Abstract
Corepressor N-CoR (nuclear receptor corepressor) and the highly related protein SMRT (silencing mediator of retinoid and thyroid hormone receptor) play important roles in different biological processes including proliferation, differentiation, and development. Understanding the biological function of these corepressors requires identification and characterization of their interacting proteins. Here we report the characterization of a novel N-CoR-interacting protein, JMJD2A (previously known as KIAA0677). JMJD2A is an evolutionarily conserved nuclear protein containing many functionally unknown domains. JMJD2A directly interacts with the N-terminal region of N-CoR through a small NID (N-CoR interaction domain) both in vitro and in vivo. Despite its copurification with N-CoR, JMJD2A is not a core subunit of the stable multiprotein N-CoR complex and is not required for N-CoR-mediated repression by thyroid hormone receptor. By chromatin immunoprecipitation cloning, we identified the human achaete scute-like homologue 2 (ASCL2/Hash2) gene as a gene regulated by JMJD2A. ASCL2 is a basic helix-loop-helix transcription factor whose mouse homolog is encoded by an imprinted gene highly expressed during the development of extraembroynic trophoblast lineages but repressed in other tissues and is essential for proper placental development. We demonstrated that JMJD2A selectively represses the expression of the ASCL2 gene but not other imprinted genes in the same imprinted locus in HeLa cells and that this repression required a functional N-CoR complex and the tandem Tudor domain of JMJD2A. Like N-CoR, JMJD2A is widely expressed in various mouse tissues. Our data indicate that JMJD2A makes use of the N-CoR complex to repress transcription and suggest that JMJD2A together with N-CoR could play a role in repressing ASCL2 expression in various tissues.
In eukaryotic cells, DNA is wrapped around histone octamers that consist of two molecules of each of the core histones H2A, H2B, H3, and H4 to form chromatin. Although chromatin was once considered to serve only as a structural component for the storage of genomic information, it is now widely accepted that chromatin also plays essential roles in the regulation of multiple processes including DNA replication, transcription, and repair (2, 8, 47, 49). Numerous proteins and protein complexes regulate transcription by posttranslationally modifying histones. These modifications include acetylation, methylation, phosphorylation, ubiquitylation, and sumoylation (2, 39, 54). It is well established that increased histone acetylation generally correlates with transcription activation and that deacetylation correlates with repression. Although the precise mechanism that links acetylation to transcriptional regulation is still not fully understood, histone acetylation catalyzed by histone acetyltransferases are thought to weaken DNA/histone interactions and affect higher-order chromatin folding to modulate the accessibility of promoter sequences to transcription factors and basal transcriptional machinery (12, 36). In addition, histone acetylation can serve as a specific code for binding of transcriptional regulatory proteins (40, 42). On the other hand, histone deacetylation catalyzed by HDACs (histone deacetylases) antagonizes acetyltransferase activity and favors the tight compaction of the histone-DNA complex.
Numerous HDAC-containing repression complexes have been identified (3, 4, 9, 11, 48). Among them are N-CoR (nuclear receptor corepressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptor) complexes. SMRT and N-CoR are two highly related proteins originally isolated and characterized by their ability to interact with the unliganded forms of nuclear hormone receptors and confer transcriptional repression (18, 34, 35). Recent studies have implicated N-CoR and SMRT in repression by diverse transcription factors including Mad/Mxi, BCL6/LAZ3, ETO, CBF, and REST/NRSF (7, 11, 22, 23, 32, 33, 43, 44). Mouse N-CoR is required for early embryonic development, and N-CoR null mice show defects in neural cell differentiation and developmental progression of specific erythrocytes and thymocytes (17, 21). To understand the mechanism by which SMRT and N-CoR mediate transcriptional repression, several groups have attempted isolation and characterization of steady-state SMRT and N-CoR complexes. These efforts collectively demonstrate that both SMRT and N-CoR exist in large protein complexes with an estimated size of 1.5 to 2 MDa and are primarily associated with TBL1 and TBLR1, two highly related WD-40 repeat proteins, GPS2, a cellular signaling protein, and HDAC3 (13, 27, 50, 51, 53). As these proteins appear to stably associate with SMRT and N-CoR in stoichiometric levels (13, 27, 50, 53), they can be viewed as the core subunits of SMRT and N-CoR complexes. Consistent with this idea, recent studies indicate that HDAC3 is required for SMRT/N-CoR-mediated repression by unliganded thyroid hormone receptor (TR) (20, 50), whereas TBL1 and TBLR1 are redundantly essential for this process (52).
In addition to the aforementioned core subunits, the diverse functions of SMRT and N-CoR are likely to be carried out by other proteins that interact transiently and/or differentially with SMRT and N-CoR. In this study, we have characterized a novel protein, KIAA0677, which was copurified with N-CoR (51). KIAA0677 is a highly evolutionally conserved protein containing several interesting domains including a Jumonji domain (JmjN) and a related JmjC domain, a PHD domain, and a tandem Tudor domain (Fig. 1A) (24). Although the function of these domains is not clear, the proteins harboring these domains have often been implicated in transcriptional regulation. Due to the presence of the JmjN and JmjC domains, this protein was recently renamed JMJD2A, a member of the proposed JMJD2 family proteins (24). We show here that JMJD2A physically interacts with N-CoR but is not a core subunit of the N-CoR complex. We identified the human achaete-scute homolog 2 (ASCL2, also called Hash2) gene, encoding a basic helix-loop-helix (bHLH) transcription factor, as a target gene repressed by JMJD2A. Mouse ASCL2, also called Mash2, is a transcriptional factor essential for placental development. Mash2−/− embryos die from placental failure at 10 days postcoitum (15). The ASCL2 gene has been mapped to human chromosome 11p15.5, a region containing a cluster of imprinted genes, including the insulin-2, insulin-like growth factor 2, and H19 genes (1, 14). We provide evidence that JMJD2A is specifically involved in the repression of the ASCL2 gene and does not appear to affect the transcriptional activity of the other genes in this imprinted region. Furthermore, this repression requires a functional N-CoR complex, suggesting a working model in which JMJD2A targets the N-CoR complex to repress ASCL2 gene expression. Our data also suggest that JMJD2A and N-CoR may be involved in the regulation of fetoplacental development through repression of ASCL2.
FIG. 1.
JMJD2A interacts with N-CoR but is unlikely to be a core subunit of the N-CoR complex. (A) Diagram showing the structural organization of the 1,064-amino-acid JMJD2A protein. The JmjN, JmjC, PHD, and tandem Tudor domains are indicated. (B) Reciprocal coimmunoprecipitation of JMJD2A and N-CoR from HeLa nuclear extracts. The immunoprecipitation (IP) and Western blotting (WB) were as indicated. For a control, IP with rabbit anti-mouse IgG was used. (C) Transiently expressed FLAG-tagged JMJD2A (F-JMJD2A) also coimmunoprecipitated with N-CoR. (D) A small fraction of JMJD2A cofractionated with N-CoR in a gel filtration experiment. HeLa cell nuclear extracts were passed through a Superose 6 column, and different fractions were collected and resolved by 7.5% SDS-PAGE. Western blots were conducted with either anti-JMJD2A or anti-N-CoR antibodies as indicated. The peak fractions for molecular weight markers were as indicated. (E) Northern analysis showing the expression of JMJD2A and N-CoR in various mouse tissues. The levels of GADPH mRNA served as a loading control.
MATERIALS AND METHODS
Preparation of whole-cell lysates and immunoprecipitation.
HeLa cells were washed with phosphate-buffered saline (PBS) two times and lysed with EBC buffer (20 mM Tris-HCl, pH 8.0, 125 mM NaCl, 2 mM EDTA, and 0.5% NP-40) with protease inhibitors. After removing cell debris by high-speed centrifugation, cell lysates were precleared with protein A-conjugated Sepharose beads for 2 h at 4°C with gentle agitation. Then specific antibodies were added and incubated overnight at 4°C with rotation. Immunoprecipitation was done by adding protein A-conjugated Sepharose beads and incubating at 4°C for another 2 h with rotation. Beads were washed five times with EBC buffer, 10 min each. Sodium dodecyl sulfate (SDS) buffer (2×) was added directly to the beads and boiled for 5 min before samples were loaded and separated by 7.5% SDS-polyacrylamide gel electrophoresis (PAGE). For multiple rounds of blotting, the membranes were stripped in stripping buffer (100 mM β-mercaptolethanol, 2% SDS, and 62.5 Tris-Cl, pH 6.7) for 30 min at 50°C.
Northern blot and immunostaining.
The nylon membrane with mRNA from different mouse tissues was purchased from Ambion Inc. (Austin, TX) and probed with 32P-labeled specific probes (amplified by reverse transcription-PCR [RT-PCR]) against either mouse JMJD2A, N-CoR, or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) according to the manufacturer's instructions. For immunostaining, cells were grown on glass cover slides overnight and fixed with 3.5% formaldehyde for 15 min. Then, cells were permeabilized with 0.02% NP-40 for 1 min and blocked with 5% goat serum for 1 h. Next, cells were incubated with the anti-JMJD2A-specific antibody for 2 h, followed by a secondary donkey anti-rabbit immunoglobulin G (IgG) conjugated with Alexa Fluor 594 for 2 h. Images were visualized by conventional microscopy.
Plasmids and transient transfection.
The various N-CoR constructs used for the in vitro transcription/translation reaction were as described previously (27, 50), and the SMRTe construct (35) was kindly provided by Don Chen (University of Massachusetts). The DNA constructs used in the Gal4 fusions were made by cloning amplified PCR products into modified pCMX-Gal mammalian expression vectors. Inverse PCR was used to generate different JMJD2A deletions. For glutathione S-transferase (GST) pull-down, the DNA encoding the peptide (597 to 720 amino acids) that contains the N-CoR binding domain of JMJD2A was amplified by RT-PCR and cloned into pGEX-4T (Pharmacia), creating an in-frame fusion to GST. The same GST fusion peptide was used for the generation of polyclonal anti-JMJD2A antibodies. To generate the ASCL2P-luc reporter, the DNA fragment corresponding to −1523 to +1 of the ASCL2 promoter was PCR amplified and cloned into the pGL3-basic reporter. The various ASCL2 promoter deletion mutants were constructed through PCR based on ASCL2P-Luc. All constructs were verified by sequencing. Transient transfections of DNA were done using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen Inc.). Most of the transient-transfection experiments were performed with triplicates and repeated at least three times.
GST pull-down assay.
GST pull-down assays were done as previously described (50). Both GST and GST-JMJD2A (597-720) fusion proteins were expressed in BL21 Escherichia coli cells, and equal amounts of each protein were immobilized onto glutathione-Sepharose beads. The beads were incubated overnight with 35S-labeled SMRT, N-CoR, other proteins, or N-CoR fragments synthesized using a TNT in vitro transcription/translation kit (Promega) according to the manufacturer's instruction. The beads were washed five times with washing buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40 with protease inhibitors). The samples were then separated by SDS-PAGE and visualized by autoradiography.
Cell culture and siRNA.
HeLa cells were routinely maintained in Dulbecco's modified Eagle medium (Invitrogen Inc.) supplemented with 10% fetal bovine serum and 1% antibiotics at 37°C under 5% CO2. For the small interference RNA (siRNA) experiments, HeLa cells were seeded the night before transfection at such a density that cells reach about 30 to 40% confluence by the time of transfection. The siRNAs against N-CoR and HDAC3 were previously described (50). The siRNA against JMJD2A was the combination of two siRNAs synthesized by Dharmacon Research: siJMJD2-I (AACACAGUUAUUGACCAUACU) and JMJD2A-II (AAGUUGAGGAUGGUCUUACCU). A combination of 20 nM of each siJMJD2A was used for transfection using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfected cells were continued in culture for 3 days before harvesting for further analyses. The efficiency of the siRNA knockdown was determined by Western analysis using corresponding specific antibodies.
Total RNA extraction and RT-PCR.
Total RNA from cultured cells was purified using the RNeasy kit (QIAGEN) according to the manufacturer's instructions. All RNA extracts were treated with RNase-free DNase to eliminate genomic DNA contamination. RT-PCR was conducted using a QIAGEN kit. Primers for the ASCL2 amplification were 5′-AGCCCGGCTCCCCGCGTTCCGCCTACT-3′ and 5′-TAGCCCCCTAACCAGCTGGAGAAGTCG-3′. Primers for the GAPDH amplification were 5′-ACAGCCTCAAGATCATCAGCAA and 5′-ACCACTGACACGTTGGCAGT-3′. Primers for the IGF-2 amplification were 5′-TGCAGTAGTTCTCCAGCTGGTAGA-3′ and 5′-AGCCTTTGTGAACCAACACCTGT-3′. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
ChIP and ChIP cloning.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (51) with the following modifications. In brief, to detect the association of JMJD2A and N-CoR with the endogenous ASCL2 promoter, approximately 2 × 109 HeLa cells in 150-mm dishes were treated with 1% formaldehyde in PBS for 10 min at room temperature with gentle agitation. The cells were rinsed twice with PBS and then incubated in 100 mM Tris-HCl (pH 9.4)-10 mM dithiothreitol (DTT) at 30°C for 15 min. After centrifugation at 3,000 rpm for 1 min, the cells were rinsed once with cold PBS and resuspended in 600 μl of solution A (10 mM HEPES, pH 7.9, 0.5% NP-40, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT). The cell lysates were centrifuged at 3,000 rpm for 5 min, and the supernatant was discarded. The nuclear pellets were resuspended in solution B (20 mM HEPES, pH 7.9, 25% glycerol, 0.5% NP-40, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA) containing protease inhibitors by vigorous pipetting to release nuclear proteins. After centrifugation at 13,000 rpm for 30 min, the nuclear pellets were resuspended in IP buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl with protease inhibitors) and sonicated to break chromatin into fragments with an average length of 0.5 to 1.0 kb. ChIP assays were then carried out as described previously (51) using the antibodies indicated in the figures.
For the identification of JMJD2A target genes, ChIP cloning was first conducted with the JMJD2A antibody essentially as described above. After the final wash, DNA-protein complexes were eluted twice in 50 μl of 10 mM DTT at 37°C for 10 min. The eluted DNA was combined and diluted 20× with dilution buffer (2 mM EDTA, 100 mM NaCl, 20 mM Tris-HCl, pH 8.1, and 0.5% Triton X-100). The N-CoR antibody was added to conduct the second round of ChIP. The resulting DNA was purified using a Qiaquick kit (QIAGEN Sciences), blunt-ended, cloned, and sequenced. The 171-bp JAR sequence is available upon request.
RESULTS
Characterization of JMJD2A as an N-CoR-associated protein.
We previously showed that KIAA0677 copurified with N-CoR (51). Recently, KIAA0677 was categorized as a member of the JMJD2 (Jumonji domain-containing 2) family and named JMJD2A (24). JMJD2A was mapped to human chromosome 1p34.3. The full-length JMJD2A cDNA is more than 4.7 kb, with an open reading frame encoding a protein of 1,064 amino acids.
As a first step to verify that JMJD2A truly interacts with N-CoR, we raised a JMJD2A-specific antibody against amino acids 592 to 720 of JMJD2A. HeLa nuclear extracts were first immunoprecipitated with an N-CoR- or JMJD2A-specific antibody and then analyzed by Western blotting. The result in Fig. 1B showed that N-CoR coprecipitated with JMJD2A and vice versa. Under the same condition, a control rabbit anti-mouse IgG did not precipitate either N-CoR or JMJD2A. To further confirm the interaction between these two proteins, a DNA construct encoding FLAG-tagged JMJD2A was transfected into HeLa cells. Whole-cell lysates were prepared from the transfected cells and used for immunoprecipitation with antibodies against either FLAG or N-CoR. Again, FLAG-JMJD2A coimmunoprecipitated with N-CoR (Fig. 1C). Together, these data provide strong evidence that JMJD2A interacts with N-CoR.
To examine the extent to which JMJD2A associates with N-CoR, HeLa nuclear extract was subjected to gel filtration analysis using a Superose 6 column and the resulting fractions were analyzed by Western blotting. Consistent with our previous data (27), N-CoR migrated as a 1.5- to 2-MDa complex(es) and peaked between fractions 9 and 11 (Fig. 1D). On the other hand, only a small fraction of JMJD2A cofractionated with N-CoR (peak 11) and the majority of JMJD2A existed either in a very small complex(es) or alone (fractions 17 to 21). This is unlikely as a result of JMJD2A’s being much more abundant than N-CoR, since depletion of JMJD2A from HeLa nuclear extracts could not deplete N-CoR and vice versa (data not shown). Thus, under our experimental conditions, much of the cellular JMJD2A protein did not appear to associate with the N-CoR complex(es), suggesting that JMJD2A is unlikely a core subunit of the N-CoR complex(es).
Expression patterns of JMJD2A.
JMJD2A is not only part of a newly identified protein family with a few conserved domains but also shows a high degree of conservation in evolution, with homologues from yeast to humans (24; our unpublished data). Somewhat surprisingly, little is known about the functions of this protein family. Given its conservation in evolution, we were interested in its expression pattern in mice. A tissue blot was analyzed by Northern hybridization, and results in Fig. 1E show that JMJD2A is broadly expressed, although thymus, ovary, and heart have relatively higher levels of expression. Noticeably, JMJD2A and N-CoR have similar patterns of expression, and both are also highly expressed in embryos.
JMJD2A is a nuclear protein and has repression activity.
We next examined the subcellular localization of JMJD2A by immunostaining. The result (Fig. 2A) showed that in HeLa cells JMJD2A was detected primarily in nuclei by using our JMJD2A-specific antibody raised against amino acids 592 to 720 of JMJD2A. In addition, the Flag-tagged JMJD2A transiently expressed in HeLa cells was also found primarily in nuclei as revealed by immunostaining using a Flag-specific antibody (data not shown). Together these results indicate that JMJD2A in HeLa cells is a nuclear protein, consistent with the fact that JMJD2A copurified with N-CoR from HeLa nuclear extracts.
FIG. 2.
JMJD2A is a nuclear protein that possesses transcriptional-repression activity. (A) Immunostaining revealed that JMJD2A is primarily a nuclear protein in HeLa cells. Also shown is DAPI (4′,6′-diamidino-2-phenylindole) staining of nuclei. (B) JMJD2A functioned as a repressor when tethered to a target reporter. The full-length JMJD2A was fused to the Gal4 DNA binding domain (Gal-JMJD2A), and repression was measured by cotransfection with the 4xUAS-TK-luc reporter. The expression of Gal-JMJD2A was confirmed by Western analysis, as shown at the bottom of panel. (C) Repression by Gal-JMJD2A was correlated with recruitment of N-CoR and SMRT corepressor complexes. HeLa cells were transfected with reporter alone or reporter plus Gal(DBD) or Gal-JMJD2A, and, after overnight incubation, ChIP assays were carried out to determine the recruitment of SMRT and N-CoR and their associated HDAC3 to the reporter.
As a nuclear protein that copurified and coimmunoprecipitated with N-CoR (Fig. 1B and 1C), we next tested whether JMJD2A possesses repression activity when tethered to DNA through a heterologous DNA binding domain (Gal-DBD). A luciferase reporter driven by a minimal thymidine kinase (TK) promoter containing four Gal binding sites (4xUAS) was cotransfected with either a Gal-DBD or Gal-JMJD2A expression construct into HeLa cells. After overnight incubation, relative luciferase activity was measured from the transfected cells. The results in Fig. 2B showed that Gal-JMJD2A actively repressed transcription in comparison to the Gal-DBD alone.
To analyze whether the repression by Gal-JMJD2A is related to the interaction between JMJD2A and N-CoR, we used ChIP assays to test whether the N-CoR complex was recruited by JMJD2A to the 4xUAS-TK-luc reporter. For this purpose, HeLa cells were transfected with the reporter alone or the reporter plus either the Gal-DBD or Gal-JMJD2A expression construct. After overnight incubation, the cells were treated with 1% formaldehyde to cross-link proteins to DNA and processed for ChIP assay as described in detail in Materials and Methods. The results in Fig. 2C showed that, while both Gal-DBD and Gal-JMJD2A bound to the reporter as expected, N-CoR was detected only in the presence of Gal-JMJD2A. This result indicates that targeting Gal-JMJD2A to the reporter resulted in the recruitment of N-CoR. Furthermore, a ChIP assay using a SMRT-specific antibody also revealed the recruitment of SMRT by Gal-JMJD2A. This result indicates that JMJD2A can interact with both SMRT and N-CoR. Consistent with the fact that HDAC3 is a core subunit of the SMRT/N-CoR complexes, HDAC3 was also recruited by Gal-JMJD2A. Together, these ChIP data provide strong evidence that JMJD2A associates with SMRT/N-CoR in vivo and also reveal that JMJD2A is likely to repress transcription by recruiting the SMRT/N-CoR complex.
JMJD2A interacts with N-CoR through a 41-amino-acid domain located in the middle of the molecule.
The above ChIP data indicate that tethering JMJD2A to DNA results in recruitment of the N-CoR complex. We next made use of this ChIP assay to map the N-CoR interaction domain in JMJD2A. As shown in Fig. 1A, JMJD2A contains the JmjC, JmjN, Tudor, and PHD domains, which are conserved among the JMJD2 family members (24). To map the N-CoR interaction domain(s), we first divided the JMJD2A protein into three fragments, the N-terminal (amino acids 1 to 370), the middle (amino acids 347 to 720), and the C-terminal (amino acids 680 to 1064) fragments. Each fragment was fused with the Gal4-DBD, and their ability to recruit N-CoR was analyzed by ChIP assay. This analysis revealed that the middle portion of JMJD2A (fragment 347-720) strongly recruited N-CoR (Fig. 3A). Fragment 1-370 could also recruit N-CoR, although it was at least five times weaker than fragment 347-720 (Fig. 3A). On the other hand, no detectable recruitment was observed for fragment 680-1064 (Fig. 3A). Thus, JMJD2A may contain two N-CoR interaction domains, a weak one in fragment 1-370 and a strong one in the region between amino acids 347 and 720. Note that this experiment did not exclude the possibility that JMJD2A indirectly recruits N-CoR via interaction with other subunits in the N-CoR complex.
FIG. 3.
Mapping the NID of JMJD2A to a small 41-amino-acid NID region. (A to D) Various JMJD2A fragments as indicated were fused to the Gal-DBD, and their ability to recruit N-CoR to the 4xUAS-TK-luc reporter was determined by ChIP assay. In each case, a ChIP assay using a Gal-DBD-specific antibody was also performed to confirm that each Gal fusion protein was expressed and bound to the reporter. ChIP with IgG served as a negative control. JΔNID represents JMJD2A deleted of the NID fragment (amino acids 597 to 637). (E) Summary of the interaction between various JMJD2A constructs and N-CoR.
We next focused our attention on further defining the N-CoR interaction domain within fragment 347-720. A series of new Gal fusion constructs were generated (Fig. 3E), and ChIP assays were employed to analyze their ability to recruit N-CoR. The result in Fig. 3B shows that fragment 597-720 was still capable of recruiting N-CoR. Further, the experiment in Fig. 3C shows that a 41-amino-acid fragment (597-637) was necessary and sufficient for the recruitment of N-CoR. Most importantly, the result in Fig. 3D shows that the full-length JMJD2A deleted of this 41-amino-acid fragment (Gal-597-638) was not able to recruit N-CoR. As controls, both Gal fusions of full-length JMJD2A and the 41-amino-acid fragment were able to recruit N-CoR under the same condition. Thus, in the full-length context, the 41-amino-acid fragment from 597 to 638 is necessary for the recruitment of N-CoR, despite the presence of a weak N-CoR interaction domain in fragment 1-370. Taken together, we conclude that the 41-amino-acid region from 597 to 638 of JMJD2A is necessary and sufficient for interaction with N-CoR. The result of these interaction assays is summarized in Fig. 3E.
Identification of a JMJD2A interaction domain in N-CoR.
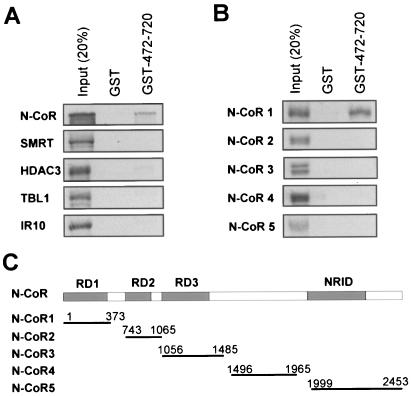
We next used an in vitro GST pulldown assay to test whether JMJD2A directly interacts with N-CoR. A GST fusion containing the JMJD2A N-CoR interaction domain was expressed in E. coli and purified. This GST fusion (GST-472-720), together with control GST alone, was used to pull down in vitro-translated, [35S]Met-labeled N-CoR, SMRT, HDAC3, TBL1, and IR10, the components of the N-CoR/SMRT complexes (50). The result in Fig. 4A showed that, among these proteins, only N-CoR bound significantly to GST-472-720. This result indicates that JMJD2A most likely interacts directly with N-CoR but not other core subunits of N-CoR. Given that SMRT was recruited by full-length JMJD2A in the ChIP assay (Fig. 2C), the lack of SMRT binding in the GST pulldown assay in Fig. 4A suggests that SMRT may interact with JMJD2A through another unidentified region.
FIG. 4.
JMJD2A directly interacts with N-CoR but not other subunits in the N-CoR complex. (A) A GST fusion of JMJD2A 472-720 containing the NID was expressed in E. coli and purified. Full-length N-CoR, SMRT, HDAC3, TBL1, and IR10 proteins were individually translated, [35S]Met labeled in vitro using the TNT transcription/translation kit, and used for GST pulldown assays. GST protein was included as a negative control. (B) The N-CoR protein was divided into five fragments, and binding to JMJD2A was analyzed by GST pulldown as in panel A. (C) Diagram showing the five N-CoR fragments used in panel B.
We further mapped the JMJD2A interaction domain in N-CoR using a series of N-CoR constructs as indicated in Fig. 4C by GST pulldown assay. The results in Fig. 4B showed that only the N-terminal domain (amino acids 1 to 373), not other regions of N-CoR, interacted with the N-CoR binding domain of JMJD2A. Thus, we conclude that JMJD2A directly interacts with the N-terminal fragment of N-CoR containing the previously identified repression domain 1 (RD1) (16).
With the knowledge that JMJD2A interacts with N-CoR through a 41-amino-acid fragment from 597 to 637, we next analyzed whether this interaction correlates with JMJD2A's repression activity. The result in Fig. 5A showed that fragment 1-370, containing the JmjC and JmjN domains, had no repression activity. To our satisfaction, the fragment containing the N-CoR interaction domain (Gal-347-720) repressed transcription from 4xUAS-TK-luc as efficiently as the full-length JMJD2A. On the other hand, the C-terminal region containing PHD and Tudor domains had a weaker but consistent repression activity. Since this region did not interact with N-CoR by ChIP assay (Fig. 3A), we suggest that it may repress transcription by interacting with another corepressor protein(s). We also tested whether the N-CoR interaction domain of JMJD2A alone was sufficient to repress transcription. The result in Fig. 5B showed that Gal-597-637 was able to repress transcription as efficiently as the full-length JMJD2A. Interestingly, deletion of the N-CoR interaction domain (Gal-597-637) did not significantly affect its repression activity, presumably because JMJD2A is capable of interacting with other corepressor proteins (Fig. 5A).
FIG. 5.
Correlation of N-CoR interaction with transcriptional repression activity of JMJD2A. (A) The ability of the N-terminal (Gal-1-370), the middle (Gal-347-720), and C-terminal (Gal-680-1064) regions of JMJD2A to repress transcription was analyzed by cotransfection with a 4xUAS-TK-luc reporter. The repression activity was compared to the control Gal-DBD (set as 1). Note that Gal-347-720 containing the NID repressed transcription as effectively as the full-length Gal fusion (Gal-JMJD2A). (B) The repression activity of the 41-amino-acid NID alone (Gal-JNID) and JMJD2A deleted of the NID (Gal-JΔNID) was analyzed as in panel A. Note that Gal-JNID alone was able to repress transcription similarly to the full-length JMJD2A. The Gal-JΔNID could also repress transcription, presumably as a result of interacting with other corepressor protein(s). Experiments in both panels A and B were repeated at least three times, and the results were statistically significant (P < 0.05).
Identification of ASCL2 as a JMJD2A target gene.
Although the Jumonji protein, a protein that also contains JmjC and JmjN domains, can function as a DNA binding transcription factor (26), JMJD2A does not appear to have a DNA binding domain based on sequence analysis. Based on the fact that JMJD2A has the ability to recruit N-CoR and represses transcription when tethered to a reporter, we hypothesized that JMJD2A could function as a cofactor that mediates the interaction between unknown transcription factors and N-CoR, although the possibility that JMJD2A possesses a sequence-specific DNA binding activity could not been excluded. Given its significant evolution conservation (24), we were specifically interested in the identification of JMJD2A-regulated genes. Toward this end, we used a ChIP cloning approach (45) to identify DNA sequences in HeLa cells that were bound by both JMJD2A and N-CoR. Specifically, HeLa cells were treated with 1% formaldehyde to cross-link chromatin-associated proteins with DNA. The chromatin was then sheared into 200- to 300-bp fragments by extensive sonication. The resulting chromatin fragments were immunoprecipitated with the JMJD2A antibody, eluted, and reprecipitated with an N-CoR-specific antibody. After reversal of cross-linking, DNA was purified, blunt ended, and cloned. Among 80 clones sequenced, 76 clones were found to contain various repetitive sequences, most likely reflecting their abundance in the genome and therefore nonspecific. Among four clones with unique sequences, a clone was found to contain a 171-bp DNA fragment that completely matched the sequence upstream of the human ASCL2 gene transcriptional starting site (Fig. 6A). The other three clones contained sequences very far away from known transcription start sites and thus were not further pursued.
FIG. 6.
Identification of the ASCL2 gene as a JMJD2A-repressed gene by ChIP cloning. (A) Gene structure of the human ASCL2 gene. The 171-bp region identified by ChIP cloning is shown as the JAR. (B) Knocking down JMJD2A in HeLa cells by siRNA led to increased expression of ASCL2. HeLa cells were transfected with siRNA specific for JMJD2A or a scrambled siRNA. Two days after transfection, cells were collected and the levels of JMJD2A protein were analyzed by Western blotting and the levels of ASCL2 mRNA were determined by RT-PCR. SRC-3 served as a loading control for protein, and GAPDH served as a control for RT-PCR. The RT-PCR was repeated at least three times, and a representative result was quantified. The value for the control was set as 1. (C) ChIP assays demonstrated the association of JMJD2A and N-CoR with the ASCL2 distal promoter but not the coding region. HeLa cells were first treated with the scrambled control siRNA or siRNA against JMJD2A and processed for ChIP assays 2 days later. The antibodies used for ChIP assays were as indicated, and rabbit anti-mouse IgG served as a negative control. DNA purified from the ChIP assays was amplified with specific primers flanking either the JAR or coding region. The experiments were repeated at least three times, and a representative result was quantified. The value for the control in each ChIP was set as 1.
ASCL2 is a mammalian member of the achaete-scute family, whose genes encode basic-helix-loop-helix transcription factors, and is strongly expressed in the extraembryonic trophoblast lineage (29, 31). The mouse ASCL2 gene is localized to an imprinted region including the insulin-2 (INS-2), insulin-like growth-factor 2 (IGF-2), and H19 genes on the distal end of chromosome 7 (14). Furthermore, mouse ASCL2 is essential for adequate differentiation of the trophoblast (15). In humans, the ASCL2/Hash2 gene has been mapped to 11p15.5, a region corresponding to the imprinted region in mouse chromosome 7 (30). In light of our ChIP cloning result, we next focused on investigating whether ASCL2 is regulated by JMJD2A and N-CoR.
To examine whether ASCL2 is regulated by JMJD2A, we first designed siRNA specific for JMJD2A and established the condition in which transfection of this siRNA substantially knocked down JMJD2A level in HeLa cells (Fig. 6B), whereas transfection of a control scramble siRNA had no effect. Control Western analysis using an SRC-3-specific antibody confirmed equal loading of the two samples (Fig. 6B). Under this condition, we found that the expression level of the ASCL2 gene was significantly elevated in the HeLa cells transfected with siRNA against JMJD2A. This result indicates that JMJD2A is actively engaged in the repression of the ASCL2 gene.
We also performed ChIP assays to confirm that JMJD2A binds specifically to the 171-bp fragment upstream of the ASCL2 promoter identified by our ChIP cloning experiment. To ensure that the signal derived from ChIP using JMJD2A antibody is not nonspecific, we performed ChIP experiments using HeLa cells treated with control siRNA or the JMJD2A-specific siRNA. The results in Fig. 6C showed that both JMJD2A and N-CoR could be detected on the ASCL2 promoter from the control siRNA-treated cells. However, knocking down JMJD2A by siRNA led to a substantial reduction of the signal for JMJD2A, indicating that the ChIP signal indeed reflected the binding of JMJD2A. Note that knocking down JMJD2A by siRNA also led to a reduced association of N-CoR, indicating that the association of N-CoR with the ASCL2 promoter is dependent on the presence of JMJD2A. Consistent with the result that knocking down JMJD2A resulted in elevated expression of ASCL2, a ChIP assay using an acetylated histone H3 antibody showed that the level of acetylated histone H3 increased. In the same ChIP experiment, we found that JMJD2A and N-CoR were not associated with the coding region of the ASCL2 gene. Altogether, our RT-PCR and ChIP data provide a working model that JMJD2A recruits the N-CoR complex to the JMJD2A promoter region (presumably by binding to the 171-bp DNA fragment) and represses ASCL2 gene expression, at least in part through histone deacetylation.
Repression by JMJD2A requires its tandem Tudor domain.
To further investigate the mechanism by which JMJD2A represses ASCL2 gene expression, we cloned the ASCL2 promoter region including the 171-bp fragment (referred as JMJD2A-associated region [JAR]) that we identified in the ChIP cloning into the pGL3-Basic vector (promoter-less luciferase reporter). Using this reporter, we found that cotransfection of a JMJD2A expression construct resulted in dose-dependent repression (Fig. 7A). To examine the role of the 171-bp JAR fragment in repression by JMJD2A, we made three additional reporters with partial or complete deletion of the JAR sequence (Fig. 7B, upper panel). By transient-transfection experiments, we found that the repression by JMJD2A was dependent on the presence of the 171-bp JAR fragment identified by our ChIP cloning, because the reporter with this fragment deleted was no longer sensitive to the repression by JMJD2A (Fig. 7B, compare bars A and D). In addition, we found that the first 79 bp of the 171-bp fragment was necessary and sufficient to mediate repression of ASCL2 promoter by JMJD2A (Fig. 7B, compare bars A and B), whereas deletion of the first 64 bp from the reporter abolished the repression by JMJD2A (Fig. 7B, compare bars A and C). Together, these results provide strong evidence that JMJD2A binds directly or indirectly to the first 79 bp of the 171-bp JAR fragment to repress ASCL2 gene expression.
FIG. 7.
Repression of the ASCL2 promoter by JMJD2A requires the tandem Tudor domain of JMJD2A and is N-CoR dependent. (A) JMJD2A repressed transcription driven by the ASCL2 promoter in a dose-dependent manner. The ASCL2 promoter including the JAR was cloned into the pGL3-basic reporter, and the ability of JMJD2A to repress this reporter was assayed by transient transfection. The expression of JMJD2A is shown at the bottom of the panel by Western blotting. (B) The first 79 bp of the JAR fragment is necessary and sufficient to mediate repression by JMJD2A. The deletion reporter constructs (bars B, C, and D) were cotransfected with or without JMJD2A, and the relative level of repression was compared to the ASCL2P-Luc reporter. The results were based on two independent experiments. (C) The tandem Tudor domain of JMJD2A is essential for its ability to repress transcription from the ASCL2P-Luc reporter. HeLa cells were cotransfected with wild-type or mutant JMJD2A and the ASCL2P-luc reporter. The relative luciferase activities from triplicates of two experiments are presented. (D) Knocking down N-CoR or HDAC3 impaired repression by JMJD2A. HeLa cells were first treated with siRNAs as indicated and 2 days later were transfected with a JMJD2A expression construct and the ASCL2P-luc reporter. The relative luciferase activities from triplicates of two experiments are presented. The effect of siRNAs was confirmed by Western blotting results shown in the lower panel.
With the result that transfection of JMJD2A was sufficient to repress transcription driven by the ASCL2 promoter containing the JAR fragment, we next wished to determine the functional domain(s) of JMJD2A important for this repression. For this deletion, we generated JMJD2A expression constructs deleting JmjN, PHD, or the tandem Tudor domains (Fig. 7C, lower panel). The results in Fig. 7C showed that deletion of the JmjN or PHD domain did not affect repression. However, deletion of the tandem Tudor domain completely abolished its repression activity. Western analysis confirmed the equal expression of the Tudor domain deletion mutant (data not shown). Thus, we conclude that the tandem Tudor domain is critically important for repression of the ASCL2 promoter by JMJD2A.
Repression by JMJD2A is N-CoR and HDAC3 dependent.
We next investigated whether the ability for JMJD2A to repress the ASCL2 promoter-driven reporter was dependent on N-CoR and its associated HDAC3. For this purpose, HeLa cells were first transfected with the control scramble siRNA or siRNA against JMJD2A, N-CoR, or HDAC3. Two days later, the cells were transfected with the ASCL2P-luc reporter and the JMJD2A expression plasmid. After overnight incubation, cells were harvested for luciferase activity assay. The results in Fig. 7D showed that, like siRNA against JMJD2A, siRNA against N-CoR and HDAC3 also substantially relieved repression by JMJD2A. The Western analysis (Fig. 7D, lower panel) confirmed the effect and specificity of each siRNA. Taken together, we conclude that JMJD2A represses transcription driven by the ASCL2 promoter in an N-CoR/HDAC3-dependent manner.
JMJD2A is not required for repression by unliganded TR.
The above results suggest that both JMJD2A and N-CoR are required for the repression of the ASCL2 gene. As a protein that copurified with N-CoR, we next wished to investigate whether JMJD2A is generally required for the repression function mediated by N-CoR. Toward this end, we compared the effect of siRNA against JMJD2A and N-CoR on genes repressed by unliganded TR, because the unliganded TR has been extensively shown to repress transcription through N-CoR. HeLa α2 cells, which constitutively express human TRα1 (10), were used for this experiment. The result in Fig. 8A shows that, while siRNA against N-CoR induced expression of ADRE, a previously identified TR target gene (52), siRNA against JMJD2A did not affect the expression of ADRE. On the other hand, both siRNAs led to increased expression of ASCL2, consistent with the result in Fig. 7 that both N-CoR and JMJD2A are required for the repression of ASCL2 gene. This result indicates that JMJD2A is not essential for N-CoR-mediated repression by TR. In addition, this result is consistent with the idea that JMJD2A is not a core subunit of the N-CoR complex.
FIG. 8.
JMJD2A is neither involved in regulation of imprinted genes in 11p15.5 nor required for N-CoR-mediated repression of a TR target gene. (A) Knocking down JMJD2A did not affect the repression of TR target gene ADRE. As a positive control, knocking down N-CoR resulted in increased expression of ADRE. The RT-PCR was repeated three times, and a representative result was quantified. For each gene, the value for the control was set as 1. (B) Ideogram of human chromosome 11. The ASCL2 gene is mapped to 11p15.5, a region of a cluster of imprinted genes: in order, a telomere (Tele) and the H19, IGF2, INS2, ASCL2, and KCNQ1 genes. (C) The involvement of JMJD2A in the expression of the imprinted KCNQ and IGF2 genes was assessed after knocking down JMJD2A by siRNA. Although the level of ASCL2 mRNA increased, the levels of the KCNQ and IGF2 genes did not change, as revealed by RT-PCR. The RT-PCR was repeated twice, and a representative result was quantified. The value for the control in each gene was set as 1. The knocking down of JMJD2A was confirmed by Western analysis.
JMJD2A is not involved in the regulation of other imprinted genes in human chromosome 11p15.5.
Given that the ASCL2 gene is located in an imprinted region at human chromosome 11p15.5 containing the KCNQ1, IGF2, and H19 genes (1, 14) (Fig. 8B), we next investigated whether JMJD2A is also involved in the transcriptional regulation of these imprinted genes. For this purpose, HeLa cells were treated with control siRNA or siRNA against JMJD2A and the levels of IGF2 and KCNQ1 were analyzed by RT-PCR. We expected that only the maternal alleles of the IGF2 and KCNQ1 genes were expressed and paternal alleles of these genes silenced. We reasoned that the expression of IGF2 and KCNQ1 would be enhanced if JMJD2A were involved in the imprinting of this locus. The results in Fig. 8C revealed that knocking down JMJD2A did not affect the expression levels of IGF2 and KCNQ1 by quantitative PCR. However, as expected, knocking down JMJD2A did lead to increased expression of ASCL2. In addition, Western analysis using our JMJD2A-specific antibody confirmed knocking down of JMJD2A by its specific siRNA. This result suggests that JMJD2A is specifically involved in the regulation of the ASCL2 gene but not other imprinted genes such as the IGF2 and KCNQ1 genes.
DISCUSSION
Corepressor N-CoR and the highly related protein SMRT play important roles in different biological processes including proliferation, differentiation, and development. Although numerous proteins (primarily transcription factors) have been shown to interact with N-CoR by various approaches including yeast and/or mammalian two-hybrid assays and in vitro and in vivo interaction or functional assays (7, 11, 22, 23, 32, 33, 43, 44), biochemical purification of SMRT and N-CoR tends to identify the proteins that associate stably and abundantly with SMRT and N-CoR. Indeed, HDAC3, GPS2, TBL1, and TBLR1 were all identified based on biochemical purification and considered as the core subunits of the SMRT/N-CoR complexes (13, 27, 50, 51, 53). Additional proteins identified by a biochemical approach that are abundantly associated with N-CoR include the methyl CpG binding protein Kaiso and the actin-binding protein IR10 (50, 51). Here we show that JMJD2A interacts directly with N-CoR, associates with only a fraction of N-CoR, and is not generally required for repression by N-CoR. In this regard, JMJD2A represents the first biochemically identified N-CoR interacting protein that is not abundantly associated with N-CoR.
Besides its copurification with N-CoR, we present both in vitro and in vivo evidence that JMJD2A interacts with N-CoR. First, JMJD2A and N-CoR could be reciprocally coimmunoprecipitated (Fig. 1B and C). Second, a fraction of JMJD2A in HeLa nuclear extracts was found to cofractionate with N-CoR in gel filtration analysis (Fig. 1D). Third, tethering JMJD2A to a reporter DNA resulted in recruitment of the N-CoR complex and repression of transcription (Fig. 2). Finally, JMJD2A interacted with N-CoR in in vitro pulldown assays (Fig. 4). By using a combination of in vitro and in vivo interaction assays, we were able to map the N-CoR interaction domain (NID) to a small 41-amino-acid region in the middle portion of JMJD2A and the JMJD2A interaction to the N-terminal RD1 region of N-CoR. Interestingly, the NID region is enriched with acidic amino acids, suggesting that its interaction with N-CoR may be electrostatically dependent. In addition, the NID region is not as evolutionarily conserved as other noticeable domains such as the JmjN, JmjC, PHD, and Tudor domains in JMJD2A. Nevertheless, this small region is highly conserved among higher eukaryotic cells (our unpublished data). Thus, it is possible that only in higher eukaryotic cells does JMJD2A make use of N-CoR complexes for repression of its target gene expression.
Despite strong evidence that JMJD2A copurifies and interacts with N-CoR, JMJD2A is unlikely a core subunit of the N-CoR complex for the following reasons. First, only a small fraction of JMJD2A was found to cofractionate with N-CoR in gel filtration experiments and much of the JMJD2A behaved as free protein (Fig. 1D). Under the same conditions, HDAC3, TBL1, and TBLR1 were previously shown to cofractionate extensively with N-CoR and SMRT. Second, although JMJD2A and N-CoR could be coimmunoprecipitated (Fig. 1B and C), we could not deplete N-CoR from HeLa nuclear extracts with our JMJD2A-specific antibody and vice versa. This is in contrast to the cases of core subunits such as HDAC3, TBL1, and Kaiso, whose codepletion with N-CoR from HeLa nuclear extracts with their specific antibodies was shown previously (27, 51). Third, JMJD2A was not required for N-CoR-mediated repression of TR target genes (Fig. 8C). On the other hand, JMJD2A repressed the ASCL2 promoter-driven transcription in an N-CoR complex-dependent manner (Fig. 7 and 8). Altogether, our data indicate that JMJD2A is unlikely a core subunit of the N-CoR complex but represses transcription by making use of the N-CoR complex.
As shown in Fig. 1A, the JMJD2A protein contains several well-conserved domains including the JmjN and JmjC domains in the N terminus and the PHD and Tudor domains in the C terminus. The function of each of these domains is not well understood. The JmjC and JmjN domains have been identified in over 100 eukaryotic and bacterial proteins, including several putative chromatin-associated proteins, especially the jumonji family of eukaryotic transcription factors (6). Although the proteins containing JmjC and JmjN domains are generally believed to be metalloenzymes, the mammalian jumonji family proteins including JMJD2A are predicted to be enzymatically inactive because of the lack of a few key conserved amino acids in this domain that are necessary for enzymatic activity (6). Recent studies show that the Jumonji, a member of the JMJD2 family proteins, can regulate transcription either as a sequence-specific DNA binding transcription factor or by inhibiting other transcription factor activity (25, 26). However, by using either in vitro-translated JMJD2A or JMJD2A purified from mammalian cells, we have not been able to detect a direct binding of JMJD2A protein to the 171-bp fragment upstream of the ASCL2 promoter by gel mobility shift assay (data not shown). A close comparison of the Jumonji transcription factor and JMJD2A revealed that the sequence involved in DNA binding of Jumonji is not present in JMJD2A. Thus, although JMJD2A is capable of repressing both endogenous ASCL2 (Fig. 6) and a transfected luciferase reporter driven by the ASCL2 promoter (Fig. 7), JMJD2A may not bind directly to the 171-bp fragment and its repressive effect may be mediated through another transcription factor(s) that binds to this fragment. To look for the putative transcription factor(s) that mediates JMJD2A action, we have analyzed the potential transcription factor binding sites in the 171-bp JAR fragment (our unpublished data) by using the Consite program with a cutoff of 80% (http://mordor.cgb.ki.se/cgi-binl). According to this program, the first 79 bp of the JAR region may contain binding sites for nuclear factor Y (NF-Y), repressor snail, E74A, and Ets family member ELF-2. Future work will test if NF-Y, snail, and ELF-2 interact directly with JMJD2A.
Tudor and its related domains have been identified in many proteins (28). Recent studies on the Tudor domain of the “survival of motor neurons” protein (SMN) indicate that it is required for interaction with the spliceosomal Sm proteins. Specifically, the Tudor domain of SMN interacts with symmetrically dimethylated arginine residues in an RG-rich region of Sm proteins (5, 38, 46). In addition, a recent report showed that the tandem Tudor domain of mammalian checkpoint protein 53BP1 binds specifically to lysine 79-methylated histone H3 and plays a role in double-strand DNA break repair (19). Interestingly, methylation of histone H4 lysine 20 in yeast controls recruitment of Crb2, the yeast homolog of 53BP1, to sites of DNA damage, although it is yet to be demonstrated whether Crb2 binds directly to the lysine 20-methylated H4 (37). In this study, we show that the tandem Tudor domain of JMJD2A is required for JMJD2A to repress the ASCL2 gene promoter (Fig. 7C). It is possible that the tandem Tudor domain mediates the interaction of JMJD2A with the putative transcription factor(s) that binds to the ASCL2 promoter.
The identification of the ASCL2 gene as a gene regulated by JMJD2A and N-CoR is of biological significance. The ASCL2 gene encodes a basic helix-loop-helix transcriptional factor. Mouse ASCL2, also called Mash2, is required for embryonic development due to its essential role in placental development (15). This placental mutant phenotype can be rescued by constructing chimeras with tetraploid wild-type embryos which provide extraembryonic tissues. Under this condition, the Mash2−/− embryos developed normally and adult Mash2−/− mice were viable, demonstrating that Mash2 has no major role in the embryo itself (15). Interestingly, expression of the ASCL2 gene is tightly regulated and tissue specific. Both mouse and human ASCL2s are only strongly expressed in the extraembryonic trophoblast lineage but repressed in other tissues (1, 15, 31). Given our data that JMJD2A represses ASCL2 expression and that both JMJD2A and N-CoR are broadly expressed in various mouse tissues (Fig. 1E), we suggest that JMJD2A and N-CoR could contribute to the restricted expression of ASCL2.
The mouse ASCL2 gene is located to the distal region of chromosome 7 within a cluster of imprinted genes, including the KCNQ1, IGF2, and H19 genes (14). The human ASCL2 gene is mapped to chromosome 11p15.5 (1, 30), the region syntenic with mouse chromosome 7, indicating that this imprinted region is highly conserved in evolution. Although the ASCL2 gene in mice is clearly an imprinted gene, human ASCL2 may escape imprinting (29). Nevertheless, our data that JMJD2A and N-CoR are involved in repression of ASCL2 raise the question as to whether JMJD2A and N-CoR could also affect other imprinted genes in the region. Since knocking down JMJD2A had little effect on KCNQ1 and IGF2 expression, whereas the same treatment released the repression of the ASCL2 gene (Fig. 8), it is unlikely that JMJD2A and N-CoR are involved in the regulation of the gene imprinting. Together, our data are consistent with a previous study showing that the allele-specific chromatin configuration could be involved in the transcriptional repression of mouse Mash2 within the imprinted cluster (41). It will be interesting to determine whether JMJD2A and N-CoR are involved in imprinted expression of Mash2 in mice in the future.
Acknowledgments
We are grateful to B. O'Malley, M. Tsai, and S. Tsai for helpful discussion and M. Stewart for critical reading of the manuscript. We also thank Celina Montemayor for transcription factor binding site analysis.
This work was supported by NIH grants DK56324 and DK58679.
REFERENCES
- 1.Alders, M., M. Hodges, A. K. Hadjantonakis, J. Postmus, I. van Wijk, J. Bliek, M. de Meulemeester, A. Westerveld, F. Guillemot, C. Oudejans, P. Little, and M. Mannens. 1997. The human Achaete-Scute homologue 2 (ASCL2,HASH2) maps to chromosome 11p15.5, close to IGF2 and is expressed in extravillus trophoblasts. Hum. Mol. Genet. 6:859-867. [DOI] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 3.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 4.Bowen, N. J., N. Fujita, M. Kajita, and P. A. Wade. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta 1677:52-57. [DOI] [PubMed] [Google Scholar]
- 5.Brahms, H., L. Meheus, V. de Brabandere, U. Fischer, and R. Luhrmann. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clissold, P. M., and C. P. Ponting. 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem. Sci. 26:7-9. [DOI] [PubMed] [Google Scholar]
- 7.Dhordain, P., O. Albagli, R. J. Lin, S. Ansieau, S. Quief, A. Leutz, J. P. Kerckaert, R. M. Evans, and D. Leprince. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94:10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 271:2335-2349. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Q., and Y. Zhang. 2003. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274:269-290. [DOI] [PubMed] [Google Scholar]
- 10.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 12.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 13.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 14.Guillemot, F., T. Caspary, S. M. Tilghman, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, D. J. Anderson, A. L. Joyner, J. Rossant, and A. Nagy. 1995. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9:235-242. [DOI] [PubMed] [Google Scholar]
- 15.Guillemot, F., A. Nagy, A. Auerbach, J. Rossant, and A. L. Joyner. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371:333-336. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 17.Hermanson, O., K. Jepsen, and M. G. Rosenfeld. 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419:934-939. [DOI] [PubMed] [Google Scholar]
- 18.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 19.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406-411. [DOI] [PubMed] [Google Scholar]
- 20.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 22.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 23.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh, M. 2004. Identification and characterization of JMJD2 family genes in silico. Int. J. Oncol. 24:1623-1628. [PubMed] [Google Scholar]
- 25.Kim, T. G., J. Chen, J. Sadoshima, and Y. Lee. 2004. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 24:10151-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, T. G., J. C. Kraus, J. Chen, and Y. Lee. 2003. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J. Biol. Chem. 278:42247-42255. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber, and C. P. Ponting. 2003. The Tudor domain ‘royal family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28:69-74. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto, T., S. Hasuike, Y. Jinno, H. Soejima, K. Yun, K. Miura, M. Ishikawa, and N. Niikawa. 2002. The human ASCL2 gene escaping genomic imprinting and its expression pattern. J. Assist. Reprod. Genet. 19:240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto, T., Y. Jinno, T. Sasaki, Y. Ikeda, H. Masuzaki, N. Niikawa, and M. Ishikawa. 1996. Genomic cloning and localization to chromosome 11p15.5 of the human achaete-scute homolog 2 (ASCL2). Cytogenet. Cell Genet. 73:312-314. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, H., Y. Liu, S. Stifani, and J. C. Cross. 1997. Developmental restriction of Mash-2 expression in trophoblast correlates with potential activation of the notch-2 pathway. Dev. Genet. 21:21-30. [DOI] [PubMed] [Google Scholar]
- 32.Nomura, T., M. M. Khan, S. C. Kaul, H. D. Dong, R. Wadhwa, C. Colmenares, I. Kohno, and S. Ishii. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ordentlich, P., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 34.Ordentlich, P., M. Downes, W. Xie, A. Genin, N. B. Spinner, and R. M. Evans. 1999. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl. Acad. Sci. USA 96:2639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, E. J., D. J. Schroen, M. Yang, H. Li, L. Li, and J. D. Chen. 1999. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc. Natl. Acad. Sci. USA 96:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 37.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 38.Selenko, P., R. Sprangers, G. Stier, D. Buhler, U. Fischer, and M. Sattler. 2001. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8:27-31. [DOI] [PubMed] [Google Scholar]
- 39.Sims, R. J., III, K. Nishioka, and D. Reinberg. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19:629-639. [DOI] [PubMed] [Google Scholar]
- 40.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, M., M. Puchyr, M. Gertsenstein, K. Harpal, R. Jaenisch, J. Rossant, and A. Nagy. 1999. Parental origin-specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech. Dev. 87:129-142. [DOI] [PubMed] [Google Scholar]
- 42.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 43.Wang, J., T. Hoshino, R. L. Redner, S. Kajigaya, and J. M. Liu. 1998. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 95:10860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., and S. W. Hiebert. 2001. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene 20:3716-3725. [DOI] [PubMed] [Google Scholar]
- 45.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26:37-47. [DOI] [PubMed] [Google Scholar]
- 46.Whitehead, S. E., K. W. Jones, X. Zhang, X. Cheng, R. M. Terns, and M. P. Terns. 2002. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 277:48087-48093. [DOI] [PubMed] [Google Scholar]
- 47.Wolffe, A. P., and D. Guschin. 2000. Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129:102-122. [DOI] [PubMed] [Google Scholar]
- 48.Wolffe, A. P., F. D. Urnov, and D. Guschin. 2000. Co-repressor complexes and remodelling chromatin for repression. Biochem. Soc. Trans. 28:379-386. [PubMed] [Google Scholar]
- 49.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 50.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon, H. G., D. W. Chan, A. B. Reynolds, J. Qin, and J. Wong. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12:723-734. [DOI] [PubMed] [Google Scholar]
- 52.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]