Abstract
Aneuploidy is a common feature of human tumors, often correlating with poor prognosis. The mitotic spindle checkpoint is thought to play a major role in aneuploidy suppression. To investigate the role of the spindle checkpoint in tumor suppression in vivo, we developed transgenic mice in which thymocytes express a dominant interfering fragment of Bub1, a kinase regulator of the spindle checkpoint. We report that, despite high-level expression of dominant-negative Bub1 (Bub1DN), a protein known to inhibit spindle checkpoint activity in cultured cells, thymocytes show no evidence of spindle checkpoint impairment. Transgenic animals also failed to show an increased predisposition to spontaneous tumors. Moreover, the Bub1DN transgene failed to alter the timing or characteristics of thymic lymphoma development in p53 heterozygous or homozygous null backgrounds, indicating that the lack of tumorigenesis is not due to suppression by p53-dependent checkpoints. These results indicate that overexpression of a Bub1 N-terminal fragment is insufficient to impair the spindle checkpoint in vivo or to drive tumorigenesis in the highly susceptible murine thymocyte system, either alone or in combination with G1 checkpoint disruption.
Genomic instability and aneuploidy are hallmarks of human cancer and frequently correlate with poor prognosis (33). Despite this common observation, the mechanisms by which aneuploidy arises and its role in tumorigenesis remain subjects of intense interest and debate (20). The spindle checkpoint suppresses chromosome segregation errors and aneuploidy by ensuring that all chromosomes achieve proper alignment and bipolar attachment to the mitotic spindle prior to anaphase onset (1, 27). The spindle checkpoint controls anaphase entry by blocking the activity of the anaphase-promoting complex (APC), a multisubunit ubiquitin ligase that degrades anaphase-blocking substrates such as Securin proteins (11, 27). Several genes are required for spindle checkpoint function in the yeast Saccharomyces cerevisiae, including BUB1, BUB3, MAD1, MAD2, MAD3, and MPS1, and homologs in metazoans appear to play conserved roles (reviewed in reference 6).
Several observations suggest that spindle checkpoint deregulation contributes to tumorigenesis. First, dominant-negative mutations or decreased expression of spindle checkpoint genes has been found in some human colon cancers and colon or other carcinoma cell lines, suggesting a role in the etiology of chromosome instability in those tumors (8, 34, 36, 41, 42). Second, several cases of the rare familial disorder mosaic variegated aneuploidy, which presents with developmental abnormalities, widespread aneuploidy, and increased risk of tumorigenesis, displayed biallelic mutations of the spindle checkpoint gene BubR1 (18). Third, mice with heterozygous mutations of the spindle checkpoint gene Mad2, BubR1, Bub3, or Rae1 are prone to spontaneous or carcinogen-induced tumorigenesis (3, 13, 29). Fourth, dominant-negative mutations in the Bub1 or BubR1 gene have been identified in thymic lymphomas from mice homozygous for a truncating mutation in the Brca2 gene (26). Finally, the Tax protein from human T-cell leukemia virus may drive chromosomal instability in human adult T-cell leukemia cells by interacting with Mad1 (21). Despite this evidence, mutations in known spindle checkpoint genes are rare in a variety of human carcinomas (7, 19, 30, 44), suggesting that genomic instability in such tumors either results from mutations in presently unknown spindle checkpoint genes or occurs through alternative mechanisms such as epigenetic reduction of gene expression. Thus, additional studies are required to define the contribution of spindle checkpoint defects in tumorigenesis.
In light of previous studies implicating spindle checkpoint impairment in thymic lymphomas in Brca2 mutant mice (26), and given the highly proliferative status of thymocytes and the high susceptibility of mouse thymocytes to tumorigenesis, we reasoned that the thymus would provide a good model system in which to test the role of spindle checkpoint impairment in tumorigenesis. Overexpression of N-terminal fragments of the Bub1 kinase impairs the spindle checkpoint in yeast and mammalian cell lines, suggesting that a similar approach could be used for dominant checkpoint inhibition in vivo (16, 26, 38, 43). Here we report the results of thymocyte overexpression of a dominant-negative Bub1 in transgenic mice alone and in combination with p53 mutation.
MATERIALS AND METHODS
Transgenic animals.
All animal studies were carried out in strict compliance with federal and local guidelines for animal care and use. The LBDN (Lck-Bub1-dominant-negative) transgene was produced by cloning the Bub1 cDNA region encoding amino acids 1 to 331 (Bub1DN) into the BamHI site of the p1017 vector (10) (a gift from Lishan Su, University of North Carolina, Chapel Hill). The Bub1DN fragment was amplified from mouse testis cDNA by PCR. A primary reaction was performed using primers Bub1NPCR (5′-TGGTGGGGTATTTCGCCGCT-3′) and Bub1CPCR (5′-TGGCTGCACTTTGCATGGTG-3′), followed by a second round of amplification using primers Bub1UBam (5′-TGCGCGGGATCCGCCACCATGGACAACCTAGAAAATGTCT-3′) and Bub1LBam (5′-AGATCTGGATCCTCACTGGGAGCAAGTATTTTGTCCAAC-3′). The resulting product was digested with BamHI and inserted into the vector. The Myc-tagged version of the transgene was produced by amplifying the Bub1DN coding region from the LBDN vector with primers that incorporated the Myc tag in place of the initiating methionine. A primary amplification was performed with primers Bub1Myc1 (5′-CTCATCTCAGAAGAGGACCTGTCTAGAGACAACCTAGAAAATGTC-3′) and Bub1LBam, followed by a second round of PCR using Bub1Myc2-2 (5′-GCCTGAAGATCTGCCACCATGGAACAAAAACTCATCTCAGAAGAGGACCT-3′) and Bub1LBam. The MycBub1DN product was digested with BamHI and BglII and cloned into the BamHI site of a modified p1017 vector in which the native XbaI site was destroyed by Klenow treatment and ligation. A single clone with the insert in the wrong orientation was subsequently digested with BstYI and religated to produce a clone with the correct orientation. Transgenes were removed from vector sequences by digestion with SacII. Transgenic animals were generated by pronuclear injection in the B6D2F1/J genetic background. Transgenic animals were identified by PCR of toe-derived DNA using primers LckFPCR1 (5′-GGCTGAGGCTGAGGGTTGACTCTAA-3′) and BubRPCR2 (5′-AAACTGATGACGGTCGCTGTTGTACT-3′). Cycling parameters were 94°C for 4 min; 35 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s; and a final incubation at 72°C for 10 min.
Antibody production.
The Bub1DN fragment was cloned into the pET-His-3E vector. His-tagged Bub1DN protein was expressed in BL21(DE3) bacteria and purified on His-Bind resin (Novagen). Purified protein was used to generate a polyclonal rabbit antiserum (Pocono Rabbit Farm and Laboratory).
Immunoblotting.
Tissues were flash frozen in liquid nitrogen and stored at −80°C. Proteins were extracted in radioimmunoprecipitation buffer and quantified by the Bradford method (Bio-Rad). Ten micrograms of protein per sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. Membranes were probed with an unpurified Bub1 polyclonal antiserum or the anti-Myc monoclonal antibody 9E10. Immunocomplexes were detected using enhanced chemiluminescence (Amersham). Equal loading was verified using an anti-beta-actin antibody (Santa Cruz Biotechnology; SC-8432). In some cases, extracts were prepared by solubilizing thymocytes in sodium dodecyl sulfate sample buffer. A lysate from equivalent cell numbers was loaded in each lane of the gel.
Flow cytometry.
Thymus, spleen, and lymph nodes were dissociated by crushing through a 70-μm screen and resuspended in FWB (phosphate-buffered saline with 1% fetal bovine serum and 0.1% [wt/vol] sodium azide). Spleen samples were treated with erythrocyte lysis solution (8.3 g NH4Cl, 1g KHCO3 per liter of water), spun down, and resuspended in FWB. Cells were counted in a hemocytometer, and 1 × 106 cells were stained with saturating amounts of a phycoerythrin-conjugated anti-CD4 (αCD4) and a fluorescein isothiocyanate-conjugated αCD8 antibody (BD Bioscience). Samples were analyzed on a FACScan flow cytometer.
Thymocyte culture.
Thymocytes were dissociated as described above and cultured in cDMEM (DMEM-H with 10% fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES, 50 μM β-mercaptoethanol, 1× penicillin-streptomycin) with 10 ng/ml interleukin-1α (IL-1α) (PeproTech) and 20 ng/ml IL-2 (R&D Systems or PeproTech).
Mitotic checkpoint challenge.
Thymocytes were cultured for 5 to 7 days in cDMEM with interleukin-1α and interleukin-2 and were subsequently treated for 8 h with 200 ng/ml nocodazole (Sigma). Cells were fixed with 1% formaldehyde in phosphate-buffered saline for 20 to 30 min at room temperature, followed by 70% ethanol overnight at −20°C. Cells were rehydrated and stained with an anti-phosphohistone H3 (PH3) antibody and propidium iodide as described elsewhere (14), except that we used 1 μg anti-PH3 (Upstate), 10 μg/ml Alexa 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes), and 100 μg/ml propidium iodide. Mitotic indices were measured by flow cytometry on a FACScan by analyzing the percentage of PH3-positive cells with 4N DNA content as described elsewhere (14). Cell cycle profiles were modeled from DNA width-versus-DNA area plots with the ModFit LT program (Verity Software House).
Survival and tumorigenesis analysis.
Animals were monitored regularly for signs of illness up to the age of 2 years. Overall and tumor-free survival times and P values were determined by Kaplan-Meier and log rank analyses (Wilcoxon P values are also shown in some cases for comparison). Animals that were tumor free at necropsy were censored in survival analyses. Animals that died of unknown causes were also censored in lymphoma-free survival analysis but not in overall survival analyses. Lymphoma diagnoses were based on histological confirmation of tumors. Comparison of tumor frequencies was done by Fisher's exact test with one-tailed P values. All statistical analyses were performed with JMP IN software (SAS Institute, Inc.).
RESULTS
Thymic overexpression of dominant-negative Bub1.
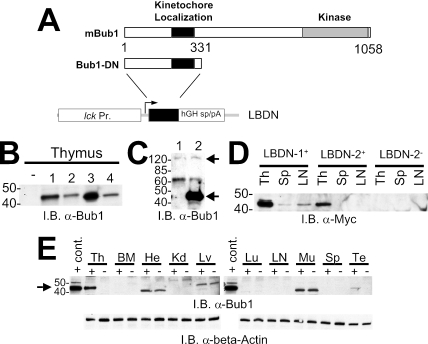
We used a transgenic approach to express a dominant-negative fragment of the Bub1 kinase (Bub1DN) specifically in the mouse thymocytes. Using cell culture systems, others have reported that expression of an N-terminal Bub1 fragment dominantly impairs the spindle checkpoint (2, 16, 26, 38). This fragment includes the conserved domain responsible for kinetochore localization and Bub3 binding and is hypothesized to interfere with checkpoint function via competitive removal of wild-type Bub1 from the kinetochore. Overexpression of a similar Bub1 fragment also impairs chromosome segregation in Saccharomyces cerevisiae (43). The transgene construct consisted of the lck proximal promoter, the Bub1DN cDNA, and splicing/polyadenylation signals from the human growth hormone gene (Fig. 1A). Two similar constructs were generated, one of which included an N-terminal Myc epitope tag on the Bub1DN fragment while the second lacked the Myc tag. Seven transgenic mouse lines (four with the Myc tag, three without) were derived on a B6D2F1/J genetic background via pronuclear injection. Strong expression of the Bub1DN protein was detected in thymus extracts from four founder lines by immunoblotting (lines 1 and 2 with the Myc tag, lines 3 and 4 without) (Fig. 1B and data not shown). Importantly, the Bub1DN protein was expressed in the thymus at much higher levels than the ∼120-kDa protein presumed to be endogenous Bub1, which was difficult to detect under standard conditions (Fig. 1C). Weak Bub1DN expression was also sometimes detected in spleen and lymph node extracts, consistent with previous reports of transgenes driven by the lck proximal promoter (Fig. 1D and E) (5, 35). To determine the specificity of transgene expression, several tissues from an animal of founder line 1 and a nontransgenic littermate were similarly examined (Fig. 1E). Bub1DN expression was detected only in the thymus and testes, while nonspecific bands were detected in the heart, liver, and muscle. Based on these results, founder lines 1 and 2 were chosen for further analyses. These animals are referred to here as transgenic LBDN (Tg-LBDN) animals.
FIG. 1.
Design and expression of the LBDN transgene in mice. (A) Schematic of the mouse Bub1 gene, the dominant-negative Bub1 fragment, and the LBDN transgene. lck Pr., lck proximal promoter; hGH, human growth hormone gene; sp/pA, splicing/polyadenylation signals. (B) Anti-Bub1 immunoblot (I.B.) showing Bub1DN expression in thymuses from four founder lines. Positions of 40- and 50-kDa protein markers are indicated. (C) Anti-Bub1 immunoblot comparing thymocyte expression levels of Bub1DN (lower arrow) and endogenous Bub1 (120 kDa; upper arrow). Lane 1, nontransgenic; lane 2, Tg-LBDN-1+. Molecular size marker positions are indicated on the left. (D) Anti-Myc tag immunoblot showing Bub1DN expression in thymus, spleen, and lymph node samples from animals of lines 1 and 2. Both lines show strong expression in the thymus, with weaker expression in the spleen and lymph node. Results for a nontransgenic control animal are shown on the right. (E) (Top) Anti-Bub1 immunoblot of thymus (Th), bone marrow (BM), heart (He), kidney (Kd), liver (Lv), lung (Lu), lymph node (LN), skeletal muscle (Mu), spleen (Sp), and testis (Te) lysates from a Tg-LBDN line 1 transgenic mouse (+) and a nontransgenic littermate (−). A lysate from Bub1DN-transfected 293 cells was used as a positive control (cont.). (Bottom) Anti-beta-actin immunoblot demonstrating equal protein loading.
Bub1DN effects on thymocyte development.
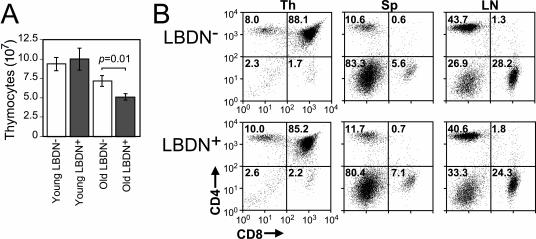
We analyzed transgenic animals to determine if Bub1DN expression induces overt thymic or T-cell phenotypes. Thymocyte counts from young animals showed no difference between transgenic animals and controls. However, 3- to 6-month-old transgenic animals displayed a modest reduction in total thymocytes (Fig. 2A). Flow cytometry analyses demonstrated that thymocyte subsets were not significantly different between transgenic and nontransgenic littermates at the ages of 2.5 (Fig. 2B) and 6.5 (data not shown) months. Similarly, no significant differences were observed in B- and T-cell percentages found in spleen and lymph node samples (Fig. 2B). These data indicate that overexpression of the Bub1DN fragment does not cause overt disruption of T-cell development but does cause modest reductions in thymocyte numbers as animals age.
FIG. 2.
The LBDN transgene causes reduced thymocyte numbers in aged animals but does not alter thymocyte and peripheral T-cell populations. (A) Mean thymocyte numbers from young (<3 months; n = 4 per genotype) and older (3 to 6.5 months; n = 13 Tg-LBDN+, 10 Tg-LBDN−) animals of LBDN line 1 and controls. Error bars, standard errors of the means. (B) Representative flow cytometry analyses of CD4 and CD8 expression in thymus, spleen, and lymph node samples from 2.5-month-old Tg-LBDN-positive and -negative animals. Percentages of cells are indicated in each quadrant.
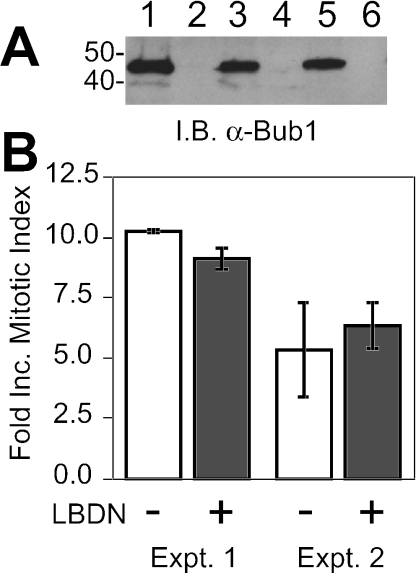
While the Bub1DN transgenic animals did not display dramatic T-cell phenotypes, it remained possible that spindle checkpoint impairment occurred in these animals. If so, transgenic thymocytes should display reduced mitotic accumulation upon treatment with the spindle inhibitor nocodazole. To test this hypothesis, thymocytes were cultured in vitro with IL-1α and IL-2, and mitotic indices were compared for mock-treated versus nocodazole-treated (8-h) samples. Importantly, IL-1α and IL-2 promoted robust proliferation while retaining Bub1DN expression (Fig. 3A). In two separate experiments, mitotic indices increased 5- to 10-fold in nocodazole-treated samples. Surprisingly, thymocytes from LBDN+ animals did not show any reduction in mitotic arrest relative to that in nontransgenic controls (Fig. 3B). These data indicate that the spindle checkpoint is intact in LBDN+ thymocytes, despite the observation that Bub1DN protein is highly expressed.
FIG. 3.
LBDN+ thymocytes have a normal spindle checkpoint response. (A) Anti-Bub1 immunoblot (I.B.) demonstrating Bub1DN expression in cultured thymocytes. Thymocytes from a Tg-LBDN+ (odd-numbered lanes) or a nontransgenic (even-numbered lanes) mouse were cultured in IL-1α plus IL-2 for 0 (lanes 1 and 2), 3 (lanes 3 and 4), or 5 (lanes 5 and 6) days. Lysates were probed with an anti-Bub1 antibody. (B) Spindle checkpoint responses of cultured thymocytes. Mitotic indices were determined for mock-treated or nocodazole-treated (8 h) thymocytes of the genotypes indicated. Fold increase (Inc.) represents treated/untreated mitotic index. Data are means ± standard errors of the means for two experiments. Two animals were used per genotype per experiment. The apparent difference in experiment 1 was not statistically significant (P = 0.11).
In performing the nocodazole challenge experiments, we noted that thymocytes from 6.5-month-old LBDN+ animals proliferated poorly relative to those from their nontransgenic counterparts (data not shown). Similar defects were not observed in thymocytes from young animals, suggesting a possible link to the reduced thymocyte numbers observed in these older mice. The basis of this proliferation defect is presently unclear, and it remains possible that the reduced proliferation is a result of chromosome segregation errors or other mitotic defects that are more pronounced as animals age. We attempted to address this question by analyzing aneuploidy rates of transgenic and nontransgenic thymocytes after 5 to 10 days of unperturbed culture. No gross differences were observed, although aneuploidy rates of both nontransgenic and transgenic cells differed widely among experiments, likely reflecting limitations associated with in vitro thymocyte culture (data not shown). Hence, while the results described above clearly indicate that the Bub1DN fragment fails to impair the spindle checkpoint response to nocodazole, it is possible that mitotic dynamics are impaired in ways not detectable by standard in vitro assays.
Bub1DN expression does not alter survival or tumorigenesis.
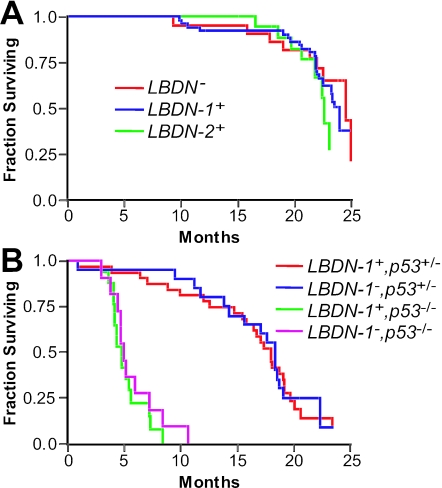
Aberrant chromosome segregation and aneuploidy have been proposed to contribute to tumorigenesis (20). According to this hypothesis, if the Bub1DN transgene increases chromosome segregation errors and aneuploidy in even a subset of thymocytes in vivo, such events would be expected to increase the risk of tumorigenesis. To test this proposal, survival analysis was performed using cohorts of Bub1DN transgenic animals from founder lines 1 and 2 and controls. Animals were sacrificed when obviously ill or when they reached the age of 22 to 24 months. Animals sacrificed at the study end point were censored in survival data if they did not display overt abnormalities or tumors. Overall survival of transgenic animals and controls was equivalent (Fig. 4A), and animals from all cohorts displayed similar incidences of age-related abnormalities such as adenomas and carcinomas of the liver and lung (data not shown). Because the LBDN transgene is expressed only in lymphocytes, the most likely tumor expected from transgene activity would be thymic or disseminated lymphoma. We noted one thymic lymphoma and one possible disseminated lymphoma in transgenic animals of Tg-LBDN line 1 (Tg-LBDN-1) (2/53; 4%), one possible disseminated lymphoma in an animal from line 2 (1/19; 5%), and no lymphomas among 22 control animals. However, these tumor frequency differences are not statistically significant (P = 0.44 for combined Tg-LBDN+ versus nontransgenic animals), and censoring survival data by the presence or absence of lymphomas also failed to show a statistically significant survival difference (data not shown). Together, these data suggest that thymic overexpression of the Bub1DN fragment has no significant effect on survival or tumorigenesis.
FIG. 4.
The LBDN transgene has no effect on survival. (A) Kaplan-Meier analysis of overall survival in mice from Tg-LBDN line 1 (n = 54), Tg-LBDN line 2 (n = 19), and nontransgenic littermate controls (n = 22). (B) Kaplan-Meier analysis of overall survival among p53-heterozygous or homozygous null mice with or without the LBDN (line 1) transgene (Tg-LBDN-1+ p53+/− [n = 32], Tg-LBDN-1− p53+/− [n = 20], Tg-LBDN-1+ p53−/− [n = 17], and Tg-LBDN-1− p53−/− [n = 11]).
p53 checkpoint inactivation does not reveal a Bub1DN effect.
Because the cellular consequences of spindle checkpoint impairment and chromosome missegregation are not well defined, we considered the possibility that p53-dependent cell cycle checkpoints might override tumorigenic effects of Bub1DN expression. That is, cells with chromosomal aberrations may be removed from the system by p53-dependent arrest or apoptosis. If so, the introduction of p53 mutations in Tg-LBDN transgenic animals would be expected to cooperate with Bub1DN expression in tumorigenesis. Additionally, if Bub1DN expression increases the rate of chromosome missegregation in vivo, Tg-LBDN mice on a p53 heterozygous background should be more likely to experience loss of the wild-type p53 allele in thymocytes or T cells. Loss of the wild-type p53 allele has been proposed to be a rate-limiting step in irradiation-induced tumorigenesis of p53 heterozygous mice (22). To test these hypotheses, survival analysis was performed on cohorts of Tg-LBDN-1 transgenic animals and nontransgenic controls generated on p53 heterozygous and homozygous null backgrounds. Contrary to the hypothesis, mice harboring the LBDN transgene on a p53 heterozygous background did not display any overall survival difference from Tg-LBDN-negative animals (Fig. 4B) (P = 0.89). Censoring survival data based on the presence or absence of lymphomas revealed a trend toward shorter lymphoma-free survival in Tg-LBDN+; p53+/− animals, but this trend also failed to reach statistical significance (P = 0.18 by the log rank test; P = 0.06 by the Wilcoxon test) (survival curve not shown). While a higher proportion of Tg-LBDN+; p53+/− animals than of Tg-LBDN−; p53+/− animals developed lymphoma (25% versus 10%, respectively), this difference also failed to reach statistical significance (P = 0.17). Similarly, the presence or absence of the LBDN transgene had no significant effect on survival or tumor spectrum in p53-null animals. Approximately 82% of Tg-LBDN+; p53−/− and 73% of Tg-LBDN−; p53−/− animals developed thymic lymphomas, and tumor latencies were very similar (Fig. 3B) (P = 0.56). Together, these data do not support a role for p53-dependent suppression of tumorigenesis in Tg-LBDN mice, nor do they support the hypothesis that the Bub1DN fragment contributes to thymic tumorigenesis in vivo.
DISCUSSION
Since the discovery of Bub1 mutations in colorectal tumor cell lines several years ago (8), considerable interest has focused on the role of spindle checkpoint impairment in human cancer. Several subsequent studies found that mutations in known spindle checkpoint genes are rare in a variety of tumors or tumor cell lines, although checkpoint impairment by epigenetic mechanisms was observed in some cases (7, 19, 25, 30, 32, 34, 36, 41, 42, 44). These studies cumulatively call into question the role of spindle checkpoint impairment in cancer.
We examined the consequences of overexpressing a dominant-negative Bub1 fragment in mouse thymocytes in vivo to determine if spindle checkpoint impairment contributes to lymphomagenesis. Surprisingly, we did not detect any evidence that the BubDN protein impaired the spindle checkpoint, as transgenic thymocytes cultured in vitro arrested normally in response to nocodazole treatment. The only phenotypes observed in Tg-LBDN mice were (i) a modest reduction in thymocyte numbers with increased age and (ii) impaired in vitro proliferation of thymocytes from aged animals. Tg-LBDN+ mice did not show significantly increased tumor susceptibility. Heterozygous or homozygous mutation of the p53 gene also failed to uncover significant tumorigenic effects of the transgene that might have been suppressed by p53-dependent checkpoints. Together, our data indicate that thymic overexpression of Bub1DN has no significant effect on thymic development or tumorigenesis.
The Bub1DN fragment used in the present work has been used extensively in cultured cells, inducing significant spindle checkpoint impairment (2, 16, 26, 38). For example, HeLa cells expressing Bub1DN displayed accelerated mitotic passage under normal growth conditions and decreased mitotic arrest and apoptosis after nocodazole treatment (38). Bub1DN expression reversed growth arrest and transformed Brca2-deficient mouse embryonic fibroblasts, rendering them resistant to nocodazole-induced mitotic arrest (26). There are several possible explanations for why expression of Bub1DN did not elicit the same spindle checkpoint effects in thymocytes. First, it is possible that cell lines used in previous in vitro studies have accumulated changes in additional factors, “relaxing” the mitotic checkpoints and making them abnormally sensitive to Bub1 inhibition. Thus, the dominant-negative Bub1 fragment may not be sufficient for checkpoint deregulation in primary cells. However, the same Bub1DN protein was recently shown to cause accelerated passage through meiosis I and to disrupt nocodazole-induced arrest in meiosis II when expressed in mouse oocytes (39). These results suggest that Bub1DN can have similar effects in primary cells cultured in vitro, although effects on meiosis and mitosis could be distinct. Thymocytes could also be resistant to the dominant-negative effects via differences in spindle checkpoint regulation or in expression levels of spindle checkpoint components. The lack of effect could also be due to technical challenges associated with the animal studies described here. For example, it is possible that Bub1DN expression levels achieved in Tg-LBDN mice are insufficient to completely override endogenous Bub1 function in thymocytes. However, we have shown that the Bub1DN fragment is expressed at significantly higher levels than endogenous Bub1. It has been noted that this fragment displays dominant-negative activity in cell culture only when the cells are challenged with spindle inhibitors (2). Nevertheless, that observation differs from other reports in which the same fragment caused shortened mitosis in untreated HeLa cells (16, 38). Such differences could reflect distinct spindle checkpoint regulation in distinct cell types, different levels of Bub1DN expression, or other unknown, variables between studies. It is noteworthy that an amino-terminal fragment of yeast Bub1 supports partial spindle checkpoint activity in vivo, independently of the C-terminal kinase domain. Nevertheless, overexpression of full-length wild-type or N-terminal Bub1 fragments in yeast caused chromosome segregation errors, supporting a dominant-negative effect due to overexpression (43). Thus, while the evidence supporting dominant-negative activities of Bub1 N-terminal fragments is compelling in yeast and cell culture systems, the behavior of these fragments in distinct cell types in vivo will need to be carefully evaluated to determine if dominant activities are induced.
The equivalent survival of p53 heterozygous mice with or without the LBDN transgene supports the conclusion that the Bub1DN fragment does not induce significant genomic instability in vivo. p53 heterozygous mice have a shortened life span and high rates of thymic lymphomagenesis in response to irradiation or chemical mutagenesis, both of which elicit increased genomic instability (22). The majority of irradiation-induced tumors in p53 heterozygous mice display loss of the wild-type p53 allele (p53LOH), suggesting that p53 loss plays a key role in tumor development. If thymic expression of the Bub1DN protein in Tg-LBDN mice induced significant genomic instability, the frequency of p53LOH and subsequent tumorigenesis should be greater in Tg-LBDN+ p53+/− animals than in controls. The absence of such cooperativity in our experimental cohorts supports the conclusion that the Bub1DN protein does not elicit significant genomic instability in vivo.
A potential caveat in interpreting tumorigenesis data in this study concerns the use of the lck proximal promoter for thymic expression of Bub1DN. It is possible that Tg-LBDN+ mice do not develop tumors because the transgene is not expressed in cell types relevant to the transformation process. The originating cell type of most cancers is controversial, but significant evidence suggests a stem cell origin for many cancers (31). It is thus possible that spindle checkpoint impairment and/or chromosome missegregation in hematopoietic stem or progenitor cells would initiate tumorigenesis, while similar effects in more-differentiated thymocytes would not. Even within the thymus, lesions may need to occur in specific thymocyte subsets, most likely the proliferative subsets, to drive tumorigenesis. Thymocytes pass through several developmental stages marked by expression of specific cell surface markers and by regulated proliferation (15, 17). Early thymocyte subsets that lack CD4 and CD8 expression (double-negative [DN] cells) represent only 1 to 2 percent of total thymocytes but account for a significant portion of thymocyte proliferation. DN thymocytes are divided into four subtypes (DN1 to DN4) based on expression of the CD44, CD25, and CD117 surface markers. The DN3 (CD44− CD25+ CD117−) and DN4 (CD44− CD25− CD117−) subsets make up approximately 90% of DN thymocytes and display high proliferative activity that gives rise to the large numbers of CD4+ CD8+ (double-positive [DP]) thymocytes (15). Thus, LBDN transgene expression in the proliferative DN3 and DN4 thymocyte subsets would provide the greatest opportunity for chromosome missegregation events that may facilitate tumor formation. Our data suggest that the LBDN transgene is expressed in DN thymocytes, since Bub1DN expression was maintained in cells cultured in IL-1α and IL-2. These conditions produced a predominant DN population after 1 week of culture (data not shown), consistent with published reports that IL-1α and IL-2 stimulate DN thymocyte proliferation (37). Our results are also consistent with a study of four transgenic mouse lines in which the same lck promoter and RNA processing signals used here drive green fluorescent protein (GFP) expression. All four lines of lck-GFP transgenic mice initiated GFP expression by the DN3 stage, with some lines expressing GFP as early as the DN1 stage (5). This expression pattern is also consistent with the mRNA expression of the endogenous lck gene, which is detectable in all DN subsets (5). We also note that several transgenes driven by the lck proximal promoter do elicit tumors in transgenic animals, indicating that the cells targeted by this promoter can be transformed in vivo (9, 12, 23, 28, 40). Thus, while we cannot rule out the possibility that Bub1DN would need to be expressed in stem cells to elicit tumorigenesis, the simplest explanation for our data is that the Bub1DN protein does not display sufficient dominant-negative activity to impair the spindle checkpoint in thymocytes.
The decreases in proliferation and overall thymocyte numbers in aged LBDN+ animals are puzzling given the lack of any apparent spindle checkpoint phenotype. Flow cytometry profiles of stimulated cells showed lower percentages of blasts as evidenced by forward and side scatter properties, suggesting that transgenic cells may be defective in responding to stimulation and/or cell cycle entry (data not shown). It is unclear how the LBDN transgene might cause such a defect and why it would be observed only in older mice. Additional analyses will be needed to understand the basis of this modest phenotype. Nevertheless, our data strongly indicate that the LBDN transgenic mice are not a suitable model for spindle checkpoint disruption in thymocytes.
Thus, additional mouse models will be required in order to definitively address the roles of bub1 mutation, spindle checkpoint impairment, and chromosome instability in tumorigenesis. The caveats involved in interpreting dominant-negative data such as those described here suggest the need for care in developing such models. While mice heterozygous for Mad2, BubR1, Rae1, and Bub3 display some increased propensity for tumorigenesis (3, 13, 29), it remains unclear whether the observed tissue specificity reflects distinct spindle checkpoint regulation or special sensitivity in those tissues. A conditional knockout strategy represents the best initial strategy to define the role for the spindle checkpoint in specific cell types in vivo. The availability of transgenic mice expressing the cre recombinase from a variety of promoters will allow tests in specific cell types in vivo. However, because the spindle checkpoint may play an essential role in normal cell division, it is possible that complete ablation will also be incompatible with tumorigenesis. In line with this hypothesis, a recently described hypomorphic mutation in the BubR1 gene caused aneuploidy and premature aging phenotypes without increasing tumorigenesis (4), and reduction of Mad2 and BubR1 expression by RNA interference in human cancer cell lines was lethal (24). It is noteworthy that spindle checkpoint mutations identified in human tumors to date are all heterozygous mutations suggested to confer dominant-negative activities (8, 26, 32). Such dominant mutations may confer sufficient checkpoint impairment for tumorigenesis while allowing enough residual activity for cell survival. Thus, conditional knock-in models of tumor-derived spindle checkpoint mutants may ultimately prove to be the best way to fully investigate the role of spindle checkpoint impairment in tumorigenesis in various tissues in vivo.
In conclusion, our results indicate that thymic overexpression of an amino-terminal Bub1 fragment does not disrupt the spindle checkpoint or promote tumorigenesis in mouse thymocytes, despite evidence from yeast and cell culture systems that the Bub1 fragment used in this study induces checkpoint impairment. This suggests that distinct cell cycle control mechanisms or redundancy may limit the effects of some dominant inhibitors in vivo, suggesting the need for caution in the use of such reagents. Our data also suggest that chromosome missegregation phenotypes observed in cell culture, such as those elicited by this reagent, may not accurately predict in vivo relevance.
Acknowledgments
Financial support for this work came from The Leukemia and Lymphoma Society of America (Fellow Award 5408-02 to D.O.C.) and NIH/National Cancer Institute grant RO1CA065773 (to T.V.D.).
We thank the University of North Carolina (UNC) animal models core facility, Hua Wu, and Lucy Lu for help with production of transgenic mice; the UNC flow cytometry facility for assistance with flow cytometry; Virginia Godfrey for assistance with pathology; Lisa Livanos of the UNC chromosome imaging core facility for help with thymocyte karyotype analysis; Lishan Su for the p1017 vector; the UNC histopathology core facility; and the UNC division of lab animal medicine.
REFERENCES
- 1.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 2.Anand, S., S. Penrhyn-Lowe, and A. R. Venkitaraman. 2003. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell 3:51-62. [DOI] [PubMed] [Google Scholar]
- 3.Babu, J. R., K. B. Jeganathan, D. J. Baker, X. Wu, N. Kang-Decker, and J. M. van Deursen. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, D. J., K. B. Jeganathan, J. D. Cameron, M. Thompson, S. Juneja, A. Kopecka, R. Kumar, R. B. Jenkins, P. C. de Groen, P. Roche, and J. M. van Deursen. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744-749. [DOI] [PubMed] [Google Scholar]
- 5.Buckland, J., D. J. Pennington, L. Bruno, and M. J. Owen. 2000. Co-ordination of the expression of the protein tyrosine kinase p56(lck) with the pre-T cell receptor during thymocyte development. Eur. J. Immunol. 30:8-18. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D. J. 2000. Complexity in the spindle checkpoint. Curr. Opin. Genet. Dev. 10:26-31. [DOI] [PubMed] [Google Scholar]
- 7.Cahill, D. P., L. T. da Costa, E. B. Carson-Walter, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 1999. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics 58:181-187. [DOI] [PubMed] [Google Scholar]
- 8.Cahill, D. P., C. Lengauer, J. Yu, G. J. Riggins, J. K. Willson, S. D. Markowitz, K. W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392:300-303. [DOI] [PubMed] [Google Scholar]
- 9.Canela, A., J. Martin-Caballero, J. M. Flores, and M. A. Blasco. 2004. Constitutive expression of tert in thymocytes leads to increased incidence and dissemination of T-cell lymphoma in Lck-Tert mice. Mol. Cell. Biol. 24:4275-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaffin, K. E., C. R. Beals, T. M. Wilkie, K. A. Forbush, M. I. Simon, and R. M. Perlmutter. 1990. Dissection of thymocyte signaling pathways by in vivo expression of pertussis toxin ADP-ribosyltransferase. EMBO J. 9:3821-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 12.Cleverley, S. C., P. S. Costello, S. W. Henning, and D. A. Cantrell. 2000. Loss of Rho function in the thymus is accompanied by the development of thymic lymphoma. Oncogene 19:13-20. [DOI] [PubMed] [Google Scholar]
- 13.Dai, W., Q. Wang, T. Liu, M. Swamy, Y. Fang, S. Xie, R. Mahmood, Y. M. Yang, M. Xu, and C. V. Rao. 2004. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 64:440-445. [DOI] [PubMed] [Google Scholar]
- 14.Doherty, S. C., S. R. McKeown, V. McKelvey-Martin, C. S. Downes, A. Atala, J. J. Yoo, D. A. Simpson, and W. K. Kaufmann. 2003. Cell cycle checkpoint function in bladder cancer. J. Natl. Cancer Inst. 95:1859-1868. [DOI] [PubMed] [Google Scholar]
- 15.Fehling, H. J., and H. von Boehmer. 1997. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 9:263-275. [DOI] [PubMed] [Google Scholar]
- 16.Geley, S., E. Kramer, C. Gieffers, J. Gannon, J. M. Peters, and T. Hunt. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153:137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haks, M. C., M. A. Oosterwegel, B. Blom, H. M. Spits, and A. M. Kruisbeek. 1999. Cell-fate decisions in early T cell development: regulation by cytokine receptors and the pre-TCR. Semin. Immunol. 11:23-37. [DOI] [PubMed] [Google Scholar]
- 18.Hanks, S., K. Coleman, S. Reid, A. Plaja, H. Firth, D. Fitzpatrick, A. Kidd, K. Mehes, R. Nash, N. Robin, N. Shannon, J. Tolmie, J. Swansbury, A. Irrthum, J. Douglas, and N. Rahman. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 36:1159-1161. [DOI] [PubMed] [Google Scholar]
- 19.Haruki, N., H. Saito, T. Harano, S. Nomoto, T. Takahashi, H. Osada, Y. Fujii, and T. Takahashi. 2001. Molecular analysis of the mitotic checkpoint genes BUB1, BUBR1 and BUB3 in human lung cancers. Cancer Lett. 162:201-205. [DOI] [PubMed] [Google Scholar]
- 20.Jallepalli, P. V., and C. Lengauer. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer 1:109-117. [DOI] [PubMed] [Google Scholar]
- 21.Jin, D. Y., F. Spencer, and K. T. Jeang. 1998. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell 93:81-91. [DOI] [PubMed] [Google Scholar]
- 22.Kemp, C. J., T. Wheldon, and A. Balmain. 1994. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat. Genet. 8:66-69. [DOI] [PubMed] [Google Scholar]
- 23.Knudson, C. M., G. M. Johnson, Y. Lin, and S. J. Korsmeyer. 2001. Bax accelerates tumorigenesis in p53-deficient mice. Cancer Res. 61:659-665. [PubMed] [Google Scholar]
- 24.Kops, G. J., D. R. Foltz, and D. W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA 101:8699-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langerod, A., M. Stromberg, K. Chin, V. N. Kristensen, and A. L. Borresen-Dale. 2003. BUB1 infrequently mutated in human breast carcinomas. Hum. Mutat. 22:420. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H., A. H. Trainer, L. S. Friedman, F. C. Thistlethwaite, M. J. Evans, B. A. Ponder, and A. R. Venkitaraman. 1999. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol. Cell 4:1-10. [DOI] [PubMed] [Google Scholar]
- 27.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 28.Mende, I., S. Malstrom, P. N. Tsichlis, P. K. Vogt, and M. Aoki. 2001. Oncogenic transformation induced by membrane-targeted Akt2 and Akt3. Oncogene 20:4419-4423. [DOI] [PubMed] [Google Scholar]
- 29.Michel, L. S., V. Liberal, A. Chatterjee, R. Kirchwegger, B. Pasche, W. Gerald, M. Dobles, P. K. Sorger, V. V. Murty, and R. Benezra. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409:355-359. [DOI] [PubMed] [Google Scholar]
- 30.Myrie, K. A., M. J. Percy, J. N. Azim, C. K. Neeley, and E. M. Petty. 2000. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett. 152:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Pardal, R., M. F. Clarke, and S. J. Morrison. 2003. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer 3:895-902. [DOI] [PubMed] [Google Scholar]
- 32.Ru, H. Y., R. L. Chen, W. C. Lu, and J. H. Chen. 2002. hBUB1 defects in leukemia and lymphoma cells. Oncogene 21:4673-4679. [DOI] [PubMed] [Google Scholar]
- 33.Sen, S. 2000. Aneuploidy and cancer. Curr. Opin. Oncol. 12:82-88. [DOI] [PubMed] [Google Scholar]
- 34.Shichiri, M., K. Yoshinaga, H. Hisatomi, K. Sugihara, and Y. Hirata. 2002. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res. 62:13-17. [PubMed] [Google Scholar]
- 35.Shimizu, C., H. Kawamoto, M. Yamashita, M. Kimura, E. Kondou, Y. Kaneko, S. Okada, T. Tokuhisa, M. Yokoyama, M. Taniguchi, Y. Katsura, and T. Nakayama. 2001. Progression of T cell lineage restriction in the earliest subpopulation of murine adult thymus visualized by the expression of lck proximal promoter activity. Int. Immunol. 13:105-117. [DOI] [PubMed] [Google Scholar]
- 36.Shin, H. J., K. H. Baek, A. H. Jeon, M. T. Park, S. J. Lee, C. M. Kang, H. S. Lee, S. H. Yoo, D. H. Chung, Y. C. Sung, F. McKeon, and C. W. Lee. 2003. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 4:483-497. [DOI] [PubMed] [Google Scholar]
- 37.Suda, T., R. Murray, C. Guidos, and A. Zlotnik. 1990. Growth-promoting activity of IL-1α, IL-6, and tumor necrosis factor-alpha in combination with IL-2, IL-4, or IL-7 on murine thymocytes. Differential effects on CD4/CD8 subsets and on CD3+/CD3− double-negative thymocytes. J. Immunol. 144:3039-3045. [PubMed] [Google Scholar]
- 38.Taylor, S. S., and F. McKeon. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89:727-735. [DOI] [PubMed] [Google Scholar]
- 39.Tsurumi, C., S. Hoffmann, S. Geley, R. Graeser, and Z. Polanski. 2004. The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J. Cell Biol. 167:1037-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virgilio, L., C. Lazzeri, R. Bichi, K. Nibu, M. G. Narducci, G. Russo, J. L. Rothstein, and C. M. Croce. 1998. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc. Natl. Acad. Sci. USA 95:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., D. Y. Jin, R. W. Ng, H. Feng, Y. C. Wong, A. L. Cheung, and S. W. Tsao. 2002. Significance of MAD2 expression to mitotic checkpoint control in ovarian cancer cells. Cancer Res. 62:1662-1668. [PubMed] [Google Scholar]
- 42.Wang, X., D. Y. Jin, Y. C. Wong, A. L. Cheung, A. C. Chun, A. K. Lo, Y. Liu, and S. W. Tsao. 2000. Correlation of defective mitotic checkpoint with aberrantly reduced expression of MAD2 protein in nasopharyngeal carcinoma cells. Carcinogenesis 21:2293-2297. [DOI] [PubMed] [Google Scholar]
- 43.Warren, C. D., D. M. Brady, R. C. Johnston, J. S. Hanna, K. G. Hardwick, and F. A. Spencer. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13:3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi, K., K. Okami, K. Hibi, S. L. Wehage, J. Jen, and D. Sidransky. 1999. Mutation analysis of hBUB1 in aneuploid HNSCC and lung cancer cell lines. Cancer Lett. 139:183-187. [DOI] [PubMed] [Google Scholar]