Abstract
Creb3l4 belongs to the CREB/ATF family of transcription factors that are involved in mediating transcription in response to intracellular signaling. This study shows that Creb3l4 is expressed at low levels in all organs and in different stages of embryogenesis but is present at very high levels in the testis, particularly in postmeiotic male germ cells. In contrast to CREB3L4 in the human prostate, of which specific expression was detected, Creb3l4 transcripts in the mouse prostate could be detected only by RT-PCR. To identify the physiological function of Creb3l4, the murine gene was inactivated by replacement with the gene encoding green fluorescent protein. Surprisingly, Creb3l4-deficient mice were born at expected ratios, were healthy, and displayed normal long-term survival rates. Despite a significant reduction in the number of spermatozoa in the epididymis of Creb3l4−/− mice, the breeding of mutant males with wild-type females was productive and the average litter size was not significantly altered in comparison to wild-type littermates. Further analyses revealed that the seminiferous tubules of Creb3l4−/− mice contained all of the developmental stages, though there was evidence for increased apoptosis of meiotic/postmeiotic germ cells. These results suggest that Creb3l4 plays a role in male germ cell development, but its loss is insufficient to completely compromise the production of spermatozoa.
Spermatogenesis is the developmental program that guides progenitor male germ cells or spermatogonia to differentiate into haploid spermatozoa. The developmental process begins in the basal layers of the seminiferous tubules by mitotic division of spermatogonia to give rise to diploid spermatocytes. These, in turn, undergo two meitotic divisions, resulting in haploid spermatids. Differentiation of spermatids into spermatozoa involves complex morphological changes, including acrosome formation, nuclear condensation, and tail formation. The correct balance between spermatogonial renewal and terminal differentiation is integral to normal spermatogenesis and can be impaired by a variety of stress conditions, such as heat shock and drugs (18, 22).
The mammalian CREB/ATF family represents a large group of basic-region leucine zipper (bZIP) transcription factors with rather diverse physiological functions. However, despite their diverse activities, CREB/ATF family members share an ability to respond to a variety of stress conditions and maintain cellular homeostasis (9, 25). The CREB/ATF factors modulate gene expression through binding to a regulatory DNA sequence known as the cAMP-responsive element (CRE) (6, 20). Upon extracellular stresses or hormonal stimulation, a signal cascade leads to induction of expression and/or the phosphorylation of a number of CREB/ATF transcription factors that bind to the CRE and then exert positive or negative effects on the transcription of cAMP-responsive genes (7, 14, 19, 33). The CREB/ATF family is composed of both ubiquitously expressed and tissue-restricted members, with the latter controlling cell-specific patterns of gene transcription. Among the numerous transcription factors of the CREB/ATF family, Crem shows high expression during spermatogenesis (7). Crem accumulates in round and elongated spermatids and is required for transcriptional activation of several postmeiotic genes, the promoters of which contain CREs. Targeted disruption of the Crem gene results in arrest of spermatogenesis in the spermatid stage. The lack of spermatid maturation leads to germ cell apoptosis (2, 21).
Creb3l4, also referred to as Aibzip and Atce1, has been recently cloned and characterized for the mouse and human (26, 31). Alignment of the amino acid sequence of Creb3l4 revealed that the protein belongs to the CREB/ATF subfamily containing CREB-H and CREB3 (17, 23). This subgroup is characterized by the presence of a putative transmembrane domain. Localization experiments using immunohistochemistry and Creb3l4-green fluorescent protein (GFP) fusion protein revealed that Creb3l4 is primarily a cytoplasmic protein. However, deletion of the transmembrane domain results in localization of the recombinant protein in the nucleus (26). Existing reports on the expression of Creb3l4 in murine and human tissues are somewhat controversial. In the mouse, the expression of Creb3l4 is restricted to the testis (31). In contrast to male germ cell-specific expression in the mouse, human CREB3L4 transcripts are detected exclusively in the prostate and in breast and prostate cancer cell lines. An immunoassay of prostate carcinoma revealed that CREB3L4 protein levels were low in noncancerous cells compared to adjacent cancerous cells. These results suggest that CREB3L4 might contribute to development of prostate cancer (26).
In the present study, we have investigated the expression of the Creb3l4 during embryonic development and in different adult tissues of the mouse. In addition, we examined the consequences of inactivation of Creb3l4 on testis and prostate development in vivo. Our results demonstrate that Creb3l4 does not play an essential role during development of either tissue, since mice lacking Creb3l4 are fertile. No gross morphological defects in spermatogenesis or in the prostate were observed.
MATERIALS AND METHODS
Generation of Creb3l4-deficient mice.
λ phage clones carrying the mouse Creb3l4 were isolated from a 129/Sv genomic mouse library (Stratagene, La Jolla, Calif.) by using the mouse Creb3l4 cDNA. For determination of the restriction map of the Creb3l4 locus and localization of the exon sequences, the isolated phage clones were examined by Southern blot analysis. The Creb3l4-targeting vector was constructed using the plasmid vector pPNT (37). To generate the Creb3l4/GFP-targeting construct, the 5.2-kb EcoRI fragment containing the sequence of exons 5 to 10 was isolated and ligated with the EcoRI-digested pPNT (clone Creb3l4-1). The 2.8-kb NotI/XhoI fragment (NotI site from polylinker of phage clone) containing the 5′-flanking region of the Creb3l4 was isolated and inserted into the NotI/XhoI-digested clone Creb3l4-1 to generate clone Creb3l4-2. Finally, the 0.9-kb BamHI/AflII fragment, which contains the coding sequence of the green fluorescent protein and the SV40 polyadenylation signal, was isolated from the pEGFP-N1 vector (Clontech, Palo Alto, CA) and inserted in the XhoI-digested Creb3l4-2 clone by blunt-end ligation (see Fig. 2A). The resulting Creb3l4/GFP-targeting vector was linearized with SalI (see Fig. 2A) and then transfected into RI embryonic stem (ES) cells (38), and colonies resistant to G418 (300 μg/ml) and ganciclovir (2 μM) were selected.
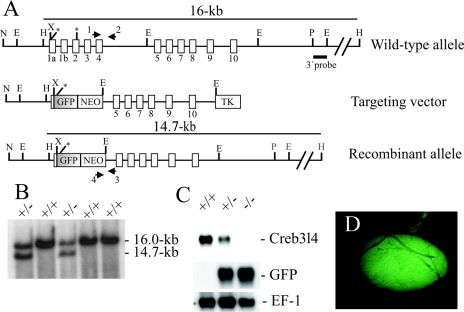
FIG. 2.
Targeted disruption of the Creb3l4 gene. (A) Structure of the wild-type, targeted vector, and recombinant allele are shown together with the relevant restriction sites. The numbers under the rectangles indicate the 11 exons of Creb3l4. Predicted translation start sites (ATG) in the Creb3l4α and Creb3l4β isoforms are indicated by single stars. The Creb3l4α isoform is transcribed from exons 1a and 2 to 10, and its predicted translation start site is the first ATG. The Creb3l4β isoform contains the sequence of exons 1b and 2 to 10, and its open reading frame starts at the second ATG. A 4.0-kb XhoI/EcoRI fragment containing exon 1a to exon 4 was replaced by a GFP gene and a pgk-neo selection cassette (NEO). The 3′ external probe used and the predicted length of HindIII restriction fragments in Southern blot analysis are shown. The primers 1, 2, 3, and 4 used to amplify the wild-type and mutant allele by PCR are also indicated. Abbreviations: TK, thymidine kinase cassette; E, EcoRI; H, HindIII; N, NotI; P, PstI; X, XhoI. (B) Southern blot analysis of the recombinant ES cell clones. Genomic DNA extracted from ES cell clones was digested with HindIII and probed with the 3′ probe shown in panel A. The Creb3l4 wild-type allele generated a 16.0-kb HindIII fragment, whereas the targeted allele yielded a 14.7-kb HindIII fragment, as indicated in panel A. (C) Northern blot analysis. Total RNA of Creb3l4−/−, Creb3l4+/− and Creb3l4+/+ was hybridized with Creb3l4 and a GFP probe. Rehybridization of blots with human elongation factor-1 cDNA (EF-1) revealed the integrity of RNA loading. (D) GFP expression in the testis of a Creb3l4−/− male. The GFP fluorescence is restricted to the seminiferous tubules.
Genomic DNA extracted from individual drug-resistant clones was screened for homologous recombination by Southern blot analysis. DNA was digested with HindIII, separated on a 0.8% agarose gel, and transferred to nylon membranes (Amersham, Braunschweig, Germany). The 0.8-kb EcoRI/PstI fragment lying externally 3′ of the targeting vector (see Fig. 2A) was radioactively labeled and used to probe the Southern blots. Hybridization was carried out at 65°C overnight in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.1% sodium dodecyl sulfate, and denatured salmon sperm DNA (100 μg/ml). Filters were washed twice at 65°C at a final stringency of 0.2× SSC/0.1% sodium dodecyl sulfate. Cells from two recombinant ES clones were injected into C57BL/6J blastocysts, and these blastocysts were transferred into DBA/BL6 pseudopregnant females. Germ line-transmitting chimeric males obtained from both lines were backcrossed to C57BL/6J and 129/Sv females, and the resulting F1 offspring were genotyped by PCR analyses.
To genotype the mice, genomic DNA was extracted from tails and analyzed by PCR. Thermal cycling was carried out for 35 cycles with denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The following primers were used to discriminate wild-type and mutant alleles: primer 1 (Creb3l4 sense), 5′-AAACAGACACCTGGGAATCC-3′, and primer 2 (Creb3l4 antisense), 5′-CCGTCGAGAAGATATCTTCTG-3′, which were designed to amplify wild-type loci. Primer 3 (Neo sense), 5′-CCTTCTATCGCCTTCTTGACG-3′, and primer 4 (Creb3l4 antisense), 5′-TTGGTGAGGGGCAGGTGAGAG-3′, were designed to amplify the targeted locus (see Fig. 2A). A 650-bp fragment of the wild-type allele was amplified with primers 1 and 2, whereas primers 3 and 4 amplified a 900-bp fragment of the mutant allele.
All animal experimentations were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Göttingen.
Northern blots and RT-PCR.
Total RNA was extracted from tissues using a QIAGEN RNA kit (QIAGEN, Hilden, Germany). For Northern blot analysis, 15 μg of RNA was electrophoresed in 1.2% agarose gels containing 2.2 M formaldehyde, transferred to nylon membranes, and hybridized with a 32P-labeled probe at the same conditions as used for Southern blot hybridizations. The following probes used for Northern blot hybridization were obtained by reverse transcription (RT)-PCR using testis RNA as template: protamine 2 (Prm2, a 258-bp RT-PCR fragment amplified with the 5′ primer 5′-AAG ACC ATG AAC GCG AGG AGC-3′ and the 3′primer 5′-GCC TCC TAC ATT TCC TGC ACC-3′), Atfa (a 379-bp fragment amplified with the 5′ primer 5′-AGA GCG GAA CTA CAC AAG ACC-3′ and the 3′ primer 5′-CTG AGG CAC TCG AAC TGT AAC-3′), Creb1 (a 367-bp cDNA fragment amplified with the 5′ primer 5′-TGT TGT TCA AGC TGC CTC AGG-3′ and the 3′ primer 5′-TTT CTG GTT GTG CCA AGC CAG-3′). The 250-bp Tnp2, 186-bp Tnp1, 520-bp Smcp, 1.2-kb Acr, and 700-bp Crem cDNA fragments were prepared from cDNA clones (1, 7, 15, 39).
RT-PCR assays were performed using 2 μg of total RNA and a One Step RT-PCR kit (QIAGEN). Primers to amplify Creb3l4, and Hprt transcripts were 5′-GTGGACTGCCCTCCGATTCG-3′ and 5′-GTCTGGGCAGCTCTGCTGGA-3′, and 5′-GTCAAGGGCATATCCAACAACAAAC-3′ and 5′-CCTGCTGGATTACATTAAAGCACTG-3′, respectively.
In situ hybridization.
Testes were fixed in 4% paraformaldehyde and embedded in paraffin. Digoxigenin (DIG)-labeled in vitro transcripts were generated from the mouse Creb3l4 cDNA according to the manufacturer's instructions (RNA labeling kit; Roche). Sense and antisense transcripts were hybridized at 37°C overnight in 50% formamide, 4× SSC, 1× Denhardt's solution, 5% dextran sulfate, 0.25 mg/ml yeast tRNA, and 0.5 mg/ml salmon sperm DNA. The posthybridization procedure includes RNase digestion and several washes in SSC solutions with a gradual reduction of the ionic strength. Labeled transcript was detected with the fluorochrome Cy3 by using the TSA amplification kit (DuPont/NEN). In brief, DIG-labeled RNA was detected by an antibody linked to biotin. Biotin was detected by streptavidin linked to horseradish peroxidase, and the signal was amplified by biotin-tyramide. This biotin-tyramide precipitate was detected by streptavidin-Cy3 (Devitron).
Fertility test.
To investigate the fertility of the Creb3l4-deficient males on a hybrid background (129/Sv × C57BL/6J) and on a 129/Sv genetic background, 10 sexually mature male Creb3l4−/− mice from each genetic background were intercrossed, each with two wild-type CD1 females, for 3 months. Females were checked for the presence of vaginal plugs and/or pregnancy. Pregnant females were removed to holding cages to give birth. The numbers and sizes of litters sired by each group of males were determined for a 3-month mating period.
Sperm analysis.
The epididymides from Creb3l4−/− and Creb3l4+/+ male mice of the hybrid genetic background were collected and dissected in Tyrode's medium. The sperm number in the cauda epididymis was determined using a Neubauer counting chamber. To investigate the acrosome reaction, spermatozoa were capacitated for 1.5 h in Tyrode's medium and then incubated for 5 min at 37°C in 5% CO2 with Tyrode's medium plus 20 μM calcium ionophore A23187 (Sigma-Aldrich Chemie). For the determination of the percentage of sperm that have undergone an acrosome reaction, sperm were fixed and stained with Coomassie brilliant blue R250 as described previously (35). At least 200 spermatozoa from each male were examined for the presence of the characteristic blue acrosomal crescent. Motility was analyzed by a CEROS computer-assisted semen analysis system (version 10; Hamilton Thorne Research, Beverly, Mass.). Epididymides of Creb3l4−/− and Creb3l4+/+ mice were dissected in in vitro fertilization medium (Medi-Cult, Jyllinge, Denmark). Sperm were allowed to swim out of the epididymides and were incubated for 1.5 h at 37°C. Aliquots (5 μl) of sperm suspension were placed into a disposable counting chamber, which was set at a temperature of 37°C. Six thousand to 10,000 spermatozoa from each of four Creb3l4−/− and Creb3l4+/+ mice were then analyzed using the following parameters: negative phase-contrast optics; recording, 60 frames/s; minimum contrast, 60; minimum cell size, 6 pixels; straightness (STR) threshold, ≥50%; cutoffs for the average path velocity (VAP) and straight line velocity (VSL), 25 and 30 μm/s, respectively; minimum progressive average path velocity (VAP), 75 μm/s. Curvilinear velocity (VCL) represents the total distance passed through by a sperm in a time unit.
Histology and immunohistochemistry.
For staining with hematoxylin-eosin and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) immunohistochemistry, tissues were fixed in Bouin's and 3.8% paraformaldehyde solution, respectively, paraffin embedded, and sectioned at a thickness of 5 μm. For routine histology, sections were stained with hematoxylin-eosin according to standard protocols. Apoptotic cells were detected using an ApopTag Peroxidase in situ apoptosis detection kit (Intergen Company, Germany) according to the instructions of the manufacturer. For quantitation of apoptosis in testes, sections of testes derived from three 12-week-old Creb3l4−/− mice and from two wild-type mice were subjected to a TUNEL assay. The numbers of TUNEL-positive and TUNEL-negative tubules were determined, and TUNEL-positive cells in each tubule were counted. The average for the 10 to 20 fields of each testis was used, and standard deviations (SD) also were determined from the examined fields.
Statistical analysis.
Paired comparisons of different sperm parameters and the apoptotic index in testis among Creb3l4−/− and Creb3l4+/+ mice were performed for statistical significance by calculating means ± SD and Student's t test.
RESULTS
Murine Creb3l4 is highly expressed in male germ cells.
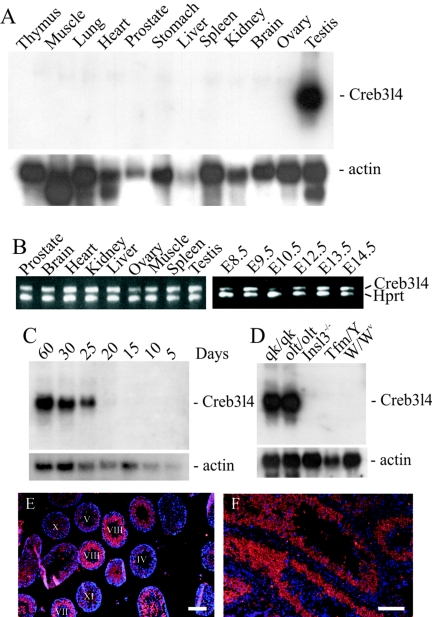
To elucidate the physiological functions of Creb3l4 in mouse, we first analyzed the structure of cDNAs. We have isolated several cDNA clones that contain different 5′ sequences. Alignments of cDNA with the genomic sequence revealed that the Creb3l4 transcribes two splicing variants, Creb3l4α and Creb3l4β. The coding sequences of the two splicing isoforms are located at the same reading frame, but Creb3l4α has an additional 49 amino acids at the N terminus. The Creb3l4α is encoded by exon 1a and 2 to 10, whereas Creb3l4β contains the coding sequence of exons 1b and 2 to 10 (see Fig. 2A). To study the expression pattern of Creb3l4, Northern blot and RT-PCR analyses were performed with total RNA from different tissues of mice. A Northern blot analysis showed that Creb3l4 was expressed exclusively in the testis; no expression in the prostate was detected (Fig. 1A). However, Creb3l4 transcripts could be detected by RT-PCR assay at different embryonic stages (embryonic day 8.5 [E8.5] to E14.5) and in different postnatal tissues (Fig. 1B). To evaluate the expression of Creb3l4 during testis development and to determine the testicular cell types expressing the Creb3l4 transcripts, we performed Northern blot and in situ hybridization analyses. As shown in Fig. 1C, the Creb3l4 transcripts could first be detected at day 20 of postnatal development. Thereafter, an increasing level of Creb3l4 expression was observed. During mouse spermatogenesis, the first wave of spermatogonia enters meiosis and gives rise to spermatocytes at ∼10 days after birth. The primary spermatocytes undergo two meiotic divisions at ∼17 days of age. The correspondence of these events with the appearance of the Creb3l4 transcripts suggests that Creb3l4 is specifically expressed in haploid germ cells. We also examined the expression of Creb3l4 in the testes of mouse mutants, in which spermatogenesis is arrested at different stages. As expected, the Creb3l4 transcripts were present in testes of olt/olt and qk/qk mutant mice (in which spermatogenesis is arrested at the spermatid stage) (Fig. 1D), whereas no transcripts could be detected in the testes of W/Wv mutant mice (which lack all germ cells), in the testes of Tfm/Y mice (in which spermatogenesis is arrested at the primary spermatocyte stage), or in the cryptorchid testes of Insl3−/− mutant mice (in which spermatogenesis is arrested at the stage of pachytene spermatocytes). The results of Northern blots were confirmed by in situ hybridization. With the fluorescence antisense probe, the Creb3l4 transcripts were barely detectable in Sertoli and Leydig cells. The most intense fluorescence signals were localized to haploid germ cells from stage VIII and are visible in elongated spermatids up to stage X (Fig. 1E and F). No staining was detected using the Creb3l4 sense probe (data not shown).
FIG. 1.
Expression analyses of the Creb3l4 gene. Northern blots with total RNA from different tissues of adult nice (A), from testis of 5-, 10-, 15, 20-, 25-, and 60-day-old mice (C) and from testis of different mutant mice (D) were hybridized with the Creb3l4 cDNA probe. Rehybridization with the β-actin cDNA revealed the level of RNA loading. (B) RT-PCR analysis with total RNA and Creb3l4-specific primers showed the presence of a 420-bp amplified fragment at all examined tissues and all embryonic stages. Production of the control Hprt products was observed throughout, demonstrating the presence of intact loaded RNA. (E and F) In situ hybridization of DIG-labeled Creb3l4 antisense transcripts to testis sections. The labeled antisense RNA was detected by streptavidin-Cy3 after signal amplification, and DNA was counterstained with DAPI (4′,6′-diamidino-2-phenylindole). The merged image shows the nuclei (blue) and the cytoplasmic localization of Creb3l4 transcripts (red). Bars, 100 μm.
Targeted disruption of Creb3l4.
The Creb3l4 gene was disrupted by homologous recombination in RI ES cells using a replacement targeting strategy (Fig. 2A). A Creb3l4/GFP-targeting construct was designed to replace a 4.0-kb XhoI/EcoRI genomic fragment containing exons 1a to 4 by GFP and neomycin-resistance gene (neoR) under the control of the Pgk promoter. The gene coding for green fluorescence protein (GFP) was inserted 25 bp upstream of the translation initiation codon ATG and thereby placed under control of the endogenous Creb3l4 promoter. Following electroporation and drug selection, homologous recombinants were detected by Southern blot analysis of HindIII-restricted genomic DNA using a 3′ external probe (Fig. 2A). The external probe detected a 16.0-kb wild-type fragment and a 14.7-kb recombinant fragment (Fig. 2B). Two Creb3l4+/− ES cell clones injected into C57BL/6J blastocysts gave rise to chimeric mice that transmitted the Creb3l4 mutation into the germ line. Chimeric mice were intercrossed with C57BL/6J or 129/Sv females to establish the Creb3l4-disrupted allele on a C57BL/6J × 129/Sv hybrid and on a 129/Sv inbred genetic background. The resulting progeny from the heterozygous intercrosses displayed a normal Mendelian ratio of Creb3l4+/+, Creb3l4+/−, and Creb3l4−/− animals, indicating that Creb3l4 is not essential for embryonic development. Creb3l4−/− mice were indistinguishable from their wild-type littermates in survival rates, appearance, and gross behavior. To confirm that the engineered disruption of the Creb3l4 had generated a null mutation, we examined the expression of Creb3l4 at the level of mRNA in testes of the three genotypes. Northern blot analysis revealed no expression of Creb3l4 in Creb3l4−/− testes. In contrast, expression of GFP in the Creb3l4-deficient testes was detected, while the level of GFP transcripts in the testes of heterozygous animals was approximately half of that detected with their Creb3l4−/− littermates (Fig. 2C). These results demonstrate the efficient expression of the GFP reporter gene under the Creb3l4 promoter and suggest that reporter gene activity can be used to track Creb3l4 expression in whole-mount tissues of Creb3l4+/− embryos and adult mice. No GFP fluorescence was detected in blastocysts or in tissues of embryos at 7.5, 10.5, and 14.5 days postcoitum (data not shown). Strong GFP fluorescence in adult testes, in which the GFP fluorescence was restricted to the seminiferous tubules, was observed (Fig. 2D). No other tissue, including different parts of the prostate, exhibited any GFP fluorescence. These results confirm that the testis is the main Creb3l4-expressing organ in mouse.
Creb3l4-deficient mice are fertile.
To investigate the consequences of the Creb3l4 gene disruption on male fertility, we intercrossed 10 Creb3l4−/− males on the C57BL/6J × 129Sv mixed and 129/Sv inbred background, each with two wild-type females, for 3 months. All matings of the Creb3l4−/− males on both genetic backgrounds were productive, and the average litter size was not significantly altered compared to that for the breeding of wild-type littermates with wild-type females (Table 1).
TABLE 1.
Fertility of Creb314+/+, Creb314+/−, and Creb314−/− mice on the genetic background C57B1/6JX129/SV and 129/Sv
| Genotype of male | No. of mice borna | No. of mice in average litter |
|---|---|---|
| C57BL/6J × 129/Sv | ||
| +/+ | 312 | 7.8 |
| −/− | 284 | 7.1 |
| 129/Sv | ||
| +/+ | 204 | 5.1 |
| −/− | 184 | 4.6 |
Ten males and 20 females were mated.
Analyses of several mutant lines, in which the spermatogenic genes expressed are inactivated, demonstrated that male mice are classified as fertile even if they produced <20% of the normal number of mature sperm or have a moderate defect in sperm motility (3). To verify whether the Creb3l4 deficiency resulted in such defects, we investigated the sperm number in the epididymis. As shown in Table 2, a significant reduction was found in the mean number of spermatozoa collected from the cauda epididymides of Creb3l4−/− males (P < 0.001). We further examined the spermatozoa by light microscopy. No morphological abnormalities in the sperm of Creb3l4−/− males could be detected. These results suggest that spermatogenesis is qualitatively normal but quantitatively reduced in Creb3l4−/− males.
TABLE 2.
Sperm analysis of Creb314+/+ and Creb314−/− micea
| Parameter | No. with indicated genotypea
|
|
|---|---|---|
| +/+ | −/− | |
| No. of sperm in cauda epididymis (107) | 2.78 ± 0.2 (4)b | 1.02 ± 0.53 (8)b |
| Sperm motility (%) | 60.1 ± 6.0 (5) | 57 ± 10.9 (5) |
| Progressive motility (%) | 48.5 ± 8.4 (5) | 48 ± 12.4 (5) |
| Sperm undergoing acrosome reaction (%) | 81.5 ± 5.0 (3) | 79 ± 7 (3) |
Data for sperm analyses are the means ± SD of the number of individual measurements indicated in parentheses.
The value for this parameter for Creb314−/− mice is significantly different from that for Creb314+/+ mice (P < 0.001, Student's t test).
Reduction of spermatogenesis and increased apoptosis of germ cells in Creb3l4−/− testis.
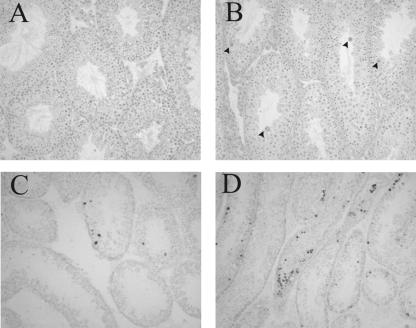
To study the basis of the reduced spermatogenesis in Creb3l4−/− males, histological sections of testes from 3-month-old mice were examined. Spermatogonia A and B, preleptotene spermatocytes, pachytene spermatocytes, and spermatids can be recognized in the tubules of Creb3l4−/− testis (Fig. 3A and B). However, several multinucleated giant cells were present in the seminiferous tubules of Creb3l4-deficient testes (Fig. 3B). These giant cells were rarely observed in wild-type mice of less than 6 months of age (32).
FIG. 3.
Spermatogenesis in the testes of Creb3l4−/− mice. Histological sections from the testes of 12-week-old wild-type (A and C) and Creb3l4−/− (B and D) mice are shown. (A and B) Staining with hematoxylin and eosin reveals the presence of all stages of spermatogenesis and the appearance of occasional multinucleated giant cells (arrows) in Creb3l4−/− tubules. In situ TUNEL staining in the seminiferous tubules of wild-type (C) and Creb3l4−/− (D) males revealed an increase in adluminal cells, with dark-stained nuclei in a subset of the tubules in mutant testes (D), while apoptotic cells in wild-type testes were most frequently located close to the basement membrane of seminiferous tubules (C). Magnification, ×200.
To determine if apoptosis contributed to the abnormality in testes of Creb3l4−/− males, a TUNEL assay was performed with testis sections. In Creb3l4−/− males, apoptotic cells were most commonly observed in adluminal regions of the seminiferous tubule (Fig. 3D). In contrast, apoptotic germ cells in the wild-type were most frequently located close to the basement membrane of seminiferous tubules (Fig. 3C). The frequency of TUNEL-positive cells is variable among tubules, but overall there were significantly more apoptotic cells in the seminiferous tubules of Creb3l4−/− males than in those of wild-type littermates (Fig. 3C and D). Quantification of apoptosis is summarized in Table 3 and is representative of testis section analysis from three Creb3l4−/− and two wild-type mice. We speculate that the increased apoptosis of germ cells in the Creb3l4−/− testis contributes to the decrease in epididymal sperm number found with Creb3l4−/− mice. Multinucleated giant cells, however, were not TUNEL positive.
TABLE 3.
Quantification of TUNEL-positive cells from testis sections
| Genotype | No. of:
|
|||
|---|---|---|---|---|
| Positive tubules | Positive cells | Negative tubules | Positive cells/tubule ± SD | |
| Creb314+/+ | 98 | 404 | 142 | 1.8 ± 1.3a |
| Creb314−/− | 128 | 737 | 70 | 4.6 ± 0.7a |
The value for this parameter for Creb314−/− mice is significantly different from that for Creb314+/+ mice (P < 0.05, Student's t test).
Sperm motility and the acrosome reaction.
To determine if Creb3l4 inactivation affected sperm motility, a computer-assisted sperm analyzer was used. Spermatozoa were isolated from the cauda epididymis and capacitated in in vitro fertilization medium for 1.5 h. No differences in the proportion of immotile, locally motile, or forwardly motile spermatozoa could be detected (Table 2). The nonlinearly and linearly motile spermatozoa, VCL, VAP, and VSL, also did not differ between wild-type and Creb3l4-deficient mice (data not shown).
Some reports indicate that sperm cAMP content increases under conditions that promote spontaneous acrosome reactions (34). Therefore, we also considered the probability that alterations in the composition of proteins in mutant sperm, which are encoded by Creb3l4 targeting genes, might affect the acrosome reaction. We found no differences in the proportions of wild-type and mutant sperm that had undergone the acrosome reaction. These results indicate that Creb3l4-deficient sperm are not significantly different from wild-type sperm when comparison of their ability to release the acrosomal contents.
Expression of potential CREB/ATF-targeted genes and CREB/ATF genes in Creb3l4−/− testes.
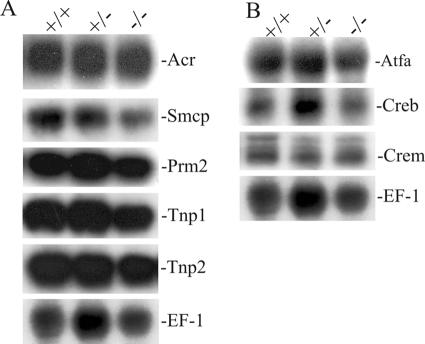
The binding of CREB/ATF transcription factors to the CRE in promoters of targeting genes mediates the activation or repression of gene expression. A significant number of germ cell-specific genes, such as acrosin (Acr), transition protein 1 (Tnp1) and Tnp2, protamine 2 (Prm2), and Smcp, contain a CRE in their regulatory promoter region (8, 24, 28, 30, 36). To examine whether the absence of Creb3l4 would modify the expression of these genes, we performed Northern blot analyses with testicular RNA. Normal expression of these genes was found with Creb3l4−/− testes (Fig. 4A), implying that they are not regulated by Creb3l4.
FIG. 4.
Analysis of gene expression in testes of Creb3l4−/− mice. Total RNA was isolated from testes of Creb3l4−/−, Creb3l4+/−, and Creb3l4+/+ mice, and 15 μg was analyzed by Northern blotting and sequential hybridization with radioactively labeled cDNA probes for the genes indicated. (A) Expression of germ cell-specific genes, which contain CREs in their promoters. (B) Expression of ATF/CREB genes. Rehybridization of blots with human elongation factor-1 cDNA (EF-1) revealed the integrity of RNA loading.
Three members of the CREB/ATF family have been reported to express in male germ cells. We examined the expression of these genes in Creb3l4−/− testes, because upregulation of these gene could, theoretically, compensate for the elimination of Creb3l4 expression. Testicular RNA derived from Creb3l4+/+ and Creb3l4−/− animals were probed with Crem, Atfa, and Creb1 cDNA (5, 19, 27). No change in the expression of these genes was detected in the testes of Creb3l4-deficient mice (Fig. 4B). These results suggest that the loss of Creb3l4 expression in testis of Creb3l4-deficient mice is not compensated for by a detectable increase of the expression of Crem, Atfa, and Creb1.
DISCUSSION
The preferential expression of Creb3l4 in haploid spermatids suggests a possible role for spermatid differentiation and/or sperm function. We show here that targeted disruption of the Creb3l4 mildly compromises germ cell development in mice, but their sperm counts and sperm function remained at sufficient levels to maintain normal fertility, thereby producing no overt consequences.
The expression analyses of Creb3l4 in adult animals confirm and extend earlier Northern blotting results that showed a high expression of Creb3l4 in the testes of mice. However, a RT-PCR assay detected the Creb3l4 expression also in different stages of embryogenesis as well as in different adult tissues. On the basis of this result, we have introduced the GFP gene under control of the endogenous Creb3l4 promoter to enable us to document Creb3l4 expression during pre- and postnatal development of heterozygous mutants. Histological analysis failed to detect the GFP fluorescence in fetal tissues. In adult tissues, strong fluorescence signals were detected only in the testis, where GFP fluorescence was restricted to germ cells. In contrast to the specific expression of CREB3L4 in the human prostate (26 and our unpublished results), Creb3l4 transcripts could be detected only by RT-PCR in mouse prostate. The absence of prostate tumors in Creb3l4-deficient mice after more than 3 years of follow-up is consistent with surveys from own institution that failed to implicate genetic alteration of CREB3L4 in prostate carcinoma (our unpublished data).
It was anticipated that altered expression of postmeiotically expressed genes containing the CRE in their promoter, would be found in Creb3l4-deficient testes. However, our analysis demonstrated normal expression of five postmeiotic germ cell markers in Creb3l4-deficient testes, suggesting that Creb3l4 does not regulate the expression of these genes. Therefore, these observations confirm previous results which showed a failure of Creb3l4 to bind to the CRE (31).
Creb3l4 belongs to a multiprotein family with functional redundancy (9). Thus, the relatively subtle phenotype of Creb3l4−/− mice may be attributable to the fact that other members of the CREB/ATF family can substitute, to some extent, for Creb3l4 function. Therefore, the role of Creb3l4 within developing germ cells remains unknown. However, given that a significant amount of apoptosis occurred among germ cells within Creb3l4−/− testes and given observations showing that mouse models with disrupted Crem, Atf3, and Atf4 function result in increased tissue-specific apoptosis (2, 11, 12, 13, 21), we are led to suggest that Creb3l4 might regulate the expression of genes required for germ cell survival. Expression analysis of Crem, Atfa and Creb in the testes of Creb3l4 mutants showed no evidence for compensatory induction of these genes. Thus, it seems that these members may play a function distinct from that of Creb3l4 in male germ cell development.
It has been reported that CREB/ATF family members mediate diverse cellular activities. For example, Creb1 and Atf1 are involved in mediating transcription in response to intracellular cAMP concentration (4). Atf2 and Atf3 have been implicated in transcription control of stress response genes (10, 40). Atf3 Atf4, Atf6, and Oasis are involved in endoplasmic reticulum stress responses (11, 12, 16, 29). Considering that many CREB/ATF family members are stress-inducible genes, Creb3l4 might play a role in adaptation to environmental stress conditions. Therefore, the lack of Creb3l4 did not dramatically affect mouse spermatogenesis, at least under laboratory conditions. However, Creb3l4 deficiency might affect sperm maturation within the testis under stress conditions. Continued efforts to understand Creb3l4 function under such conditions should provide more information about the versatile role of Creb3l4 in stress response during the progress of spermatogenesis.
Acknowledgments
We thank M. Schindler, H. Riedesel, and S. Wolf for help in the generation and breeding of knockout mice. We also thank C. Müller for help with particular experiments. We thank R. M. Sharpe (MRC, Edinburgh, Scotland) for critical reading of the manuscript.
This study was supported by grant AD129/2 from Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aho, H., M. Schwemmer, D. Tessman, D. Murphy, G. Mattei, W. Engel, and I. M. Adham. 1996. Isolation, expression, and chromosomal localization of the human mitochondrial capsule selenoprotein gene (MCSP). Genomics 32:184-190. [DOI] [PubMed] [Google Scholar]
- 2.Blendy, J. A., K. H. Kaestner, G. F. Weinbauer, E. Nieschlag, and G. Schutz. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380:162-165. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, P. E., O. Chisholm, R. J. Arceci, E. R. Stanley, and J. W. Pollard. 1996. Absence of colony-stimulating factor-1 in osteopetrotic (csfmop/csfmop) mice results in male fertility defects. Biol. Reprod. 55:310-317. [DOI] [PubMed] [Google Scholar]
- 4.De Cesare, D., and P. Sassone-Corsi. 2000. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 64:343-369. [DOI] [PubMed] [Google Scholar]
- 5.De Graeve, F., A. Bahr, B. Chatton, and C. Kedinger. 2000. A murine ATFa-associated factor with transcriptional repressing activity. Oncogene 19:1807-1819. [DOI] [PubMed] [Google Scholar]
- 6.Deutsch, P. J., J. P. Hoeffler, J. L. Jameson, J. C. Lin, and J. F. Habener. 1988. Structural determinants for transcriptional activation by cAMP-responsive DNA elements. J. Biol. Chem. 263:18466-18472. [PubMed] [Google Scholar]
- 7.Foulkes, N. S., B. Mellstrom, E. Benusiglio, and P. Sassone-Corsi. 1992. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature 355:80-84. [DOI] [PubMed] [Google Scholar]
- 8.Ha, H., A. J. van Wijnen, and N. B. Hecht. 1997. Tissue-specific protein-DNA interactions of the mouse protamine 2 gene promoter. J. Cell. Biochem. 64:94-105. [PubMed] [Google Scholar]
- 9.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Hai, T., C. D. Wolfgang, D. K. Marsee, A. E. Allen, and U. Sivaprasad. 1999. ATF3 and stress responses. Gene Expr. 7:321-335. [PMC free article] [PubMed] [Google Scholar]
- 11.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 12.Hartman, M. G., D. Lu, M. L. Kim, G. J. Kociba, T. Shukri, J. Buteau, X. Wang, W. L. Frankel, D. Guttridge, M. Prentki, S. T. Grey, D. Ron, and T. Hai. 2004. Role for activating transcription factor 3 in stress-induced β-cell apoptosis. Mol. Cell. Biol. 24:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hettmann, T., K. Barton, and J. M. Leiden. 2000. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev. Biol. 222:110-123. [DOI] [PubMed] [Google Scholar]
- 14.Iordanov, M., K. Bender, T. Ade, W. Schmid, C. Sachsenmaier, K. Engel, M. Gaestel, H. J. Rahmsdorf, and P. Herrlich. 1997. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 16:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm, U., W. M. Maier, S. Tsaousidou, I. M. Adham, K. Willison, and W. Engel. 1990. Mouse preproacrosin: cDNA sequence, primary structure and postmeiotic expression in spermatogenesis. Differentiation 42:160-166. [DOI] [PubMed] [Google Scholar]
- 16.Kondo, S., T. Murakami, K. Tatsumi, M. Ogata, S. Kanemoto, K. Otori, K. Iseki, A. Wanaka, and K. Imaizumi. 2005. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7:186-194. [DOI] [PubMed] [Google Scholar]
- 17.Lu, R., P. Yang, P. O'Hare, and V. Misra. 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meistrich, M. L., G. Wilson, G. Shetty, and G. A. Shuttlesworth. 2003. Restoration of spermatogenesis after exposure to toxicants: genetic implications. Adv. Exp. Med. Biol. 518:227-237. [DOI] [PubMed] [Google Scholar]
- 19.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 20.Montminy, M. R., and L. M. Bilezikjian. 1987. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328:175-178. [DOI] [PubMed] [Google Scholar]
- 21.Nantel, F., L. Monaco, N. S. Foulkes, D. Masquilier, M. LeMeur, K. Henriksen, A. Dierich, M. Parvinen, and P. Sassone-Corsi. 1996. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380:159-162. [DOI] [PubMed] [Google Scholar]
- 22.Neuer, A., S. D. Spandorfer, P. Giraldo, S. Dieterle, Z. Rosenwaks, and S. S. Witkin. 2000. The role of heat shock proteins in reproduction. Hum. Reprod. Update 6:149-159. [DOI] [PubMed] [Google Scholar]
- 23.Omori, Y., J. Imai, M. Watanabe, T. Komatsu, Y. Suzuki, K. Kataoka, S. Watanabe, A. Tanigami, and S. Sugano. 2001. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oosterhuis, J. H., and F. A. van der Hoorn. 1999. Testis-specific TTF-D binds to single-stranded DNA in the c-mos and Odf1 promoters and activates Odf1. J. Biol. Chem. 274:11708-11712. [DOI] [PubMed] [Google Scholar]
- 25.Persengiev, S. P., and M. R. Green. 2003. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis 8:225-228. [DOI] [PubMed] [Google Scholar]
- 26.Qi, H., C. Fillion, Y. Labrie, J. Grenier, A. Fournier, L. Berger, M. El-Alfy, and C. Labrie. 2002. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 62:721-733. [PubMed] [Google Scholar]
- 27.Ruppert, S., T. J. Cole, M. Boshart, E. Schmid, and G. Schutz. 1992. Multiple mRNA isoforms of the transcription activator protein CREB: generation by alternative splicing and specific expression in primary spermatocytes. EMBO J. 11:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulten, H. J., K. Nayernia, K. Reim, W. Engel, and P. Burfeind. 2001. Assessment of promoter elements of the germ cell-specific proacrosin gene. J. Cell. Biochem. 83:155-162. [DOI] [PubMed] [Google Scholar]
- 29.Shen, J., E. L. Snapp, J. Lippincott-Schwartz, and R. Prywes. 2005. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol. Cell. Biol. 25:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steger, K., T. Klonisch, K. Gavenis, R. Behr, V. Schaller, B. Drabent, D. Doenecke, E. Nieschlag, M. Bergmann, and G. F. Weinbauer. 1999. Round spermatids show normal testis-specific H1t but reduced cAMP-responsive element modulator and transition protein 1 expression in men with round-spermatid maturation arrest. J. Androl. 20:747-754. [PubMed] [Google Scholar]
- 31.Stelzer, G., and J. Don. 2002. Atce1: a novel mouse cyclic adenosine 3′,5′-monophosphate-responsive element-binding protein-like gene exclusively expressed in postmeiotic spermatids. Endocrinology 143:1578-1588. [DOI] [PubMed] [Google Scholar]
- 32.Takano, H., and K. Abe. 1987. Age-related histologic changes in the adult mouse testis. Arch. Histol. Jpn. 50:533-544. [DOI] [PubMed] [Google Scholar]
- 33.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 34.Tash, J. S., and A. R. Means. 1983. Cyclic adenosine 3′,5′ monophosphate, calcium and protein phosphorylation in flagellar motility. Biol. Reprod. 28:75-104. [DOI] [PubMed] [Google Scholar]
- 35.Thaler, C. D., and R. A. Cardullo. 1995. Biochemical characterization of a glycosylphosphatidylinositol-linked hyaluronidase on mouse sperm. Biochemistry 34:7788-7795. [DOI] [PubMed] [Google Scholar]
- 36.Topaloglu, O., G. Schlueter, K. Nayernia, and W. Engel. 2001. A 74-bp promoter of the Tnp2 gene confers testis- and spermatid-specific expression in transgenic mice. Biochem. Biophys. Res. Commun. 289:597-601. [DOI] [PubMed] [Google Scholar]
- 37.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 38.Wurst, W., and A. L. Joyner. 1993. Production of targeted embryonic stem cell clones, p. 33-61. In A. L. Joyner (ed.), Gene targeting: a practical approach. IRL Press, Oxford, England.
- 39.Yelick, P. C., C. Kozak, Y. K. Kwon, M. F. Seldin, and N. B. Hecht. 1991. The mouse transition protein 1 gene contains a B1 repetitive element and is located on chromosome 1. Genomics 11:687-694. [DOI] [PubMed] [Google Scholar]
- 40.Zoumpourlis, V., P. Papassava, S. Linardopoulos, D. Gillespie, A. Balmain, and A. Pintzas. 2000. High levels of phosphorylated c-Jun, Fra-1, Fra-2 and ATF-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene 19:4011-4021. [DOI] [PubMed] [Google Scholar]