Abstract
Dbf4/Cdc7 is required for DNA replication in Saccharomyces cerevisiae and appears to be a target in the S-phase checkpoint. Previously, a 186-amino-acid Dbf4 region that mediates interactions with both the origin recognition complex and Rad53 was identified. We now show this domain also mediates the association between Dbf4 and Mcm2, a key Dbf4/Cdc7 phosphorylation target. Two conserved sequences, the N and M motifs, have been identified within this Dbf4 region. Removing motif M (Dbf4ΔM) impairs the ability of Dbf4 to support normal cell cycle progression and abrogates the Dbf4-Mcm2 association but has no effect on the Dbf4-Rad53 interaction. In contrast, deleting motif N (Dbf4ΔN) does not affect the essential function of Dbf4, disrupts the Dbf4-Rad53 interaction, largely preserves the Dbf4-Mcm2 association, and renders the cells hypersensitive to genotoxic agents. Surprisingly, Dbf4ΔM interacts strongly with Orc2, while Dbf4ΔN does not. The DBF4 allele dna52-1 was cloned and sequenced, revealing a single point mutation within the M motif. This mutant is unable to maintain interactions with either Mcm2 or Orc2 at the semipermissive temperature of 30°C, while the interaction with Rad53 is preserved. Furthermore, this mutation confers increased resistance to genotoxic agents, which we propose is more likely due to a role for Dbf4 in the resumption of fork progression following checkpoint-induced arrest than prevention of late origin firing. Thus, the alteration of the M motif may facilitate the role of Dbf4 as a checkpoint target.
The initiation of DNA replication requires both the formation of prereplicative complexes (pre-RCs) at origins and the activity of specific protein kinases in the G1 phase of the cell cycle (reviewed in references 2 and 23). In recent years, it has been firmly established that a number of replication factors also play key roles during the S-phase checkpoint (reviewed in references 30 and 36). Studies with the budding yeast Saccharomyces cerevisiae, as well as with fission yeast, frogs, and mammals, have implicated one of the replicative kinase complexes, Dbf4/Cdc7, as a target of the S-phase checkpoint response to genotoxic agents (reviewed in references 21 and 25). Levels of the Cdc7 kinase are relatively constant throughout the cell cycle, while those of its regulatory subunit, Dbf4, are low from the end of mitosis until a burst of new synthesis occurs in late G1 phase (6, 36, 52). Work with Xenopus laevis cell extracts has demonstrated that the binding of Cdc7 to chromatin requires the presence of Dbf4 (22), suggesting that it targets Cdc7 to pre-RCs. Chromatin fractionation assays with S. cerevisiae indicate that Cdc7 is chromatin bound throughout the cell cycle (52), while both Dbf4 and Cdc7 have been shown to interact with origin recognition complex (ORC) subunits (13, 18). Presently it is not known whether Dbf4 relocates Cdc7 to origins in budding yeast in late G1 phase, analogous to the findings with Xenopus, or may instead simply associate with and activate Cdc7 that is already ORC bound. Once Dbf4/Cdc7 becomes active, it is believed to phosphorylate Mcm2, a subunit of the heterohexameric MCM complex and a pre-RC component. This phosphorylation may result in local DNA unwinding (16), which is consistent with the putative MCM complex role as a replicative helicase (20, 27). In budding yeast, the initiation of replication is choreographed, with individual origins categorized as early, middle, or late firing (40). Dbf4/Cdc7 is required throughout S phase for initiation events to occur, as inactivation of Cdc7 during this stage of the cell cycle prevents further origins from firing (3, 10). Following exposure to the ribonucleotide reductase inhibitor hydroxyurea (HU), which results in replication fork stalling and triggers the S-phase checkpoint, Dbf4 is phosphorylated and removed from chromatin. Significantly, both events require the activity of the Rad53 checkpoint kinase (37, 52), which has been shown to physically interact with Dbf4 (9, 13). Similar mechanisms have been reported for Schizosaccharomyces pombe (4) and Xenopus laevis (7). The preservation of unfired origins may aid in the efficient resumption of DNA synthesis once the checkpoint is lifted, while Dbf4/Cdc7 may assist in reactivating replication at stalled forks (12). Additionally, Dbf4/Cdc7 has recently been shown to function in translesion synthesis, part of the checkpoint response following exposure to DNA damaging agents, such as UV radiation or methyl methanesulfonate (MMS) (38).
We have previously demonstrated that a 186-amino-acid Dbf4 region mediates interactions with both the ORC and Rad53. This suggests a mechanism whereby Rad53 directly binds to and/or phosphorylates the Dbf4 N terminus when the S-phase checkpoint is triggered, resulting in its displacement from unfired replication origins (13). This notion is supported by the finding that the inhibition of further origin firing, characteristic of the S-phase checkpoint, fails to occur in rad53 mutant strains (42, 46).
Dbf4 orthologs have been identified in organisms as diverse as fruit flies, frogs, mice, and humans (26, 31). Sequence comparisons between the different species have uncovered three relatively well-conserved amino acid stretches that have been termed the N, M, and C motifs (31). Studies with S. pombe have demonstrated that both motifs N and C are involved in the response to genotoxic agents but are not required for normal cell cycle progression (14, 34). Less is known about motif M, although it appears to be important for cell proliferation, since a mutant with combined deletions of motifs N and M cannot support growth in fission yeast (34).
Despite the obvious importance of these conserved regions for the replicative and checkpoint functions of Dbf4, the key interactions they mediate have not thus far been investigated. Here we show that deletion of motif M results in a disruption of the interaction between Dbf4 and Mcm2, while association with the Rad53 kinase remains intact. Similarly, a motif M point mutation in the DBF4 allele, which is temperature sensitive for normal growth, disrupts interactions with both Orc2 and Mcm2, while the interaction with Rad53 is preserved. Furthermore, this allele confers increased resistance to genotoxic agents. Thus, mutation of motif M may favor the association of Dbf4 with Rad53, thereby facilitating its role as an S-phase checkpoint target.
MATERIALS AND METHODS
Yeast strains.
DY-1 (MATa ade2-1 can1-100 trp1-1 his3-11,-15 ura3-1 leu2-3,-112 pep4::LEU2) was used for all two-hybrid analyses. DY-2 (MATa can1-11 ura3-52 dna52-1) was the host for the Dbf4 fragment complementation assay. Both strains were used in the MMS and HU growth and viability assays. The diploid strain DY-27 (MATa/MATa, his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ +/met15Δ ura3Δ/ura3Δ) was used to create DY-75 (MATa/MATa his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ +/met15Δ ura3Δ/ura3Δ dbf4ΔM/+) and DY-76 (MATa/MATa his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ +/met15Δ ura3Δ/ura3Δ dbf4ΔN/+). DY-76 was then sporulated and dissected to obtain DY-77 (MATa his3Δ1 leu2Δ0 ura3Δ) and DY-78 (MATa his3Δ1 leu2Δ0 ura3Δ dbf4ΔN).
Plasmid construction.
pEG-Dbf4-FL, -296, and -Δ110-296, pCM190-Dbf4-FL, (1-296), and -Δ110-296, pJG-Orc2, and -FHA1 have been previously described (13). pEG-Dbf4ts-FL was constructed by PCR amplification of the entire coding sequence of the DBF4 allele (dna52-1) from DY-2 genomic DNA, using forward and reverse primers with incorporated EcoRI and BamHI restriction endonuclease sites, respectively. The resultant PCR product was digested with these enzymes and cloned into the two-hybrid bait vector pEG-202 (1) that had also been digested with EcoRI and BamHI, creating an in-frame fusion with LexA coding sequence. pEG-Dbf4ΔM was created by amplifying the first 786 bp (encoding amino acids 1 to 262) of DBF4 using DY-1 genomic DNA as template, a forward primer with an added EcoRI site, and a reverse primer with a HindIII site, created in part by a silent base change from adenine to guanine at position 786. Base pairs 928 to 2112 (encoding amino acids 310 to 704) were then amplified using a forward primer which contained a HindIII restriction site created, in part, by changing bases 928 to 930 from ATA to CTT, causing a conservative amino acid change from isoleucine to leucine, and a reverse primer which included a NcoI restriction site. Following digestion with the appropriate restriction enzymes, the two DBF4 fragments were ligated together at their respective HindIII restriction sites and cloned into pEG-202, which had been digested with EcoRI and NcoI, creating an in-frame fusion with a LexA coding sequence. pEG-Dbf4ΔN was created by PCR amplification of bp 1 to 405 (encoding amino acids 1 to 135) using a forward primer with an EcoRI restriction site and a reverse primer containing a SalI site created by a silent base change from adenine to cytosine at position 402 and a base change from guanine to cytosine, resulting in a conservative amino acid change from glutamic acid to aspartic acid at position 135 of the protein product. Base pairs 537 to 2112 (amino acids 179 to 704) were amplified using forward and reverse primers with added SalI and NcoI sites, respectively. Following digestion with the appropriate restriction enzymes, the two fragments were ligated through their respective SalI sites and cloned into pEG-202, which had been digested with EcoRI and NcoI, creating an in-frame fusion with a LexA coding sequence. pCM190-Dbf4ts-FL, -ΔM, and -ΔN were created by excising the region between bp 170 and 1972 from pEG-Dbf4ts-FL, -ΔM, and -ΔN, respectively, using the natural BglII and Kpn2I sites within DBF4. These fragments were then cloned into the pCM190-Dbf4-FL vector, which had been digested with BglII and Kpn2I, thus replacing the DBF4 region between these restriction sites. Full-length MCM2 was PCR amplified from DY-1 genomic DNA using forward and reverse primers with added NcoI and XhoI restriction sites, respectively, and was cloned into the pJG 4-6 two-hybrid prey vector (a derivative of pJG4-5 [17], with an extended polylinker insert; gift of R. Brent) at these sites, resulting in an in-frame fusion with a hemagglutinin (HA) coding sequence and the creation of pJG-Mcm2. pJG-Cdc7 and -Rad53 were generated by PCR amplification of full-length CDC7 and RAD53 coding sequences using DY-1 genomic DNA as template, with forward and reverse primers incorporating EcoRI and BglII sites, respectively. Following digestion with EcoRI and BglII, the PCR products were cloned into the pJG-4-6 vector at these sites, resulting in an in-frame fusion with HA coding sequence.
Gene replacement.
DBF4ΔM and DBF4ΔN were amplified from pEG-DBF4ΔM and pEG-DBF4ΔN, respectively, using a forward primer containing an EcoRI restriction site and a reverse primer containing a BamHI restriction site. pRS406 (47) and the PCR products were digested with EcoRI and BamHI and ligated to create pRS-DBF4ΔM and pRS-DBF4ΔN. Gene replacement techniques were performed according to the methods of Rothstein (41), using diploid strain DY-27 to create DY-75 and DY-76 diploids, respectively. Sporulation and tetrad dissection were carried out according to the methods of Burke et al. (5).
Two-hybrid and Western analyses.
Liquid culture two-hybrid analyses were performed as previously described (1). The lacZ reporter plasmid pSH18-34, pEG-202-derived bait, and pJG4-6-derived prey plasmids were transformed into DY-1. Cultures were initially grown to a concentration of 2 × 107 cells/ml in 10 ml SD medium (5) without uracil, histidine, and tryptophan. Cells were washed and then induced for 6 h in 20 ml of 2% galactose-1% raffinose medium (BD Bioscience) lacking uracil, histidine, and tryptophan. Using a total of 5 × 106 cells, the interactions between fusion proteins were detected by the quantitative β-galactosidase (β-Gal) assay on permeabilized cells. Reaction tubes were centrifuged at 16,000 × g for 5 min, and the supernatant was collected to measure the A420. The β-Gal activity was calculated with the following formula: β-Gal activity = 1,000 × A420/(t × V × A600), where t = time of reaction (in minutes) and V = volume of culture used in the assay (in milliliters). The remaining culture was then centrifuged (2,000 × g, 3 min). All subsequent steps were performed at 4°C. The pellet was resuspended in 400 μl of ice-cold lysis buffer (10 mM Tris-Cl, pH 8.0-140 mM NaCl-1 mM EDTA-1% Triton X-100, with protease inhibitors) and lysed with a bead beater by using 0.5 g of 0.5-mm glass beads (BioSpec Products, Inc.) per sample. Lysate was centrifuged (10,000 × g, 30 s), and the supernatant was called whole-cell extract (WCE). Protein concentrations were quantified by Bradford assay, and protein expression was analyzed by Western blotting. The LexA-tagged bait proteins were detected using a rabbit polyclonal anti-LexA antibody (Invitrogen), and the HA-tagged prey proteins were detected using a mouse monoclonal anti-HA antibody (Sigma). Alexa Fluor 647 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse polyclonal secondary antibodies (Molecular Probes) were used. All detections were performed on a Typhoon 9400 laser scanning system (Amersham Bioscience).
Coimmunoprecipitation.
DY-1 cells were transformed with pCM190- and pJG4-6-derived expression vectors and were initially grown to 1 × 107 cells/ml in 10 ml SD medium lacking uracil and tryptophan and supplemented with 5 μg/ml doxycycline (Dox). Cells were then centrifuged (2,000 × g, 3 min), the pellet was resuspended in 20 ml of 2% galactose-1% raffinose medium (BD Bioscience) lacking uracil, tryptophan, and Dox, grown for 6 h, and centrifuged (2,000 × g, 3 min). All subsequent steps were performed as described previously (13).
Complementation assays.
DY-2 cells were transformed with the doxycycline-repressible plasmids pCM-190-Dbf4-FL, -ΔM, - ΔN, -Δ110-296, and 1-296 or unmodified pCM190 (15) and grown to saturation in SD medium lacking uracil (SD-Ura), supplemented with Dox (6 μg/ml). A total of 5 × 106 cells of each were washed in distilled H2O, resuspended in SD-Ura without Dox, streaked on SD-Ura plates, and grown at 23°C, 30°C, or 37°C for 3 days. A control SD-Ura plate supplemented with Dox (6 μg/ml) was streaked with the same transformed strains and grown at 23°C. The above yeast transformants were then transformed with pJG 4-6 and pJG-Cdc7 and grown to saturation in SD medium lacking uracil and tryptophan (SD-Ura-Trp) and supplemented with Dox (6 μg/ml). Cells were washed twice in distilled H2O, streaked onto plates of SD-Ura-Trp supplemented with Dox (6 μg/ml) and galactose-raffinose lacking uracil and tryptophan (Gal/Raf-Ura-Trp), and grown at 23°C.
Growth assays.
DY-1 and DY-2 were transformed with pCM190, pCM190-Dbf4-FL, or pCM190-Dbf4ts-FL. Cultures of DY-1 and DY-2 transformants were grown to saturation (∼1 × 108 cells/ml), and serial 10-fold dilutions, ranging from 1 × 107 cells/ml to 1 × 104 cells/ml, were prepared for both strains. A 5-μl volume from each cell suspension was spotted onto a series of plates in decreasing concentrations from the top of each plate to the bottom. SD lacking uracil (to select for plasmid maintenance) was supplemented with various concentrations of HU ranging from 0 to 100 mM or MMS ranging from 0 to 0.015%. Each series was spotted in duplicate, with one plate series being incubated at 23°C and the other at 30°C for 3 days.
Viability assay.
DY-1 and DY-2 cells were diluted to a concentration of 1 × 106 cells/ml in YPD medium. Ten-milliliter culture volumes were treated with MMS (0.013%, 0.0195%, or 0.026%) or HU (50 mM, 100 mM, or 150 mM) and incubated at 30°C with shaking (215 rpm) in parallel with untreated control cultures. After 4 h of incubation, 50-μl culture aliquots were removed and diluted 200-fold. Fifty-microliter aliquots of each dilution suspension were spread onto YPD plates in triplicate. Plates with DY-1 cells were then incubated for 2 days at 30°C, while those with DY-2 cells were incubated at 23°C for 2 days. The triplicate series of plates were counted using a Quebec colony counter. The colony counts for each triplicate series were averaged and recorded. In each case, the percentage of viable colonies was calculated relative to untreated control plate count averages. Results presented are the averages of three independent experiments along with standard deviations.
RESULTS
Dbf4 is targeted to chromatin in the late G1 phase of the cell cycle, when its activation of Cdc7 is required to trigger the initiation of DNA replication (reviewed in reference 44). We had previously identified a 186-amino-acid region of Dbf4 that mediates interactions with both ORC, through the Orc2 subunit, as well as the Rad53 checkpoint kinase, via its FHA1 and FHA2 domains (13). Given the likelihood that the Dbf4-ORC interaction is important for targeting Dbf4/Cdc7 activity to origins of replication, we decided to investigate the effect of removing a DBF4 sequence encoding amino acids 110 to 296 on plasmid-based growth complementation of yeast strain DY-2 (dna52-1), which expresses temperature-sensitive Dbf4 (Dbf4ts-FL). DY-2 cells were transformed with doxycycline-repressible pCM190-derived (15) constructs, expressing either full-length Dbf4 (pCM190-Dbf4-FL), Dbf4 lacking amino acids 110 to 296 (pCM190-Dbf4 Δ110-296) (Fig. 1), or empty pCM190 vector. For both pCM190-Dbf4-FL and pCM190-Dbf4Δ110-296, a sequence encoding a C-terminal Myc18 tag was incorporated into the construct. While expression of wild-type Dbf4 restored growth to DY-2 cells at 37°C, expression of Dbf4 Δ110-296 did not (data not shown). This suggests that the missing Dbf4 region (amino acids 110 to 296) is indeed required for normal cell proliferation.
FIG. 1.
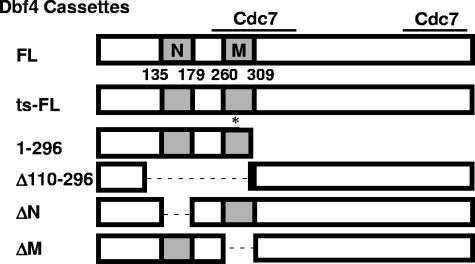
Dbf4 mutants. Full-length Dbf4 (Dbf4FL) and mutants used in this study are depicted. The N and M motifs, which are highly conserved among eukaryotes, are illustrated in gray. Numbers below indicate corresponding amino acid position, and the two regions that bind Cdc7 (19) are indicated by solid bars. The single amino acid change from proline to leucine at position 277 resulting from the dna52-1 point mutation is indicated by an asterisk (Dbf4ts-FL). DNA cassettes encoding these Dbf4 variants were used for the construction of both pEG-202-based (1) and pCM190-based (15) expression vectors.
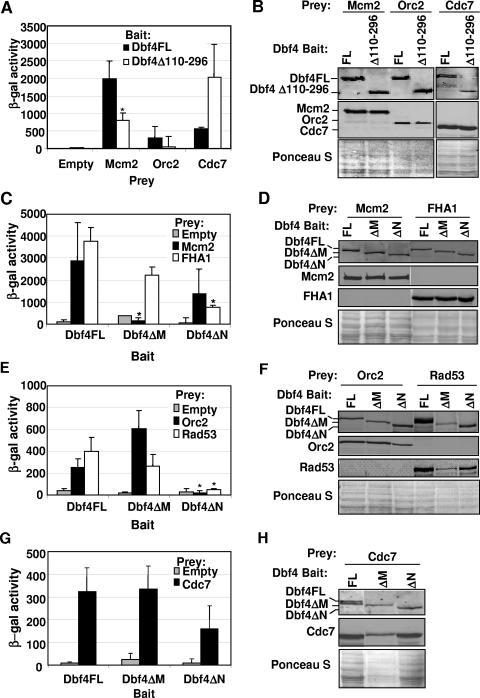
The previous observation that removal of these amino acids abrogates the interaction between Dbf4 and Orc2 suggests one mechanism whereby DNA replication may be inhibited (13). We were nevertheless curious to determine whether this region might also mediate the previously reported physical interaction between Dbf4 and the Dbf4-dependent kinase (DDK) substrate Mcm2 (28). A disrupted Dbf4-Mcm2 interaction would provide another explanation of how cell cycle progression is impeded by deletion of Dbf4 amino acids 110 to 296. In order to investigate this possibility, two-hybrid assays were conducted using pEG-202-derived (1) bait plasmids expressing either full-length Dbf4 (pEG-Dbf4-FL) or Dbf4 Δ110-296 (pEG-Dbf4Δ110-296), in combination with pJG4-6-derived (see Materials and Methods) prey plasmids expressing full-length Mcm2 (pJG-Mcm2), Orc2 (pJG-Orc2), or Cdc7 (pJG-Cdc7). As shown in Fig. 2A, removal of Dbf4 amino acids 110 to 296 reduced the interaction with Mcm2 to approximately half its normal level. This closely parallels what is observed with Orc2 prey (Fig. 2A), as previously reported (13). Immunoblot analysis indicated that levels of full-length Dbf4 and Dbf4Δ110-296 bait were comparable (Fig. 2B), ruling out decreased steady-state levels of the latter as a cause for the weakened two-hybrid signal generated by the deletion mutant with either Mcm2 or Orc2. Similarly, the levels of Mcm2 and Orc2 preys were not affected by the particular bait construct being expressed. In contrast, Dbf4Δ110-296 demonstrated a robust association with Cdc7 (Fig. 2A), consistent with previous data indicating that the Cdc7 interacting regions of Dbf4 span amino acids 241 to 416 and 573 to 695 (Fig. 1). The fact that the two-hybrid signal with the Cdc7 prey was higher with the Dbf4Δ110-296 bait than with the Dbf4FL bait may indicate that residues within the deleted region partially mask one or both of the Cdc7 binding regions in the folded full-length protein.
FIG. 2.
Two-hybrid analysis of Dbf4 variant interactions with replication and checkpoint factors. (A) Two-hybrid assays using bait constructs pEG-Dbf4-FL and pEG-Dbf4Δ110-296 along with pJG-Mcm2, pJG-Orc2, pJG-Cdc7, and empty pJG 4-6 as prey vectors were carried out as described in Materials and Methods. Relative β-galactosidase activity is shown and represents the average of at least three trials. Error bars represent standard deviations. The asterisk indicates a statistically significant difference (P < 0.05; paired Student's t test) in the signal for Dbf4Δ110-296 relative to Dbf4FL. Insufficient trials were conducted to perform this analysis for the Cdc7 prey set. (B) To confirm that all baits and preys were properly expressed, culture aliquots were removed just prior to the measurement of β-galactosidase activity, and whole-cell extracts were prepared and subjected to immunoblot analysis. Bait proteins were detected with rabbit polyclonal anti-LexA antibody (Invitrogen), and prey proteins were detected with mouse monoclonal anti-HA antibody (Sigma). Ponceau S staining of the membrane to confirm equal loading of whole-cell extracts is shown. (C, E, and G) Two-hybrid assays were carried out as described above by using pEG-Dbf4FL, pEG-Dbf4ΔM, and pEG-Dbf4ΔN as bait, with pJG 4-6, -Mcm2, -FHA1, -Orc2, -Rad53, and -Cdc7 preys. Relative β-galactosidase activities are shown and in each case represent the average of three trials. Error bars represent standard deviations. Asterisks indicate those two-hybrid interactions with significant differences at a P level of <0.05. In each case the values obtained with Dbf4ΔM or Dbf4ΔN are compared to the equivalent interaction using Dbf4FL as bait. (D, F, and H) Confirmation of protein expression as described above.
Given that the deletion of Dbf4 amino acids 110 to 296 eliminated all or most of conserved motifs N and M (Fig. 1), we decided to examine how more precise excisions of these regions would influence the ability of Dbf4 to interact with replication and checkpoint factors. Additional two-hybrid bait vectors were therefore constructed, expressing Dbf4 lacking either motif N (pEG-Dbf4ΔN, amino acids 136 to 178 removed [Fig. 1]) or motif M (pEG-Dbf4ΔM, amino acids 263 to 309 removed [Fig. 1]). When these were tested in combination with the prey constructs pJG-Mcm2, pJG-Orc2, pJG-Rad53, and pJG-FHA1, a variety of deletion-dependent effects were observed. The removal of motif N resulted in a sharp drop in the two-hybrid signal between Dbf4 and either full-length Rad53 or its FHA1 domain alone (Fig. 2C and E), consistent with the observation in S. pombe that deletion of this region from its Dbf4 ortholog, Dfp1, results in sensitivity to a broad range of genotoxic agents (14). In contrast, the interaction with Mcm2 was preserved to a greater extent with Dbf4ΔN (Fig. 2C), consistent with the finding that removal of motif N does not prevent Dfp1 from carrying out its essential function (14). For Dbf4ΔM, the results were essentially reversed, the interaction with Mcm2 was completely abrogated while removal of motif M had only a modest effect on the strength of the two-hybrid signal with full-length Rad53 or its FHA1 domain alone (Fig. 2C and E). Indeed, the modest reduction in two-hybrid signal between Dbf4ΔM and Rad53 may be due to lower levels of both proteins in this transformant (Fig. 2F). Surprisingly, when Orc2 was used as prey in conjunction with Dbf4FL, Dbf4ΔN, and Dbf4ΔM baits, the absence of motif N rather than motif M reduced the relative strength of interaction (Fig. 2E). Finally, we monitored the interaction between the same three Dbf4 baits and Cdc7 and found similar two-hybrid signal strengths in each case (Fig. 2G). For all trials, the levels of bait and prey proteins were monitored by immunoblot analysis (Fig. 2B, D, F, and H).
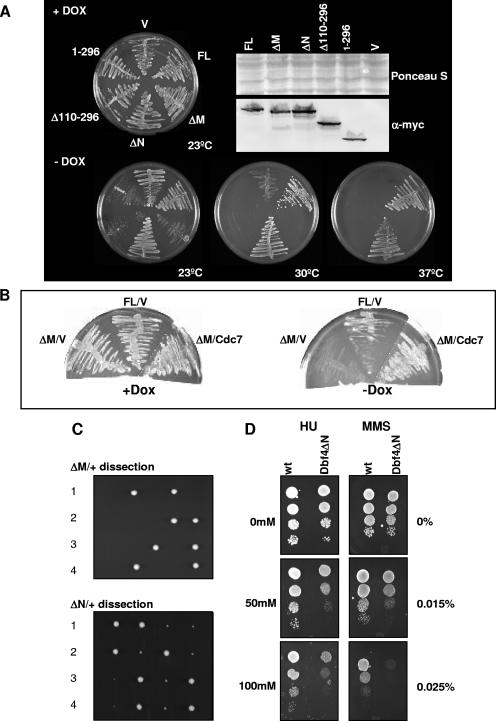
In light of our determination that removal of conserved Dbf4 regions leads to reduced interactions with specific cell cycle factors, we decided to evaluate whether one or both of motifs N and M are required for Dbf4 to carry out its role in normal cell cycle progression. This was of particular interest, given previous studies with S. pombe showing that removal of the N motif does not prevent Dfp1 from fulfilling its essential function while additional deletion of the M motif was found to eliminate the ability of Dfp1 to support cell cycle progression (14, 34). In order to evaluate the requirement of these two conserved Dbf4 regions for S. cerevisiae growth, we prepared further doxycycline-repressible pCM190 constructs, expressing Dbf4 variants lacking either motif N (pCM190-Dbf4ΔN, with amino acids 136 to 178 removed [Fig. 1]) or motif M (pCM190-Dbf4ΔM, with amino acids 263 to 309 removed [Fig. 1]) and tested their ability to complement growth in DY-2 cells. While pCM190-Dbf4ΔN was able to complement growth as efficiently as pCM190-Dbf4-FL at both semipermissive (30°C) and restrictive (37°C) temperatures, pCM190-Dbf4ΔM was not able to do so (Fig. 3A). Moreover, pCM190-Dbf4ΔM acted in a dominant-negative fashion, inhibiting growth at 23°C relative to cells transformed with empty vector. A plausible explanation for this phenomenon is that overexpressed Dbf4ΔM, which is impaired for association with Mcm2, may sequester Cdc7, thereby reducing its interaction with endogenous Dbf4 and the subsequent phosphorylation of Mcm2. The same effect would be expected for overexpression of pCM190-Δ110-296, since it produces a Dbf4 variant lacking most of motif M (Fig. 1) while the corresponding two-hybrid bait demonstrates a strong interaction with Cdc7 (Fig. 2A) and, indeed, such a dominant-negative effect was observed (Fig. 3A). In further support of this model, we found that coexpression of plasmid-encoded Cdc7 with Dbf4ΔM was able to restore growth at 23°C (Fig. 3B). As an additional control, pCM190-Dbf4(1-296), expressing only the first 296 amino acids of Dbf4, also inhibited growth at 23°C, as previously reported (13). In order to establish that Dbf4ΔN is able to support growth in the absence of any other Dbf4 variant, we generated a +/ΔN diploid strain. Following sporulation and tetrad dissection, four viable spores were consistently obtained (Fig. 3C). In nearly every case, two of the four spore-derived colonies were considerably smaller than the two others. We determined that the smaller colonies corresponded to the Dbf4ΔN segregants (not shown). Thus, despite the fact that the Dbf4ΔN haploid cells are viable, they display some growth impairment relative to isogenic wild-type cells, although this was less apparent when the cells were spotted following exponential growth in liquid medium (Fig. 3D). Tetrad dissection was similarly conducted with +/ΔM diploids, and in each case only two of four spores were viable (Fig. 3C), confirming the requirement of motif M for regular cell cycle progression.
FIG. 3.
Dbf4 motif M is required for normal cell growth, while motif N confers resistance to both HU and MMS. (A) DY-2 cells were transformed with pCM190-Dbf4-FL, -Dbf4ΔM, -Dbf4ΔN, -Dbf4Δ110-296, -Dbf4(1-296), or empty vector pCM190 (V). A complementation assay was performed as described in Materials and Methods. Cells were grown either in the presence (+Dox) or absence (-Dox, entire row) of Dox, which represses pCM190-based expression, at the indicated temperatures. Western blot analysis to confirm plasmid-based expression was conducted using a mouse monoclonal anti-Myc antibody (Sigma) and Alexa Fluor 488 goat anti-mouse immunoglobulin G polyclonal secondary antibody (Invitrogen). (B) DY-2 cells transformed with pCM190-Dbf4-FL (FL) or pCM190-Dbf4ΔM (ΔM) as described above were further transformed with either pJG 4-6 (V) or pJG-Cdc7 (Cdc7) and grown in the presence or absence of Dox, as indicated, at 23°C. (C) Tetrad dissection of DY-75 (ΔM/+) and DY-76 (ΔN/+) strains. Gene replacement, sporulation, and tetrad analysis were performed as described in Materials and Methods. Growth of spores following dissection of four tetrads corresponding to each strain is shown. (D) A 10-fold dilution series of the Dbf4ΔN and isogenic wild-type haploid strains at a starting concentration of 1 × 107 cells/ml was spotted onto YPD plates containing the indicated concentrations of MMS and HU. Plates were incubated at 30°C for 2 days.
Since the removal of Dbf4 motif N abrogates its association with Rad53 (Fig. 2C and E), we examined whether Dbf4 ΔN haploids are hypersensitive to agents that normally trigger the S-phase checkpoint. Growth of the Dbf4 ΔN cells relative to isogenic haploid wild-type cells was inhibited in the presence of either MMS, an alkylating agent that causes DNA lesions, or HU, a ribonucleotide reductase inhibitor that causes replication fork stalling (Fig. 3D).
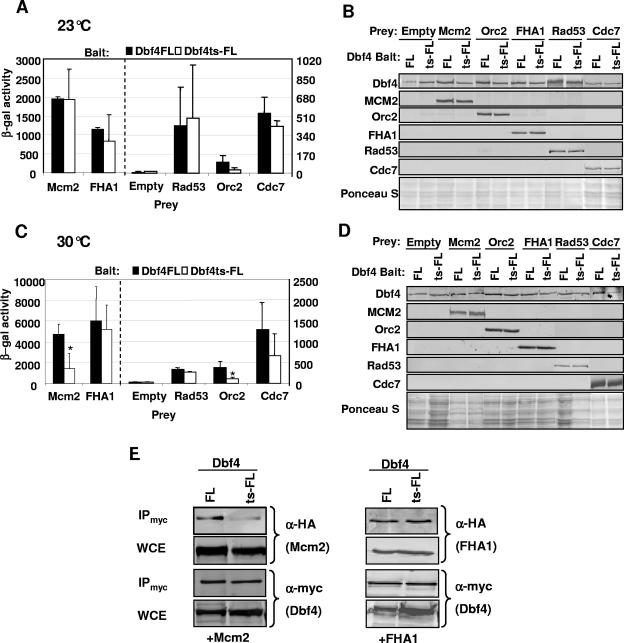
A number of temperature-sensitive (ts) DBF4 mutant strains have been identified which demonstrate a cell cycle arrest just prior to the initiation of DNA replication at restrictive temperature (reviewed in reference 31). We cloned and sequenced the Dbf4 ts allele dna52-1 (11, 48) from strain DY-2 and determined that it has a single point mutation, resulting in a change from proline to leucine at position 277 within motif M. This is identical to the previously reported sequence for the dbf4-1 allele (24). Given our finding that the elimination of motif M disrupts the interaction between Dbf4 and the DDK target Mcm2, we decided to test the effect of the P277L mutation on this association. We therefore prepared an additional two-hybrid bait construct by cloning the dna52-1 allele into pEG-202 to produce pEG-Dbf4ts-FL (Fig. 1). When the assay was carried out at permissive temperature for the dna52-1 allele (23°C), no significant difference was observed between Dbf4FL (wild type) and Dbf4ts-FL with respect to the strength of the two-hybrid signal for interactions with Mcm2, the FHA1 domain of Rad53, or full-length Rad53, Orc2, or Cdc7 (Fig. 4A and B). At 30°C (semipermissive temperature for DY-2), a sharp drop in association between Dbf4ts-FL and Mcm2 was observed, while very little difference was seen when Dbf4ts-FL or Dbf4FL was used as bait in combination with either full-length Rad53 or its FHA1 domain alone (Fig. 4C and D). Orc2 also had a weakened association with Dbf4ts-FL relative to Dbf4FL at 30°C. Assays at nonpermissive temperature (37°C) were attempted, but regardless of whether the bait used was wild-type or mutant Dbf4, the signals were not appreciably above background values obtained with empty vectors, indicating that the assay is suboptimal at this temperature. Finally, to confirm the effect of the Dbf4 P277L mutation on the interaction with Mcm2 at 30°C, coimmunoprecipitation experiments were carried out, and they revealed that the Dbf4 mutation greatly impaired its ability to interact with Mcm2, while its association with the Rad53 FHA1 domain remained unchanged (Fig. 4E).
FIG. 4.
Dbf4ts-FL has weakened interactions with both Orc2 and Mcm2. (A and C) Two-hybrid assays were carried out as described in Materials and Methods using bait constructs pEG-Dbf4-FL and pEG-Dbf4ts-FL. pJG-Mcm2, -Orc2, -FHA1, -Rad53, and -Cdc7 were used as prey, along with pJG 4-6 (empty). Induction of prey expression was carried out at either 23°C (A) or 30°C (C). Relative β-galactosidase activity is shown and in each case represents the average of at least three separate trials, with the error bars indicating standard deviations. Asterisks indicate a significant difference (P < 0.05; paired Student's t test) in the signal for Dbf4ts-FL relative to Dbf4-FL with equivalent preys. The dashed line indicates a separation of prey interactions for the left and right scales on the y axes. (B and D) Immunoblot analyses to verify bait and prey expression were carried out as described in the legend for Fig. 2. (E) Coimmunoprecipitation (IP) experiments were carried out as described in Materials and Methods. Shown are immunoblots of IP and input WCEs detected with either monoclonal anti-HA (for Mcm2 and FHA1 detection) or anti-Myc antibodies (for Dbf4FL and Dbf4ts-FL detection). In each case, 20 μg of the input WCE (determined by Bradford assay) and one-fourth of the final bead suspension were blotted.
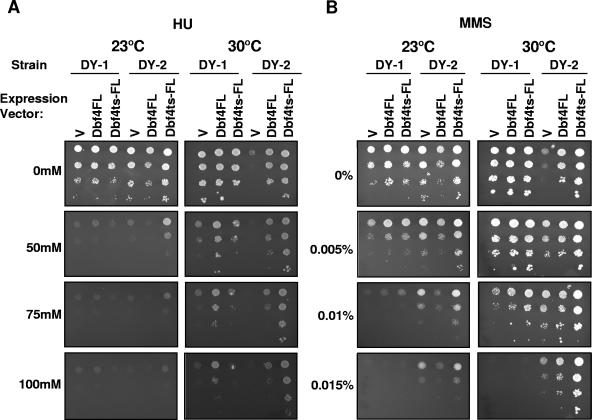
Since Dbf4ts-FL has weakened interactions with replication factors Mcm2 and Orc2 at 30°C, relative to Dbf4FL, but little change in its association with Rad53, we were interested in determining whether this mutant would be resistant to agents that normally provoke a checkpoint response. We therefore compared the ability of wild-type strain DY-1 and dna52-1 strain DY-2 to withstand various levels of either MMS or HU (Fig. 5). Serial dilutions of each strain, transformed with empty pCM190 vector, pCM190-Dbf4-FL (Fig. 1) (13), or pCM190-Dbf4ts-FL (see Materials and Methods), were spotted on a series of SD-Ura medium plates with various concentrations of either HU or MMS. At all concentrations tested, DY-1 and DY-2 cells transformed with empty vector seemed equally sensitive to HU at 23°C (Fig. 5A). In contrast, at this same temperature, empty vector DY-2 transformants actually grew better on elevated concentrations of MMS than did wild-type transformants (Fig. 5B). Remarkably, at semipermissive temperature for the dna52-1 allele, this growth advantage over the wild-type strain on MMS was maintained (30°C) (Fig. 5B). Plasmid-based expression of Dbf4ts-FL and, to a lesser degree, Dbf4FL enhanced growth on MMS for both DY-1 and DY-2 cells at 30°C. The most dramatic effect was observed with the combination of DY-2 cells transformed with pCM190-Dbf4ts-FL (Fig. 5B). A particularly surprising, yet reproducible, result from this analysis was that DY-2 cells transformed with empty vector actually grew better at 30°C on medium containing low concentrations of MMS than medium with no MMS. This was true for untransformed DY-2 cells exposed to MMS as well but was not observed with HU or UV treatment (Fig. 5A and data not shown). Intriguingly, the effects of Dbf4 expression for cells spotted on HU medium were allele specific, with DY-1 (wild-type) and DY-2 (dna52-1) cells benefiting most from overexpression of Dbf4FL and Dbf4ts-FL, respectively (Fig. 5A).
FIG. 5.
Dbf4ts-FL confers resistance to both MMS and HU. DY-1 and DY-2 cells, transformed with pCM190-Dbf4-FL, pCM190-Dbf4ts-FL, or empty pCM190 (V), were spotted in 10-fold dilution series onto selective media plates containing the indicated concentrations of MMS and HU, as described in Materials and Methods. Each plate series was incubated at 23°C or 30°C for 3 days, as indicated. It should be noted that reduced DY-1 cell growth at 23°C relative to 30°C likely reflects a slower cell cycle (see 0 mM HU and 0% MMS panels) rather than increased sensitivity to genotoxic agents at the lower temperature.
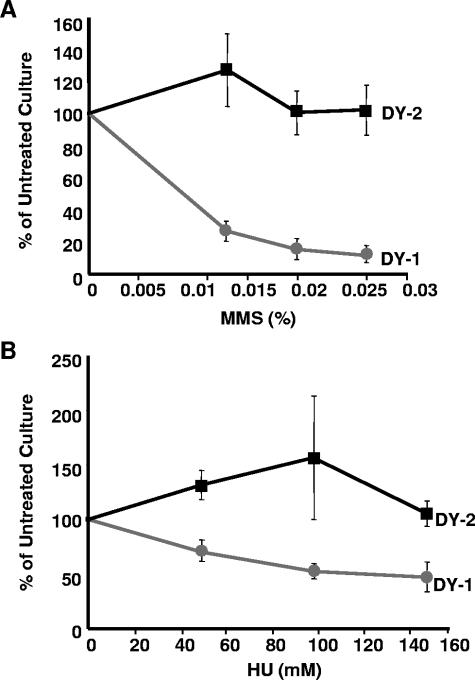
Finally, to evaluate the viability of DY-1 and DY-2 cells following shorter HU and MMS exposures, cultures were grown in media supplemented with various concentrations of these reagents for 4 hours and then plated on media without the supplements. Relative to untreated controls, the DY-2 cells maintained high levels of viability at all concentrations of HU and MMS tested, whereas the DY-1 cells fared much worse, demonstrating a clear dose-dependent drop in survival (Fig. 6).
FIG. 6.
Cells expressing Dbf4ts-FL are resistant to short-term exposure to either MMS or HU. DY-1 and DY-2 cells were treated with three concentrations of MMS (0.013%, 0.0195%, and 0.026%) (A) or HU (50 mM, 100 mM, and 150 mM) (B) for 4 h, plated in triplicate, and incubated as described in Materials and Methods. Colonies were counted, and the percentages were calculated relative to untreated control plated cells. Each data point represents the average of three independent experiments. Error bars represent standard deviations.
DISCUSSION
The M and N motifs of Dbf4 mediate specific interactions with replication and checkpoint factors.
Sequence comparisons of Dbf4 and its orthologs in other eukaryotes have revealed a number of conserved regions, including the M and N motifs (31, 49). For the first time, we have linked these motifs and their replication or checkpoint roles to previously identified physical interactions with replication and checkpoint factors. Our data demonstrate that deletion of the M motif abolishes the essential function of Dbf4 (Fig. 3A and C) and strongly suggest that this is due to the abrogation of its interaction with Mcm2 (Fig. 2C). Interestingly, while the original deletion of Dbf4 amino acids 110 to 296 reduced the interaction with Mcm2 considerably (Fig. 2A), the excision of motif M (removal of amino acids 263 to 309) resulted in a more dramatic effect on this association (Fig. 2C). This suggests that amino acid residues 297 to 309 make an important contribution to the Dbf4-Mcm2 interaction. While Cdc7 has also been shown to interact with Mcm2, this association was much weaker than that observed for Dbf4 and Mcm2 (28). Thus, it is very likely that disruption of Dbf4 motif M is sufficient to prevent effective DDK interaction with its critical substrate. By extending our analysis to the Dbf4 P277L mutant encoded by the temperature-sensitive dna52-1 allele, we showed that a single amino acid change within motif M is sufficient to dramatically impair the Dbf4-Mcm2 interaction at 30°C, but not at 23°C (Fig. 4A and C). Significantly, several other dbf4 alleles have been sequenced from temperature-sensitive strains, including dbf4-1 (which is identical to dna52-1 based on our sequence data), dbf4-2, and dbf4-3, all of which result in a change of one of two conserved motif M proline residues (Pro277 and Pro308), while dbf4-4 and dbf4-5 produce severely truncated proteins with intact M motifs (24). It has been shown previously that lysates from dbf4-1 cells fail to phosphorylate Mcm2 in an in vitro kinase assay (35), consistent with the notion that the Dbf4 P277L mutation disrupts the Dbf4-Mcm2 association. An alternative explanation is that this mutation instead abrogates the Dbf4-Cdc7 interaction, thereby preventing kinase activation. However, our data show that at 30°C the interaction between the mutated Dbf4 and Mcm2 is greatly impaired, whereas its interaction with Cdc7 is not significantly reduced (Fig. 4C). Deletion of Dbf4 motif N produced a strongly reduced two-hybrid signal for interaction with the checkpoint kinase Rad53 (Fig. 2C and E) and rendered haploid cells hypersensitive to both MMS and HU (Fig. 3D). These results are consistent with the previously proposed role for Dbf4 as a checkpoint target (21, 12, 39). Interestingly, it was the elimination of motif N and not motif M that had the greatest effect on the Dbf4-Orc2 interaction. It has been previously established that following exposure to HU, S. cerevisiae Dbf4 is phosphorylated and displaced from origins which are then prevented from firing, in a Rad53-dependent fashion (37, 42, 52). This may be important in allowing cells to efficiently recover once the checkpoint has been lifted (see below). Therefore, it is proposed that under conditions of genotoxic stress, Rad53 interacts with and phosphorylates the Dbf4 N motif, disrupting its interaction with Orc2 and releasing Dbf4 from origins. Consistent with this model, deletion of an eight-amino-acid stretch in the N motif of S. pombe Dfp1, including a number of potential Ser and Thr phosphorylation sites, or mutations of these residues result in sensitivity to a wide range of DNA-damaging agents (14, 49). However, this is hard to reconcile with the fact that plasmid-based expression of Dbf4ΔN was able to complement the growth defect in DY-2 cells at restrictive temperature as effectively as Dbf4FL (Fig. 3A), which suggests that the interaction between Dbf4 and Orc2 may not be essential for targeting Dbf4 to origins. While previous work has shown that the chromatin association of Dbf4 is greatly reduced in an orc2-1 mutant at restrictive temperature, there was nevertheless a proportion of total cellular Dbf4 that remained associated with the chromatin pellet in cell fractionation assays (36). These two models can be reconciled if the interaction with Orc2 is only one of several that helps to efficiently tether Dbf4 to origins. This would also explain the partial growth defect observed with the Dbf4ΔN strain (Fig. 3C). Phosphorylation of Dbf4 by Rad53 may cause a conformational change that disrupts interactions with additional ligands, such as other ORC subunits or MCM proteins. Interestingly, even at permissive temperature, the association of Orc2 with Dbf4ts was compromised relative to the strength of the two-hybrid signal for wild-type Dbf4-Orc2 (Fig. 4A). This further suggests that a tight association between the two proteins is not strictly required for cell cycle progression, which is consistent with the observed delocalization of Dbf4 foci in an orc2-1 background at 23°C as judged by immunofluorescence (36). Given the robust Dbf4ΔM-Orc2 association (Fig. 2E), the effects of the Dbf4ts 277 mutation appear to extend beyond motif M.
A single amino acid change within Dbf4 motif M confers resistance to genotoxic agents.
It has been previously shown that cdc7ts mutants are sensitive to genotoxic agents (32), and we have shown here that this is also true for the Dbf4ΔN mutant (Fig. 3D). It was therefore interesting to observe that Dbf4 P277L (Dbf4ts-FL), encoded by the dna52-1 allele, maintains a strong interaction with Rad53, while associations with both Orc2 and Mcm2 are compromised (Fig. 4). Furthermore, DY-2 cells are more resistant to MMS than DY-1 (wild-type) cells (Fig. 5 and 6), and in either strain the overexpression of Dbf4ts-FL, and to a lesser extent Dbf4FL, confers improved growth in the presence of MMS (Fig. 5B). What is the explanation for this effect? One possibility is that, due to its weakened interaction with replicative factors, Dbf4ts-FL is more easily displaced from origins by Rad53 during an MMS-provoked checkpoint response. This may improve the efficiency with which further origin firing is suppressed. However, the prevention of late origin firing is unlikely to lead to greater viability in the face of MMS exposure. A comparison of two mec1 mutants that fail to suppress late origin firing revealed that mec1Δ cells are hypersensitive to MMS while mec1-100 cells are not. Furthermore, the mec1Δ strain was found to have a 10-fold-higher rate of fork catastrophe than the mec1-100 strain (50). Thus, another explanation for the improved growth observed with the DY-2 cells is that the efficient Rad53-Dbf4 association stabilizes Dbf4 and, following checkpoint adaptation, Dbf4/Cdc7 aids in the resumption of normal replication fork progression through modification of a previously identified fork-associated substrate(s), such as Mcm2 (28), Cdc45 (33), and/or the polα-primase complex (52). Indeed, this may help to explain why DY-2 cells actually grow better at 30°C in the presence of lower concentrations of MMS (0.005% and 0.01%, but not 0.015%) (Fig. 5B). Although clearly speculative, it is possible that the deleterious effects of MMS are offset by the ability of Dbf4 to help to restart paused forks, mediated by its interaction with activated Rad53. Since replication fork stalling occurs even during an unperturbed S phase (8, 51), this activity of Dbf4 might aid recovery from both natural and lesion-induced impediments to fork progression. Although this effect was not observed with HU and UV exposure (Fig. 5A and data not shown), it is possible that the optimal doses of each to see such a growth enhancement were not used. Alternatively, the contrasting observations may be due to agent-specific differences in the checkpoint response (29). Given that Dbf4/Cdc7 has been shown to function in translesion synthesis (38), some additional candidate substrates include alternative polymerases and/or accessory proteins which are thought to permit replication fork progression through damaged DNA.
Another potential role for Dbf4 is that its interaction with Rad53 serves as a signal for downstream checkpoint effectors, consistent with the cell cycle arrest observed when the Dbf4 N terminus is overexpressed (13). In this scenario, the efficient initial establishment of the checkpoint might maintain the viability of a greater number of cells, which then successfully reenter the cell cycle following adaptation.
Plasmid-expressed Dbf4ts-FL is more beneficial than plasmid-expressed Dbf4FL for both DY-1 and DY-2 cells exposed to MMS (Fig. 5B). Curiously, this is not the case for cells grown on HU medium, as episomal and genomic expression of the same form of Dbf4 (i.e., episomal Dbf4FL for the DY-1 strain and episomal Dbf4ts-FL for the DY-2 strain) results in the best growth (Fig. 5A). Since Dbf4 and Cdc7 have been shown to form oligomers (45), it is possible that complexes with either Dbf4FL or Dbf4ts-FL are more effective than those with a mix of both at dealing with the consequences of HU-induced fork slowing, while such complexes or their composition may not be important for recovery from MMS-induced fork blockage
Conservation of eukaryotic DDK function.
Our finding that Dbf4 motif M is required for normal cell cycle progression, while motif N is most likely involved in checkpoint responses, adds to the growing body of evidence that DDK function is conserved among eukaryotes (reviewed in references 12 and 25), since similar conclusions have been arrived at from studies of Dbf4 ortholog motifs in both fission yeast (14, 34, 49) and humans (25, 43). In the present report, we have advanced the understanding of how Dbf4 fulfills both roles by establishing important functional connections between these motifs and specific interactions with DNA replication and checkpoint factors. Our finding that the conserved M motif mediates the Dbf4-Mcm2 interaction is supported by the fact that MCM proteins, and Mcm2 in particular, have been shown to be DDK substrates in eukaryotes as diverse as yeast and humans (reviewed in references 12 and 25). A role for Dbf4 in preventing the firing of late origins during the S-phase checkpoint also appears to be a common feature of eukaryotes (reviewed in reference 12). Despite this conserved function, we favor a model whereby the Dbf4 motif N-Rad53 interaction guards against the effects of genotoxic agents by aiding in the recovery from replication fork arrest. This is supported by the work with mec1 mutants showing that the failure to suppress late origin firing per se does not render cells hypersensitive to genotoxic insult, whereas increased rates of fork catastrophe do (50). As stated above, an attractive idea that merits further study is the notion that an interaction with Rad53 may stabilize Dbf4 during the checkpoint and thereby facilitate a role in resuming replication, potentially through the phosphorylation of DDK fork substrates (12).
In the present report we have, for the first time, made important functional connections between conserved Dbf4 regions and cell cycle factors associated with the roles these motifs play. It will be of considerable interest to determine whether these mechanisms can now be extended to all eukaryotes.
Acknowledgments
We thank David Schnurr for technical assistance, Susan Gasser (FMI, Basel) and Philippe Pasero (CNRS, Montpellier) for yeast strains and plasmids, and Grant Brown (University of Toronto) for critical reading of the manuscript.
This research was funded by a grant from the Natural Sciences and Engineering Research Council of Canada. A.E.V. is the recipient of an Ontario Graduate scholarship, and B.P.D. is a Research Scientist of the Canadian Cancer Society.
REFERENCES
- 1.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed., p. 13.53-13.61. Wiley, New York, N.Y.
- 2.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 3.Bousset, K., and J. F. Diffley. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, G. W., and T. J. Kelly. 1999. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc. Natl. Acad. Sci. USA 96:8443-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Cheng, L., T. Collyer, and C. F. Hardy. 1999. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19:4270-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman, and J. Gautier. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11:203-213. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030-1033. [DOI] [PubMed] [Google Scholar]
- 9.Dohrmann, P. R., G. Oshiro, M. Tecklenburg, and R. A. Sclafani. 1999. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics 151:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson, A. D., W. L. Fangman, and B. J. Brewer. 1998. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 12:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumas, L. B., J. P. Lussky, E. J. McFarland, and J. Shampay. 1982. New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol. Gen. Genet. 187:42-46. [DOI] [PubMed] [Google Scholar]
- 12.Duncker, B. P., and G. W. Brown. 2003. Cdc7 kinases (DDKs) and checkpoint responses: lessons from two yeasts. Mutat. Res. 532:21-27. [DOI] [PubMed] [Google Scholar]
- 13.Duncker, B. P., K. Shimada, M. Tsai-Pflugfelder, P. Pasero, and S. M. Gasser. 2002. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA 99:16087-16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung, A. D., J. Ou, S. Bueler, and G. W. Brown. 2002. A conserved domain of Schizosaccharomyces pombe dfp1+ is uniquely required for chromosome stability following alkylation damage during S phase. Mol. Cell. Biol. 22:4477-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 16.Geraghty, D. S., M. Ding, N. H. Heintz, and D. S. Pederson. 2000. Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J. Biol. Chem. 275:18011-18021. [DOI] [PubMed] [Google Scholar]
- 17.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, C. F. 1996. Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol. Cell. Biol. 16:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy, C. F., and A. Pautz. 1996. A novel role for Cdc5p in DNA replication. Mol. Cell. Biol. 16:6775-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimi, Y. 1997. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272:24508-24513. [DOI] [PubMed] [Google Scholar]
- 21.Jares, P., A. Donaldson, and J. J. Blow. 2000. The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO Rep. 1:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jares, P., M. G. Luciani, and J. J. Blow. 2004. A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication. BMC Mol. Biol. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 24.Kihara, M., W. Nakai, S. Asano, A. Suzuki, K. Kitada, Y. Kawasaki, L. H. Johnston, and A. Sugino. 2000. Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. Its protein kinase activity is regulated by Rad53p. J. Biol. Chem. 275:35051-35062. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J. M., M. Yamada, and H. Masai. 2003. Functions of mammalian Cdc7 kinase in initiation/monitoring of DNA replication and development. Mutat. Res. 532:29-40. [DOI] [PubMed] [Google Scholar]
- 26.Landis, G., and J. Tower. 1999. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126:4281-4293. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. K., and J. Hurwitz. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei, M., Y. Kawasaki, M. R. Young, M. Kihara, A. Sugino, and B. K. Tye. 1997. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 11:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J. S., S. R. Kuo, and T. Melendy. 2003. Comparison of checkpoint responses triggered by DNA polymerase inhibition versus DNA damaging agents. Mutat. Res. 532:215-226. [DOI] [PubMed] [Google Scholar]
- 30.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532:41-58. [DOI] [PubMed] [Google Scholar]
- 31.Masai, H., and K. Arai. 2000. Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem. Biophys. Res. Commun. 275:228-232. [DOI] [PubMed] [Google Scholar]
- 32.Njagi, G. D., and B. J. Kilbey. 1982. cdc7-1, a temperature sensitive cell-cycle mutant which interferes with induced mutagenesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 186:478-481. [DOI] [PubMed] [Google Scholar]
- 33.Nougarede, R., S. F. Della, P. Zarzov, and E. Schwob. 2000. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 20:3795-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino, K., T. Takeda, E. Matsui, H. Iiyama, C. Taniyama, K. Arai, and H. Masai. 2001. Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J. Biol. Chem. 276:31376-31387. [DOI] [PubMed] [Google Scholar]
- 35.Oshiro, G., J. C. Owens, Y. Shellman, R. A. Sclafani, and J. J. Li. 1999. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol. 19:4888-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasero, P., B. P. Duncker, E. Schwob, and S. M. Gasser. 1999. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 13:2159-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasero, P., K. Shimada, and B. P. Duncker. 2003. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle. 2:568-572. [PubMed] [Google Scholar]
- 38.Pessoa-Brandao, L., and R. A. Sclafani. 2004. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics 167:1597-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pike, B. L., N. Tenis, and J. Heierhorst. 2004. Rad53 kinase activation-independent replication checkpoint function of the N-terminal forkhead-associated (FHA1) domain. J. Biol. Chem. 279:39636-39644. [DOI] [PubMed] [Google Scholar]
- 40.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 41.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 42.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 43.Sato, N., M. Sato, M. Nakayama, R. Saitoh, K. Arai, and H. Masai. 2003. Cell cycle regulation of chromatin binding and nuclear localization of human Cdc7-ASK kinase complex. Genes Cells 8:451-463. [DOI] [PubMed] [Google Scholar]
- 44.Sclafani, R. A. 2000. Cdc7p-Dbf4p becomes famous in the cell cycle. J. Cell Sci. 113:2111-2117. [DOI] [PubMed] [Google Scholar]
- 45.Shellman, Y. G., I. E. Schauer, G. Oshiro, P. Dohrmann, and R. A. Sclafani. 1998. Oligomers of the Cdc7/Dbf4 protein kinase exist in the yeast cell. Mol. Gen. Genet. 259:429-436. [DOI] [PubMed] [Google Scholar]
- 46.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solomon, N. A., M. B. Wright, S. Chang, A. M. Buckley, L. B. Dumas, and R. F. Gaber. 1992. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 8:273-289. [DOI] [PubMed] [Google Scholar]
- 49.Takeda, T., K. Ogino, E. Matsui, M. K. Cho, H. Kumagai, T. Miyake, K. Arai, and H. Masai. 1999. A fission yeast gene, him1+/dfp1+, encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol. 19:5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y., M. Vujcic, and D. Kowalski. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21:4938-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinreich, M., and B. Stillman. 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18:5334-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]