Abstract
Rho GTPases regulate multiple cellular processes affecting both cell proliferation and cytoskeletal dynamics. Their cycling between inactive GDP- and active GTP-bound states is tightly regulated by guanine nucleotide exchange factors and GTPase-activating proteins (GAPs). We have previously identified CdGAP (for Cdc42 GTPase-activating protein) as a specific GAP for Rac1 and Cdc42. CdGAP consists of an N-terminal RhoGAP domain and a C-terminal proline-rich region. In addition, CdGAP is a member of the impressively large number of mammalian RhoGAP proteins that is well conserved among both vertebrates and invertebrates. In mice, we find two predominant isoforms of CdGAP differentially expressed in specific tissues. We report here that CdGAP is highly phosphorylated in vivo on serine and threonine residues. We find that CdGAP is phosphorylated downstream of the MEK-extracellular signal-regulated kinase (ERK) pathway in response to serum or platelet-derived growth factor stimulation. Furthermore, CdGAP interacts with and is phosphorylated by ERK-1 and RSK-1 in vitro. A putative DEF (docking for ERK FXFP) domain located in the proline-rich region of CdGAP is required for efficient binding and phosphorylation by ERK1/2. We identify Thr776 as an in vivo target site of ERK1/2 and as an important regulatory site of CdGAP activity. Together, these data suggest that CdGAP is a novel substrate of ERK1/2 and mediates cross talk between the Ras/mitogen-activated protein kinase pathway and regulation of Rac1 activity.
RhoA, Rac1, and Cdc42, the best-characterized members of the Rho family of small GTPases, are critical regulators of many cellular activities, such as cell dynamics, cell growth, intracellular membrane trafficking, gene transcription, cell cycle progression, and apoptosis (4, 36). The Rho proteins operate as molecular switches that cycle between an active GTP-bound and an inactive GDP-bound state. This GDP/GTP cycle is tightly regulated by three families of proteins: the guanine nucleotide exchange factors, which activate Rho GTPases by inducing the exchange of GDP for GTP (5); the GTPase-activating proteins (GAPs), which enhance the intrinsic GTPase activity, leading to the inactive state of the GTPase (14); and the guanine nucleotide dissociation inhibitors, which sequester Rho GTPases in their GDP-bound, inactive state (28).
More than 40 RhoGAP family members have been characterized in eukaryotes ranging from yeast to humans (23). Recent analysis of the human genome unraveled 66 different genes encoding potential RhoGAP domain-containing proteins, far outnumbering the existing 23 mammalian Rho GTPases (2, 30). In most cases, GAP proteins are large proteins containing multiple signaling modules that mediate the cross talk between GAPs and other signaling pathways or serve to regulate the GAP activity (14). GAP proteins act not only as negative regulators but also function as downstream effectors of Rho GTPases. For instance, n-chimaerin can induce actin reorganization independently of its RhoGAP domain (16), and TCGAP plays a direct role in insulin-stimulated glucose transport (6). Given that Rho GTPases are implicated in a large number of biological responses, each GAP protein may selectively regulate a specific Rho GTPase signaling pathway. In addition, the overabundance of GAP proteins strongly suggests a tight regulation of their activity in a spatial and temporal fashion. Indeed, accumulating evidence reveals that GAPs are regulated by lipid interaction, protein-protein interaction, phosphorylation, and proteolytic degradation (1, 13, 31, 34).
CdGAP (Cdc42 GTPase-activating protein) is a serine- and proline-rich RhoGAP protein showing GAP activity against both Cdc42 and Rac1 but not Rho A (17). In addition to its N-terminal GAP domain, CdGAP contains a central domain and a C-terminal proline-rich domain (PRD) harboring five consensus Src homology 3 (SH3)-binding sites whose functions are still unclear. We have recently shown that the endocytic protein intersectin interacts with CdGAP through a subset of its SH3 domains and negatively regulates CdGAP's activity, providing evidence of a direct regulation through protein-protein interaction (13). Here we report that CdGAP is highly phosphorylated on serine and threonine residues in the proline-rich region. We found that CdGAP interacts with members of the mitogen-activated protein kinase (MAPK) signaling pathway, RSK-1 and extracellular signal-regulated kinase 1/2 (ERK1/2), and is phosphorylated in vitro by these Ser/Thr kinases. Mutation of key residues in the ERK docking site of CdGAP reduces both ERK binding and phosphorylation of CdGAP. In Swiss 3T3 fibroblasts, endogenous CdGAP is phosphorylated in response to platelet-derived growth factor (PDGF), and this in vivo phosphorylation of CdGAP is reduced in the presence of the MEK1 inhibitor PD98059. We identified Thr776 in the proline-rich domain of CdGAP as a major in vivo phosphorylation site of ERK-1, and amino acid substitution of this threonine for alanine significantly affects the GAP activity of CdGAP. We propose that CdGAP mediates cross talk between the MAPK and Rac1 signaling pathways and that phosphorylation of CdGAP by ERK1/2 participates to negatively regulate CdGAP activity.
MATERIALS AND METHODS
Reagents and antibodies.
Recombinant activated rat RSK-1 and human ERK-1 proteins, PD98059, myelin basic protein (MBP), and long S6 kinase substrate peptide (KRQEQIAKRRRLSSLRASTAKSGGSQK) were obtained from Upstate Biotechnology, Inc. (Lake Placid, NY). Lipofectamine was obtained from Invitrogen (Carlsbad, CA). Human recombinant PDGF-BB was obtained from Calbiochem. Protease inhibitor cocktail tablets were from Roche Applied Science (Indianapolis, IN). A protein assay kit (micro-BCA) was from Pierce Chemical (Rockford, IL). Trypsin-TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone) was purchased from Sigma. Protein G- and A-Sepharose were obtained from Pharmacia Biotech. [γ32P]ATP (3,000 Ci/mmol) and [32P]orthophosphate (3,000 μCi/ml) were purchased from Perkin-Elmer. Anti-Rsk-1 and -Erk1/2 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and phospho-specific antibodies recognizing RSK-1 and ERK1/2 were from Upstate Biotechnology. Anti-CdGAP antibodies were obtained by immunization of rabbits with CdGAP proline-rich domain (amino acids [aa] 515 to 820) fused to glutathione S-transferase (GST) and affinity purified on a CH-Sepharose column (Pharmacia Biotech) covalently coupled to the CdGAP peptide ESQGASQPKPSTPQESLGAG (aa 601 to 620). Anti-Rac1 antibodies were purchased from Upstate Biotechnology. Myc-tagged proteins were detected by using the 9E10 anti-Myc monoclonal antibody (MAb), which was kindly provided by Nicole Beauchemin (McGill University, Montreal, Canada).
DNA constructs and site-directed mutagenesis.
The threonine 612, 769, and 776 and phenylalanine 677 and 679 residues located in the proline-rich region of CdGAP in pRK5myc vector (13) were substituted for alanine residues by using a PCR oligonucleotide-directed mutagenesis approach according to standard protocols. The following primers were used: T612A (5′-GAAGCCAGCGCGCCACAAGAGAGCCTCGGG-3′ and 3′-TCGGTCGGCTTCGGGTCGCGCGGTGTTCTC-5′), T769A (5′-CTCTCTCCCCCTCTTGCTCCTGCTCCTCCTCCC-3′ and 3′-CCTTAGAGAGAGGGGGAGAACGAGGACGAGG-5′), T776A (5′-CCTCCTCCCCCCGCTCCTCTGGAGGAGGAGCCT-3′ and 3′-GAATGAGGACGAGGAGGGGGGCGAGGAGACCTC-5′), T769AT776A (5′-CCCCCTCTTGCTCCTCCTCCCCCCGCTCCTCTGGAGGAGGAG-3′ and 3′-GAGAGAGGGGGAGAACGAGGACGAGGAGGAGGGGGGCGAGGAGACCTC-5′), and FFAA (5′-CAGCCCCAGCTCCCGCTCCAGAAGCCCCTGG-3′ and 3′-GGACTCAGTTCGGGTCGGGGTCGAGGGCGAGGTCTT-5′). The PCR products were digested by using NheI/XbaI restriction enzymes and then inserted into pRK5mycCdGAP-s, digested with NheI/XbaI, and dephosphorylated by using calf intestine phosphatase (NEB). The proline-rich domain of CdGAP (aa 516 to 820) wild type or threonine and phenylalanine mutations were subcloned into pTrcHisA vector by PCR with pRK5mycCdGAP-s, -T612A, -T769A, -T776A, -T769AT776A, or -FFAA as templates and 5′-CGGATCCCAAGGTTCAGAGAGTGG-3′ and 3′-CGGGTTGAACAAATAACG-5′, respectively, as forward and reverse primers. Both PCR products and pTrcHisA vector were digested with BamHI/EcoRI and ligated together. The pRK5myc CdGAP deletion mutants CdGAP-ΔGAP, CdGAP-GAP, CdGAP-ΔPRD, and CdGAP-PRD were produced as described previously (13). To produce the pRK5mycCdGAP-l, the mouse cDNA clone mpf00743 containing the longer C-terminal tail of mouse CdGAP (mCdGAP) was obtained from the Kazuka DNA Research Institute (Chiba, Japan) and was cloned into pRK5mycCdGAP-s by PCR with 5′-CATGCCATGGCACAAGGTTCAGAGAGTGG-3′ and 3′-AGTGGGAGAGCAGATAGAATGATCTAGAG-5′ as forward and reverse primers. The resulting PCR product and pRK5mycCdGAP-s were digested with NheI and XbaI and ligated together.
Expression of recombinant proteins.
Recombinant Rac1 and PAK (aa 56 to 272) were produced in Escherichia coli strain DH5α as GST fusion proteins and purified on glutathione-Sepharose beads as described previously (13, 18). pTrcHisA containing hexahistidine fusion proteins—CdGAP-PRD, CdGAP-PRD-T612A, CdGAP-PRD-T769A, CdGAP-PRD-T776A, CdGAP-PRD-T769AT776A, and CdGAP-PRD-FFAA—were transformed into E. coli DH5α strain and grown in 100 ml of LB medium at 30°C overnight. The next day, 900 ml of M9 medium 1× was added before induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma-Aldrich) for 1 h. Bacteria pellets were lysed and sonicated in 10 ml of lysis buffer containing 50 mM HEPES, 300 mM NaCl, and protease inhibitor cocktail (Sigma-Aldrich), followed by the addition of 10% Triton X-100 and centrifugation for 45 min at 4,000 × g. Then, 20 mM imidazole pH 8.0 was added to the supernatant, and His-tagged recombinant proteins were purified by adding 500 μl (50:50) of Ni-nitrilotriacetic acid beads (Amersham) per liter of culture and rotating for 3 h. After a quick spin, the beads were washed twice with buffer A (50 mM HEPES, 300 mM NaCl, 1% Triton X-100, 20 mM imidazole [pH 8.0]) and twice in buffer B (50 mM HEPES, 300 mM NaCl, 1% Triton X-100, 20 mM imidazole [pH 6.5]). The proteins were eluted by incubating beads three times in 3 ml of buffer C (50 mM HEPES, 300 mM NaCl, 1% Triton X-100, 200 mM imidazole [pH 6.5]) for 30 min each time. Eluates were pooled and concentrated in Centricon (Millipore) and washed twice with 4 ml of 1× cold phosphate-buffered saline to remove the imidazole. Protein concentration and purity were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining.
Preparation of mouse tissues.
Murine tissue samples were obtained from adult male BALB/c mice, aged 7 to 9 weeks. Tissues were collected and homogenized in radioimmunoprecipitation assay buffer (1× phosphate-buffered saline [pH 7.4], 0.1% SDS, 1% Triton X-100, 12 mM deoxycholic acid, and protease inhibitor cocktail according to the manufacturer [Roche]). Total tissue lysates were centrifuged at 5,000 × g for 10 min. The protein concentration in the resulting supernatant was determined by using the BCA protein assay kit (Pierce Chemical), and 200-μg portions of proteins from each tissue were loaded onto a SDS-7.5% PAGE gel and transferred onto a nitrocellulose membrane. CdGAP was detected by using affinity-purified polyclonal anti-CdGAP antibodies. Competition assay was performed by incubating the membrane with 10 μg of GST-tagged CdGAP-PRD or with GST alone as a control.
Cell transfection, immunoprecipitation, and immunoblotting.
COS-7, HEK293, and Swiss 3T3 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS; Invitrogen) and antibiotics and maintained at an atmosphere of 10% CO2 at 37°C. COS-7 cells were transfected by using DEAE-dextran as described previously (29). Briefly, 5 μg of pRK5myc-CdGAPs, -CdGAP-l, -CdGAP-ΔPRD, -CdGAP-PRD, -CdGAP-GAP, -CdGAP-ΔGAP, -CdGAPT769A, -CdGAPT769AT776A, -CdGAPT612A, -CdGAPT776A, or -CdGAPFFAA was used per 100-mm dish. Then, 48 h posttransfection, cells were lysed in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100) containing 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 20 mM NaF, and 1 mM sodium orthovanadate, followed by centrifugation for 15 min at 1,000 × g. Then, 1 mg of the resulting postnuclear supernatant was incubated overnight at 4°C with 5 μg of anti-Myc antibodies and 20 μl of 50% protein G-Sepharose. Samples were washed three times in lysis buffer and subjected to SDS-PAGE, followed by immunoblotting analysis with anti-Myc, -ERK1/2, or -RSK-1 antibodies and visualized by enhanced chemiluminescence (Perkin-Elmer). Endogenous CdGAP-l was immunoprecipitated from protein lysates obtained from Swiss 3T3 cells treated or not with PDGF (5 ng/ml) or 20% FBS for 5 and 30 min as described above with anti-CdGAP antibodies and protein A-Sepharose. Western blotting was performed with anti-CdGAP antibodies.
In vivo [32P]orthophosphate labeling.
Confluent Swiss 3T3 fibroblasts grown on 150-mm dishes were serum starved for 24 h. Then, cells were washed once in phosphate-free medium and then incubated in the same medium for 1 h prior to labeling. Cells were incubated for 3 h in the presence of 0.5 mCi of [32P]orthophosphate/ml. The cells were either unstimulated or stimulated with dialyzed FBS (20% for 30 min) or PDGF (5 ng/ml for 5 min) or pretreated with PD98059 (50 μM for 1 h) prior to stimulation with PDGF. Endogenous CdGAP was immunoprecipitated from cell lysates, and proteins were separated by SDS-PAGE. Radiolabeled proteins were detected by autoradiography with an enhancing screen and Biomax MS film (Sigma-Aldrich) at −80°C. COS-7 cells grown onto 100-mm dishes were transfected with Myc-tagged CdGAP or different protein mutants as described above. At 48 h posttransfection, cells were washed once in phosphate-free medium supplemented with 1% serum and incubated for 1 h in the same medium prior to labeling as described above.
In gel kinase assay.
After immunoprecipitation of Myc-tagged CdGAP from COS-7 cells, proteins were resolved by SDS-PAGE with a 10% acrylamide resolving gel containing 0.5 mg of recombinant His-tagged CdGAP-PRD/ml. After electrophoresis, the gel was washed twice with 30 mM Tris-HCl (pH 7.5)-20% isopropanol. The gel was then washed twice (30 min each) in 30 mM Tris-HCl-2 mM dithiothreitol (pH 7.5) and incubated for 45 min in the same buffer containing 6 M urea. The proteins were then subjected to renaturation by three washes of 45 min each in 30 mM Tris-HCl-2 mM DTT (pH 7.5) containing 0.05% Tween 20 and, respectively, 3, 1.5, and 0.75 M urea and then washed for 2 h in 30 mM Tris-HCl-2 mM DTT (pH 7.5)-0.05% Tween 20. The gel was then incubated for 30 min in the kinase reaction buffer (30 mM Tris-HCl, 2 mM DTT, 10 mM MgCl2, 10 mM MnCl2; pH 7.5). Phosphorylation was carried out by incubating the gel in the same buffer containing 10 μCi of [γ-32P]ATP/ml and 100 μM ATP for 45 min at room temperature. The gel was washed extensively in 5% (vol/vol) trichloroacetic acid-1% (wt/vol) sodium pyrophosphate for 24 h. The gel was dried, and the radiolabeled bands were visualized by autoradiography.
In vitro kinase assays.
COS-7 cells were transfected with pRK5myc or pRK5mycCdGAP as described above. Proteins were immunoprecipitated, and the pellets were washed three times with lysis buffer and twice with kinase buffer (20 mM morpholinepropanesulfonic acid [MOPS; pH 7.2], 25 mM β-glycerophosphate, 5 mM EGTA, 1 mM sodium orthovanadate, 1 mM DTT). Immunoprecipitates were incubated at 30°C for 10 min in 50 μl of kinase reaction buffer (kinase buffer plus 10 mM MgCl2, 100 μM ATP, and 10 μCi of [γ-32P]ATP/ml). The reaction was stopped by the addition of Laemmli sample buffer, and the phosphorylated proteins were separated by SDS-PAGE and visualized by autoradiography. His-tagged CdGAP-PRD wild type and threonine and phenylalanine protein mutants were phosphorylated with 20 ng of recombinant activated ERK-1 (Upstate) or 20 ng of recombinant activated RSK-1 (Upstate) in reaction buffer. MBP (0.1 mg/ml; Sigma) or long S6 substrate peptide (20 μM; Upstate) were used as positive controls for ERK-1 and RSK-1, respectively.
Phosphoamino acid analysis.
Phosphoamino acid analysis was performed as described previously (3). Briefly, CdGAP wild-type or protein mutants overexpressed in COS-7 cells were radiolabeled and immunoprecipitated as described above. His-tagged CdGAP-PRD was phosphorylated in vitro by recombinant activated ERK-1 or RSK-1. Samples were resolved by SDS-PAGE and electroblotted onto polyvinylidene difluoride membrane. Bands corresponding to 32P-labeled CdGAP proteins were excised, placed in a 1.5-ml screw-cap tube, and washed extensively with distilled water. Acid hydrolysis was performed by incubating samples in 200 μl of 6 N HCl, followed by heating at 110°C for 60 min, lyophilization, and dissolution in 10 μl of pH 1.9 buffer (88% formic acid, glacial acetic acid, H2O; 2.5:7.8:89.7 [vol/vol/vol]) containing 0.5 μl of 2 mg of combined unlabeled phosphoamino acid standards (phosphoserine, -threonine, and -tyrosine)/ml. The phosphoamino acids were first separated by high voltage (1.5 kV) electrophoresis at pH 1.9 for 20 min by using a Hunter thin-layer electrophoresis system (CBS Scientific, Del Mar, CA), followed by a second-dimension electrophoresis in pH 3.5 buffer (glacial acetic acid, pyridine, H2O; 5:0.5:94.5 [vol/vol/vol]). The standards were visualized by spraying a 0.25% (wt/vol) ninhydrin acetone solution, followed by incubation at 65°C for 10 min. The radiolabeled amino acids were detected by autoradiography with an enhancing screen and Kodak Biomax film at −80°C.
Tryptic phosphopeptide mapping.
Radiolabeled proteins were resolved by SDS-PAGE and electroblotted onto nitrocellulose membrane. Corresponding CdGAP bands were excised and digested with 10 μg of TPCK-treated trypsin for 4 h at 37°C. Peptides were diluted in 500 μl of water and lyophilized by using a Speed-Vac. The peptides were then oxidized in 50 μl of performic acid for 60 min on ice, diluted to 500 μl with deoinized water before lyophilization. Pellets were dissolved in pH 1.9 buffer (88% formic acid, glacial acetic acid, H2O; 2.5:7.8:89.7 [vol/vol/vol]) and were first separated by electrophoresis in pH 1.9 buffer 25 min at 1.0 kV by using a Hunter thin-layer electrophoresis system (CBS Scientific, Del Mar, CA), followed by a second-dimension separation by ascending chromatography in phospho-chromatography buffer (glacial acetic acid, pyridine, n-butanol, H2O; 7.5:25:37.5:30 [vol/vol/vol/vol]). The radiolabeled phosphopeptides were detected by autoradiography.
In vitro GAP assay.
COS-7 cells were transfected with pRK5myc, pRK5mycCdGAP, pRK5mycCdGAP-T769A, pRK5mycCdGAP-T776A, and pRK5mycCdGAP-T769AT776A as described above. At 48 h posttransfection, Myc-tagged proteins were immunoprecipitated as described above with anti-Myc antibodies. Samples were then washed three times in 20 mM Tris-HCl (pH 7.5)-150 mM NaCl-1% Triton X-100 and twice in 20 mM Tris-HCl (pH 7.5) and used for the in vitro GAP assay as follows. The amount of immunoprecipitated CdGAP is estimated on Coomassie blue staining by comparison with different amounts of purified bovine serum albumin. According to this estimation, immune complexes corresponding to 1 μg of immunoprecipitated CdGAP were resuspended in 24 μl of 20 mM Tris-HCl (pH 7.5), 0.1 mM DTT, 1 mM GTP, and 0.86 mg of bovine serum albumin/ml. At the same time, 2 μg of recombinant Rac1 was incubated with 5 μCi of [γ-32P]GTP (30 Ci/mmol) in 20 μl of 20 mM Tris-HCl (pH 7.5), 25 mM NaCl, 0.1 mM DTT, and 5 mM EDTA for 10 min at 30°C. GTP-loaded Rac1 was kept on ice after addition of 20 mM MgCl2. A total of 3 μl of [γ-32P]GTP-loaded Rac1 was incubated at 20°C with the immune complexes. After 0 and 6 min of incubation, 4-μl mixtures were diluted in 1 ml of cold buffer A (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM MgCl2) and filtered through prewetted nitrocellulose filters. Filters were washed with 10 ml of cold buffer A, dried, and counted.
Rac activation assay.
HEK293 cells grown onto 100-mm dishes were transfected with Lipofectamine according to the manufacturer's procedures. Briefly, cells were transfected with 2 μg of pRK5myc, pRK5mycCdGAP, pRK5mycCdGAP-T769A, pRK5mycCdGAP-T776A, and pRK5mycCdGAP-T769AT776A, together with 1.5 μg of pRK5mycRac1 and 1 μg of pRK5mycRasV12. Cells were then serum starved overnight and lysed the next day in lysis buffer B (25 mM HEPES [pH 7.5], 100 mM NaCl, 1% NP-40, 5% glycerol) containing 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 20 mM NaF, and 1 mM sodium orthovanadate, followed by centrifugation for 15 min at 1,000 × gmax. The amounts of GTP-loaded Rac1 in the supernatant were measured by using a pull-down assay with GST-CRIB domain of PAK as described previously (18).
RESULTS
CdGAP isoforms are differentially expressed in mouse tissues.
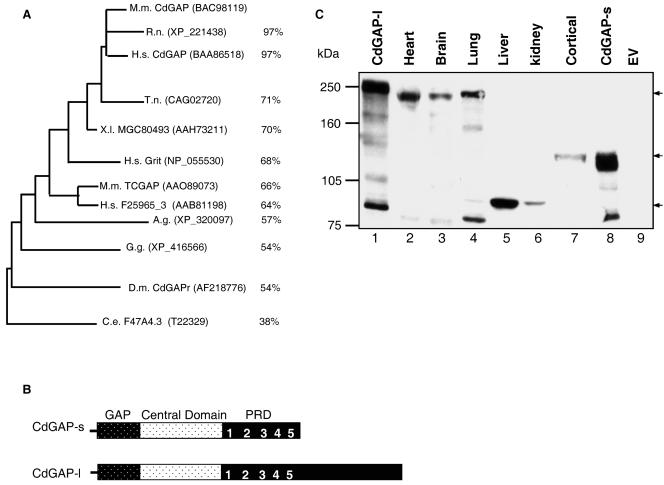
We have previously identified and characterized mCdGAP, a protein of 820 amino acids. Since then, CdGAP-related genes have been found in both vertebrate and invertebrate organisms. In addition to the previously characterized ortholog of mCdGAP in Drosophila melanogaster (32), a single gene encoding a putative protein with high degree of homology to CdGAP within the RhoGAP domain is found in at least six other organisms, including rat, fish, frog, chicken, and worm (Fig. 1A). In Homo sapiens, there are at least three different genes encoding proteins that are highly homologous to mCdGAP. One encodes a previously characterized protein named Grit (also known as p200RhoGAP, RICS, p250GAP, and GC-GAP) (22, 26, 27, 35, 38), which shares 68% homology to mCdGAP within the rhoGAP domain and one uncharacterized cDNA, F25965_3, which is the human ortholog of mouse TCGAP protein (6). The cDNA KIAA1204 is predicted to encode a protein of 1,444 aa that shares 97% homology to mCdGAP within the rhoGAP domain and 84% homology within the entire protein sequence. The longer C-terminal tail of human CdGAP suggested that a similar protein might also exist in Mus musculus. Indeed, BLASTP analysis at the National Center for Biotechnology Information GenBank databases revealed a novel mouse cDNA identical to that of CdGAP but with an additional 1,815 bp at the 3′ end (GenBank accession number BAC98119). This cDNA is predicted to encode a protein of 1,425 amino acids with a predicted molecular mass of 155,724 Da (Fig. 1B). We have named these two isoforms: CdGAP long (CdGAP-l) and CdGAP short (CdGAP-s). When overexpressed into fibroblasts, we found that both CdGAP-s and CdGAP-l migrate higher than their expected molecular masses of 90 and 155 kDa, respectively (Fig. 1C, lanes 1 and 8). In fact, CdGAP-s migrates at 125 kDa and CdGAP-l migrates at 250 kDa. These mobility shifts may be in part due to posttranslational modifications such as phosphorylation in addition to unidentified modifications of the proteins. Using affinity-purified polyclonal antibodies against the third proline-rich sequence of CdGAP, three major bands of 250, 125, and 90 kDa could be detected in lysates of different mouse tissues and primary mouse cortical neurons (Fig. 1C). These bands were not recognized in Western blots with preimmune sera (data not shown). The band of 250 kDa, which migrates at the same level as overexpressed CdGAP-l (lane 1), is present in the heart, brain, and lung tissues (lanes 2, 3, and 4). The 90-kDa band, which corresponds to the expected molecular mass of CdGAP-s, is very abundant in liver and present in kidney tissues (lanes 5 and 6). We hypothesized that in these tissues posttranslational modifications of CdGAP-s are absent or different than the overexpressed CdGAP-s protein migrating at a higher molecular mass of 125 kDa. Interestingly, cortical neurons express a CdGAP protein of 125 kDa (lane 7) corresponding to the overexpressed CdGAP-s (lane 8). All three bands of 250, 125, and 90 kDa were also detected by using affinity-purified polyclonal antibodies to the first proline-rich sequence of CdGAP (data not shown). In addition, preadsorption of antibodies with GST-tagged CdGAP-PRD reduced significantly their ability to recognize all three bands (data not shown). We conclude that at least three abundant endogenous CdGAP proteins exist in mouse.
FIG. 1.
Tissue distribution of CdGAP proteins. (A) Phylogenetic analysis of the RhoGAP domain of CdGAP and CdGAP-related proteins generated with Treeview after CLUSTAL W analysis. GenBank accession numbers are indicated in parentheses. The percent identities to mCdGAP are shown at the right. M.m., Mus musculus; H.s., Homo sapiens; R.n. Rattus norvegicus; X.l., Xenopus laevis; C.e., Caenorhabditis elegans; D.m., Drosophila melanogaster; G.g., Gallus gallus; A.g., Anopheles gambiae; T.n., Tetraodon nigroviridis. (B) Structure of short and long mCdGAP protein. (C) Total protein cell lysates from mouse tissues, primary cortical neurons, or COS-7 cells overexpressing CdGAP-s and CdGAP-l were resolved by SDS-PAGE, and CdGAP was revealed by immunoblotting analysis with affinity-purified polyclonal anti-CdGAP antibodies. Lanes: 1, 8, and 9, 5 μg; 2 and 3, 150 μg; 4 to 6, 50 μg; 7, 100 μg. Arrows indicate the three major CdGAP proteins. EV, empty vector.
CdGAP-s and CdGAP-l are highly phosphorylated in fibroblast cells.
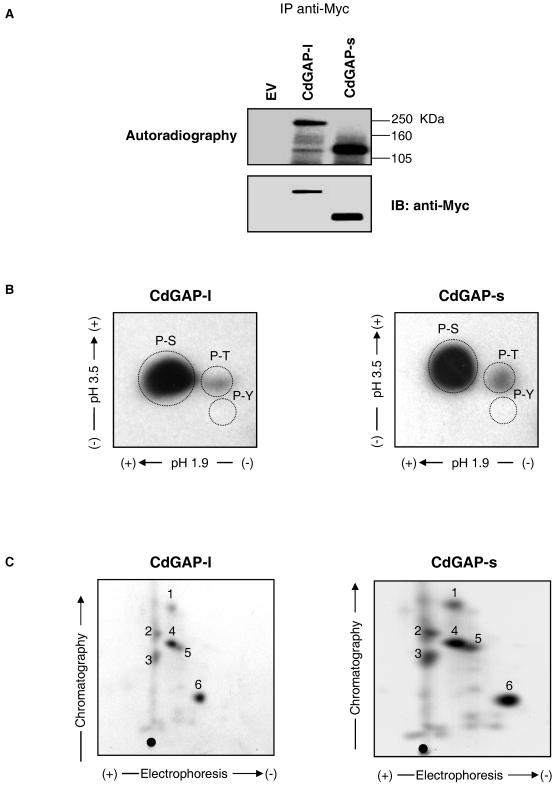
To examine whether CdGAP-s and CdGAP-l are phosphorylated in vivo, Myc-tagged CdGAP-s and CdGAP-l were expressed in COS-7 fibroblasts that were then incubated in phosphate-free medium supplemented with [32P]orthophosphate for 2 h prior to lysis. As shown in Fig. 2A, both immunoprecipitated CdGAP proteins are phosphorylated in vivo to a similar extent in COS-7 cells. To assess the content of phosphorylated residues on CdGAP-s and CdGAP-l, a phosphoamino acid analysis of immunoprecipitated CdGAP-s and CdGAP-l was performed and showed that both CdGAP proteins are highly phosphorylated on serine residues and to a lesser extent on threonine and not on tyrosine (Fig. 2B). The absence of tyrosine phosphorylation was also confirmed by immunoblotting with anti-phosphotyrosine antibodies (data not shown). The tryptic phosphopeptide maps of both immunoprecipitated CdGAP-s and CdGAP-l show a similar pattern of phosphorylation with six major phosphopeptides (Fig. 2C), suggesting that most of the phosphorylation sites are present in the short form of CdGAP. Thus, these results show that CdGAP-s and CdGAP-l are phosphorylated on serine and threonine residues in fibroblasts.
FIG. 2.
CdGAP-s and CdGAP-l have similar phosphorylation profiles. (A) COS-7 cells expressing Myc-tagged CdGAP long (l) or short (s) forms were metabolically labeled with 0.5 mCi of [32P]orthophosphate/ml. The proteins were immunoprecipitated (IP) with anti-Myc antibodies. The samples were resolved by SDS-PAGE, and radiolabeled proteins were identified by autoradiography (upper panel). The membrane was immunoblotted (IB) with anti-Myc antibodies to show the total amount of immunoprecipitated CdGAP (bottom panel). EV, empty vector. (B and C) The phosphorylated protein bands corresponding to CdGAP-l and CdGAP-s were cut and subjected to phosphoamino acid analysis (B) or tryptic phosphopeptide mapping (C). Phosphopeptides or phosphoamino acids were resolved by thin-layer chromatography and detected by autoradiography. Migration of phosphoamino acid standards is indicated with dashed circles: phosphoserine (P-S), phosphothreonine (P-T), and phosphotyrosine (P-Y). Numbers 1 to 6 on phosphopeptide maps represent the six most abundant tryptic phosphopeptides. •, origin of migration.
CdGAP is predominantly phosphorylated in the proline-rich domain.
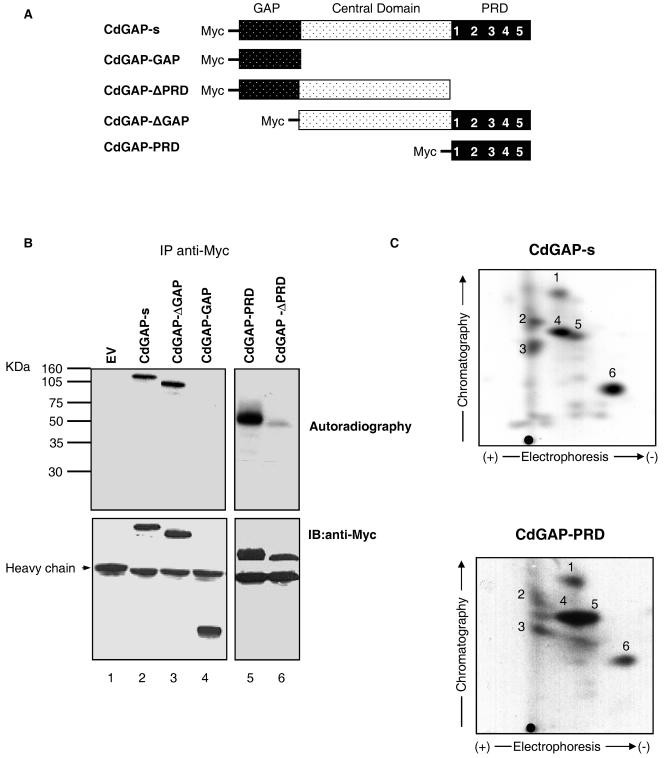
CdGAP-s consists of an N-terminal GAP domain, a central domain, and a C-terminal PRD harboring five consensus SH3-binding motifs. To determine which region of CdGAP-s is phosphorylated in vivo, we investigated the phosphorylation status of various deletion mutants of CdGAP (Fig. 3A) expressed in COS-7 cells. As shown in Fig. 3B, CdGAP lacking the GAP domain (CdGAP-ΔGAP [lane 3]) is as well phosphorylated as the wild-type protein (lane 2). Indeed, the GAP domain by itself is not phosphorylated in vivo (CdGAP-GAP [lane 4]). Interestingly, the CdGAP-PRD protein (lane 5) is phosphorylated at levels similar to those of the wild-type CdGAP and removing the PRD significantly decreases the phosphorylation levels of the mutant protein CdGAP-ΔPRD (lane 6). To confirm that the PRD contains the majority of the phosphorylation sites, we performed a two-dimensional tryptic phosphopeptide mapping of immunoprecipitated CdGAP and CdGAP-PRD. We found that the proline-rich domain comprises the six major phosphopeptides also found in the wild-type CdGAP protein (Fig. 3C). These findings indicate that the majority of the phosphorylation sites are present in the proline-rich domain of CdGAP.
FIG. 3.
Phosphorylation analysis of CdGAP deletion mutants. (A) Schematic representation of CdGAP-s, CdGAP-GAP, CdGAP-ΔPRD, CdGAP-ΔGAP, and CdGAP-PRD constructs. (B) COS-7 cells transfected with empty vector (EV) or pRK5myc encoding CdGAP-s or various protein mutants were labeled with [32P]orthophosphate for 3 h. Proteins were immunoprecipitated (IP) from cell lysates by using anti-Myc antibodies. The samples were resolved by SDS-PAGE, and the radiolabeled proteins were identified by autoradiography (upper panel). The membrane was immunoblotted (IB) with anti-Myc antibodies to show the total amount of immunoprecipitated CdGAP proteins (bottom panel). (C) Comparison of tryptic phosphopeptide patterns between CdGAP-s and CdGAP-PRD. The phosphorylated protein bands corresponding to CdGAP-s and CdGAP-PRD were cut and subjected to two-dimensional tryptic phosphopeptide mapping. •, origin of migration.
CdGAP-s associates with ERK1/2 and RSK-1.
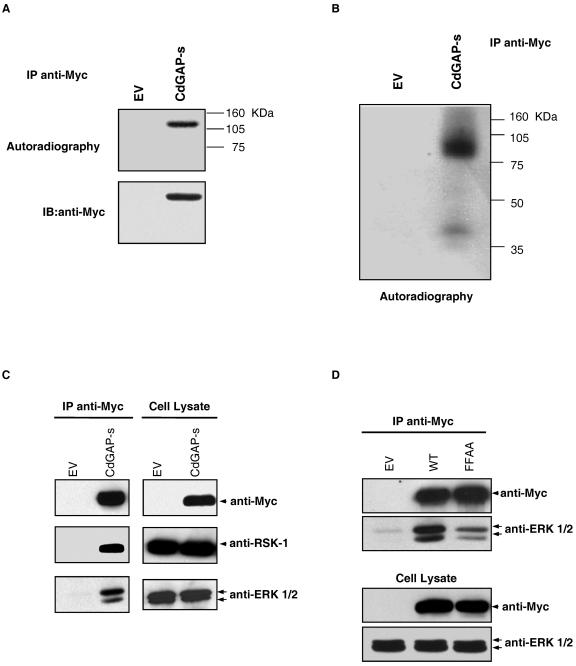
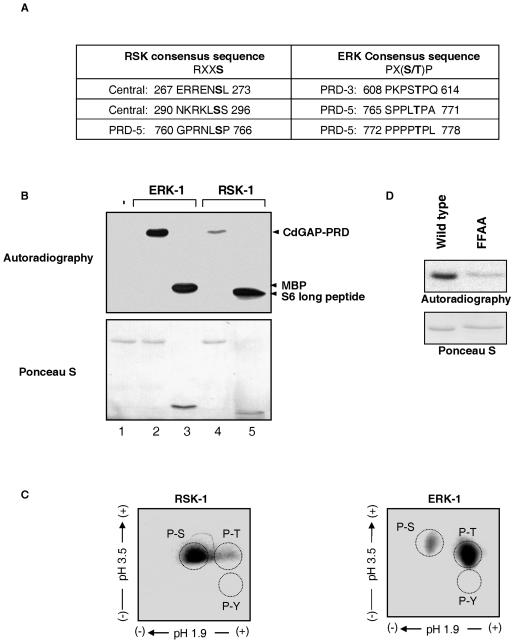
To identify the putative kinase(s) involved in CdGAP phosphorylation, we searched for kinase activities present in CdGAP immunoprecipitates. For this purpose, Myc-tagged CdGAP-s was immunoprecipitated from COS-7 cell lysates and incubated in kinase buffer containing [γ-32P]ATP. Interestingly, CdGAP-s was phosphorylated even without the addition of exogenous kinases (Fig. 4A), suggesting that kinases coimmunoprecipitate with CdGAP-s. To further characterize the kinases associated with CdGAP-s, we performed an in-gel kinase assay in which purified His-tagged CdGAP-PRD was used as a substrate embedded in the polyacrylamide gel. The proteins were renatured, and the gel was incubated with [γ-32P]ATP. As shown in Fig. 4B, two major kinases corresponding to the molecular masses of 90 and 40 kDa were able to renature efficiently and to phosphorylate CdGAP-PRD in the polyacrylamide gel. Curiously, this in-gel phosphorylation pattern is similar to those obtained by at least two other groups who used different proteins as substrates (8, 24). In both cases, the authors showed that the 40-kDa kinases represented p42 ERK-2 and p44 ERK-1, whereas the 90-kDa kinase appeared to be p90 RSK-1. We have also investigated this possibility since the protein sequence of CdGAP contains a significant number of ERK1/2 and RSK-1 consensus phosphorylation sites PX(S/T)P and RXXS, respectively (see Fig. 6A). As shown in Fig. 4C, both ERK 1/2 and RSK-1 coimmunoprecipitate with CdGAP expressed in COS-7 cells. Members of the MAPK family of proteins are known to interact with substrates through consensus docking motifs (12). In the case of ERK1/2, two docking sequences have been identified: the D domain and the DEF domain (12). The D domain consists of a cluster of basic amino acids adjacent to a cluster of hydrophobic residues. Many MAPK family members are able to bind to the D domain, whereas the DEF domain comprising the amino acid sequence FXFP is specific to ERK1/2 (12). The proline-rich sequence of CdGAP-s contains a consensus DEF domain (FPFP). To determine whether this DEF domain is required for the interaction between ERK1/2 and CdGAP-s, we generated an FPFP-to-APAP mutant in which the phenylalanine residues at positions 677 and 679 were substituted with alanines. As shown in Fig. 4D, the ability of the FFAA mutant to bind ERK1/2 was significantly reduced compared to the wild-type protein, suggesting that the FPFP sequence of CdGAP is required for interacting with ERK1/2.
FIG. 4.
ERK1/2 and RSK interact with CdGAP-s. (A) COS-7 cells were transfected with empty vector (EV) or pRK5-myc CdGAP-s. Proteins were immunoprecipitated (IP) by using anti-Myc antibodies. An in vitro kinase assay was performed by incubating immunoprecipitates in kinase buffer containing [γ-32P]ATP. The samples were resolved by SDS-PAGE, and radiolabeled proteins were identified by autoradiography (upper panel). The membrane was immunoblotted (IB) with anti-Myc antibodies to show the total amount of immunoprecipitated CdGAP (bottom panel). (B) An in-gel kinase assay was performed by resolving immunoprecipitated Myc-CdGAP on SDS-PAGE containing purified His-tagged CdGAP-PRD protein. The kinases were renatured, and the acrylamide gel was incubated in kinase buffer containing [γ-32P]ATP. (C and D) Immunoprecipitated CdGAP-s and CdGAP-s-FFAA from COS-7 cell lysates was resolved by SDS-PAGE, and proteins were transferred on nitrocellulose membrane for immunoblotting (IB) with anti-Myc, polyclonal anti-ERK1/2, and RSK-1 antibodies. Protein expression levels are shown as input controls (cell lysate) in the right panel (C) and the bottom panel (D).
FIG. 6.
RSK-1 and ERK-1 phosphorylate CdGAP in vitro. (A) Consensus phosphorylation sites of RSK and ERK in CdGAP protein sequence. The potential phosphorylation site within the consensus motif is shown in bold. (B) An in vitro kinase assay was performed by incubating recombinant His-tagged CdGAP-PRD without (lane 1) or with activated ERK-1 (lane 2) or activated RSK-1 (lane 4) in the presence of 10 μCi of [γ-32P]ATP/ml. As positive controls, ERK-1 was incubated with MBP (lane 3) and RSK-1 with long S6 kinase substrate peptide (lane 5). The products were resolved by SDS-PAGE, and phosphorylated substrates were detected by autoradiography (upper panel). Comparable amounts of substrate are shown by Ponceau-S staining (lower panel). (C) His-tagged CdGAP-PRD proteins phosphorylated by either ERK-1 or RSK-1 were subjected to phosphoamino acid analysis. (D) In vitro kinase assay with His-tagged CdGAP-PRD and CdGAP-PRD-FFAA and activated ERK-1 (autoradiograph, upper panel). Protein loading was determined by Ponceau-S staining (bottom panel).
CdGAP-l is phosphorylated in vivo in response to serum and PDGF stimulation via the MAPK pathway.
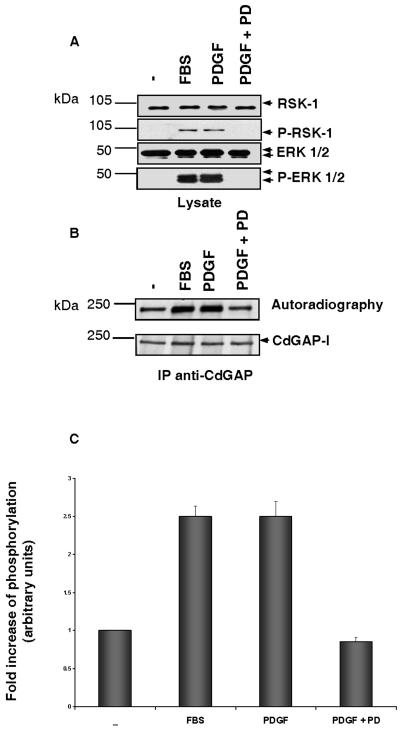
To examine whether endogenous CdGAP is phosphorylated in response to activation of the MAPK pathway, Swiss 3T3 fibroblasts were serum starved and then incubated in phosphate-free medium supplemented with [32P]orthophosphate for 2 h prior to stimulation with FBS or PDGF for 30 and 5 min, respectively. As expected, both RSK-1 and ERK1/2 are activated after stimulation of the fibroblasts by serum or PDGF (Fig. 5A). The activation of these kinases is inhibited in cells incubated with the MEK-1 inhibitor PD98059 prior to stimulation with PDGF (Fig. 5A). CdGAP-l, the most abundant CdGAP isoform in Swiss 3T3 cells, shows a 2.5-fold increase in the level of phosphorylation in response to FBS and PDGF, and this is inhibited in the presence of the MEK-1 inhibitor PD98059 (Fig. 5B and C). These results suggest that endogenous CdGAP is phosphorylated in vivo in response to activation of the MAPK pathway.
FIG. 5.
The MEK-1 inhibitor PD98059 blocks PDGF-stimulated CdGAP phosphorylation. Serum-starved Swiss 3T3 cells were labeled with 0.5 mCi of [32P]orthophosphate/ml for 2 h and then were either left unstimulated (−) or stimulated with 20% FBS, PDGF (5 ng/ml) alone, or PDGF (5 ng/ml) after treatment with 50 μM PD98059 for 1 h. (A) Protein cell lysates resolved by SDS-PAGE were immunoblotted with anti-ERK1/2 and anti-RSK-1 antibodies or with polyclonal anti-P-ERK1/2 and anti-P-RSK1 antibodies to show MAPK pathway activation during similar conditions to CdGAP phosphorylation. (B) CdGAP was immunoprecipitated (IP) by using anti-CdGAP antibodies and subjected to SDS-PAGE, and the radiolabeled proteins were identified by autoradiography (upper panel). The membrane was immunoblotted with anti-CdGAP antibodies to show the total amount of immunoprecipitated CdGAP (lower panel). (C) Quantitative analysis of CdGAP phosphorylation. The fold increase in CdGAP phosphorylation was determined by densitometry, and the values correspond to the average of at least three independent experiments.
The proline-rich domain of CdGAP-s is phosphorylated in vitro by ERK-1 and RSK-1.
ERK1/2 is a proline-directed kinase that phosphorylates serine and threonine residues that precede proline residues within the Pro-X-(Ser/Thr)-Pro consensus motif. On the other hand, RSK-1 phosphorylates serines and threonines within the Arg-X-X-Ser/Thr consensus motif (9). The amino acid sequence of CdGAP contains three consensus phosphorylation motifs for ERK1/2 located within the proline-rich sequence and three consensus RSK-1 phosphorylation sites: two in the central domain and one in the proline-rich sequence (Fig. 6A). To examine the ability of ERK-1 and RSK-1 to directly phosphorylate CdGAP, in vitro kinase assays were performed with purified His-tagged CdGAP-PRD incubated with recombinant activated ERK-1 or RSK-1. Figure 6B shows that CdGAP-PRD is efficiently phosphorylated by ERK-1 in vitro. In fact, we found that at similar protein concentrations, MBP and CdGAP are equally good substrates for ERK-1. RSK-1 is also able to phosphorylate CdGAP-PRD, although to a lower extent than ERK-1. To determine the content of phosphorylated residues on CdGAP-PRD, we performed a phosphoamino acid analysis on His-tagged CdGAP-PRD, phosphorylated in vitro by either ERK-1 or RSK-1. As expected from the consensus phosphorylation motifs (Fig. 6A), ERK-1 phosphorylates CdGAP mainly on threonine residues, whereas CdGAP is predominantly phosphorylated on serine residues by RSK-1 (Fig. 6C). We also investigated whether the FFAA mutant is phosphorylated by ERK1/2 in vitro. As shown in Fig. 6D, ERK-1 fails to phosphorylate the FFAA mutant, indicating that the FPFP motif is not only important for ERK1/2 binding but also for efficient phosphorylation by ERK-1.
ERK-1 phosphorylates CdGAP on Thr769 and Thr776 in vitro.
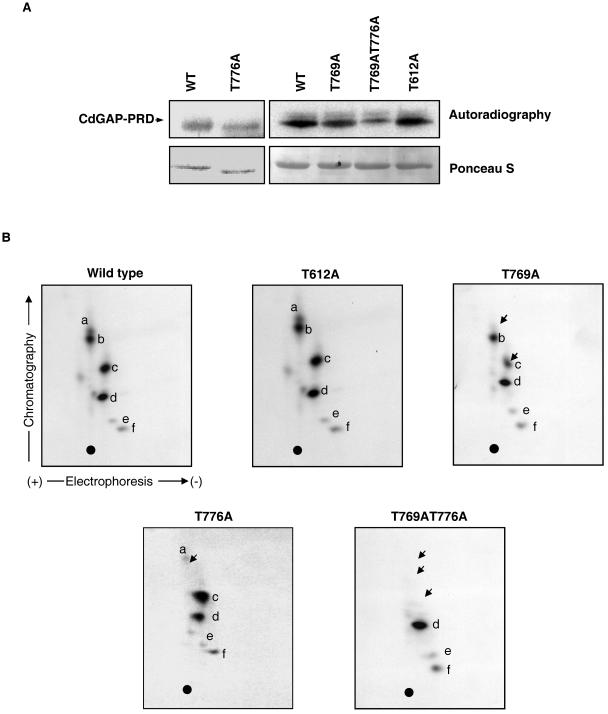
To identify the phosphorylation sites for ERK-1, each threonine of the three potential ERK-1 phosphorylation sites of CdGAP was substituted for an alanine residue. As shown in Fig. 7A, CdGAP-PRD containing the T612A amino acid substitution is phosphorylated by ERK-1 at levels comparable to those of the wild-type protein, and the tryptic phosphopeptide map does not show any noticeable differences (Fig. 7B). However, the phosphorylation of CdGAP-PRD (T769A) and the double mutant CdGAP-PRD (T769AT776A) by ERK-1 are significantly reduced compared to wild-type CdGAP-PRD (Fig. 7A). In fact, the tryptic phosphopeptide map of the T769A mutant protein reveals that phosphorylation of peptide a is absent and phosphorylation of peptide c is reduced compared to the wild-type protein (Fig. 7B). In the phosphopeptide map of the double mutant T769AT776A, we found that phosphorylation of peptides a, b, and c has disappeared (Fig. 7B). Since Thr769 and Thr776 are located on the same tryptic peptide, these results suggest that phosphopeptides a, b, and c are the result of partial tryptic digestion. Although total phosphorylation of the T776A mutant protein by ERK-1 is slightly reduced (Fig. 7A), tryptic phosphopeptide mapping (Fig. 7B) clearly indicates that phosphopeptide a is missing and that phosphopeptide c is increased compared to the wild-type protein. These data suggest that in the absence of threonine 776, threonine 769 becomes hyperphosphorylated by ERK-1. Altogether, these findings demonstrate that Thr769 and Thr776 are phosphorylation target sites of ERK-1 in vitro.
FIG. 7.
Thr769 and Thr776 in the proline-rich domain of CdGAP are phosphorylated in vitro by ERK-1. (A) In vitro phosphorylation of His-tagged CdGAP-PRD wild type or the indicated alanine mutants (T612A, T769A, T776A, and T769AT776A) by recombinant activated ERK-1 (upper panel). The bottom panel corresponds to Ponceau-S staining, indicating that equal amounts of protein were used. Wild type and T776A are on a separate gel containing lower amounts of proteins. (B) Two-dimensional separation of tryptic phosphopeptides derived from wild-type CdGAP-PRD, -T612A, T769A, -T776A, or -T769AT776A. Letters a to f designate the six most abundant phosphopeptides present in CdGAP-PRD wild type. Equal counts of samples were applied onto the thin-layer chromatography plates, and autoradiographs with the same exposure times are presented for each sample. Arrows in panel B indicate the reduction or absence of phosphopeptides a, b, and c. •, origin of migration.
Thr776 is an in vivo phosphorylation site of CdGAP-s.
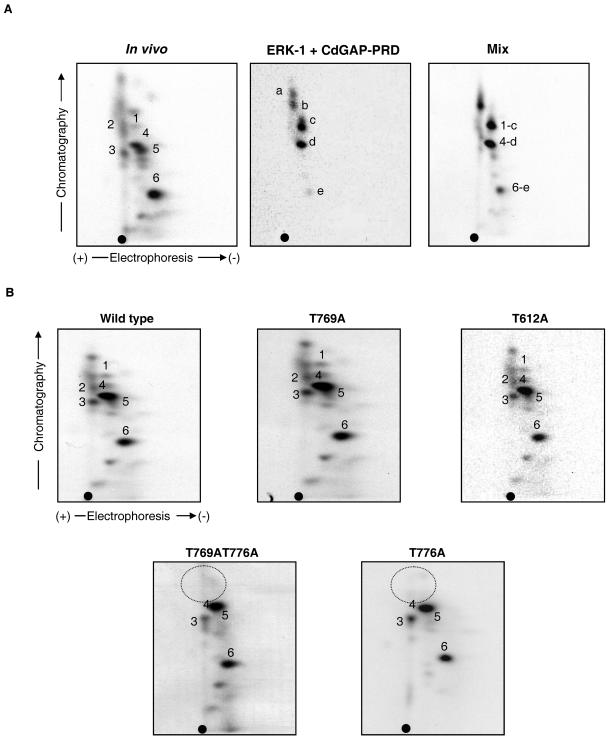
To determine whether CdGAP is a physiological substrate for ERK-1, we mixed the phosphopeptides obtained by tryptic digestion of recombinant His-tagged CdGAP-PRD phosphorylated with ERK-1 in vitro with those obtained from immunoprecipitated CdGAP phosphorylated in vivo in COS-7 cells. As shown in Fig. 8A, phosphopeptides 1, 4, and 6 from the in vivo phosphopeptide map comigrate with phosphopeptides c, d, and e from the in vitro map, suggesting that ERK-1 phosphorylates CdGAP in vitro and in vivo on similar sites. We then compared the in vivo phosphopeptide patterns of CdGAP mutants to that of wild-type CdGAP expressed in COS-7 cells. We found that the in vivo phosphorylation patterns of both T769A and T612A CdGAP protein mutants are very similar to that of the wild-type protein (Fig. 8B). However, phosphopeptides 1 and 2 are absent in the in vivo map of both T776A and the double mutant T769AT776A. Together, these results strongly suggest that CdGAP is an in vivo substrate of ERK-1 and that Thr776 is a phosphorylation site in vivo.
FIG. 8.
Thr776 is an in vivo phosphorylation site of CdGAP. (A) Comparison of the tryptic phosphopeptide maps between in vivo- and in vitro-phosphorylated CdGAP by ERK-1. Tryptic phosphopeptide map derived from in vivo 32P-labeled Myc-CdGAP-s immunoprecipitated from COS-7 cell lysate (left panel). Tryptic phosphopeptide map of His-tagged recombinant CdGAP-PRD phosphorylated in vitro by activated ERK-1 (middle panel). Tryptic phosphopeptide map derived from a mixture of the in vitro and in vivo phosphopeptides (right panel). Equal counts of samples were applied onto the thin-layer chromatography plates, and autoradiographs with the same exposure times are presented for each sample. Phosphopeptides from the in vitro map that comigrate with the in vivo phosphopeptides are indicated in right panel (1-c, 4-d, and 6-e). (B) In vivo phosphopeptide mapping of 32P-labeled Myc-CdGAP wild type or the indicated protein mutants. Dashed circle indicates the missing phosphopeptides. •, origin of migration.
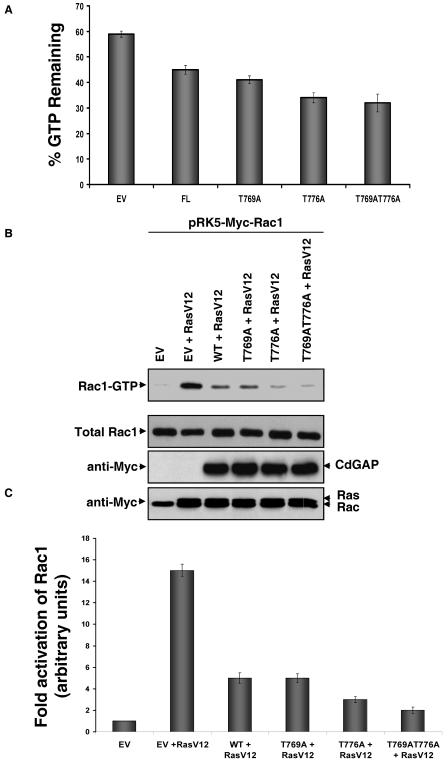
T776A and T769AT776A mutant proteins have increased GAP activity toward Rac1 in response to oncogenic Ras.
To assess whether threonine phosphorylation of CdGAP is involved in the regulation of its GAP activity, [32P]GTP-loaded Rac1 was incubated with myc-tagged CdGAP or with myc-tagged CdGAP threonine mutant proteins immunoprecipitated from COS-7 cell lysates. The GTPase stimulating activity of CdGAP was measured by the estimation of the ratio of Rac1-bound GTP in the absence versus the presence of CdGAP as described previously (13). As shown in Fig. 9A, 45% of GTP remains bound to Rac1 after a 6-min incubation in the presence of immunoprecipitated CdGAP compared to 59% of GTP bound to Rac1 incubated with immune complexes from lysates of COS-7 cells transfected with empty vector. However, the GAP activity of the T776A and T769AT776A mutant proteins shows a significant 1.4- and 1.5-fold increase compared to the wild-type protein, respectively. The GAP activity of the T769A mutant protein appears to be only weakly affected compared to the wild-type protein. To further support these findings, we investigated the in vivo GAP activity of CdGAP wild type and threonine mutant proteins toward Rac1 activated by RasV12 in serum-starved HEK 293 cells. The GTPase stimulating activity of CdGAP was measured by the amount of GTP-bound Rac1 obtained in a pull-down assay using GST-CRIB domain of PAK. As shown in Fig. 9B and C, RasV12 stimulated a 15-fold increase in the level of activated Rac1. In the presence of wild-type CdGAP or the T769A mutant proteins, the levels of activated Rac1 show a threefold reduction compared to RasV12 alone. However, both T776A and T769AT776A mutant proteins show a higher GAP activity resulting in a 5- and 7.5-fold reduction in the levels of activated Rac1 in response to RasV12, respectively. These results correlate well with the in vitro GAP assays and indicate that phosphorylation of at least one threonine residue in the CdGAP protein sequence downstream of the Ras/ERK signaling pathway is sufficient to modulate the intrinsic GTPase stimulating activity toward Rac1. We conclude that phosphorylation of Thr776 plays an important regulatory role in the GAP activity of CdGAP.
FIG. 9.
Phosphorylation of Thr776 reduces CdGAP activity in vitro and in vivo. (A) [γ-32P]GTP-loaded Rac1 was incubated at 20°C with anti-Myc immune complexes from lysates of COS-7 cells transfected with empty vector (EV) or pRK5 encoding Myc-tagged CdGAP-s (FL), -T769A, -T776A, or T769AT776A, and a GAP assay was performed. Equal amounts of immunoprecipitated CdGAP and CdGAP protein mutants were estimated in each sample by Coomassie blue-stained SDS-PAGE by comparison with different amounts of purified bovine serum albumin. (B) HEK293 cells were transfected with empty vector (EV) or pRK5 encoding Myc-tagged CdGAP-s (WT), -T769A, -T776A, or T769AT776A, together with Myc-Rac1 and Myc-RasV12. GTP-loaded Rac1 proteins were pulled down from cell lysates by using GST-PAK1 (aa 56 to 272). GTP-bound Rac1 and total Rac1 in protein cell lysates were detected by Western blotting with anti-Rac1 antibodies. The amounts of CdGAP and myc-tagged RasV12 in protein cell lysates were revealed by Western blotting with anti-myc antibodies. (C) Quantitative analysis of Rac1 activation by RasV12 in the absence or presence of CdGAP wild type and threonine protein mutants. Error bars represent standard deviations relative to three separate experiments.
DISCUSSION
In the present study, we show that the proline-rich domain of CdGAP is phosphorylated in vivo at multiple sites containing serine and threonine residues. We also demonstrate that CdGAP is phosphorylated downstream of the MEK-ERK pathway in response to serum or PDGF stimulation of fibroblasts. In particular, we find that CdGAP interacts with both ERK1/2 and RSK-1 and is directly phosphorylated by ERK-1 and RSK-1 in vitro. Site-directed mutagenesis reveals that threonine 776 of CdGAP is a phosphorylation site for ERK-1 and is an important regulatory site of CdGAP activity.
The incredibly large number of RhoGAP proteins strongly suggests a tight regulation of their activities at specific sites within the cell. Indeed, accumulating evidence indicates that RhoGAP activities are regulated by a wide variety of mechanisms, including phosphorylation. For example, tyrosine phosphorylation of p190RhoGAP by Src is necessary for its association with p120RasGAP and activation of its rhoGAP activity in vivo (11, 31). On the other hand, the in vitro GAP activity of RICS, a GTPase-activating protein for Cdc42 and Rac1, is inhibited by phosphorylation from Ca2+/calmodulin-dependent protein kinase II (27). Interestingly, MgcRacGAP, known to be involved in cytokinesis and a GAP for Rac1 and Cdc42, is functionally converted to a GAP for RhoA after serine phosphorylation by Aurora B kinase (20). In the present study, we found that the replacement of threonine 776 by an alanine within the proline-rich domain of CdGAP is sufficient to induce a 1.5- and 2.0-fold increase in the in vitro and in vivo CdGAP activity toward Rac1, respectively. These findings suggest that phosphorylation of threonine 776 is an important regulatory site of CdGAP activity and may lead to a conformational change affecting the enzymatic activity. However, it is clear that CdGAP contains additional phosphorylation sites within the proline-rich domain that may affect the GAP activity as well. Future identification of these phosphorylation sites will help us to understand better the mechanism of regulation of CdGAP activity. In addition, it is more than likely that phosphorylation of the proline-rich domain may affect the localization of the protein or alter protein-protein interactions. Indeed, CdGAP contains five consensus SH3-binding sites. However, we have not yet been able to identify any SH3 domain-containing proteins binding to these motifs. Interestingly, Thr776 is located directly in the fifth proline-rich sequence PPLTPAPPPPTP. Therefore, it is possible that phosphorylation of serine or threonine residues within the proline-rich domain causes a conformational change that negatively regulates their ability to bind SH3 domains. Similarly, it has been reported that phosphorylation of the proline-rich sequence of SOS is important to modulate its interaction with the SH3 domain-containing adaptor molecule Grb2 (7).
To identify potential Ser/Thr kinases that interact with CdGAP, we performed an in-gel kinase assay. The success of this technique depends greatly on the ability of kinases to renature in the polyacrylamide gel. Several lines of evidence have demonstrated that RSK and ERK1/2 are able to efficiently recover their kinase activity after in-gel renaturation (8, 24). We found striking similarities between the in-gel kinase profile reported in these previous studies and our own results. In fact, we found that the two kinase activities recovered from the in-gel kinase are indeed ERK1/2 and RSK-1. Consistent with these results, we observed that CdGAP is present in a ternary protein complex including ERK1/2 and RSK-1. Moreover, we present evidence that the proline-rich domain of CdGAP is directly phosphorylated by ERK-1 and RSK-1 in vitro on distinct sites. These results suggest that both enzymes can phosphorylate CdGAP independently of each other. However, since RSK-1 phosphorylates CdGAP significantly less than ERK-1 in vitro, it remains to be determined whether prephosphorylation of CdGAP by ERK-1 or another kinase leads to a better substrate for RSK-1. It is also possible that efficient phosphorylation of CdGAP by RSK-1 requires the full-length protein and not only the proline-rich sequence. In fact, although the PRD domain of CdGAP contains a minimal RSK recognition phosphorylation site, the two additional ones in the central domain are preceded by arginine residues, which could make these sites more favorable for RSK-1. The levels of kinase activity obtained in the in-gel kinase experiment suggested that RSK-1 is more efficient than ERK-1 in phosphorylating the PRD of CdGAP, but the data obtained from the in vitro kinase assay with recombinant activated kinases suggests the opposite. These conflicting results can be explained by at least two possibilities. First, it is possible that RSK-1 recovers more efficiently its kinase activity after renaturation than ERK1/2. Second, RSK-1 seems to be more abundant than ERK-1/2 in coimmunoprecipitation assays with CdGAP.
In the present study, we have shown that the majority of the in vivo phosphorylation sites of CdGAP are on serine residues. Since RSK-1 interacts with and phosphorylates CdGAP on serine residues, it is likely that RSK-1 is responsible for most of the in vivo serine phosphorylation. In particular, treatment of Swiss 3T3 cells with the MEK inhibitor that blocks both ERK1/2 and RSK-1 activation also completely abolishes PDGF-induced CdGAP phosphorylation. We are currently investigating the role of RSK-1 phosphorylation on CdGAP, and future studies will provide valuable knowledge on this issue. Nevertheless, our studies clearly demonstrate that, although the extent of threonine phosphorylation in vivo is weak, the importance of this phosphorylation on CdGAP activity is significant.
The carboxy-terminal tail of CdGAP contains a number of putative ERK phosphorylation sites containing the consensus sequence P-X-S/T-P (10). The in vitro and in vivo phosphopeptide mapping of CdGAP protein mutants strongly support the conclusion that ERK-1 phosphorylates CdGAP on Thr776 in vivo. However, amino acid substitution of both Thr769 and Thr776 to alanine did not completely abolish ERK-1 phosphorylation of CdGAP in vitro. In addition to the three ERK putative phosphorylation sites mutated in the present study, the proline-rich domain of CdGAP contains 12 S/T-P motifs containing the minimum consensus motif for ERK phosphorylation (33). Interestingly, two of these sites are adjacent to a putative DEF domain containing the FPFP motif known to be an ERK docking site (12). Indeed, mutation of this motif alters CdGAP binding to ERK1/2 and leads to a loss of CdGAP phosphorylation by ERK-1 in vitro.
CdGAP belongs to a novel family of RhoGAP proteins that are phylogenetically well conserved among different species. Up to three human genes encode for CdGAP-related proteins which consist of a RhoGAP domain at the N terminus and multiple SH3-binding motifs at the C terminus of the proteins. Interestingly, the DEF domain and the sequence surrounding Thr776 are found only in CdGAP and not in the closely related Grit or TCGAP. This suggests that ERK may exclusively interact with and phosphorylate CdGAP. In fact, among all of the characterized RhoGAP proteins, only mCdGAP and human DLC-1 proteins contain a DEF domain. Interestingly, we observed the differential expression of at least two major isoforms of CdGAP in specific mouse tissues. CdGAP-l (250 kDa) is predominantly expressed in the brain, lung, and heart, whereas CdGAP-s (90 kDa) is predominantly expressed in the liver and kidney. Whether the differential expression of CdGAP leads to a tissue-specific function for each isoform will require further investigation. We also found that both overexpressed CdGAP-s and CdGAP-l migrates higher than their expected molecular masses of 90 and 155 kDa, respectively. It will be of great interest to investigate the posttranslational modifications responsible for this impressive mobility shift.
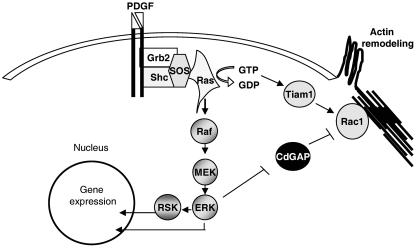
Thus far, one of the most exciting roles attributed to RhoGAP proteins is their implication in the cross talk between members of the Rho family of small GTPases. For example, p120RasGAP interacts with and regulates p190RhoGAP activity, suggesting a possible interplay between Ras and Rho GTPases (25). The connection between the Ras/MAPK pathway and the effects on cytoskeletal dynamics becomes more evident with the identification of a number of cytoskeleton-related proteins as ERK and RSK substrates (15, 19, 21, 37). Here we have demonstrated that phosphorylation of Thr776 by ERK affects CdGAP activity both in vitro and in vivo. One possibility is that mitogenic signal regulates Rac1 through phosphorylation and downregulation of CdGAP activity by ERK, leading to Rac1 activation and cytoskeletal remodeling (Fig. 10). In conclusion, we provide evidence that CdGAP is a novel ERK substrate and may play roles in the connection between the Ras/MAPK and Rac1 pathways.
FIG. 10.
CdGAP mediates cross talk between the Ras/MAPK pathway and the regulation of Rac1 activity. Upon PDGF stimulation, Ras activates the MAPK pathway, leading to gene expression and phosphorylation of many cytoplasmic and membrane proteins. In addition, Ras causes cytoskeleton remodeling by activating a guanine nucleotide exchange factor (Tiam-1) for Rac1. We propose that ERK downregulation of CdGAP activity is an additional mechanism by which Ras can maintain active Rac1 in response to growth factor stimulation.
Acknowledgments
We thank Louise Larose and Fiona Bedford for critically reading the manuscript and for helpful discussions. We also thank Louise Larose for support with phosphoamino and phosphopeptide analysis. We especially thank Josephine Aho for the cloning of CdGAP-l.
This study was supported by the Canadian Cancer Society through the National Cancer Institute of Canada. E.I.D is supported by a Canada Graduate Scholarship from the CIHR. N.L.-V. is a recipient of a CIHR new investigator award.
REFERENCES
- 1.Ahmed, S., J. Lee, R. Kozma, A. Best, C. Monfries, and L. Lim. 1993. A novel functional target for tumor-promoting phorbol esters and lysophosphatidic acid. The p21rac-GTPase activating protein n-chimaerin. J. Biol. Chem. 268:10709-10712. [PubMed] [Google Scholar]
- 2.Bernards, A., and J. Settleman. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14:377-385. [DOI] [PubMed] [Google Scholar]
- 3.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 4.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116:167-179. [DOI] [PubMed] [Google Scholar]
- 5.Cerione, R. A., and Y. Zheng. 1996. The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, S. H., J. Hwang, M. Legendre, M. Zhang, A. Kimura, and A. R. Saltiel. 2003. TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 22:2679-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbalan-Garcia, S., K. R. Degenhardt, and D. Bar-Sagi. 1996. Insulin-induced dissociation of Sos from Grb2 does not contribute to the down regulation of Ras activation. Oncogene 12:1063-1068. [PubMed] [Google Scholar]
- 8.Douville, E., and J. Downward. 1997. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15:373-383. [DOI] [PubMed] [Google Scholar]
- 9.Erikson, R. L. 1991. Structure, expression, and regulation of protein kinases involved in the phosphorylation of ribosomal protein S6. J. Biol. Chem. 266:6007-6010. [PubMed] [Google Scholar]
- 10.Gonzalez, F. A., D. L. Raden, and R. J. Davis. 1991. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266:22159-22163. [PubMed] [Google Scholar]
- 11.Hu, K. Q., and J. Settleman. 1997. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 16:473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13:163-175. [PMC free article] [PubMed] [Google Scholar]
- 13.Jenna, S., N. K. Hussain, E. I. Danek, I. Triki, S. Wasiak, P. S. McPherson, and N. Lamarche-Vane. 2002. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J. Biol. Chem. 277:6366-6373. [DOI] [PubMed] [Google Scholar]
- 14.Jenna, S., and N. Lamarche-Vane. 2003. The superfamily of Rho GTPase activating proteins, p. 68-87. In M. Symons (ed.), Rho GTPases. Kluwer Academic Press/Plenum Press, New York, N.Y.
- 15.Klemke, R. L., S. Cai, A. L. Giannini, P. J. Gallagher, P. de Lanerolle, and D. A. Cheresh. 1997. Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 137:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozma, R., S. Ahmed, A. Best, and L. Lim. 1996. The GTPase-activating protein n-chimaerin cooperates with Rac1 and Cdc42Hs to induce the formation of lamellipodia and filopodia. Mol. Cell. Biol. 16:5069-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamarche-Vane, N., and A. Hall. 1998. CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J. Biol. Chem. 273:29172-29177. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., E. Saint-Cyr-Proulx, K. Aktories, and N. Lamarche-Vane. 2002. Rac1 and Cdc42 but not RhoA or Rho kinase activities are required for neurite outgrowth induced by the Netrin-1 receptor DCC (deleted in colorectal cancer) in N1E-115 neuroblastoma cells. J. Biol. Chem. 277:15207-15214. [DOI] [PubMed] [Google Scholar]
- 19.Miki, H., M. Fukuda, E. Nishida, and T. Takenawa. 1999. Phosphorylation of WAVE downstream of mitogen-activated protein kinase signaling. J. Biol. Chem. 274:27605-27609. [DOI] [PubMed] [Google Scholar]
- 20.Minoshima, Y., T. Kawashima, K. Hirose, Y. Tonozuka, A. Kawajiri, Y. C. Bao, X. Deng, M. Tatsuka, S. Narumiya, W. S. May, Jr., T. Nosaka, K. Semba, T. Inoue, T. Satoh, M. Inagaki, and T. Kitamura. 2003. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 4:549-560. [DOI] [PubMed] [Google Scholar]
- 21.Mitsushima, M., A. Suwa, T. Amachi, K. Ueda, and N. Kioka. 2004. Extracellular signal-regulated kinase activated by epidermal growth factor and cell adhesion interacts with and phosphorylates vinexin. J. Biol. Chem. 279:34570-34577. [DOI] [PubMed] [Google Scholar]
- 22.Moon, S. Y., H. Zang, and Y. Zheng. 2003. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J. Biol. Chem. 278:4151-4159. [DOI] [PubMed] [Google Scholar]
- 23.Moon, S. Y., and Y. Zheng. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13:13-22. [DOI] [PubMed] [Google Scholar]
- 24.Moor, A. N., and L. Fliegel. 1999. Protein kinase-mediated regulation of the Na+/H+ exchanger in the rat myocardium by mitogen-activated protein kinase-dependent pathways. J. Biol. Chem. 274:22985-22992. [DOI] [PubMed] [Google Scholar]
- 25.Moran, M. F., P. Polakis, F. McCormick, T. Pawson, and C. Ellis. 1991. Protein-tyrosine kinases regulate the phosphorylation, protein interactions, subcellular distribution, and activity of p21ras GTPase-activating protein. Mol. Cell. Biol. 11:1804-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, T., M. Komiya, K. Sone, E. Hirose, N. Gotoh, H. Morii, Y. Ohta, and N. Mori. 2002. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 22:8721-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okabe, T., T. Nakamura, Y. N. Nishimura, K. Kohu, S. Ohwada, Y. Morishita, and T. Akiyama. 2003. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-d-aspartate receptor signaling. J. Biol. Chem. 278:9920-9927. [DOI] [PubMed] [Google Scholar]
- 28.Olofsson, B. 1999. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 11:545-554. [DOI] [PubMed] [Google Scholar]
- 29.Olson, M. F., A. Ashworth, and A. Hall. 1995. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 269:1270-1272. [DOI] [PubMed] [Google Scholar]
- 30.Peck, J., G. t. Douglas, C. H. Wu, and P. D. Burbelo. 2002. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 528:27-34. [DOI] [PubMed] [Google Scholar]
- 31.Roof, R. W., M. D. Haskell, B. D. Dukes, N. Sherman, M. Kinter, and S. J. Parsons. 1998. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol. Cell. Biol. 18:7052-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sagnier, T., A. Grienenberger, M. Mariol, H. Berenger, J. Pradel, and Y. Graba. 2000. Dynamic expression of d-CdGAPr, a novel Drosophila melanogaster gene encoding a GTPase activating protein. Mech. Dev. 94:267-270. [DOI] [PubMed] [Google Scholar]
- 33.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, A. J. DeMaggio, M. F. Hoekstra, J. Blenis, T. Hunter, and L. C. Cantley. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su, L., J. M. Agati, and S. J. Parsons. 2003. p190RhoGAP is cell cycle regulated and affects cytokinesis. J. Cell Biol. 163:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taniguchi, S., H. Liu, T. Nakazawa, K. Yokoyama, T. Tezuka, and T. Yamamoto. 2003. p250GAP, a neural RhoGAP protein, is associated with and phosphorylated by Fyn. Biochem. Biophys. Res. Commun. 306:151-155. [DOI] [PubMed] [Google Scholar]
- 36.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 37.Woo, M. S., Y. Ohta, I. Rabinovitz, T. P. Stossel, and J. Blenis. 2004. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol. Cell. Biol. 24:3025-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, C., H. Ma, E. Bossy-Wetzel, S. A. Lipton, Z. Zhang, and G. S. Feng. 2003. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J. Biol. Chem. 278:34641-34653. [DOI] [PubMed] [Google Scholar]