Abstract
The ability of p53 to promote apoptosis and cell cycle arrest is believed to be important for its tumor suppression function. Besides activating the expression of cell cycle arrest and proapoptotic genes, p53 also represses a number of genes. Previous studies have shown an association between p53 activation and down-regulation of c-myc expression. However, the mechanism and physiological significance of p53-mediated c-myc repression remain unclear. Here, we show that c-myc is repressed in a p53-dependent manner in various mouse and human cell lines and mouse tissues. Furthermore, c-myc repression is not dependent on the expression of p21WAF1. Abrogating the repression of c-myc by ectopic c-myc expression interferes with the ability of p53 to induce G1 cell cycle arrest and differentiation but enhances the ability of p53 to promote apoptosis. We propose that p53-dependent cell cycle arrest is dependent not only on the transactivation of cell cycle arrest genes but also on the transrepression of c-myc. Chromatin immunoprecipitation assays indicate that p53 is bound to the c-myc promoter in vivo. We report that trichostatin A, an inhibitor of histone deacetylases, abrogates the ability of p53 to repress c-myc transcription. We also show that p53-mediated transcriptional repression of c-myc is accompanied by a decrease in the level of acetylated histone H4 at the c-myc promoter and by recruitment of the corepressor mSin3a. These data suggest that p53 represses c-myc transcription through a mechanism that involves histone deacetylation.
p53 is a sequence-specific DNA binding transcription factor that is expressed as an unstable protein. In response to abnormal proliferative signals and many stress signals, including DNA damage, p53 is stabilized and posttranslationally modified. Once activated, p53 regulates the expression of a number of target genes that collectively contribute to p53-dependent cellular responses. p53 can induce cells to undergo a transient arrest in G1 to allow time for repair of damaged DNA that could otherwise lead to mutations and genomic instability. Activated p53 can also eliminate cells through mechanisms that involve prolonged arrest in G1 or apoptosis. The elimination of damaged, stressed, or abnormally proliferating cells by p53 is considered to be the principal means by which p53 mediates tumor suppression.
Aside from its transcriptional activation function, p53 can also act as a transcriptional repressor. There is accumulating evidence to show that the repression of certain genes by p53 may be important for its ability to carry out its functions. For instance, ectopic expression of various p53-repressed genes, including Bcl-2 (2, 48), survivin (13, 29), MAP4 (32), PIK3CA (40), and p202 (4), was shown to inhibit p53-dependent apoptosis. The mechanism of transrepression remains a controversial area of p53 biology and may or may not be dependent on the site-specific DNA binding activity of p53. Proposed mechanisms include interference with the function of transcriptional activators, interference with the basal transcriptional machinery, recruitment of chromatin modifying factors to reduce promoter accessibility, and recruitment of transcriptional corepressors (12). In addition, recent studies have suggested that p53-dependent transcriptional repression of certain genes occurs indirectly as a consequence of p53-dependent transactivation of p21WAF1 (10, 23, 38). Transcripts not normally expressed in G1 or G2 will appear to be repressed as a result of p21-dependent cell cycle arrest. Alternatively, the Rb-E2F complexes that fail to dissociate when Rb remains hypophosphorylated as a result of cyclin-dependent kinase inhibition by p21 function as transcriptional repressors of genes carrying E2F-responsive sites.
It has been known for some time that p53 activation is associated with c-myc down-regulation in some cells. In rat embryo fibroblasts (34) and mouse myeloid leukemia cells (22) that carry a temperature-sensitive p53 protein, temperature shift to 32.5°C activates wild-type p53 function and leads to a decrease in the level of c-myc mRNA. It is unclear, however, if p53 directly represses c-myc and whether this repression is important for p53-mediated cellular responses.
The c-myc oncoprotein is a transcription factor that promotes cell growth and proliferation, as well as apoptosis under certain conditions. Deregulated c-myc can induce aberrant proliferation, loss of terminal differentiation, abrogation of DNA damage-induced cell cycle arrest, genomic instability, and oncogenesis (33). As p53 and c-myc are involved in many of the same cellular processes, it is perhaps not surprising that they affect similar targets and modulate each other's activities. For example, p53 activates the expression of p21 (8) and gadd45 (19), while c-myc represses these genes (26, 30), consistent with the observation that c-myc can interfere with p53-induced cell cycle arrest (11, 46). The opposing effects of c-myc and p53 at the p21 locus were shown to have a profound physiological effect in HCT116 colon carcinoma cells by switching the outcome of a p53 response from cell cycle arrest to apoptosis (37). Hence, the functional interactions between p53 and c-myc may have broad implications in tumorigenesis.
We have examined the transcriptional repression of c-myc by p53 and its physiological significance to p53 function. We demonstrate that c-myc is repressed by p53 in a number of mouse and human cell lines and mouse tissues and that this repression occurs independently of p21 transactivation. We provide evidence that p53 binds to the c-myc promoter in vivo and represses the promoter through a mechanism that involves histone deacetylation. To evaluate the physiological significance of c-myc repression by p53, we show that ectopic c-myc expression interferes with the ability of p53 to induce G1 cell cycle arrest and cellular differentiation. We propose that p53 represses the expression of c-myc to promote these processes.
MATERIALS AND METHODS
Cell culture and retroviral infection.
DP16.1, BJ-T, AML3, and K562 cells were maintained in α-minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum. Baf-3 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 0.1 ng/ml interleukin-3 (IL-3), and 0.0004% β-mercaptoethanol. IL-3 was removed 2 hours before irradiation, and the unirradiated control was also maintained in the absence of IL-3 for the same period of time. For retroviral infections, the phoenix Eco packaging cells were transfected using calcium phosphate with the various retroviral constructs. Viral supernatants were collected 48 h later and incubated with DP16.1/p53ts cells along with 8 μg/ml Polybrene for 16 h. Cells were evaluated 24 to 48 h postinfection.
Northern blot analysis.
Total RNA was isolated from cells and tissues using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For each sample, 10 μg of RNA was run on a denaturing agarose gel and transferred to a positively charged nylon membrane. The blots were hybridized with 32P-radiolabeled c-myc, p21, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes, followed by standard washes. Phosphorimaging analysis was carried out, and RNA transcript levels were quantitated using ImageQuant software (Molecular Dynamics).
Cell cycle analysis.
DNA content was analyzed in live cells to facilitate the identification of green fluorescent protein-positive (GFP+; infected) and GFP− (uninfected) cells. DP16.1/p53ts cells were incubated with Hoechst 33342 to a final concentration of 3 μg/ml in α-minimal essential medium for 45 min. Cells were then washed and resuspended in phosphate-buffered saline. Cell cycle distribution was examined using the Becton Dickinson LSR II flow cytometer and FACSDiva software. The relative proportion of cells in each phase of the cell cycle was determined using the automated ModFit program (Verity Software House, Inc.).
RNAi.
The DNA sequence used for c-myc RNA interference (RNAi) was described in reference 15. The hairpin sequence, 5′-TGCTGTTGACAGTGAGCGCAGAACATCATCATCCAGGACTTAGTGAAGCCACAGATGTAAGTCCTGGATGATGATGTTCTTTGCCTACTGCCTCGGA-3′ (underlining indicates the sense and antisense sequences), was cloned into a murine stem cell virus-U6 short hairpin RNA (shRNA) retroviral vector (gift from Scott Lowe).
Differentiation assay.
Hemoglobin expression was visualized by staining cells with 2,7-diaminofluorene (DAF; Sigma) as described previously (16). Briefly, a working solution was made by mixing 1 ml of DAF stock solution (10 mg/ml in 90% acetic acid), 100 μl of 30% hydrogen peroxide, and 10 ml of 0.2 M Tris, pH 7.0. Cells were added to an equal volume of the DAF working solution, and the mixture was incubated at room temperature for 5 min. A 10-μl aliquot of the stained cells was then counted with a hemocytometer, and the proportion of hemoglobin-expressing cells was determined (number of blue-stained cells over total number of cells).
Apoptosis assay.
Apoptosis was assessed using Annexin V-phycoerythrin and 7-amino-actinomycin D (7-AAD) staining according to the manufacturer's instructions (BD PharMingen). The proportion of apoptotic cells (Annexin V positive, 7-AAD negative) was determined using a FACScan flow cytometer and CellQuest software (Becton Dickinson).
Chromatin immunoprecipitation.
The chromatin immunoprecipitation procedure was adapted from the methods of Eberhardy et al. (5). Briefly, 1.5 × 107 cells were used per immunoprecipitation reaction mixture. Cells were cross-linked in 1% formaldehyde for 10 min, followed by the addition of glycine to a final concentration of 0.125 M to stop the cross-linking reaction. Cells were washed twice in phosphate-buffered saline and lysed in cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 8.0, 85 mM KCl, 0.5% NP-40, Complete protease inhibitor cocktail (Roche)] on ice for 10 min. The nuclei were pelleted at 5,000 rpm and lysed in nuclei lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], Complete protease inhibitor cocktail [Roche]) on ice for 10 min. The chromatin was sonicated to an average length of 500 bp. The samples were precleared by incubating with blocked Staph A cells (Boehringer Mannheim; Staph A cells were blocked with 1 mg/ml herring sperm DNA and 1 mg/ml bovine serum albumin) for 15 min. The Staph A cells were then pelleted and discarded, and the protein-chromatin complexes were incubated with no antibody, anti-p53 antibody (2 μg; FL393; Santa Cruz), anti-acetylated histone H4 antibody (3 μl; catalog number 06-866; Upstate Biotechnology), anti-mSin3a antibody (2 μg; AK-11; Santa Cruz), or rabbit immunoglobulin G (IgG; 2 μg) at 4°C overnight. Each reaction mixture was then incubated with 10 μl of blocked Staph A cells for 15 min at room temperature. The Staph A cells were pelleted, and the supernatant from the no-antibody sample was used as total input chromatin (input). The Staph A pellets were washed twice in dialysis buffer (2 mM EDTA, 50 mM Tris, pH 8.0, 0.2% Sarkosyl) and four times in IP wash buffer (100 mM Tris, pH 9.0, 500 mM LiCl, 1% NP-40, 1% deoxycholic acid). The protein-chromatin complexes were eluted from the Staph A cells twice in IP elution buffer (50 mM NaHCO3, 1% SDS), followed by reverse cross-linking in 0.3 M NaCl along with 1 μg of RNase A at 67°C for 5 h. The reactions were precipitated with 2.5 volumes of ethanol at −20°C overnight. The reaction mixtures were then centrifuged at 14,000 rpm for 20 min, and the pellets were air dried and resuspended in 100 μl Tris-EDTA-proteinase K buffer (final reaction concentrations, 10 mM Tris, pH 7.5, 5 mM EDTA, 0.25% SDS, and proteinase K [1 U]) and incubated at 45°C for 2 h. Subsequently, the samples were purified by phenol-chloroform extraction. NaCl (final concentration of 0.14 M), glycogen (20 μg), and 2.5 volumes of ethanol were then added, and the samples were allowed to precipitate overnight at −20°C. The following day, samples were centrifuged at 14,000 rpm for 20 min and the pellets were air dried and resuspended in 60 μl of water. Two microliters of the purified DNA was used for each PCR. The primer sequences used for the various PCR amplicons were as follows: for the mouse c-myc gene, −1706 to −1534 (5′-TGTAGGATAAGCAAATCCCGAGG-3′ and 5′-TCCTGAATACTACGCTGTGCATTC-3′), −512 to −259 (5′-ATACGCAGGGCAAGAACACAG-3′, and 5′-TTTTTTCCTCCTCTCGCTTCC-3′), +48 to +236 (5′-AGTGAGAAGTGTCTGCCCGC-3′ and 5′-TTGGAAGAGCCGTGTGTGC-3′), +748 to +1079 (5′-AGTCAACGAATCGGTCACATCC-3′ and 5′-TCCTGAGGTCTTTGGAGAAGGG-3′); for the mouse p21 gene, −2927 to −2595 (5′-CGGAGACCAGCAGCAAAATCG-3′ and 5′-TGACACATACACACCCCAGGCAC-3′).
RESULTS AND DISCUSSION
c-myc is repressed in a p53-dependent manner in mouse and human cells and mouse tissues.
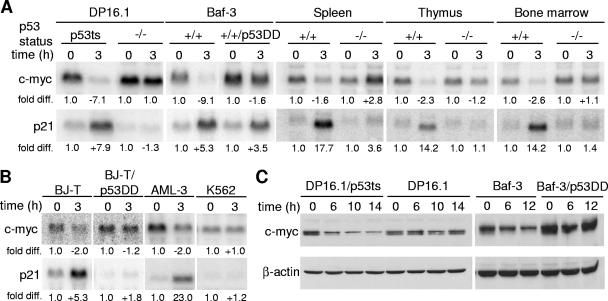
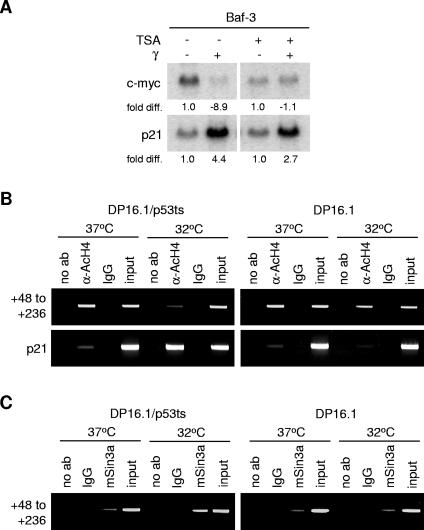
To determine if the repression of c-myc occurs in a p53-dependent manner, Northern blot analyses were performed to examine c-myc transcript levels in cells and tissues that differ with respect to p53 gene status. DP16.1/p53ts cells were derived by stable expression of a temperature-sensitive p53 allele (p53Val-135) in the p53-null mouse erythroleukemia cell line DP16.1 (16). After shifting the cells (DP16.1/p53ts) to the permissive temperature at 32°C to activate wild-type p53 function, c-myc transcript levels decreased within 3 h (Fig. 1A). This reduction occurred in a p53-dependent manner, as temperature shift had no effect on the level of c-myc mRNA in the parental DP16.1 cells. To ensure that p53-dependent c-myc repression was not restricted to DP16.1/p53ts cells, we used Baf-3 cells, a mouse pro-B-cell line with endogenous wild-type p53 that can be activated by γ-irradiation. c-myc was repressed strongly after γ-irradiation in Baf-3 cells, but not in Baf-3/p53DD cells, which stably express a dominant-negative p53 protein that interferes with the function of the endogenous wild-type p53 protein (39). c-myc transcript levels were also repressed in the spleen, thymus, and bone marrow of γ-irradiated p53+/+ mice but not p53−/− mice. To determine if p53-dependent c-myc repression also occurs in human cells, we γ-irradiated BJ-T cells (normal human BJ fibroblasts immortalized by ectopic expression of the telomerase catalytic subunit [47]). c-myc mRNA levels decreased in irradiated BJ-T cells, but not in BJ-T/p53DD cells, which carry the dominant-negative p53 allele (Fig. 1B). In addition, c-myc mRNA levels decreased in γ-irradiated AML-3 cells (a human acute myeloid leukemia cell line that expresses wild-type p53 [43]) but not in γ-irradiated p53-null K562 cells. The p53-dependent repression of c-myc transcripts is also reflected in a reduction in c-myc protein levels. Western blot analyses revealed a decrease in myc protein levels after temperature shift in DP16.1/p53ts cells and in Baf-3 cells after γ-irradiation but not in DP16.1 and Baf-3/p53DD cells (Fig. 1C). We conclude that p53-dependent repression of c-myc is conserved in multiple mouse and human cell types.
FIG. 1.
c-myc is repressed in a p53-dependent manner. (A) c-myc transcript is repressed in mouse cells and tissues in a p53-dependent manner. DP16.1/p53ts and DP16.1 cells were maintained at 37°C or shifted to 32°C. Baf-3 and Baf-3/p53DD cells, as well as p53+/+ and p53−/− mice, were left untreated or γ-irradiated (6 Gy). At 3 h after treatment, total RNA was harvested from the cells or mouse tissues (spleen, thymus, and bone marrow). Northern blot analyses were performed, and the blots were hybridized with 32P-labeled cDNA probes for c-myc, p21, and GAPDH (loading control). Signal intensities were quantified by phosphorimaging analyses. Each signal (c-myc or p21) was first normalized to the GAPDH signal within the same sample. In each cell line or tissue, the untreated sample (37°C or unirradiated) signal was assigned a value of 1.0 and the treated sample (32°C or γ-irradiated) was expressed as the fold difference relative to it. (B) p53-dependent repression of c-myc transcript in human cells. BJ-T, BJ-T/p53DD, AML-3, and K562 cells were left untreated or γ-irradiated (6 Gy). Total RNA was harvested from the cells 3 h after treatment. Northern blot analyses and signal quantifications were performed as described for panel A. (C) Western blot analyses using antibodies against c-myc (N-262; Santa Cruz) and β-actin (loading control). DP16.1/p53ts and DP16.1 cells were shifted to 32°C, and Baf-3 and Baf-3/p53DD cells were γ-irradiated (6 Gy). Proteins extracts were made at the times indicated.
c-myc repression is not dependent on p21WAF1.
Recent studies have reported that the down-regulation of many of the previously identified p53-repressed genes is mediated by p21. For instance, the repression of CHK1, stathmin, cyclin B1, cdc2, BRCA1, cyclin A2, and hTERT is abrogated in p21-deficient cells, and the expression of p21 alone is sufficient to repress these genes (10, 23, 38). To determine if the p53-dependent repression of c-myc transcription is mediated through p21, we examined c-myc transcript levels in p21−/− mice before and after γ-irradiation. As shown in Fig. 2, c -myc mRNA levels decreased in the thymus, spleen, and bone marrow of p21-deficient mice after γ-irradiation. We conclude that p53-dependent repression of c-myc is not a consequence of p53-dependent transactivation of p21.
FIG. 2.
c-myc repression is not mediated by p21. Total RNA was harvested from the thymus, spleen, and bone marrow of p21−/− mice after exposure to γ-irradiation (6 Gy) and subjected to Northern blot analysis as described in the legend for Fig. 1.
c-myc repression is required for the efficient induction of G1 cell cycle arrest and differentiation by p53.
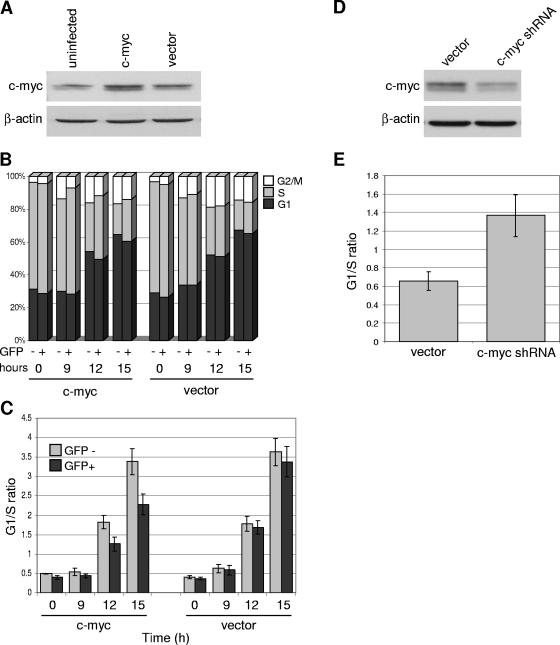
To examine the consequences of persistent c-myc expression during p53 activation, we ectopically expressed c-myc and assayed for various p53-associated functions, including G1 cell cycle arrest, differentiation, and apoptosis. DP16.1/p53ts cells were retrovirally infected with pBabe-c-myc-IRES-GFP or with vector only (pBabe-IRES-GFP). As shown in the Western blot in Fig. 3A, c -myc-infected cells expressed a slightly higher level of c-myc protein than uninfected and vector-infected cells. Infected cells were shifted to 32°C to activate wild-type p53 function, and at various times the cell cycle profiles were examined by flow cytometry after staining with Hoechst 33342 stain. We identified the infected cells on the basis of GFP expression and evaluated the cell cycle profiles of the GFP+ (infected cells) and GFP− (uninfected cells) gated populations. The latter represent cells that were not infected and, hence, serve as additional controls in these experiments; infection efficiencies ranged from 40% to 60%. The proportion of cells in each phase of the cell cycle is shown in Fig. 3B; G1/S ratios were also calculated (Fig. 3C) to portray the extent of G1 cell cycle arrest. As shown in Fig. 3C, ectopic expression of c-myc reduced the extent of G1 cell cycle arrest after p53 activation.
FIG. 3.
c-myc repression is required for the efficient induction of G1 cell cycle arrest by p53. (A) DP16.1/p53ts cells were retrovirally infected with the pBabe-c-myc-IRES-GFP (c-myc) or pBabe-IRES-GFP (vector) construct. At 48 h after infection, cells were harvested, and protein extracts were prepared and analyzed by Western blotting using an anti-c-myc (N-262; Santa Cruz) or anti-β-actin antibody. Infection efficiencies ranged from 40% to 60%. (B) Cell cycle analyses of cells expressing ectopic c-myc or vector only. DP16.1/p53ts cells infected with c-myc or empty vector (as described for panel A) were shifted to 32°C for the indicated amounts of time. To measure DNA content, cells were stained with Hoechst 33342 and analyzed by flow cytometry to determine the proportion of GFP− (uninfected) and GFP+ (infected) cells that were in the G1, S, and G2/M phases of the cell cycle. (C) G1/S ratios were calculated from the data in panel B to portray the extent of G1 cell cycle arrest. Each bar represents the average of three independent experiments ± the standard error. (D) DP16.1/p53ts cells were retrovirally infected with a c-myc shRNA construct or empty vector. At 24 h after infection, cells were harvested, and protein extracts were analyzed by Western blotting. Infection efficiencies ranged from 50% to 70%. (E) Cell cycle analyses of cells expressing c-myc shRNA or vector only. At 24 h after infection, DNA content was determined as described for panel B, and G1/S ratios were determined. Cells were maintained at 37°C throughout. Each bar represents the average of three independent experiments ± the standard error.
If the repression of c-myc were required for the induction of G1 cell cycle arrest, then artificially knocking down c-myc should promote G1 arrest even in the absence of p53 activation. We introduced c-myc shRNA by retroviral infections into DP16.1/p53ts cells. As shown in Fig. 3D, the c-myc protein level was decreased in c-myc shRNA-infected cells compared with vector-infected cells. In the absence of wild-type p53 function at 37°C, knock-down of c-myc promoted G1 cell cycle arrest as demonstrated by the increased G1/S ratio in Fig. 3E.
These data are consistent with the ability of c-myc to promote entry into S phase and oppose p53-dependent G1 arrest. c-myc was shown previously to influence the cellular response to p53 activation by inhibiting p21 expression and favoring apoptosis over cell cycle arrest (37). Hence, c-myc repression by p53 may constitute an integral component of the signaling pathway through which p53 promotes cell cycle arrest. Likewise, a recent study (42) demonstrated that the repression of cdc25c by p53 contributes to the induction of G2 arrest.
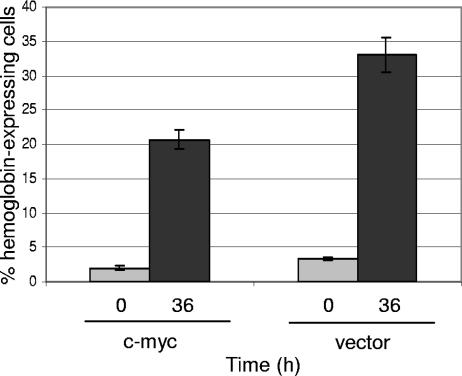
Previously we showed that incubation of DP16.1/p53ts cells at 32°C resulted in the expression of hemoglobin, a marker of erythroid differentiation (16). To determine if c-myc interferes with hemoglobin expression induced during p53 activation, c-myc- and vector-infected DP16.1/p53ts cells were maintained at 37°C or incubated at 32°C for 36 h. Cells were then stained with DAF to determine hemoglobin expression (16). As shown in Fig. 4, the percentage of hemoglobin-expressing cells was reduced in c-myc-infected samples compared with vector-infected samples. This finding is consistent with previous reports showing that c-myc inhibits differentiation in myoblasts (28), erythroleukemia (3), B-lymphoid (45), and myeloid (20) cells.
FIG. 4.
Ectopic c-myc expression interferes with p53-induced cellular differentiation. DP16.1/p53ts cells infected with c-myc or empty vector were incubated at 32°C for 0 or 36 h. Cells were stained for hemoglobin expression using DAF as described elsewhere (16). Stained cells were counted with a hemocytometer, and the proportion of hemoglobin-expressing cells was determined (number of blue cells/total number of cells). Results represent the means of three independent experiments ± the standard errors. Unsorted whole cell populations were used; infection efficiencies ranged from 40 to 60%.
c-myc repression is not required for apoptosis induction by p53.
Next, we wished to evaluate the effect of c-myc expression on p53-induced apoptosis. DP16.1/p53ts cells ectopically expressing c-myc or empty vector were kept at 37°C or shifted to 32°C for various periods of time. Cells were stained with Annexin-V as a marker for apoptosis, and the proportion of apoptotic cells was determined by flow cytometry. Ectopic c-myc expression led to an increase in apoptotic cells upon p53 activation at 32°C (Fig. 5). Hence, c-myc repression is not a prerequisite for efficient induction of apoptosis by p53. On the contrary, c-myc enhanced p53-dependent apoptosis. c-myc is known to sensitize cells to undergo apoptosis through p53-dependent as well as p53-independent pathways, possibly through the activation of ARF, stimulation of Bax activity, repression of Bcl-2 and Bcl-XL, and repression of p21 (6, 18, 37, 41). The ability of c-myc to promote entry into S phase and oppose p53-dependent cell cycle arrest (Fig. 3) may account in part for its ability to increase apoptosis upon p53 activation (Fig. 5).
FIG. 5.
Ectopic c-myc expression enhances apoptosis induction by p53. DP16.1/p53ts cells infected with c-myc or empty vector were incubated at 32°C for the indicated amounts of time. Cells for the zero-hour time point were maintained at 37°C for the duration of the experiment. Cells were stained with Annexin V-phycoerythrin and 7-AAD and analyzed by flow cytometry to determine the proportion of GFP− and GFP+ cells that were apoptotic (Annexin V positive, 7-AAD negative). Each bar represents the mean of three independent experiments ± the standard error.
p53 binds to the c-myc gene in vivo.
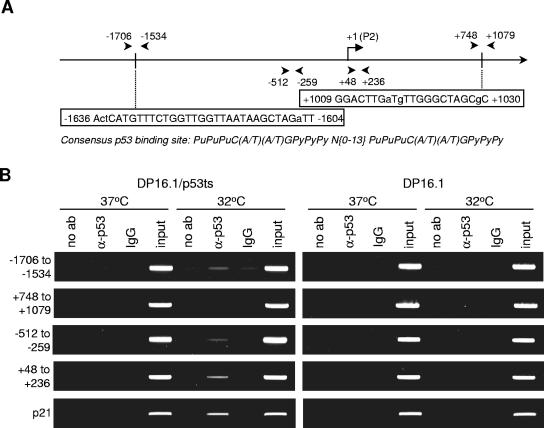
p53 has been shown to bind to certain regions within repressed genes, such as Map4 (31) and survivin (13, 29), and to be associated with the recruitment of histone deacetylases. The observed decrease in the level of acetylated histones at the target promoters could lead to a reduction in promoter accessibility, resulting in transcriptional repression. To determine if p53 binds to the c-myc promoter in vivo, chromatin immunoprecipitation assays were performed. DP16.1/p53ts and DP16.1 cells were maintained at 37°C or shifted to 32°C for 3 h. Chromatin immunoprecipitations were carried out as described elsewhere (5, 21) by using a polyclonal p53 antibody (FL-393; Santa Cruz). We scanned the mouse c-myc promoter and 5′ region (−3000 to +1200) for potential p53 binding sites (two repeats of the 10-bp motif, 5′-PuPuPuC(A/T)(A/T)GPyPyPy-3′, separated by 0 to 13 bp; Pu is a purine and Py is a pyrimidine) (7). Within the consensus sequence, the C and G residues are considered to be particularly important for p53 binding. There are at least four potential binding sequences within the region that contain two or three mismatches that are not at the C and G positions. PCR primers were designed to flank these sites, and binding was assessed by the enrichment of PCR signal in the anti-p53 sample compared to the no-antibody and control IgG reaction mixtures. At 32°C in DP16.1/p53ts cells, p53 consistently bound to the region around −1706 to −1534, which flanked one of the p53 consensus sites at −1636 to −1604 (Fig. 6A and B). This binding was not observed in DP16.1 cells lacking p53 nor in DP16.1/p53ts cells at 37°C. PCR amplification of a region (+748 to +1079) that flanks another p53 consensus site (+1009 to +1030) failed to show enrichment in binding signal in both DP16.1/p53ts and DP16.1 cells. The two other consensus sites were also examined, and weak or no enrichment in binding signal was observed (data not shown). These results indicate that p53 binds to the c-myc promoter in vivo and that this binding is restricted to certain regions of the gene. Further analysis of the c-myc promoter revealed p53 binding in the −512 to −259 and +48 to +236 regions, which lack consensus p53 binding sequences, suggesting that p53 binds through novel or noncanonical binding sites. In a previous study, p53 was shown to repress Map4 transcription by binding to the proximal promoter region, which lacks a consensus p53 binding sequence (31). Because the average size of the sheared chromatin was 500 bp in our ChIP assays, we cannot be certain whether p53 binds to one or both of these regions or whether it binds to a region close by. The ChIP findings have been reproduced three times using different cell extracts. Moreover, the enrichment in PCR signal in the immunoprecipitated sample over the no-antibody control was retained within a range of PCR amplification cycles.
FIG. 6.
p53 binds to the c-myc gene in vivo. (A) Schematic representation of the murine c-myc promoter and 5′ region. The reference point +1 is placed at the transcription start site of the P2 promoter. Arrows indicate the positions of PCR primers used in panel B. Two consensus p53 binding sites and their positions on the gene are also shown; capital letters indicate the residues that satisfy the consensus p53 binding sequence, while lowercase letters indicate mismatches. (B) DP16.1/p53ts and DP16.1 cells were maintained at 37°C or 32°C for 3 h. Cells were harvested for chromatin immunoprecipitation assays as described in Materials and Methods. Samples were immunoprecipitated with no antibody (no ab), a p53 antibody, or rabbit IgG; input controls (1/100 dilution) were also included. PCR analyses on the immunoprecipitated chromatin were carried out using primers that flank the regions as shown in panel A. Numbers on the left of the figure indicate the regions on the c-myc gene that are flanked and amplified by the primers. PCR amplification using primers against a p53 binding site at the p21 promoter was also included as a control.
To assess the regions on the c-myc promoter that are required for p53-dependent repression, we conducted a series of luciferase-reporter assays. Luciferase constructs that carry various segments of the c-myc promoter were transiently transfected along with p53 or empty vector into p53-null H1299 cells. The longest of these constructs extended from −2093 to +360 and included all the regions that bind p53. We did not observe p53-dependent repression of any of the luciferase-reporter constructs despite the ability of p53 to stimulate luciferase expression from a reporter plasmid containing the p53 binding site from p21WAF1 (data not shown). Our inability to detect p53-mediated repression of c-myc in a transient reporter assay might be explained if repression can only occur within the context of assembled chromatin.
p53 represses c-myc expression via a histone deacetylation mechanism.
As p53 has been associated with the recruitment of histone deacetylases to its repressed gene targets (13, 29, 31), a similar repression mechanism may operate at the c-myc promoter. If so, this could explain the inability of the transient luciferase assays to detect p53-mediated repression, since luciferase expression is not occurring within the context of histone assembly. To determine if histone deacetylase activity is required for c-myc repression by p53, Baf-3 cells were treated with the histone deacetylase inhibitor trichostatin A (TSA) prior to γ-irradiation. We showed earlier that c-myc repression in irradiated Baf-3 cells is dependent on p53 (Fig. 1). As shown in Fig. 7A, TSA fully abrogated the repression of c-myc mRNA in these cells. To determine if histones at the c-myc promoter are deacetylated in the presence of activated p53, chromatin immunoprecipitation assays were performed using an anti-acetylated histone H4 antibody, and extracts were prepared from DP16.1/p53ts and DP16.1 cells cultured at 37°C and 32°C (Fig. 7B). p53 activation led to a decrease in the level of acetylated histone H4 at the +48 to +236 region; this decrease was not observed in the DP16.1 cells. These results suggest that histone deacetylation occurs when p53 is activated and bound to the c-myc promoter. p53 may mediate histone deacetylation by recruitment of corepressor proteins; Murphy and coworkers (31) demonstrated that p53 represses the Map4 promoter by recruiting the corepressor mSin3a, which associates with HDAC1. Chromatin immunoprecipitations using an anti-mSin3a antibody indicated that p53 activation in the DP16.1/p53ts cells led to an increase in mSin3a association with the +48 to +236 region of the c-myc promoter (Fig. 7C). This increased association was absent in the DP16.1 cells. Together, these data support a model in which p53 mediates transcriptional repression of c-myc through a mechanism that involves binding to the c-myc promoter and promotion of histone deacetylation. The repression of c-myc transcription by c-myc autorepression, as well as through Smads in response to transforming growth factor beta, is dependent on p107 (1, 25). Pocket proteins, such as Rb, p107, and p130, have been shown to interact with and recruit histone deacetylases to repress transcription at E2F sites (9, 35). It is unclear if the reduction in histone acetylation at the c-myc promoter in response to p53 also involves p107. In preliminary experiments using p107−/− mice, we observed that c-myc was normally repressed in response to γ-irradiation, suggesting that p107 is not required for γ-induced c-myc repression in at least some tissues (data not shown).
FIG. 7.
p53-mediated transcriptional repression of c-myc involves histone deacetylation at the c-myc promoter. (A) Baf-3 cells were treated with 12.5 nM TSA for 9 h. At 6 h after drug treatment, cells were γ-irradiated where indicated. RNA was harvested 3 h later (9 h total TSA treatment time) and subjected to Northern blot analysis as described in the legend for Fig. 1. (B) Chromatin immunoprecipitations were performed as described in the legend for Fig. 6. Protein-DNA complexes were immunoprecipitated with no antibody (no ab), an antibody against acetylated histone H4 proteins (α-AcH4), or rabbit IgG. (C) Chromatin immunoprecipitations with no antibody (no ab), anti-mSin3a, or rabbit IgG in DP16.1/p53ts and DP16.1 cells.
Our findings indicate that c-myc repression by p53 is required for p53-dependent cell cycle arrest and differentiation but not for apoptosis. This raises a question regarding the importance of c-myc repression for p53-dependent tumor suppression. In some tumor models, p53 loss accelerates tumor growth through a decrease in tumor cell apoptosis, whereas in other models, p53 loss increases tumor cell proliferation rates. In the choroid plexus epithelium, for example, tumor formation correlates with the loss of apoptosis, rather than an increase in proliferation (44). Similarly, in the Eμ-myc-induced lymphomagenesis mouse model, blockage of the apoptotic pathway by the expression of bcl-2 or dominant-negative caspase 9 is sufficient to override p53 function in suppressing tumor development (36). In contrast, p53 loss accelerates tumor formation in Wnt-1 and ras transgenic mouse models, primarily due to an increase in the rate of tumor cell proliferation without affecting apoptosis (14, 17). In addition, p21−/− mice, which lack a G1 checkpoint, are prone to tumorigenesis, albeit at a slower rate than p53−/− mice (27). It is possible, therefore, that defective cell cycle control may contribute to tumor development. Thus, c-myc repression by p53 may be important for tumor suppression in certain cell types and under certain conditions.
The role of cellular differentiation in tumor suppression is not well understood, despite the pervasive views that blocked differentiation underlies many forms of leukemia and that a poorly differentiated phenotype is characteristic of malignant cell populations. Some studies have shown that induced differentiation of myeloid leukemic cells converts these malignant cells to a nonmalignant phenotype (24). In addition, retinoic acid is used as an effective cancer therapeutic to induce terminal differentiation of human promyelocytic leukemia cells carrying the PML/RARα fusion protein (24). Hence, the repression of c-myc by p53 may also contribute to its tumor suppressor function through the promotion of cellular differentiation.
Acknowledgments
We thank members of the Benchimol laboratory for useful discussions of the manuscript. We are grateful to Wissam Assaily for technical assistance with the mice work.
This work was supported by grants from the Terry Fox Foundation and the National Cancer Institute of Canada. J.S.L.H was supported by an NSERC predoctoral scholarship and a Canada Graduate Scholarship.
REFERENCES
- 1.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 2.Chiou, S. K., L. Rao, and E. White. 1994. Bcl-2 blocks p53-dependent apoptosis. Mol. Cell. Biol. 14:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppola, J. A., and M. D. Cole. 1986. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature 320:760-763. [DOI] [PubMed] [Google Scholar]
- 4.D'Souza, S., H. Xin, S. Walter, and D. Choubey. 2001. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J. Biol. Chem. 276:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Eberhardy, S. R., C. A. D'Cunha, and P. J. Farnham. 2000. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J. Biol. Chem. 275:33798-33805. [DOI] [PubMed] [Google Scholar]
- 6.Eischen, C. M., D. Woo, M. F. Roussel, and J. L. Cleveland. 2001. Apoptosis triggered by Myc-induced suppression of Bcl-XL or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 21:5063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 8.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira, R., L. Magnaghi-Jaulin, P. Robin, A. Harel-Bellan, and D. Trouche. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. USA 95:10493-10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottifredi, V., O. Karni-Schmidt, S. S. Shieh, and C. Prives. 2001. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol. Cell. Biol. 21:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermeking, H., J. O. Funk, M. Reichert, J. W. Ellwart, and D. Eick. 1995. Abrogation of p53-induced cell cycle arrest by c-Myc: evidence for an inhibitor of p21WAF1/CIP1/SDI1. Oncogene 11:1409-1415. [PubMed] [Google Scholar]
- 12.Ho, J., and S. Benchimol. 2003. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 10:404-408. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, W. H., S. Biade, J. T. Zilfou, J. Chen, and M. Murphy. 2002. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 277:3247-3257. [DOI] [PubMed] [Google Scholar]
- 14.Hundley, J. E., S. K. Koester, D. A. Troyer, S. G. Hilsenbeck, M. A. Subler, and J. J. Windle. 1997. Increased tumor proliferation and genomic instability without decreased apoptosis in MMTV-ras mice deficient in p53. Mol. Cell. Biol. 17:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung, L., and V. Kumar. 2004. Specific inhibition of gene expression and transactivation functions of hepatitis B virus X protein and c-myc by small interfering RNAs. FEBS Lett. 560:210-214. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, P., S. Chung, and S. Benchimol. 1993. Growth suppression of Friend virus-transformed erythroleukemia cells by p53 protein is accompanied by hemoglobin production and is sensitive to erythropoietin. Mol. Cell. Biol. 13:1456-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, J. M., L. Attardi, L. A. Godley, R. Laucirica, D. Medina, T. Jacks, H. E. Varmus, and L. A. Donehower. 1997. Absence of p53 in a mouse mammary tumor model promotes tumor cell proliferation without affecting apoptosis. Cell Growth Differ. 8:829-838. [PubMed] [Google Scholar]
- 18.Juin, P., A. O. Hueber, T. Littlewood, and G. Evan. 1999. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 13:1367-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 20.Larsson, L. G., I. Ivhed, M. Gidlund, U. Pettersson, B. Vennstrom, and K. Nilsson. 1988. Phorbol ester-induced terminal differentiation is inhibited in human U-937 monoblastic cells expressing a v-myc oncogene. Proc. Natl. Acad. Sci. USA 85:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 22.Levy, N., E. Yonish-Rouach, M. Oren, and A. Kimchi. 1993. Complementation by wild-type p53 of interleukin-6 effects on M1 cells: induction of cell cycle exit and cooperativity with c-myc suppression. Mol. Cell. Biol. 13:7942-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr, K., C. Moritz, A. Contente, and M. Dobbelstein. 2003. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 278:32507-32516. [DOI] [PubMed] [Google Scholar]
- 24.Lotem, J., and L. Sachs. 2002. Epigenetics wins over genetics: induction of differentiation in tumor cells. Semin. Cancer Biol. 12:339-346. [DOI] [PubMed] [Google Scholar]
- 25.Luo, Q., J. Li, B. Cenkci, and L. Kretzner. 2004. Autorepression of c-myc requires both initiator and E2F-binding site elements and cooperation with the p107 gene product. Oncogene 23:1088-1097. [DOI] [PubMed] [Google Scholar]
- 26.Marhin, W. W., S. Chen, L. M. Facchini, A. J. Fornace, Jr., and L. Z. Penn. 1997. Myc represses the growth arrest gene gadd45. Oncogene 14:2825-2834. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Caballero, J., J. M. Flores, P. Garcia-Palencia, and M. Serrano. 2001. Tumor susceptibility of p21Waf1/Cip1-deficient mice. Cancer Res. 61:6234-6238. [PubMed] [Google Scholar]
- 28.Miner, J. H., and B. J. Wold. 1991. c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol. Cell. Biol. 11:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirza, A., M. McGuirk, T. N. Hockenberry, Q. Wu, H. Ashar, S. Black, S. F. Wen, L. Wang, P. Kirschmeier, W. R. Bishop, L. L. Nielsen, C. B. Pickett, and S. Liu. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21:2613-2622. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, K. O., and W. S. El-Deiry. 1999. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 10:223-230. [PubMed] [Google Scholar]
- 31.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy, M., A. Hinman, and A. J. Levine. 1996. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 10:2971-2980. [DOI] [PubMed] [Google Scholar]
- 33.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 34.Ragimov, N., A. Krauskopf, N. Navot, V. Rotter, M. Oren, and Y. Aloni. 1993. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene 8:1183-1193. [PubMed] [Google Scholar]
- 35.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, C. A., J. S. Fridman, M. Yang, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1:289-298. [DOI] [PubMed] [Google Scholar]
- 37.Seoane, J., H. V. Le, and J. Massague. 2002. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729-734. [DOI] [PubMed] [Google Scholar]
- 38.Shats, I., M. Milyavsky, X. Tang, P. Stambolsky, N. Erez, R. Brosh, I. Kogan, I. Braunstein, M. Tzukerman, D. Ginsberg, and V. Rotter. 2004. p53-dependent downregulation of telomerase is mediated by p21/waf1. J. Biol. Chem. 65:4530-4543. [DOI] [PubMed] [Google Scholar]
- 39.Shaulian, E., A. Zauberman, D. Ginsberg, and M. Oren. 1992. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 12:5581-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, B., P. G. Reddy, A. Goberdhan, C. Walsh, S. Dao, I. Ngai, T. C. Chou, O. C. P., A. J. Levine, P. H. Rao, and A. Stoffel. 2002. p53 regulates cell survival by inhibiting PIK3CA in squamous cell carcinomas. Genes Dev. 16:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soucie, E. L., M. G. Annis, J. Sedivy, J. Filmus, B. Leber, D. W. Andrews, and L. Z. Penn. 2001. Myc potentiates apoptosis by stimulating Bax activity at the mitochondria. Mol. Cell. Biol. 21:4725-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St. Clair, S., L. Giono, S. Varmeh-Ziaie, L. Resnick-Silverman, W. J. Liu, A. Padi, J. Dastidar, A. DaCosta, M. Mattia, and J. J. Manfredi. 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol. Cell 16:725-736. [DOI] [PubMed] [Google Scholar]
- 43.Sutcliffe, T., L. Fu, J. Abraham, H. Vaziri, and S. Benchimol. 1998. A functional wild-type p53 gene is expressed in human acute myeloid leukemia cell lines. Blood 92:2977-2979. [PubMed] [Google Scholar]
- 44.Symonds, H., L. Krall, L. Remington, M. Saenz-Robles, S. Lowe, T. Jacks, and T. Van Dyke. 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703-711. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, C. B., E. H. Humphries, L. M. Carlson, C. L. Chen, and P. E. Neiman. 1987. The effect of alterations in myc gene expression on B cell development in the bursa of Fabricius. Cell 51:371-381. [DOI] [PubMed] [Google Scholar]
- 46.Vafa, O., M. Wade, S. Kern, M. Beeche, T. K. Pandita, G. M. Hampton, and G. M. Wahl. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031-1044. [DOI] [PubMed] [Google Scholar]
- 47.Vaziri, H., and S. Benchimol. 1998. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 8:279-282. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., L. Szekely, I. Okan, G. Klein, and K. G. Wiman. 1993. Wild-type p53-triggered apoptosis is inhibited by bcl-2 in a v-myc-induced T-cell lymphoma line. Oncogene 8:3427-3431. [PubMed] [Google Scholar]