Abstract
The Mus81-Eme1 endonuclease is implicated in the efficient rescue of broken replication forks in Saccharomyces cerevisiae and Schizosaccharomyces pombe. We have used gene targeting to study the function of the Mus81-Eme1 endonuclease in mammalian cells. Mus81-deficient mice develop normally and are fertile. Surprisingly, embryonic fibroblasts from Mus81−/− animals fail to proliferate in vitro. This proliferation defect can be rescued by expression of the papillomavirus E6 protein that promotes degradation of p53. When grown in culture, Mus81−/− cells have elevated levels of DNA damage, acquire chromosomal aberrations, and are hypersensitive to agents that generate DNA cross-links. In contrast to the situation in yeast, murine Mus81 is not required for replication restart following camptothecin treatment. Mus81−/− mice and cells are hypersensitive to DNA cross-linking agents. Cross-link-induced double-strand break formation is normal in Mus81−/− cells, but the resolution of repair intermediates is not. The persistence of Rad51 foci in Mus81−/− cells suggests that Mus81 acts at a late step in the repair of cross-link-induced lesions. Despite these defects, Mus81−/− mice do not show increased predisposition to lymphoma or any other malignancy in the first year of life.
The processes of homologous recombination contribute significantly to the accurate repair of genomic damage resulting both from intrinsic and exogenous sources. The role homologous recombination plays in double-strand break (DSB) repair in mammalian cells is increasingly well understood (29). However, despite evidence that mammalian mutants defective in homologous recombination are also extremely sensitive to agents that induce DNA interstrand cross-links (ICLs), the role of homologous recombination in ICL repair is less well characterized (10, 21). In addition to the damage ICLs inflict at the nucleotide level, they can block essential cellular processes such as transcription, replication, and recombination. It is postulated that ICL repair involves the formation of DSBs and subsequent repair through homologous recombination. However, the order of events and the nucleases that catalyze each step have not been identified (19, 33, 41, 50). Cross-link repair is especially important in the clinic where ICL-inducing agents, such are mitomycin C (MMC) and cis-platin, are widely used as antitumor agents. However, sources of endogenous or spontaneous ICLs are poorly characterized, and the importance of ICL repair in normal cellular processes and the maintenance of health is not clear.
The subunits of the mammalian Mus81-Eme1 endonuclease complexes were identified by sequence similarity to Mus81-Eme1 (Mms4) from Saccharomyces cerevisiae and Schizosaccharomyces pombe (1, 7, 12, 14, 44). Haploid yeast Mus81 mutants are sensitive to agents that result in replication fork collapse but not to those that generate DSBs (9, 20, 28, 36, 57). Thus, Mus81-Eme1 is specifically involved in the process of replication restart in yeast. In addition, fission yeast Mus81-Eme1 is essential for meiotic recombination (8, 24, 46, 53). In budding yeast, Mus81 and its partner, Mms4, have a lesser role in meiotic recombination (17, 18); however, as in fission yeast, Mus81-Mms4 mutants have a severe recombination-dependent meiotic defect (28, 30). In vitro, Mus81-Eme1 cleaves a number of synthetic DNA substrates, including 3′ flaps, replication forks, nicked Holliday junctions, and Holliday junctions (12, 15, 24, 30, 46). These structures are designed to mimic potential in vivo substrates; however, how closely in vitro specificity reflects in vivo function is not known. To more directly determine the function of Mus81-Eme1 in mammalian cells, we have disrupted the mouse Mus81 gene and analyzed the effect Mus81 deficiency has in vitro and in vivo. We find that Mus81-deficient mice and cells have significantly increased sensitivities to MMC-induced toxicity. Mus81 is not required for the formation of DSBs after cross-link damage, but it is required for their resolution. Unlike yeast Mus81 mutants, murine cells that lack Mus81 are relatively resistant to camptothecin (CPT), suggesting that alternative or redundant mechanisms ensure replication restart in mammalian cells. Significantly, despite increased sensitivity to agents that induce DNA interstrand cross-links and evidence that Mus81-deficient cells suffer increased chromosomal aberrations in vitro, no increase in the incidence of spontaneously occurring tumors or malignancy is seen in Mus81-deficient mice. In this respect, the data presented here differ significantly from those reported in a different Mus81−/− mouse model (39).
MATERIALS AND METHODS
Generation of Mus81 knockout mice.
Genomic clones containing MUS81 sequences were identified by screening a 129SvEvBrd-derived lambda pKOS mouse genomic library with MUS81 sequence-specific primers. A 9.2-kb genomic clone containing MUS81 exons 5 through 16 was used to generate the targeting vector. Heterozygous pLox-Mus81 flanked mice were generated by Lexicon Genetics Inc. (The Woodlands, Tex.) by introducing flanking pLox sites upstream of exon 9 and downstream of exon 12. A 1.7-kb PGK-Neo selection cassette flanked by two Frt sites was introduced between exon 12 and the downstream LoxP site. The vector was linearized by digestion with NotI and electroporated into 129SvEvBrd(LEX1) embryonic stem (ES) cells. G418-fialuridine-resistant ES cell clones were isolated and verified for homologous recombination at the MUS81 locus by Southern blot analysis. ES cell clones containing the targeting construct were injected into C57BL/6(albino) blastocysts, and resulting chimeras were mated with C57BL/6(albino) females to generate heterozygous animals containing one pLox-MUS81 allele each. These mice were subsequently crossed with protamine-Cre mice (43) that undergo recombination of the target gene in the male germ line but not in other tissues. The resulting males heterozygous for both the pLox-MUS81 allele and the protamine-Cre transgene were crossed with C57BL/6 females to obtain heterozygous mice harboring a disrupted MUS81 allele. These mice were subsequently crossed to generate all genotypes employed in the studies reported here. Mice were maintained in a pathogen-free environment. Genotyping was carried out by PCR on DNA isolated from mouse tail biopsy samples. Reactions with the primer pair 5′CTAGCCGCTTGCGTTCCACAATGT3′ and 5′GGAGCTAAGGCCTAGCGAGTACAG3′ resulted in amplification of a 400-bp product from the wild-type allele, while reactions using the primer pair 5′GGTGTGGCCCTGATGGAAGAG3′ and 5′GGAGCTAAGGCCTAGCGAGTACAG3′ amplified a 370-bp product from the disrupted allele.
Cell culture.
Mouse embryonic fibroblasts (MEFs) were isolated from mixed-genotype littermate embryos at 13.5 days postcoitum. Primary MEFs were harvested and processed for DNA content following 2 days in culture. Twenty-four hours after plating, primary MEFs were infected with LXSN retrovirus containing the coding sequence for human papillomavirus type 16 E6 and cultured in Dulbecco's modified Eagle medium supplemented with 10% calf serum and 100 μg/ml penicillin and streptomycin.
Prior to flow cytometry, cells were cultured in the presence of 10 μM bromodeoxyuridine (Calbiochem) for 30 min and fixed in 70% ethanol. Cells were stained with fluorescein isothiocyanate-anti-bromodeoxyuridine (Becton Dickinson) according to the manufacturer's protocol. After several washes, cells were resuspended in phosphate-buffered saline containing 5 μg/ml propidium iodide and 100 μg/ml RNase for 30 min before fluorescence-activated cell sorter analysis (FACSCalibur; Becton Dickinson). The cell cycle distribution of each sample was calculated using Cell Quest software.
Genomic stability assay.
To prepare metaphase spreads, cells were incubated in 500 ng/ml nocodazole (Sigma) for 4 h, and 0.1 μg/ml demecolcine (Sigma) was added to the medium for the last hour. Cells were harvested and resuspended in hypotonic buffer (0.075 M KCl) at 37°C for 10 min. After fixation in methanol:acetic acid (3:1), cells were dropped onto glass slides and stained with Giemsa stain (Sigma) for 10 min. Chromosome aberrations, including chromosome breaks, fragment and chromosomal fusions, and dicentricity, were analyzed by direct visualization using a 100× objective. Twenty-five to 30 metaphases from each cell type were scored.
Colony survival and whole-animal sensitivity assay.
To determine cellular sensitivity to DNA-damaging agents, 100 or 500 cells were seeded in six-well plates. Cells were cultured continuously in the concentrations of MMC, CPT and hydroxyurea (HU) indicated below. cis-Platin and nitrogen mustard were added to the culture media for 2 h, and the cells were then cultured in drug-free medium until colonies were visible (10 to 14 days). Where indicated, cells were exposed to UV-C light with a wavelength of 254 nm or were irradiated by use of a cesium 137 source. Following 10 to 14 days in culture, cells were fixed in methanol and stained with Giemsa stain, and colonies were counted. Triplicate cultures were scored for each treatment.
The ability of human Mus81 to complement the MMC sensitivity of Mus81−/− MEFs was assayed by transient transfection. Approximately 0.5 × 106 cells were transfected with pCDNA-3HaMus81WT (wild type); with pCDNA-3HaMus81DD, an endonuclease-dead mutant of Mus81 (12); and with a control vector expressing green fluorescent protein, pcDNAGFP. Effectene from QIAGEN was used for all transfections. Parallel transfection with pCMU-β-galactosidase was used to determine that ∼50% of cells were transfected. Twenty-four h after transfection, 100 or 500 cells were seeded in six-well plates, and an indicated concentration of MMC was added for 2 h. Cells were washed and incubated in fresh media until colonies were visible. The remaining cells were cultured for 72 h and then used to check for the expression of the Mus81 protein by use of Western blotting.
Whole-animal sensitivity studies.
Mice (12 to 18 weeks of age) were injected intraperitoneally with a single dose of MMC at 5 mg/kg of body weight and 10 mg/kg. Survival and body weight measurements were recorded daily. The percentages of survival were calculated according to the Kaplan and Meier method (31). Differences in survival between the different groups were tested for significance by the Wilcoxon-Gehan test.
Affinity purification of Mus81 antibody and immunoblotting.
Affinity-purified mouse Mus81 antibody was generated by purifying Mus81 serum (12) over GST-mus-Mus81 full-length protein that had been cross-linked to glutathione-Sepharose by use of dimethylpimelimidate (Pierce). For immunoblotting, cells were lysed in 20 mM HEPES, pH 7.4, 150 mM NaCl, 5% glycerol, 0.1% NP-40, 0.1% β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, and 5 μg/ml each of leupeptin, pepstatin, and aprotinin. Lysates were cleared by centrifugation at 10,000 × g for 10 min. Protein concentration was determined using Bradford reagent. One hundred micrograms of cell lysate was resolved on 10% acrylamide-sodium dodecyl sulfate gels. Immunoblots were incubated in phospho-Chk1 (Ser345) (1:1,000; Cell Signaling), Chk1 and tubulin (1:1,000; Santa Cruz), hemagglutinin (1:5,000; Covance), and PCNA (1:5,000; Santa Cruz). The incubations were followed by treatment with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antiserum. Chemiluminescence was used to detect the respective proteins.
Immunofluorescence.
Cells were grown on cover slides overnight before exposure to 3 μM MMC or 1 nM CPT for 1 h. Cells were washed and incubated in fresh media for the amounts of time indicated below. In situ fractionation was performed prior to fixation (40). Cells were fixed in 4% formaldehyde, permeabilized in 0.5% Triton X-100, blocked with 10% calf serum, and then incubated with primary antibody overnight in 4°C and with secondary antibody for 1 h at room temperature. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted in 70% glycerol. Primary antibody dilutions were of γH2AX (Trevigen) at 1:150 and Rad51 (Abcam) at 1:100. Alexa-fluor 488- or 546-conjugated anti-mouse or anti-rabbit immunoglobulin G (Molecular Probes) was used at a dilution of 1:1,000. Images were captured using a charge-coupled-device camera (Photometrics). Gray-scale images were processed using Adobe Photoshop 7.0. Three hundred cells were counted for each sample. Cells containing two or more distinct foci were scored as positive.
Pulsed-field gel electrophoresis.
Cells were either not treated or treated with 3 μM MMC for 1 h and then incubated in fresh media for an additional 24 h. Approximately 1 × 106 cells were collected and embedded in 0.85% agarose insert prepared with a contour-clamped homogeneous electric field disposable plug mold (Bio-Rad). The agarose inserts were incubated in proteinase K (1 mg/ml) at 50°C for 48 h and thereafter washed four times in 10 mM Tris-Cl, pH 7.5, 1 mM EDTA prior to loading onto a 0.85% agarose gel. Pulsed-field gel electrophoresis was carried out with a contour-clamped homogeneous electric field DRIII system (Bio-Rad). The gel was run at 14°C for 24 h at 4 V/cm at a 120° angle with a 60- to 120-s switch time for 24 h. DNA was visualized by ethidium bromide staining.
RESULTS
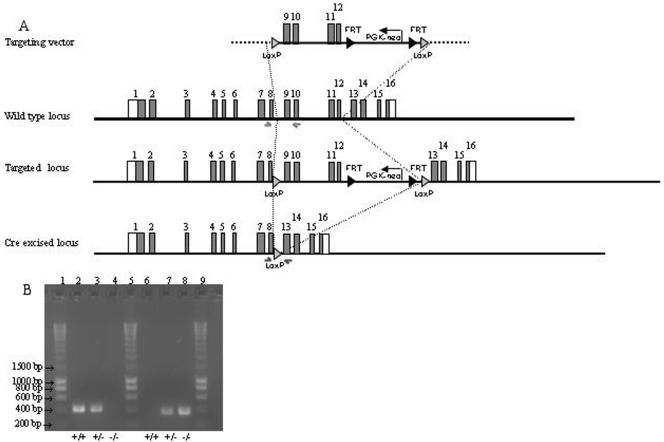
The function of mammalian Mus81 was investigated by targeted disruption of the gene in mice. A construct in which exons 9 to 12, which include the endonuclease domain, were altered by the introduction of PGK-Neo and flanked by pLox sites was introduced into mouse embryonic stem cells (Fig. 1A). pLox-Mus81-Neo mice were crossed with protamine-Cre mice that express the Cre recombinase exclusively in the male germ line (43). Resulting compound heterozygous males were crossed with C57BL/6 females. The presence of the targeting construct in embryonic stem cells and in mice was monitored by Southern blot analysis (not shown). Cre-mediated excision of the pLox-Mus81-Neo cassette was monitored by PCR (Fig. 1B). Mice and cells in which the both copies of the cassette were disrupted are designated as Mus81−/−. Mus81−/− mice were born at expected Mendelian frequencies and were indistinguishable from wild-type littermates in terms of development, growth, immune function, and fertility.
FIG. 1.
Targeted disruption of the MUS81 gene. (A) Schematic of the targeting strategy. The positions of the primers of the wild-type and disrupted alleles are indicated by arrows. (B) Genotyping was carried out by PCR on DNA isolated from mouse tail biopsy samples. The wild-type allele resulted in the amplification of a 400-bp product (lanes 2 to 4), and the disrupted allele resulted in a 370-bp product (lanes 6 to 8). Lanes 1, 5, and 9 contain molecular weight markers.
Mus81-deficient MEFs fail to proliferate in vitro.
Despite the vitality of Mus81−/− mice, repeated attempts to culture Mus81-deficient MEFs in vitro resulted in the loss of viable cells within 3 to 5 days of culture. Cells from wild-type and heterozygous animals appeared healthy and grew to confluence at the expected rates. The effect Mus81 deficiency has on the cell cycle distribution of early-passage MEFs was examined by flow cytometry. Following 2 days in culture, the Mus81-deficient cell culture showed a significant increase in the number of cells with a sub-G1 content of DNA and a significant loss of cells with 4N DNA content (Fig. 2A). We reasoned that the failure of Mus81−/− cells to proliferate might be related to the oxygen-induced stress that occurs when MEFs are cultured in vitro (47). Loss of p53 function is commonly associated with escape from damage sensitivity in MEFs. We therefore infected freshly isolated MEFs with retrovirus expressing E6, a viral protein that promotes p53 degradation (51). Using this procedure, Mus81−/− cell lines that grow under standard laboratory conditions were established. The cell culture analysis described below was carried out using E6-expressing MEFs from Mus81−/−, Mus81+/−, and Mus81+/+ mice.
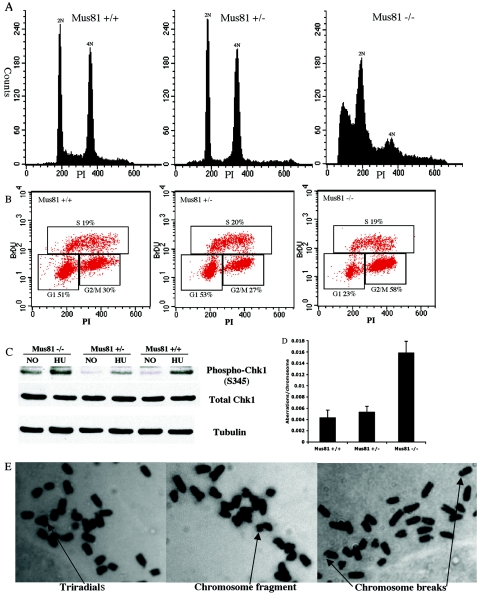
FIG. 2.
Chromosomal aberrations and checkpoint activation in Mus81−/− MEFs. (A) The DNA content of early-passage Mus81−/−, Mus81+/−, and wild-type MEFs was determined by flow cytometric analysis of propidium iodide-stained cells. (B) The cell cycle distribution of E6-expressing Mus81−/−, Mus81+/−, and wild-type cells was determined by flow cytometric analysis of bromodeoxyuridine- and propidium iodide-stained cells. (C) Phosphorylation of the Chk1 kinase was elevated in Mus81−/− cells prior to the addition of HU. The abundance of the Chk1 protein is not altered. Tubulin was monitored to ensure equal loading. (D) Chromosomal aberrations are increased in metaphase spreads from Mus81−/− MEFs. (E) Representative metaphase spreads from Mus81−/− cells.
Mus81 deficiency results in checkpoint activation.
The effect of Mus81 deficiency on cell cycle progression was analyzed by flow cytometry (Fig. 2B). The percentage of Mus81−/− cells with 4N DNA content, 58%, was approximately 30% higher than those of either wild-type or Mus81+/− cells (27% and 30%, respectively). An equivalent decrease in the number of Mus81−/− cells with 2N content was seen. The S phase populations were approximately equal in all three cultures. No increase in the number of mitotic figures was detected in Mus81−/− cultures. This cell cycle profile suggests that Mus81−/− cells replicate all, or nearly all, of their DNA with near-normal kinetics but that they delay in G2. To determine if the G2 delay involves the activation of a DNA damage checkpoint, the phosphorylation status of the checkpoint kinase Chk1 was examined. As shown in Fig. 2C, Chk1 phosphorylation was elevated in untreated Mus81−/− cells. In all three cell types, Chk1 phosphorylation increased following treatment with the replication inhibitor hydroxyurea.
The slight but constitutive checkpoint activation seen in Mus81−/− cells is suggestive of increased levels of spontaneous DNA damage. In cells with compromised p53 function, DNA damage could lead to increased levels of chromosomal abnormalities; therefore, metaphase spreads were examined for the presence of chromosome breaks, fusions, and triradials (Fig. 2D). An approximately threefold increase in the incidence of spontaneous chromosomal abnormalities was detected in cells from Mus81−/− mice compared to wild-type controls (P < 0.05). No significant difference in the incidence of chromosomal aberrations was seen in Mus81+/− MEFs compared to that in Mus81+/+ MEFs (Fig. 2E).
Damage sensitivity of Mus81-deficient MEFs.
Mus81-deficient cells and animals have previously been shown to be hypersensitive to the DNA cross-linking agent MMC (39). How Mus81-Eme1 acts to repair MMC-induced damage has not been determined. The sensitivity of Mus81−/− MEFs to MMC and the role of Mus81 in the generation and resolution of MMC-induced lesions were examined. Mus81-deficient MEFs were found to be hypersensitive to MMC, whereas Mus81+/− MEFs had the same level of resistance as wild-type cells did (Fig. 3A). Expression of wild-type human Mus81, but not of a point mutant of Mus81 that lacks endonuclease activity, partially restored resistance to MMC (Fig. 3B and C). The relative insensitivity of Mus81+/− cells could be explained if 50% expression of Mus81 protein is sufficient to protect cells from the toxic effects of DNA cross-linking agents or if the heterozygous cells compensate for the reduced gene dosage by expressing increased levels of protein. Western blot analysis was used to distinguish between these possibilities. Mus81 antibodies were purified using full-length murine Mus81 fused to glutathione S-transferase as an affinity matrix. The resulting antibodies recognized both N-terminal and C-terminal portions of Mus81 (data not shown). Western blot analysis showed that the expression of Mus81 in heterozygous cells was very similar (between 80 and 100%) to that of wild-type cells (Fig. 3D). The relative resistance of heterozygous cells to MMC and cis-platin is thus most likely explained by the fact that cells compensate for the reduced gene dosage by increased protein expression. A longer exposure of the entire Western blot did not reveal any novel bands in the lysates from either null or heterozygote cells. A protein of 36 kDa would be expected if the disrupted allele allowed expression of the 5′ exons.
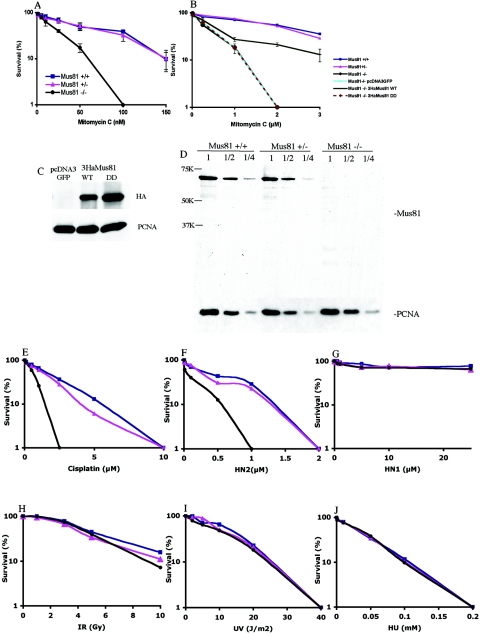
FIG. 3.
Mus81-deficient cells are sensitive to cross-linking agents. (A) Survival of wild-type, Mus81+/−, and Mus81−/− cells following chronic exposure to MMC. (B) Transient transfection of Mus81 (3HaMus81WT) but not of an endonuclease-dead mutant (3HaMus81DD) rescues the MMC sensitivity of Mus81−/− cells. Twenty-four h after transfection, cells were exposed to the indicated concentrations of MMC for 2 h, and cells were then cultured in drug-free medium for 14 days, fixed, and stained with Giemsa stain. The transfection efficiency, ∼50%, was determined by β-galactosidase staining. (C) Expression of 3HaMus81 protein 72 h following transfection. (D) Near-wild-type levels of expression of Mus81 protein in Mus81+/− cells. The abundance of endogenous Mus81 was determined by Western blot analysis using polyclonal anti-Mus81 serum against full-length protein. Serial twofold dilutions of lysates from wild-type, Mus81+/−, and Mus81−/− cells are shown. PCNA was probed to assure equal loading. No novel cross-reacting species were detected in Mus81−/− or Mus81+/− cells. Colony survival assays: Mus81−/− cells are sensitive to DNA cross-linking agent cis-platin (E) and the bifunctional nitrogen mustard HN2 (F). Mus81−/− cells are not hypersensitive to a monofunctional nitrogen mustard, HN1 (G), ionizing radiation (H), UV light (I), or chronic exposure to hydroxyurea (J).
MMC toxicity is thought to result mainly from the generation of ICLs; however, MMC also damages individual bases and causes intrastrand cross-links. To determine whether Mus81−/− cells are generally sensitive to cross-linking agents, the toxicities of cis-platin and nitrogen mustard were also assessed. Whereas MMC-induced DNA adducts cause relatively little helix distortion, cis-platin adducts cause major distortions to the DNA helix (21). Mus81-deficient MEFs were found to be hypersensitive to cis-platin (Fig. 3E). Thus, the sensitivity of Mus81-deficient cells to cross-linking agents does not depend on helix distortion. The sensitivity of Mus81 specifically to cross-links compared to its sensitivity to monoadducts was tested by assessing the toxicity of the bifunctional nitrogen mustard HN2 with its monofunctional analogue HN1. Both agents cause N-alkylpurine adducts, but only the bifunctional agent causes interstrand cross-links (19). Mus81−/− cells were hypersensitive to HN2 but not to HN1 (Fig. 3F and G). Mus81−/− MEFs have little or no increased sensitivity to γ radiation, UV light, or hydroxyurea (Fig. 3H through J).
The repair defect in Mus81−/− cells.
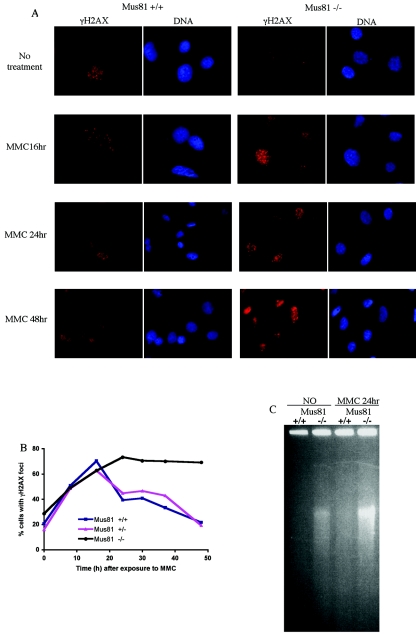
MMC and other agents that generate ICLs are extremely toxic to cells that are undergoing DNA replication, and this toxicity correlates with the generation of DSBs (2, 19, 38). However, the mechanism by which the breaks are generated is not known. The Mus81-Eme1 endonuclease efficiently cleaves 3′ flaps in vitro (14). This enzymatic activity predicts a role for Mus81 in the generation of DSBs by the direct cleavage of blocked replication forks (5, 41). To determine if Mus81 is required for the generation of DSBs in MMC-treated cells, we first looked at the kinetics of γ-H2AX focus formation by immunofluorescence. γ-H2AX, the phosphorylated form of the histone variant H2AX, is rapidly and extensively phosphorylated in large regions of chromatin adjacent to DSBs (49). The presence of γ-H2AX is widely used as a marker for DSBs. Consistent with the elevated phosphorylation of Chk1 seen in Mus81−/− MEFs, the number of Mus81-deficient cells that have γ-H2AX foci was elevated prior to treatment (28.7% ± 1% compared to 20.6% ± 0.6% in wild-type cells [P < 0.05]) (Fig. 4A). Following a 1-h treatment with MMC, the number of wild-type cells with γ-H2AX foci increased for 16 h (Fig. 4B). The kinetics of γ-H2AX focus formation is consistent with the need for MMC-exposed cells to pass through S phase to generate DSBs (2, 38). Having reached a peak in which ∼70% of cells had γ-H2AX foci, the numbers of wild-type and Mus81+/− cells with γ-H2AX foci gradually declined to basal levels within 48 h of drug treatment. Mus81−/− MEFs accumulated γ-H2AX foci at the same rate as wild-type cells did, but the percentage of cells with γ-H2AX foci remained constant at ∼70% for the duration of the experiment. Although γ-H2AX foci are widely viewed as representing DSBs, the possibility that other forms of damage might also induce H2AX phosphorylation cannot be excluded. We therefore used pulsed-field gel electrophoresis to confirm that MMC induces DSBs in Mus81−/− cells. Under the electrophoretic conditions used, intact genomic DNA is too large to enter the gel. The presence of DSBs is revealed as lower-molecular-weight DNA that is able to enter the gel. Low-molecular-weight DNA was not detected in untreated wild-type MEFs (Fig. 4C). However, a small amount of DNA from untreated Mus81−/− cells entered the gel. MMC treatment of wild-type cells resulted in detectable amounts of lower-molecular-weight DNA. MMC treatment of Mus81−/− cells also resulted in increased amounts of low-molecular-weight DNA. Taken together with the γ-H2AX focus formation data, these data suggest that Mus81 is not required for the generation of DSBs following MMC treatment.
FIG. 4.
Mus81 is not needed for DSB formation following MMC treatment. (A) γ-H2AX foci are formed following MMC exposure. MEFs were seeded onto to coverslips and grown overnight prior to exposure to 3 μM MMC for 1 h. At the indicated times, cells were extracted in situ, fixed, and immunostained for γ-H2AX. (B) Quantification of γ-H2AX foci in wild-type, Mus81+/−, and Mus81−/− cells. (C) Detection of DSB by pulsed-field gel electrophoresis. Wild-type and Mus81−/− cells were untreated (NO) or treated for 1 h with 3 μM MMC, washed and incubated in fresh media for 24 h. Plugs of 1 ×106 cells were embedded in low-melting-point agarose prior to proteinase K treatment and pulsed-field gel electrophoresis. Ethidium bromide staining was used to visualize DNA.
Rad51 foci form and persist in Mus81-deficient cells.
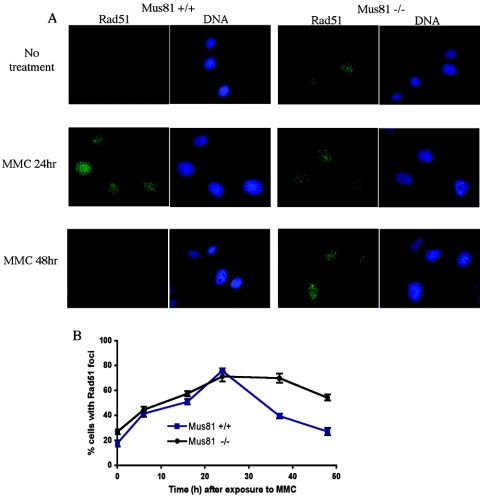
Rad51, the mammalian homologue of the bacterial RecA protein, forms the single-stranded DNA nucleoprotein filament needed for the homology search and strand invasion steps of DSB repair through homologous recombination (55). The Rad51 protein forms visible nuclear foci similar to γ-H2AX foci in response to DNA damage. However, Rad51 foci are found only in replicative or postreplicative cells. Focus formation is dependent on a number of upstream activities, and the foci are thought to represent sites of homologous recombination (26, 35, 37). A number of mutant cell lines that are extremely sensitive to MMC show delayed or defective Rad51 focus formation (6, 34). A defect in Rad51 focus formation in Mus81-deficient cells would suggest that Mus81 functions upstream of the strand invasion step in the homologous repair pathway. Following exposure to MMC, Rad51 foci were found to accumulate with nearly identical kinetics in wild-type and Mus81-deficient MEFs (Fig. 5B). Twenty-four h after treatment, the number of cells with Rad51 foci peaked in the wild-type culture, and by 48 h the number of cells with Rad51 had decreased almost to pretreatment levels. By contrast, in Mus81−/− samples, the number of cells with Rad51 foci remained elevated for the duration of the experiment. The generation and persistence of Rad51 foci suggest that Mus81−/− cells initiate but fail to complete the repair of MMC-induced damage.
FIG. 5.
Rad51 focus formation in Mus81−/− and wild-type cells. (A) MEFs were seeded onto to coverslips and grown overnight prior to exposure to 3 μM MMC for 1 h. At the indicated times, cells were extracted in situ, fixed, and immunostained for Rad51. (B) Quantification of Rad51 foci in wild-type and Mus81−/− cells.
Mus81-deficient cells are not sensitive to camptothecin.
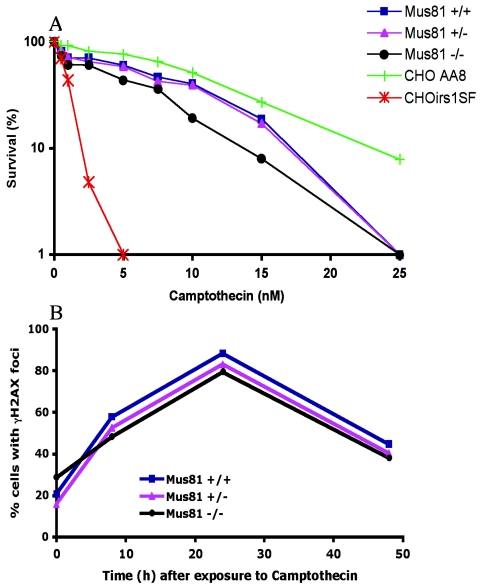
The topoisomerase inhibitor CPT stabilizes topoisomerase 1-DNA intermediates and inhibits the religation of the two DNA ends (48). It thus creates a single-strand break in which one end is free and the other is covalently coupled to topoisomerase. If present during DNA replication, CPT-induced breaks are converted into double-strand breaks by advancing replication forks (54). In both fission yeast and budding yeast, Mus81-Eme1/Mms4 mutants are extremely sensitive to camptothecin (20, 36, 57). The CPT sensitivity of Mus81-deficient murine cells was therefore examined. As shown in Fig. 6A, Mus81−/− cells are not significantly more sensitive to CPT treatment than wild-type cells are (P = 0.43). Using identical experimental conditions, CPT was found to be highly toxic to irs1SF cells (a CHO cell line with a mutation in the Xrcc3 gene [56]). Thus, the experimental conditions were appropriate for the detection of hypersensitivity. γ-H2AX foci were monitored following treatment with 1 nM CPT for 1 h. After CPT exposure, the number of wild-type cells with γ-H2AX foci increased for approximately 24 h. Having reached a peak in which 60 to 70% of cells had γ-H2AX foci, the number of cells with foci declined gradually to basal levels within 48 h of drug treatment. No significant difference was seen in the kinetics of γ-H2AX focus formation and disappearance in Mus81-deficient cells compared to wild-type or heterozygote controls (P = 0.59). Similar results were obtained when the kinetics of Rad51 foci formation was followed (data not shown). Together, these analyses suggest that the majority of CPT-induced DSBs are efficiently and appropriately repaired in Mus81−/− cells.
FIG. 6.
Mus81−/− cells are not hypersensitive to camptothecin. (A) Cell survival following chronic exposure to camptothecin. The sensitivity of the CHO (Chinese hamster ovary)-derived Irs1SF cell line was determined and compared to that of CHO-AA8 cells. (B) Quantification of γ-H2AX foci in wild-type, Mus81+/−, and Mus81−/− cells following exposure to CPT. Cells were exposed to 1 μM CPT for 1 h and incubated in drug-free medium for the indicated time prior to processing for immunostaining as described above.
Mus81-deficient mice are healthy and viable.
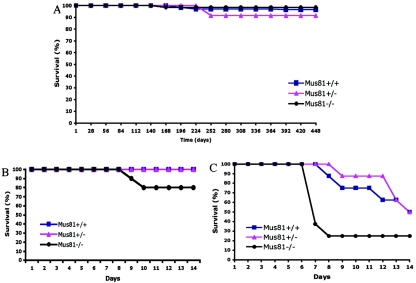
Defects in DNA repair processes are frequently associated with increased susceptibility to tumors or premature ageing. A recent study found evidence that proper biallelic expression of Mus81 was required for tumor suppression in mice (39). To determine if susceptibility to tumors is a direct consequence of Mus81 disruption, we monitored the health and survival of wild-type, heterozygous, and homozygous Mus81 mutants for more than 15 months. More than 90% of the animals in all three cohorts remained healthy and disease free for the duration of the experiment (Fig. 7A). Even in the oldest Mus81−/− mice (n = 12 at 20 months), no increased mortality or signs of sickness were detected. Mus81−/− mice were analyzed in an extensive battery of histological and clinical biochemical tests, and no differences from wild-type mice were found. These findings suggest that Mus81 expression is not required for tumor suppression in mice.
FIG. 7.
Long-term survival and cross-link sensitivity of Mus81−/− mice. (A) Analysis of survival of wild-type (n = 66), Mus81+/− (n = 23), and Mus81−/− (n = 65) mice versus age in days. To date, no evidence of lymphoma has been detected in any Mus81−/− or Mus81+/− mouse. (B) Mus81−/− mice are sensitive to MMC. Survival of mice injected intraperitoneally with (B) 5 mg/kg MMC (n = 10 for all three strains) and (C) 10 mg/kg (n = 8 for all three strains).
Mus81-deficient mice are sensitive to MMC-induced lethality.
Mus81−/− cells are extremely sensitive to MMC (Fig. 3A); however, the cells used in this study express the papillomavirus E6 protein, and the sensitivity could result from a synergistic effect of Mus81 and p53 deficiency. Thus, the sensitivities of wild-type, heterozygous, and Mus81-deficient mice were tested. Following the administration of a single dose of 5 mg/kg MMC, 3 out of 10 Mus81−/− mice died within 14 days. This was in contrast to results for both wild-type and heterozygous mice, which all survived this dose. A similar trend was also seen when a higher dose of MMC was used. Following injection of 10 mg/kg MMC, 80% of Mus81−/− mice died within 1 week, compared to the 50% of the wild-type and 50% of the heterozygous animals that died within 14 days (Fig. 7B). This pattern of MMC toxicity is similar to that reported in reference 39 and suggests that MMC tolerance is significantly compromised in the two strains of Mus81-deficient mice.
DISCUSSION
We have investigated the function of mammalian Mus81 by characterizing the phenotypes of cells and mice bearing a targeted disruption of the endonuclease domain of Mus81. We find that Mus81-deficient mice are viable and fertile. Thus, in agreement with a similar study of Mus81 function, we find that Mus81 does not play an essential role in murine development (39). In spite of this, we find that fibroblasts taken from Mus81−/− mice fail to proliferate in vitro unless they are transformed with the E6 protein. The inactivation of p53 has previously been shown to rescue proliferation defects associated with the disruption of several DSB repair genes (23, 25, 52). However, in these previous examples, disruption of the repair gene resulted in a proliferation defect that was also manifest in the whole animal. The contrast between the normal development and health of Mus81−/− mice and the failure of Mus81−/− cells to proliferate in vitro suggests that Mus81 is required in order to survive damage that occurs frequently in culture but is less common in the whole organism. We suspect that the failure of Mus81−/− MEFs to proliferate in culture relates to the increased oxidative damage of DNA that occurs when cells are grown under standard laboratory conditions (27, 47). A similar proliferation defect was not reported for Mus81-deficient or Eme1-deficient ES cells (1, 39). However, since p53 function is suppressed in ES cells (3), these cells might be expected to behave similarly to E6-expressing MEFs. Although E6-expressing Mus81−/− fibroblast cell lines grew at rates similar to those of wild-type controls, we found elevated chromosomal aberrations, and Mus81−/− cells tended to accumulate in the G2 phase of the cell cycle. Furthermore, the increased phosphorylation of Chk1 seen in untreated Mus81−/− cells suggests that they have constitutively elevated levels of DNA damage.
Mus81-deficient cells are specifically sensitive to agents that generate ICLs.
As previously reported for Eme1−/− embryonic stem cells, we find that Mus81−/− fibroblasts are hypersensitive to MMC and other agents that cause cross-links but only mildly sensitive to infrared, UV, or HU (1). We find that γ-H2AX focus formation after MMC damage occurs with the same kinetics in wild-type and Mus81−/− cells. This and the pulsed-field gel electrophoresis analysis strongly argue that the endonuclease activity of Mus81-Eme1 is not required for ICL-induced DSB formation. A role for mammalian Mus81 at a late step in ICL repair is consistent with a genetic analysis of budding yeast indicating that Mus81 acts late in recombination (22). Niedernhofer et al. showed that there was normal kinetics of DSB formation following MMC treatment of cells that lack Ercc1 (41), and these authors speculated that Mus81-Eme1 endonuclease function might be needed to generate DSBs from replication forks that have stalled because the replication templates could not be separated (41). Since MMC-induced DSB formation is not compromised in Mus81-deficient cells, the question of how they form remains to be addressed. Rothfuss and Grompe recently showed that incision of ICLs (i.e., single-strand nicking on one or both sides of the cross-link) occurs rapidly and is independent of cell cycle position (50). They also showed that subsequent DSB formation is dependent on DNA replication. Thus, in this model, the conversion of the incised ICL site into a DSB is dependent on replication forks encountering a nick; it is not actively catalyzed by an endonuclease acting on the replication fork. Identification of the enzyme(s) that makes the initial incision will be key to determining how ICLs are repaired in mammalian cells.
We also find that damage-induced focus formation of Rad51, a central molecule in homologous recombination, as well as that of γ-H2AX, is not impaired in the absence of Mus81. These data suggest that Mus81 either acts independently of Rad51 or acts at a later step in the repair process. The persistence of both γ-H2AX foci and Rad51 foci may indicate that Mus81-deficient cells are unable to process ICL lesions beyond the assembly of Rad51 filaments. However, it is also possible that in the absence of Mus81, novel, nonproductive recombination intermediates accumulate.
Mus81-deficient murine cells are not sensitive to CPT.
The mild sensitivity of Mus81-deficient cells to CPT was something of a surprise. Both budding yeast and fission yeast mutants of Mus81-Eme1/Mms4 are extremely sensitive to CPT (20, 36, 57). This sensitivity is rescued by the expression of the bacterial Holliday junction resolvase, RusA (4, 20). Thus, in the absence of Mus81 function, an exogenous Holliday junction resolvase is able to restore viability, presumably through its ability to promote productive replication restart. Therefore, the presence of a second Holliday junction-resolving activity might explain the lack of sensitivity of Mus81−/− murine cells to CPT. Two Holliday junction-resolving activities, one involving Mus81 and an undefined resolvase, known as resolvase A, have previously been shown to be present in a single human cell type (15). Thus, it is possible that murine resolvase A is able to support the repair of CPT-induced damage.
Mus81−/−: a tumor suppressor or not?
Our data concerning the longevity and lack of malignancy in Mus81−/− mice differ dramatically from those described in a recent report in which Mus81 deficiency resulted in early-onset tumors and death (39). Of the Toronto group's mice, only 27% of homozygous and 50% of heterozygous animals survived the first year of life (39). By contrast, in this study, more than 95% of the Mus81-deficient mice survived disease free for at least 15 months. The Mus81-deficient mice in this study may eventually develop tumors, but if they do, it will be with greatly delayed onset. The basis for the discrepancies in longevity and tumor susceptibility between the two studies is not clear. Strain differences are often invoked to explain differences in the rates or penetrance of phenotypes in mouse models. The mice in this study were generated from 129 ES cells, and they were initially crossed with C57BL/6 and then back with a 129 strain to allow Cre-mediated excision of the targeting construct (43). Thus, the final strain is a 129/BL6 mix. Although a different 129-derived embryonic stem-cell line was used for disruption, the Toronto group's mice are also 129/BL6 mixes (Razqallah Hakem, personal communication). Thus, although not identical, the two strains are closely related. Further studies will be needed to determine if a modifier or suppressor mutation accounts for differences in these two models of Mus81 deficiency. Given that the Mus81-deficient mice described here do not spontaneously develop tumors, it will be of interest to characterize the genetic interaction between Mus81 and p53. The cellular phenotypes described here predict a role for p53 in protecting Mus81−/− cells from chromosomal aberrations.
The targeting strategy used by the Toronto group replaced exons 3 and 4 with a selectable marker; the marker and its promoter were left in place in the final construct (39). Therefore, it is formally possible that this promoter inadvertently leads to the increased expression of a proto-oncogene. Such off-target effects, although rare, have given rise to dramatic phenotypic differences in closely related mouse models (45). Although there is no obvious proto-oncogene in the vicinity of murine Mus81, this hypothesis would account for the observation that the Mus81 heterozygous mice were found to be nearly as tumor prone as the homozygous disrupted mice, despite the fact that they have wild-type levels of resistance to DNA damage (39).
The phenotypes of Mus81−/− cells and mice are similar to the phenotypes seen following the disruption of several of the Fanconi anemia complementation (FANC) group genes. Fanconi anemia (FA), a rare autosomal recessive disease, is characterized by congenital abnormalities, progressive bone marrow failure, and cancer susceptibility (reviewed in reference [16]). FA cells are hypersensitive to agents that induce cross-links but have, at most, modestly increased sensitivities to agents that induce other forms of nucleotide damage or that induce strand breaks. Thus, the clinical manifestation of FA is perhaps the best evidence that the inability to repair naturally occurring ICLs poses a serious threat to human health.
Like Mus81 deficiency, the disruption of mouse FANC A, C, or G genes results in increased sensitivity to MMC but not to other DNA-damaging agents (11, 13, 32, 42, 58, 59). Remarkably, the disruption of these genes also does not result in increased tumor susceptibility (11, 13, 32, 42, 58, 59). Given that both the FANC and Mus81 functions are required to repair ICL but not to suppress tumors, it is tempting to suggest that ICLs do not occur frequently enough to create serious problems in mice. On the other hand, the profound anemias and cancer susceptibilities of Fanconi anemia patients suggest that humans might be more dependent on pathways that repair ICL for tumor avoidance.
Acknowledgments
We are grateful to Peiqing Sun (The Scripps Research Institute) for providing LXSN E6 retrovirus and to Paul Russell and Pierre-Henri Gaillard for encouragement and advice.
This work was funded by a National Cancer Institute grant awarded to C.H.M.
REFERENCES
- 1.Abraham, J., B. Lemmers, M. P. Hande, M. E. Moynahan, C. Chahwan, A. Ciccia, J. Essers, K. Hanada, R. Chahwan, A. K. Khaw, P. McPherson, A. Shehabeldin, R. Laister, C. Arrowsmith, R. Kanaar, S. C. West, M. Jasin, and R. Hakem. 2003. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22:6137-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkari, Y. M., R. L. Bateman, C. A. Reifsteck, S. B. Olson, and M. Grompe. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 20:8283-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., B. T. Spike, L. W. Rodewald, T. J. Hope, M. Klemm, R. Jaenisch, and G. M. Wahl. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8:145-155. [DOI] [PubMed] [Google Scholar]
- 4.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessho, T. 2003. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J. Biol. Chem. 278:5250-5254. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. K., U. Ear, A. Bhattacharyya, C. Calderone, M. Beckett, R. R. Weichselbaum, and A. Shinohara. 1998. Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem. 273:21482-21488. [DOI] [PubMed] [Google Scholar]
- 7.Blais, V., H. Gao, C. A. Elwell, M. N. Boddy, P. H. Gaillard, P. Russell, and C. H. McGowan. 2004. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell 15:552-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 9.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W.-D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldecott, K., and P. Jeggo. 1991. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 255:111-121. [DOI] [PubMed] [Google Scholar]
- 11.Chen, M., D. J. Tomkins, W. Auerbach, C. McKerlie, H. Youssoufian, L. Liu, O. Gan, M. Carreau, A. Auerbach, T. Groves, C. J. Guidos, M. H. Freedman, J. Cross, D. H. Percy, J. E. Dick, A. L. Joyner, and M. Buchwald. 1996. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 12:448-451. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X. B., R. Melchionna, C. M. Denis, P. H. Gaillard, A. Blasina, I. Van de Weyer, M. N. Boddy, P. Russell, J. Vialard, and C. H. McGowan. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8:1117-1127. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, N. C., H. J. van de Vrugt, M. A. van der Valk, A. B. Oostra, P. Krimpenfort, Y. de Vries, H. Joenje, A. Berns, and F. Arwert. 2000. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 9:1805-1811. [DOI] [PubMed] [Google Scholar]
- 14.Ciccia, A., A. Constantinou, and S. C. West. 2003. Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 278:25172-25178. [DOI] [PubMed] [Google Scholar]
- 15.Constantinou, A., X. B. Chen, C. H. McGowan, and S. C. West. 2002. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21:5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 17.de los Santos, T., N. Hunter, C. Lee, B. Larkin, J. Loidl, and N. M. Hollingsworth. 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Los Santos, T., J. Loidl, B. Larkin, and N. M. Hollingsworth. 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159:1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 21.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 22.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank, K. M., N. E. Sharpless, Y. Gao, J. M. Sekiguchi, D. O. Ferguson, C. Zhu, J. P. Manis, J. Horner, R. A. DePinho, and F. W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5:993-1002. [DOI] [PubMed] [Google Scholar]
- 24.Gaillard, P. H., E. Noguchi, P. Shanahan, and P. Russell. 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12:747-759. [DOI] [PubMed] [Google Scholar]
- 25.Gao, Y., D. O. Ferguson, W. Xie, J. P. Manis, J. Sekiguchi, K. M. Frank, J. Chaudhuri, J. Horner, R. A. DePinho, and F. W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897-900. [DOI] [PubMed] [Google Scholar]
- 26.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halliwell, B. 2003. Oxidative stress in cell culture: an under-appreciated problem? FEBS Lett. 540:3-6. [DOI] [PubMed] [Google Scholar]
- 28.Interthal, H., and W. D. Heyer. 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:812-827. [DOI] [PubMed] [Google Scholar]
- 29.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21:8981-8993. [DOI] [PubMed] [Google Scholar]
- 30.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan, E., and P. Meier. 1958. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53:457-481. [Google Scholar]
- 32.Koomen, M., N. C. Cheng, H. J. van de Vrugt, B. C. Godthelp, M. A. van der Valk, A. B. Oostra, M. Z. Zdzienicka, H. Joenje, and F. Arwert. 2002. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum. Mol. Genet. 11:273-281. [DOI] [PubMed] [Google Scholar]
- 33.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275:26632-26636. [DOI] [PubMed] [Google Scholar]
- 34.Larminat, F., M. Germanier, E. Papouli, and M. Defais. 2004. Impairment of homologous recombination control in a Fanconi anemia-like Chinese hamster cell mutant. Biol. Cell 96:545-552. [DOI] [PubMed] [Google Scholar]
- 35.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 36.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99:14970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHugh, P. J., W. R. Sones, and J. A. Hartley. 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McPherson, J. P., B. Lemmers, R. Chahwan, A. Pamidi, E. Migon, E. Matysiak-Zablocki, M. E. Moynahan, J. Essers, K. Hanada, A. Poonepalli, O. Sanchez-Sweatman, R. Khokha, R. Kanaar, M. Jasin, M. P. Hande, and R. Hakem. 2004. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science 304:1822-1826. [DOI] [PubMed] [Google Scholar]
- 40.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. Jaspers, H. B. Beverloo, J. H. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noll, M., K. P. Battaile, R. Bateman, T. P. Lax, K. Rathbun, C. Reifsteck, G. Bagby, M. Finegold, S. Olson, and M. Grompe. 2002. Fanconi anemia group A and C double-mutant mice: functional evidence for a multi-protein Fanconi anemia complex. Exp. Hematol. 30:679-688. [DOI] [PubMed] [Google Scholar]
- 43.O'Gorman, S., N. A. Dagenais, M. Qian, and Y. Marchuk. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94:14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogrunc, M., and A. Sancar. 2003. Identification and characterization of human MUS81-MMS4 structure specific endonuclease. J. Biol. Chem. 278:21715-21720. [DOI] [PubMed] [Google Scholar]
- 45.Olson, E. N., H. H. Arnold, P. W. Rigby, and B. J. Wold. 1996. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1-4. [DOI] [PubMed] [Google Scholar]
- 46.Osman, F., J. Dixon, C. L. Doe, and M. C. Whitby. 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12:761-774. [DOI] [PubMed] [Google Scholar]
- 47.Parrinello, S., E. Samper, A. Krtolica, J. Goldstein, S. Melov, and J. Campisi. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pommier, Y., P. Pourquier, Y. Fan, and D. Strumberg. 1998. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400:83-105. [DOI] [PubMed] [Google Scholar]
- 49.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 50.Rothfuss, A., and M. Grompe. 2004. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 52.Smiraldo, P. G., A. M. Gruver, J. C. Osborn, and D. L. Pittman. 2005. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 65:2089-2096. [DOI] [PubMed] [Google Scholar]
- 53.Smith, G. R., M. N. Boddy, P. Shanahan, and P. Russell. 2003. Fission yeast Mus81. Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165:2289-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strumberg, D., A. A. Pilon, M. Smith, R. Hickey, L. Malkas, and Y. Pommier. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 56.Tebbs, R. S., Y. Zhao, J. D. Tucker, J. B. Scheerer, M. J. Siciliano, M. Hwang, N. Liu, R. J. Legerski, and L. H. Thompson. 1995. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl. Acad. Sci. USA 92:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99:13669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitney, M. A., G. Royle, M. J. Low, M. A. Kelly, M. K. Axthelm, C. Reifsteck, S. Olson, R. E. Braun, M. C. Heinrich, R. K. Rathbun, G. C. Bagby, and M. Grompe. 1996. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood 88:49-58. [PubMed] [Google Scholar]
- 59.Yang, Y., Y. Kuang, R. M. De Oca, T. Hays, L. Moreau, N. Lu, B. Seed, and A. D. D'Andrea. 2001. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood 98:3435-3440. [DOI] [PubMed] [Google Scholar]