Abstract
The stability of metazoan genomes during their duplication depends on the spatiotemporal activation of origins and the progression of forks. Human rRNA genes represent a unique challenge to DNA replication since a large proportion of them exist as noncanonical palindromes in addition to canonical tandem repeats. Whether origin usage and/or fork elongation can cope with the variable structure of these genes is unknown. By analyzing single combed DNA molecules from HeLa cells, we studied the rRNA gene replication program according to the organization of canonical versus noncanonical rRNA genes. Origin positioning, spacing, and timing were not affected by the underlying rRNA gene physical structure. Conversely, fork arrest, both temporary and permanent, occurred more frequently when rRNA gene palindromes were encountered. These findings reveal that while initiation mechanisms are flexible enough to adapt to an rRNA gene structure of any arrangement, palindromes represent obstacles to fork progression, which is a likely source of genomic instability.
When cells divide, a copy of the genome must be generated with a high fidelity so that each daughter cell can be given a replica of the genetic material. Metazoa duplicate their genomes through the initiation of DNA replication at multiple sites along the length of each chromosome and by synthesizing the intervening DNA through the progression of forks. Origins that are spaced too far apart and/or forks that are interfered with are potential sources of genomic instability (9, 26). Despite its relative importance for the faithful propagation of the genome, what constitutes the spatiotemporal distribution of replication origins together with the progression of forks—the DNA replication program—remains to be elucidated.
The locus carrying the rRNA genes has been studied extensively as a model to address this issue. From yeast to humans, rRNA genes are broadly considered to be organized into hundreds of tandemly repeated units that each includes a gene followed by a nontranscribed spacer. In yeast, although a potential origin resides in every rRNA gene unit, clusters of synchronously firing origins are separated by large gaps spanning a few units whose origins are silent (32). In human rRNA genes, a consensus for the initiation site exists, specifically in the nontranscribed spacer, with a preferential zone upstream of the transcription unit (15, 24, 41, 47). However, the distribution of functional human rRNA gene origins and their timing of activation in relation to each other are not known. In other words, do human origins fire in clusters in adherence with the yeast model?
With respect to replication fork kinetics at rRNA genes, most work has focused on a fork barrier that maps to the 3′ end of the transcription unit (reviewed in reference 37). In yeast, not only is the fork barrier polar, preventing forks from entering the transcription unit against RNA polymerase I movement, but it is also nearly 100% efficient (4, 23). Multiple pause sites due to the local chromatin structure in yeast mutants deficient for the Rrm3p helicase suggest that fork obstruction might be multimodal in rRNA genes (20, 43). In human rRNA genes, the programmed fork barrier and its position are conserved (24). In contrast to the case in Saccharomyces cerevisiae, forks traveling in both directions are blocked and a significant proportion of forks traverse the boundary unaffected. Although the stability of the locus is influenced by fork-related events (36), quantification of the fork barrier efficiency and other potential blocks/pause sites in human rRNA genes have not been reported to date.
Recently, a more complex organization of human rRNA genes was described, revealing a fundamental divergence from the standard model of clusters of highly repetitive, nearly identical rRNA gene units (6). A significant proportion of the genes are organized into palindromes interspersed among the tandem repeats, yielding a mosaic of noncanonical and canonical rRNA genes. Palindromes are acknowledged sources of genomic instability, which has been proposed to result from problematic DNA replication, at least in yeast (18, 25, 44). While most rRNA gene replication studies have been carried out in yeast, rRNA gene palindromes do not exist in this organism. Tetrahymena thermophila does contain rRNA gene palindromes that undergo amplification through multiple initiation events on the same fragment of DNA and activation of a fork barrier at the center of the palindrome (48). Therefore, an account of where origins fire from and how forks travel in the context of human rRNA gene palindromes would provide clues as to how the stability of the locus is maintained in this species.
Molecular combing (3, 27) produces sufficiently large DNA molecules on which multiple active origins and progressing forks can be monitored at the level of a single molecule (2, 19, 31, 32). Since the original order of genes and flanking nontranscribed spacers is preserved during the combing process, replication can be attributed to either canonical or noncanonical rRNA genes, a prerequisite if differences between the programs of these two classes of human rRNA genes are to be delineated.
Using combed DNA from HeLa cells, this study establishes and compares key parameters of the human rRNA gene replication programs of canonical versus noncanonical rRNA genes. No significant initiation differences, both in origin spacing and in timing, were observed in relation to the underlying rRNA gene physical structure. rRNA gene palindromes, however, were associated with a greater degree of fork stalling and/or arrest than forks elongating through canonical tandem repeats. These results suggest that while initiation mechanisms in humans are amenable to rRNA gene rearrangement, replication forks are sensitive to rRNA gene palindromes, and that these inverted repeats represent obstacles to fork progression.
MATERIALS AND METHODS
DNA preparation.
HeLa cells were cultured in minimal essential medium (Eagle) with Earle's balanced salt solution, nonessential amino acids, and sodium pyruvate (BioWhittaker) supplemented with 10% fetal bovine serum. One day prior to labeling, 106 cells were seeded in a T75 flask. Twenty-four hours later, cells were pulse labeled for 30 min by replacing the normal medium with prewarmed medium containing 100 μM iododeoxyuridine (IdU). At the end of the first labeling period, the cells were washed twice with warm phosphate-buffered saline (PBS) and then pulse labeled once more for 30 min with medium containing 100 μM chlorodeoxyuridine (CldU). Cells were then harvested, and genomic DNA was extracted and combed as previously described (27).
Hybridization and fluorescence revelation.
The two EcoRI fragments covering the 5′ and 3′ ends of the human rRNA gene transcription unit were labeled with biotin and digoxigenin, respectively, by random priming. Seven hundred nanograms of each probe, 5 μg of human cot-1 DNA (Gibco BRL), and 10 μg of herring sperm DNA were precipitated and then resuspended in hybridization buffer consisting of 50% formamide, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% sodium dodecyl sulfate, 0.5% sarcosyl, 10 mM NaCl, and 33.3% blocking solution (Boehringer blocking reagent; 1% [wt/vol] in PBS, 0.05% Tween 20). Combed DNA was denatured in 1 M NaOH for 30 min, and denatured probes were then added. Hybridization took place overnight in a humid HybChamber (Genemachines) at 42°C. The following day, slides were washed with 2× SSC-50% formamide for 5 min three times and then with 2× SSC for 5 min twice. The antibodies used for fluorescence detection of probe and replication signals were as follows: (i) sheep anti-digoxigenin-fluorescein isothiocyanate (Roche) and streptavidin-Alexa Fluor 750 (Molecular Probes); (ii) donkey anti-sheep 488 (Molecular Probes) and biotin-conjugated rabbit anti-streptavidin (Rockland); (iii) streptavidin-Alexa Fluor 750, mouse anti-bromodeoxyuridine (anti-BrdU; Becton Dickinson), and rat anti-BrdU (Harlan Seralab); and (iv) goat anti-mouse-Alexa Fluor 350 (Molecular Probes) and donkey anti-rat-Texas Red (Jackson Immunochemicals). All incubations were carried out in a humid chamber at 37°C for 30 min, except for step iii, which took place for 1 h. After each step, excess antibodies were washed with 1× PBS for 5 min three times, and then slides were mounted in Slowfade light antifade reagent (Molecular Probes).
Image acquisition and analysis.
Fluorescent signals were captured with a Zeiss Axioplan 2 microscope equipped with a Photometrics HQ charge-coupled device camera using SmartCapture software (Digital Scientific). Probe and replication signals were measured with Cartographix (Institut Pasteur). Analyses were performed on DNAs from two separate labeling experiments. Since there were no significant differences between the results from the two experiments, the data were merged and are presented together here. Photos were arranged using Adobe Photoshop. Background fluorescent spots surrounding the fiber of interest were removed to highlight the signal.
RESULTS
HeLa rRNA genes make up a mosaic of canonical and noncanonical physical structures.
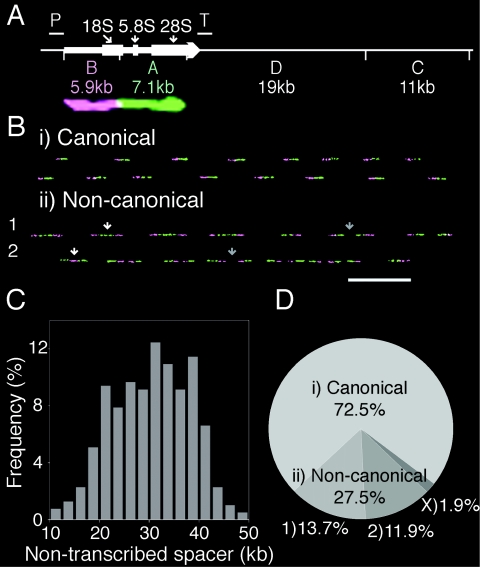
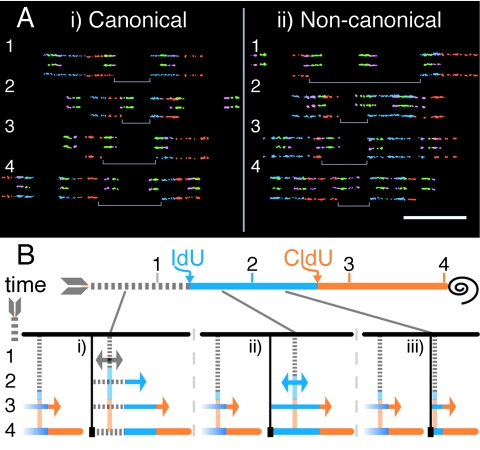
The classical structure of a single human rRNA gene unit is provided in Fig. 1A. EcoRI fragments A and B, corresponding to the transcription unit, were labeled with digoxigenin and biotin, respectively, hybridized to combed genomic HeLa DNA, and detected with green and infrared (shown as purple) coupled antibodies to reveal the physical organization of the locus. The canonical structure of tandem repeats of transcription units separated by nontranscribed spacers was confirmed (Fig. 1Bi). The variability among the nontranscribed spacers in canonical rRNA genes from HeLa cells, illustrated by the molecules in Fig. 1B, panel i, and the histogram in Fig. 1C, is similar to that reported for other cell lines (6). Clusters of rRNA genes were detected as inverted repeats or palindromes, which were collectively termed noncanonical rRNA genes (Fig. 1Bii). Type 1 palindromes have internal A probes or 3′ sequences with respect to the direction of transcription, and type 2 palindromes have internal B probes or 5′ sequences. For simplicity, the classes of noncanonical rRNA genes were pooled and considered one category when analyzing DNA replication. The relative proportions of canonical and noncanonical rRNA genes from HeLa cells are provided in Fig. 1D.
FIG. 1.
Genomic structure of human rRNA genes. (A) Classical structure of one rRNA gene unit. The horizontal arrow represents the transcribed region, with the 18S, 5.8S, and 28S rRNA coding regions shown as thicker boxes. EcoRI restriction sites, indicated by short vertical white lines, divide the 43-kb rRNA gene unit into four fragments, i.e., A, B, C, and D. Promoter and transcription termination sequences are denoted by “P” and “T,” respectively. An example of an enlarged hybridized probe signal corresponding to a single classical rRNA gene unit is provided. (Adapted from reference 17 with permission of the publisher.) (B) Physical structure of human rRNA genes based on probes hybridized to combed molecules. Canonical units for two molecules are shown in panel i, and noncanonical units for two molecules are shown in panel ii. White arrows indicate type 1 and type 2 noncanonical palindromic units. The darker arrows for both types of noncanonical units point out transcribed sequence palindromes separated by nontranscribed spacer DNA. Bar = 50 kb. (C) Histogram showing the nontranscribed spacer length distribution in HeLa canonical rRNA genes. (D) Pie chart showing the relative percentages of canonical and noncanonical rRNA genes in HeLa cells, with a breakdown of type 1 and 2 noncanonical units. Type X rRNA genes contain multiple contiguous 5′-3′ transcribed regions, but they are not shown here because their contribution to human rRNA genes is significantly reduced.
Origins fire from both the rRNA genes and the intergenic spacer.
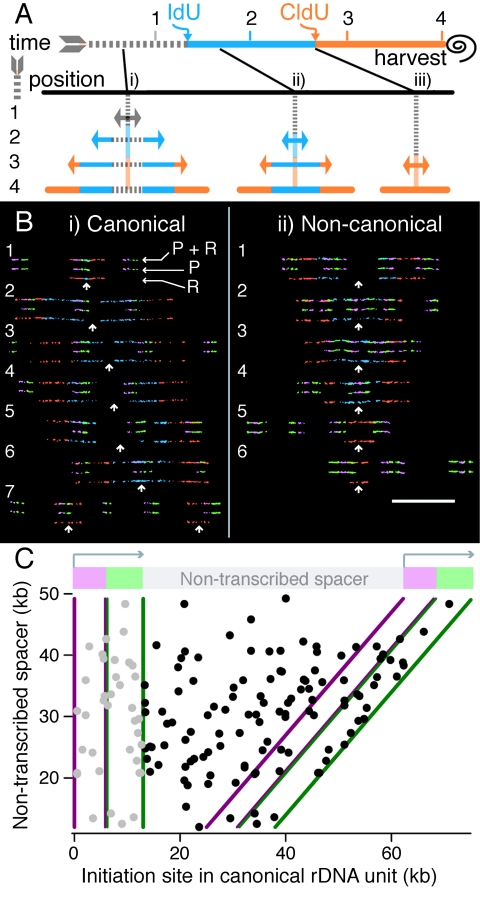
To map initiation sites on combed molecules, nascent DNA in asynchronous exponentially dividing HeLa cells was sequentially pulse labeled with iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) for 30 min each. DNA from this cell population was extracted and combed. Incorporated IdU and CldU were detected as blue and red, respectively. A scheme of expected DNA replication signals from bidirectional origins providing positional information based on this experimental paradigm is shown in Fig. 2A. When rRNA gene probes are hybridized in combination with the detection of BrdU analogues, replication analyses can be assigned to rRNA genes whenever probes superimpose on the replication tracts.
FIG. 2.
DNA replication initiation mapping at human rRNA genes. (A) Expected replication signals from bidirectional origins on combed DNA. Asynchronous cells were initially labeled with IdU (blue) followed by CldU (red) and then harvested. The black spiral denotes the end of the labeling period and the time of cell harvest. A schematic of a combed DNA molecule with the positions of three potential initiation events (i, ii, and iii) is provided. The black lines joining the time scale with the sites of initiation show when during labeling the origins fire. Initiation event i occurs prior to the incorporation of modified nucleotides. Initiation event ii occurs during the IdU pulse, and event iii occurs during the CldU pulse. The processes of replication and nucleotide incorporation are provided for the three previous time points, 1, 2, and 3, that give rise to the three types of signals used for initiation mapping. (B) Observed initiation events in the human rRNA gene locus. Fibers with probes specific for the rRNA gene transcription unit (3′ infrared, 5′ green) containing replication signals for initiation mapping are shown for canonical (i) and noncanonical (ii) rRNA genes. To facilitate understanding, four-color molecules containing both probe and replication (P + R) signals were vertically decomposed into the corresponding two-color probe (P) and replication (R) images. White arrows indicate the sites of initiation. Molecules 1 and 6 (i) and 2 to 4 (ii) contain origins that map to the rRNA coding genes. Fiber 7 in panel i contains two origins that occured during the CldU pulse, thefirst one in the transcription unit and the second in the nontranscribed spacer. The initiation sites in molecules 2 to 5 (i) and molecules 1, 5, and 6 (ii) are positioned in the intergenic spacers. Bar = 50 kb. (C) Scatter plot of origin positions in canonical rRNA genes. For canonical units, the nontranscribed spacers separating the rRNA genes vary in size. Initiation sites are shown (black circles) as a function of the nontranscribed spacer length in the rRNA gene units where the origins were found. Initiation events in the transcription unit were replotted for the preceding transcription unit (light gray circles) to help visualize whether an initiation zone at the 3′ end of the gene exists.
One hundred sixteen initiation events were mapped in canonical rRNA genes, with 37 (31.9%) localized to the transcription units and 79 (68.1%) localized to the nontranscribed spacers (for examples, see Fig. 2Bi). To determine whether an initiation preference for rRNA coding or noncoding DNA exists, the relative contributions of each to the total DNA length were quantified. An examination of 17,588-kb canonical rRNA genes revealed a composition of 30.2% (5,306 kb) transcription unit and 69.8% (12,282 kb) nontranscribed spacer DNA. Since the proportions of origins firing from the transcription units and from the nontranscribed spacers are similar to the DNA content percentages (P > 0.05 by the chi-square test), a preference for initiation was not found. To investigate whether initiation zones appear at a particular rRNA gene unit size, origin positions were plotted as a function of the nontranscribed spacer length (Fig. 2C). No initiation zones were observed, regardless of how far apart the transcription units were separated.
Thirty-one initiation events localized to noncanonical rRNA genes. Fifteen (48.4%) initiation sites mapped to the rRNA genes, and 16 (51.6%) mapped to the intergenic spacers (examples are provided in Fig. 2Bii). An analysis of the noncanonical rRNA gene content was performed to determine which sequence the active origins favored. rRNA coding and noncoding sequences made 49.1% (3,254 kb) and 50.9% (3,337 kb) contributions, respectively, to the 6,631 kb of noncanonical rRNA genes analyzed. These percentages do not differ greatly from the ratio of origins mapped to the rRNA genes and the intergenic spacers (P > 0.05 by the chi-square test). Therefore, in a similar fashion to canonical rRNA genes, an initiation preference for coding versus noncoding sequences of noncanonical rRNA genes was not observed.
Neighboring origins fire asynchronously at regular intervals.
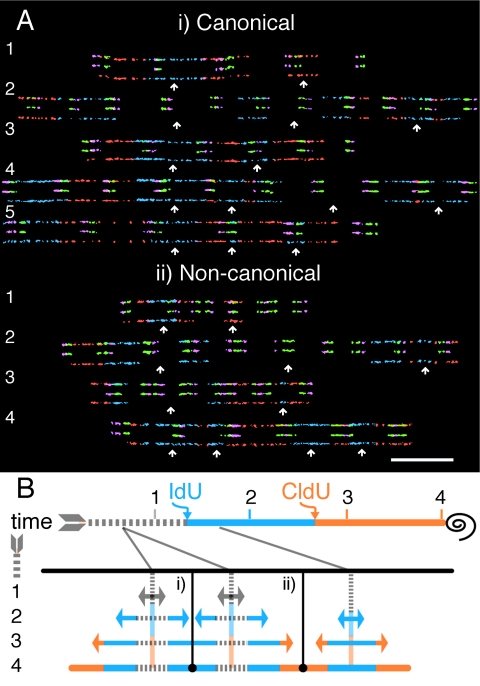
The appearance of multiple initiations on individual fibers allowed for a spatiotemporal analysis of how nearby rRNA gene origins are activated. An immediate neighbor was found for 52.6% of origins in canonical rRNA genes (61/116) and for 45.2% of origins in noncanonical rRNA genes (14/31). The fibers shown in Fig. 3A are typical examples of molecules with two or more initiation events. Since merging between oncoming forks from two origins can yield complicated replication signals, a scheme has been provided to aid in their interpretation (Fig. 3B).
FIG. 3.
Origin spacing and timing in canonical and noncanonical rRNA genes. (A) Single molecules containing multiple initiation events and fork merges. White arrows denote origin positions. Molecules were aligned according to the position of the first origin. Only two origin neighbors can be observed in molecules 1 and 3 for both panels i and ii. Three initiation events are found in the remaining fibers, except for molecule 4 in panel ii, which contains four origins. A merge between oncoming forks cannot be observed for the pair of origins in fiber 1 for both panels i and ii. A merge during the IdU pulse can be observed for the second fiber in panels i and ii. The remainder of the molecules contain signals with fork merges occurring during CldU labeling. Bar = 50 kb. (B) Expected signals of merged forks from multiple initiation sites on a single fiber. As shown in Fig. 1, the sequential labeling of IdU (blue) followed by CldU (red) and subsequent cell harvesting (black spiral) are shown. Vertical black lines represent the positions of merging forks. In panel i, a merge occurs during the IdU pulse, and in panel ii, a merge occurs during the CldU pulse, with the outcomes given at time point 4. The prior nucleotide incorporation giving rise to these two types of merge signals from three origins is provided for time points 1, 2, and 3. The timing of origin activation is shown in relation to the addition of the IdU/CldU pulse labels by gray solid lines.
Considering the type of replication tracts indicating an origin, the time of activation could be assigned prior to or during the IdU or CldU labeling period (see Fig. 2A for an explanation). As observed for the examples provided, many neighboring origins fired at different times. This result was consistent for the majority of molecules with multiple origins, regardless of which rRNA gene category was scrutinized. Therefore, although adjacent origins fired within approximately 60 min of each other, finely tuned synchrony was not observed.
With regard to origin spacing, origins fired every one to five rRNA gene units, with a mode of 2. No two functional origins occurred closer than 23.1 kb apart in canonical rRNA genes and 33.9 kb apart in noncanonical rRNA genes. Upper limits of 205.8 kb and 129.6 kb for interorigin distances were observed for canonical and noncanonical rRNA genes, respectively. No statistical difference was found between the mean interorigin distance for canonical rRNA genes (82.4 ± 42.9 kb) and that for noncanonical rRNA genes (70.6 ± 32.5 kb).
Among all observed rRNA gene initiation events, 78.9% occurred in canonical rRNA genes (116/147) and 21.1% occurred in noncanonical rRNA genes (31/147). These proportions do not differ significantly from the percentages of contribution of the different structural categories to the total length of the HeLa rRNA genes (72.5% canonical rRNA genes and 27.5% noncanonical rRNA genes; P > 0.05 by the chi-square test) (Fig. 1D). This result lends support to the interorigin data showing that the spatial distribution of active origins is not affected by the rRNA gene physical structure.
Noncanonical rRNA genes are associated with stalled forks.
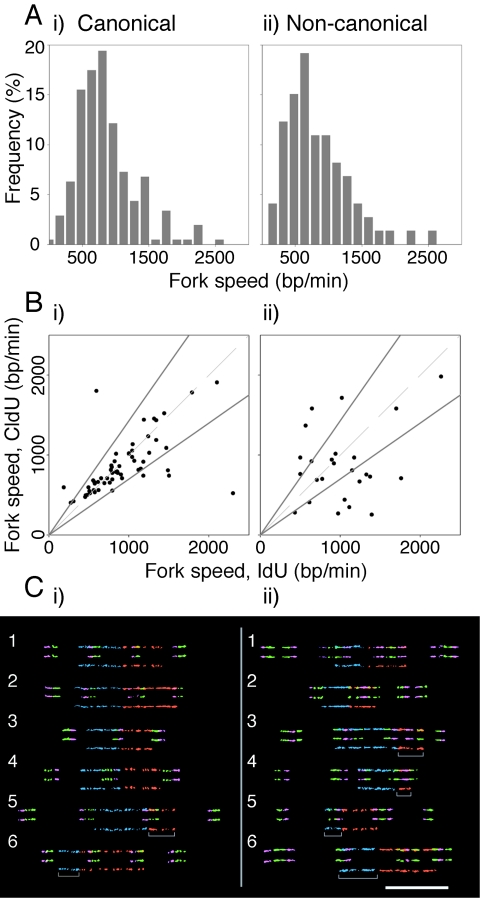
An analysis of DNA replication on combed DNA is also amenable to a study of fork kinetics. Fork speeds were calculated by dividing the length of each replication signal, blue or red, by the time of labeling for one of the nucleotides (30 min). Fork pausing would result in a decrease in the amount of DNA synthesized during the IdU/CldU labeling periods and would have a negative effect on the fork speed. To test whether forks encountered obstacles when replicating rRNA gene palindromes, the fork speeds of canonical and noncanonical rRNA genes were compared. On average, forks moved at indistinguishable speeds through canonical rRNA genes (927 ± 440.3 bp/min; n = 207) and noncanonical rRNA genes (909.4 ± 480.4 bp/min; n = 73) (see Fig. 4A for frequency distributions).
FIG. 4.
Effect of noncanonical rRNA genes on fork progression. (A) Histograms showing fork speed distributions for canonical (i) and noncanonical (ii) rRNA genes. (B) Fork speeds based on IdU and CldU tracts from single forks plotted against each other for canonical (i) and noncanonical (ii) rRNA genes. The dashed gray lines represent equal speeds calculated from IdU and CldU labels of the same length. The two solid lines represent thresholds that allow for a 30% difference in fork speed between the IdU and CldU pulses. (C) Forks with speed information available from both IdU and CldU labels. Molecules 1 to 4 for canonical rRNA genes (i) and molecules 1 and 2 for noncanonical rRNA genes (ii) contain IdU and CldU tracts of approximately the same length. Molecules 5 and 6 (i) and 3 to 6 (ii) are examples of data excluded by the thresholds established in panel B. They have IdU and CldU replication tracts from single forks that differ by >30%. The gray open rectangles indicate the regions of fork stalling.
If forks stalled infrequently, the effect on the bulk fork speed could have been insufficient to shift the population average. To increase the sensitivity of the test, speed changes at the individual fork level were analyzed. Fork speeds based on the IdU and CldU components of single replication tracts from one fork were plotted against each other (Fig. 4B). A significant positive correlation between the lengths of the blue and red signals for individual forks was found for canonical rRNA genes (R = 0.637; P < 0.001; n = 58). This statistic implies that despite the fork speed heterogeneity in canonical rRNA genes (see the fork speed frequency distribution in Fig. 4A, panel i), once a fork acquires a certain speed, the rate of movement is constant. Conversely, the correlation between the lengths of the blue and red signals for individual forks was absent for noncanonical rRNA genes (R = 0.294; P > 0.05; n = 25). The loss of this correlation for noncanonical rRNA genes suggests that while fork speeds are also heterogeneous in this case (see the fork speed frequency distribution in Fig. 4Aii), the inherent rate of movement for any given fork is not maintained. The forks in noncanonical rRNA genes are subject to periods of slowing down or pausing.
The DNA substrate over which forks decreased their speed was analyzed with the aim of delineating their relationship. Thresholds allowing a 30% difference in the blue and red replication signals were established to help identify molecules with considerable fork speed changes. Only 10.3% (6/58) of the data points were excluded by the thresholds for canonical rRNA genes, in contrast to 64% (16/25) for noncanonical rRNA genes (P < 0.001 by two-sided Fisher's exact test) (Fig. 4B). No common rRNA gene element correlated with a decreased fork speed in canonical rRNA genes (for example, see molecules 5 and 6 in Fig. 4Ci). In noncanonical rRNA genes, palindromes were not always associated with a reduced fork speed: forks could slow several kb ahead of an rRNA gene palindrome (Fig. 4Cii, molecules 5 and 6) or could even speed up while synthesizing a palindrome (molecules 3 and 6). However, for the majority of the molecules excluded by the thresholds (11/16), palindromes colocalized with the shorter replication tracts (Fig. 4Cii, molecules 3 and 4). Altogether, these observations implicate noncanonical rRNA genes in transiently perturbing fork progression, and this disruption colocalizes with the palindromic physical structures in at least half of the cases.
Unidirectional forks occur more frequently in noncanonical rRNA genes.
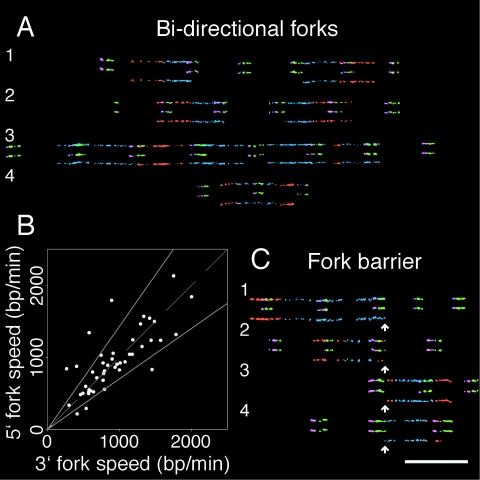
Mirror replication signals on either side of a midpoint consisting of bisymmetric IdU and CldU labels provide direct evidence of bidirectional origins in human rRNA genes (reviewed in reference 13) (Fig. 2A). Sometimes, however, the replication tract from one of the two forks was completely absent (see Fig. 5A for examples). The presence of incoming replication signals from a second nearby origin excludes the possibility that nonvisualization of the outgoing fork was due to DNA breakage. These signals, henceforth termed unidirectional forks, may be due to unidirectional initiation. Three other plausible explanations for this type of signal invoke bidirectional initiation with a subsequent blocked fork (illustrated in Fig. 5B). These models cannot be delineated by the methods used in this study. Whether forks collapse or are maintained after fork blockage also cannot be determined with the replication signals of combed DNA.
FIG. 5.
Unidirectional forks as a result of permanent fork arrest. (A) Examples of forks that are not coupled to a second fork moving away from single origins in canonical (i) and noncanonical (ii) rRNA genes. In the left part of each molecule, incoming replication signals are observed from a nearby origin in the region where an outgoing bidirectional fork counterpart is expected but absent. In panel i, molecules 3 and 4 possess complete replication signals from a neighboring origin in place of a simple incoming replication signal. Gray open rectangles denote all possible sites of fork blocking, as explained for panel B. (B) Scheme to explain three possibilities of events that give rise to a unidirectional fork. The IdU and CldU labeling periods are denoted by horizontal blue and red lines, respectively, followed by a black spiral signifying cell harvest. The horizontal black lines are divided to indicate that the three unidirectional forks occur on different molecules. Gray lines give the time of origin firing relative to the labeling periods for three origins that yield unidirectional forks. In panel i, an origin fires prior to labeling. One of the forks is blocked at some distance from the initiation site. A similar situation occurs in panel ii; however, initiation occurs during the IdU pulse. In panel iii, one fork is blocked at the origin. The replication signal outcomes are shown at time point 4 for all three cases. The processes of fork progression preceding the observed signals are given for time points 1, 2, and 3. Vertical black lines represent the positions of fork arrest.
The proportions of unidirectional forks in canonical versus noncanonical rRNA genes are a relative measure of the failure to establish a second bidirectional fork and/or fork arrest. Eleven unidirectional forks were observed in canonical rRNA genes, and nine unidirectional forks were observed in noncanonical rRNA genes. Bidirectional replication signals were observed 89 and 24 times in canonical and noncanonical rRNA genes, respectively. Therefore, 11% of forks were defective in canonical rRNA genes (11/100). This fraction significantly increased, to 27.3% (9/33), in the context of rRNA gene palindromes (P < 0.05 by two-sided Fisher's exact test), thereby implicating noncanonical rRNA gene with either faulty fork setup or fork obstruction.
The fork barrier at the transcription termination site is 10% efficient in canonical rRNA genes.
In the previous section, fork arrest was inferred by the presence of unidirectional forks. Another way to account for a fork barrier is to directly visualize premature termination of an elongating fork. According to the known position of the fork barrier in rRNA genes, shorter replication tracts in conjunction with the 3′ end of the transcription unit were expected. A comparison of the lengths of the IdU and CldU signals to analyze fork stalling, however, did not indicate a fork barrier at this site (Fig. 4). Therefore, in order to quantify the efficiency of this fork barrier, other analyses using different replication tract substrates were required.
When speed information was available for both forks moving away from a site of initiation, it was observed that the two forks moved at similar rates (Fig. 6A). Plotting both speeds of the two bidirectional forks against each other confirmed this observation by revealing a significant positive correlation (R = 0.783; P < 0.001; n = 45) (Fig. 6B). This correlation was independent of the fork speed. For example, in Fig. 6A, molecule 3 shows two highly correlated fast forks and molecule 4 shows two highly correlated slow forks. Also, the distance from the origin where labels were observed did not affect the correlation. Molecules 1 to 4 in Fig. 6A illustrate this point.
FIG. 6.
Quantifying the fork barrier efficiency in canonical rRNA genes. (A) Fibers with similar bidirectional fork speeds for single origins. For molecules 1 to 3, the speeds from the IdU label are available since initiation occurred prior to IdU labeling. Speeds from the bidirectional forks could be calculated based on the CldU labels for molecules 1, 2, and 4 since merging with forks from other origins did not occur during this labeling period. (B) Bidirectional fork speeds from single origins plotted against each other. The fork direction was determined by the 5′-3′ orientation of the transcription unit. Forks with equal speeds are denoted by the dashed line. Thresholds corresponding to a 30% difference in speeds for diverging forks are shown with solid lines. (C) Position of the fork barrier based on the site where one of the two bidirectional forks from a single origin stopped. All molecules resulted in bidirectional fork speed data excluded by the thresholds in panel B. White arrows indicate the positions of fork barriers.
For the goal of mapping fork barriers, it is the absence of this relationship that is important. Seven data points were excluded by thresholds that allowed for a 30% difference in the speeds of the two bidirectional forks (Fig. 6B). In six of the seven cases, termination of the shorter replication tract occurred at the 3′ end of the transcription unit (Fig. 6C). Premature termination of a fork may be due to a barrier, but it also can reflect a merge with a previously blocked fork. Because of this ambiguity, even though the direction of the observed blocked fork was discernible, the polarity of the fork barrier could not be ascertained. Information about the frequency of the fork barrier, however, is still accessible. Based on the replication tracts from bidirectional origins, the 3′ end of the transcription unit was crossed 59 times without stopping a fork. Therefore, the fork barrier at the transcription termination sequences is 9.2% (6/65) efficient at blocking oncoming forks.
DISCUSSION
This work investigates the human rRNA gene replication program according to the organization of the locus, namely, canonical versus noncanonical units. It represents one of the first studies of human DNA replication that provides a quantitative view of the in vivo process of replication in specific sequence tracts using a DNA-fiber technique. The method chosen for this study involves differentially labeling nascent DNA with two halogenated nucleotides, IdU followed by CldU, extending the fibers by molecular combing, and then attributing replication tracts to rRNA genes through their colocalization with probes hybridized to the rRNA coding sequences. The spatiotemporal initiation patterns, together with fork progression, were based on the orientations, lengths, and relative positions of the replication signals. Analyses of origins and forks at the single-molecule level allowed for the detection of rare events and complexities, phenomena that are often obscured in bulk studies. Another advantage of using this approach was the conservation of the in vivo order of rRNA genes during preparation of the combed DNA substrate. In this way, the effect of noncanonical rRNA genes on initiation and fork movement could be delineated. Until now, the majority of rRNA gene replication studies have been carried out in yeast, an organism in which rRNA gene palindromes do not exist. The data presented here, however, suggest that significant differences do exist in humans, depending on the order and orientation of rRNA genes which are not present as a tandem array of nearly identical units, thereby providing new insights into how human cells replicate rRNA genes.
Organization of initiation in the human rRNA gene locus.
Altogether, the data on origin firing reveal that the initiation profiles do not differ greatly between canonical and noncanonical rRNA genes (Fig. 2 and 3). Our data suggest that initiation can take place in the nontranscribed spacers and the transcription units in both structural categories of rRNA genes. Previous in vivo studies indicated that the genetic determinants of human rRNA gene origin activity are differentially defined depending on the technique used (reviewed in reference 12). Two-dimensional gels revealed scattered initiation sites restricted to the nontranscribed spacer (24). Methods using nascent strands uncovered regions of preferential initiation, specifically a primary zone upstream of the gene (15, 41, 47). A lesser-used zone downstream of the gene was also identified (15). Another study using nascent strand analysis found a significant proportion of multiple initiation sites dispersed throughout the rRNA gene repeats, including the transcription units (47). Initiation within the rRNA coding sequences was further corroborated by a later study showing extension of the primary zone to the midpoint of the gene (41). In summary, these studies suggest that there are preferential initiation zones upstream and downstream of the rRNA genes, with occasional origins located within the genes. Our data provide additional information showing that whatever origins are present in human rRNA genes, they are not likely to be influenced by rRNA gene palindromes.
Our data suggest no initiation site preference between coding and noncoding sequences in canonical and noncanonical rRNA genes (Fig. 2). Using the combed DNA approach, the positioning of replication origins at the midpoint of the nucleotide analogue signals is based on the assumption that replication forks progress uniformly and bidirectionally. However, in subsequent sections of Results, data were presented to the contrary: (i) approximately 10% of forks in canonical rRNA genes and 60% of forks in noncanonical rRNA genes changed speed during the course of pulse labeling (Fig. 4) and (ii) nearly 10% and 30% of origins in canonical and noncanonical rRNA genes, respectively, yielded unidirectional replication signals (Fig. 5). As a consequence, origin preferences between and within the nontranscribed spacer and the transcription unit, as previously reported (15, 24, 41, 47), could have been obscured. Furthermore, the variability of the nontranscribed spacer (Fig. 1) introduces additional uncertainty about the nucleotide sequence from which origins fired. Techniques that provide base pair resolution, such as two-dimensional gels and nascent strand analysis, will help determine the precise origin locations in canonical versus noncanonical rRNA genes.
Human rRNA gene replication origins are considered to be mostly restricted to transcriptionally silent DNA (reviewed in reference 12). Can this constraint be satisfied by the initiation data presented here? In human rRNA genes, 50% of the units are transcribed since about one-half of the cluster is nucleosome-free (8). Approximately 70% and 50% of origins map to the nontranscribed spacer in canonical and noncanonical rRNA genes, respectively (Fig. 2C). Therefore, a sufficient number of origins can be appropriately positioned to avoid initiation from transcriptionally active rRNA genes.
To date, the spatiotemporal distribution of multiple rRNA gene origins has been quantified for only one other organism, S. cerevisiae (32). In yeast, clusters of synchronously firing origins spanning 20 to 30 kb are separated by large 60-kb gaps. Although most adjacent human rRNA gene origins fire within 60 min of each other, the timing of initiation is staggered (Fig. 3A). Furthermore, the distance between two origins could vary from tens of kilobases to a couple hundred kilobases, with a peak of 80 kb on average. Therefore, there is little evidence that the yeast rRNA gene initiation program is conserved in humans. Rather, the regulation of human rRNA gene initiation timing and spacing is more relaxed, albeit with upper and lower limits of when and how far apart origins fire. Yet despite this relaxed definition of the spatiotemporal distribution of origins, our data suggest that initiation mechanisms are robust: human rRNA gene microrearrangement in the form of palindromes influenced neither the spacing nor the timing of neighboring origins (Fig. 3).
Interorigin distances were measured between origins inferred from combed replication signals (Fig. 2A). Based on these signals, a single initiation event had been assumed to occur at the midpoint of any two bidirectional replication tracts. However, it is possible that two or more origins lie at the center of diverging replication forks, the probability of which increases with the size of the gap between the pair of observed signals (for example, the first origin in both molecule 2 in Fig. 3Ai and molecule 3 in Fig. 3Aii). Consequently, the replicon size, the fragment of DNA replicated from a single origin, and hence the interorigin distances may have been overestimated. The probabilities of this overestimation are equal for canonical and noncanonical rRNA genes since origins were defined using the same criteria for each. Therefore, the lack of a difference in average interorigin distances between these two categories of human rRNA genes is unlikely to change should origins be found closer together by another method.
An inefficient fork barrier in canonical rRNA genes accommodates origin spacing.
In all organisms studied to date, the fork barrier at the 3′ end of the transcription unit is polar, arresting forks moving in the direction opposite to transcription (reviewed in reference 37). The only two exceptions are fission yeast and human rRNA genes, where the site is conserved but forks in both directions are blocked (24, 39). One implication of a bidirectional fork barrier is that if fork blocking is close to 100% efficient, forks essentially cannot leave the rRNA gene unit from which replication initiated. In such a scenario, each rRNA gene unit would be required to contain a functional origin to fully replicate the locus. However, adjacent origins were observed to span several rRNA gene repeats (Fig. 3A). The passive replication of the intervening units crucially depends upon a somewhat less efficient fork barrier.
In support of this hypothesis, not only was the fork barrier found to be inefficient, in agreement with a previous study (24), but it was also quantified as blocking approximately 10% of incoming forks. The low efficiency of the fork barrier means that the majority of forks can leave the rRNA gene unit from which they originated and passively replicate the next unit. In this way, canonical rRNA genes can be entirely duplicated from widely spaced origins, thereby ensuring that the initiation program is tolerated by the cell.
Roles of fork obstruction in human rRNA genes may include acting as a backup to prevent collision with transcription polymerases and/or providing substrates for recombination.
The function of the fork barrier at the 3′ end of the transcription unit is a matter of debate (36). In yeast, due to the position and polarity of the fork barrier and the simultaneous activities of transcription and replication on the same molecule (38), its suggested role was to prevent deleterious head-on collisions between transcription and replication polymerases (5). The spatiotemporal separation of rRNA gene transcription and replication domains in HeLa nuclei (33), however, suggests that human cells do not need to regulate polymerase traffic the same way that yeast does. The low-frequency fork barrier observed here might represent a safety mechanism to prevent polymerase collisions when the segregation of transcription and replication domains falters.
The finding that recombinogenic double-strand ends can form at the site of a fork barrier (28, 29) has inspired another functional role: fork arrest is involved in the homogenization, contraction, expansion, and/or extrachromosomal circle formation of the rRNA gene array (36). In human rRNA genes, the average fork speed calculated here of approximately 1 kb/min for both canonical and noncanonical rRNA genes (Fig. 4) is lower than the 1.7 kb/min reported for HeLa cells (21). This result suggests that the formation of recombinogenic substrates is equally probable from canonical and noncanonical rRNA genes. However, a detailed analysis at the single-molecule level revealed that fork pausing and/or blocking is predominant in noncanonical rRNA genes (Fig. 4 and 5). Insofar as recombinogenic substrates arise from these fork impediments (46), the genetic material in noncanonical rRNA genes initiates recombination in the majority of cases. The percentage of noncanonical rRNA genes in HeLa cells remained stable for two different cell cultures over 15 population doublings (results not shown). Therefore, the fork obstruction observed here relates to the homogenization of rRNA gene sequences either through equal gains and losses or through normal restoration of the generated double-strand ends (36).
Implications of blocked forks in rRNA gene palindromes regarding Robertsonian translocations.
Reciprocal crossovers involving inverted repeats would yield one dicentric chromosome and one acentric fragment, creating a significant threat to genomic integrity (42). Our findings that fork blockage and potential double-strand ends localize to noncanonical rRNA genes raise the possibility that inverted human rRNA gene repeats may participate in this recombination pathway. In support of this hypothesis, rRNA gene sequences mapped to the breakpoint in Robertsonian translocations involving nonhomologous acrocentric chromosomes (for example, see reference 7). However, the extreme rarity of this class of Robertsonian translocation (reviewed in reference 42), together with a profound bias towards noncrossover gene conversion during double-strand-break repair in mammalian cells (22, 35), suggests that reciprocal crossovers involving inverted rRNA gene repeats are suppressed during homologous recombination.
Mechanisms of fork obstruction in noncanonical rRNA genes.
Palindromes are proposed to adopt hairpin structures during lagging-strand synthesis, which can challenge polymerase movement and, in turn, arrest the entire replisome (14, 18, 45). These studies were carried out in bacteria or yeast with palindromes of only a few hundred base pairs or less. While direct evidence is lacking for kb-sized palindromes forming hairpins, these secondary structures were invoked to explain the rearranged products from a large 15.3-kb palindrome inserted into mice (1). It is therefore possible that the perturbed forks in human rRNA gene palindromes, whose sizes are on the same order of magnitude as the 15.3-kb mouse palindrome, may also be related to hairpin formation. Work carried out with Escherichia coli suggested that the resolution of the palindrome-associated hairpin occurs separately behind the replication fork (10). In this case, other mechanisms to explain fork obstruction in noncanonical rRNA genes are needed.
Ahead of the replication fork, DNA undergoes negative supercoiling by the action of topoisomerases to allow for the opening of the DNA double helix by the approaching replisome (reviewed in references 34 and 40). This condition favors a cruciform formation of palindrome sequences (30). Another possibility, therefore, is that an rRNA gene cruciform generated by the topoisomerase-related negative supercoiling stalls/stops the fork.
Chromatin and its remodeling complexes can influence the accessibility of factors important for replication efficiency (reviewed in references 11 and 16). Studies of the chromatin state associated with canonical and noncanonical rRNA genes and the relationship between forks and chromatin remodeling will help determine the role that chromatin plays in human rRNA gene fork progression.
Acknowledgments
We thank Bénédicte Michel, Dmitry Gordenin, and the reviewers for their helpful comments on the manuscript.
Ronald Lebofsky is supported the Natural Sciences and Engineering Council of Canada.
REFERENCES
- 1.Akgun, E., J. Zahn, S. Baumes, G. Brown, F. Liang, P. J. Romanienko, S. Lewis, and M. Jasin. 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17:5559-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385-394. [DOI] [PubMed] [Google Scholar]
- 3.Bensimon, A., A. Simon, A. Chiffaudel, V. Croquette, F. Heslot, and D. Bensimon. 1994. Alignment and sensitive detection of DNA by a moving interface. Science 265:2096-2098. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. [DOI] [PubMed] [Google Scholar]
- 6.Caburet, S., C. Conti, C. Schurra, R. Lebofsky, S. J. Edelstein, and A. Bensimon. A new paradigm for ribosomal DNA arrays incorporating a broad range of palindromic pseudogenes. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 7.Cheung, S. W., L. Sun, and T. Featherstone. 1990. Molecular cytogenetic evidence to characterize breakpoint regions in Robertsonian translocations. Cytogenet. Cell Genet. 54:97-102. [DOI] [PubMed] [Google Scholar]
- 8.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 9.Coverley, D., and R. A. Laskey. 1994. Regulation of eukaryotic DNA replication. Annu. Rev. Biochem. 63:745-776. [DOI] [PubMed] [Google Scholar]
- 10.Cromie, G. A., C. B. Millar, K. H. Schmidt, and D. R. Leach. 2000. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 154:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeret, C., Y. Vassetzky, and M. Mechali. 2001. Chromatin remodelling and DNA replication: from nucleosomes to loop domains. Oncogene 20:3086-3093. [DOI] [PubMed] [Google Scholar]
- 12.DePamphilis, M. L. 1999. Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21:5-16. [DOI] [PubMed] [Google Scholar]
- 13.Edenberg, H. J., and J. A. Huberman. 1975. Eukaryotic chromosome replication. Annu. Rev. Genet. 9:245-284. [DOI] [PubMed] [Google Scholar]
- 14.Flores, M. J., H. Bierne, S. D. Ehrlich, and B. Michel. 2001. Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J. 20:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gencheva, M., B. Anachkova, and G. Russev. 1996. Mapping the sites of initiation of DNA replication in rat and human rRNA genes. J. Biol. Chem. 271:2608-2614. [DOI] [PubMed] [Google Scholar]
- 16.Gerbi, S. A., and A. K. Bielinsky. 2002. DNA replication and chromatin. Curr. Opin. Genet. Dev. 12:243-248. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez, I. L., and J. E. Sylvester. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320-328. [DOI] [PubMed] [Google Scholar]
- 18.Gordenin, D. A., K. S. Lobachev, N. P. Degtyareva, A. L. Malkova, E. Perkins, and M. A. Resnick. 1993. Inverted DNA repeats—a source of eukaryotic genomic instability. Mol. Cell. Biol. 13:5315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrick, J., P. Stanislawski, O. Hyrien, and A. Bensimon. 2000. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 300:1133-1142. [DOI] [PubMed] [Google Scholar]
- 20.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, D. A., and A. Pombo. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linskens, M. H. K., and J. A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little, R. D., T. H. Platt, and C. L. Schildkraut. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13:6600-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobachev, K. S., B. M. Shor, H. T. Tran, W. Taylor, J. D. Keen, M. A. Resnick, and D. A. Gordenin. 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148:1507-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 27.Michalet, X., R. Ekong, F. Fougerousse, S. Rousseaux, C. Schurra, N. Hornigold, M. van Slegtenhorst, J. Wolfe, S. Povey, J. S. Beckmann, and A. Bensimon. 1997. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277:1518-1523. [DOI] [PubMed] [Google Scholar]
- 28.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel, B., M. J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuuchi, K., M. Mizuuchi, and M. Gellert. 1982. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 156:229-243. [DOI] [PubMed] [Google Scholar]
- 31.Norio, P., and C. L. Schildkraut. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science 294:2361-2364. [DOI] [PubMed] [Google Scholar]
- 32.Pasero, P., A. Bensimon, and E. Schwob. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pliss, A., K. Koberna, J. Vecerova, J. Malinsky, M. Masata, M. Fialova, I. Raska, and R. Berezney. 2005. Spatio-temporal dynamics at rDNA foci: global switching between DNA replication and transcription. J. Cell. Biochem. 94:554-565. [DOI] [PubMed] [Google Scholar]
- 34.Postow, L., N. J. Crisona, B. J. Peter, C. D. Hardy, and N. R. Cozzarelli. 2001. Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. USA 98:8219-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothstein, R., and S. Gangloff. 1999. The shuffling of a mortal coil. Nat. Genet. 22:4-6. [DOI] [PubMed] [Google Scholar]
- 37.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 38.Saffer, L. D., and O. L. Miller. 1986. Electron-microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol. Cell. Biol. 6:1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, J. A., S. M. Kim, and J. A. Huberman. 1998. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 238:220-230. [DOI] [PubMed] [Google Scholar]
- 40.Schvartzman, J. B., and A. Stasiak. 2004. A topological view of the replicon. EMBO Rep. 5:256-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott, R. S., K. Y. Truong, and J. M. Vos. 1997. Replication initiation and elongation fork rates within a differentially expressed human multicopy locus in early S phase. Nucleic Acids Res. 25:4505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaffer, L. G., and J. R. Lupski. 2000. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34:297-329. [DOI] [PubMed] [Google Scholar]
- 43.Torres, J. Z., J. B. Bessler, and V. A. Zakian. 2004. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S-cerevisiae DNA helicase Rrm3p. Genes Dev. 18:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran, H., N. Degtyareva, D. Gordenin, and M. A. Resnick. 1997. Altered replication and inverted repeats induce mismatch repair-independent recombination between highly diverged DNAs in yeast. Mol. Cell. Biol. 17:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaver, D. T., and M. L. DePamphilis. 1984. The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J. Mol. Biol. 180:961-986. [DOI] [PubMed] [Google Scholar]
- 46.Weitao, T., M. Budd, L. L. Hoopes, and J. L. Campbell. 2003. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 278:22513-22522. [DOI] [PubMed] [Google Scholar]
- 47.Yoon, Y., J. A. Sanchez, C. Brun, and J. A. Huberman. 1995. Mapping of replication initiation sites in human ribosomal DNA by nascent-strand abundance analysis. Mol. Cell. Biol. 15:2482-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Z., D. M. Macalpine, and G. M. Kapler. 1997. Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol. Cell. Biol. 17:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]