Abstract
Activation of ubiquitination occurs during spermatogenesis and is dependent on the induction of isoforms of the UBC4 family of ubiquitin-conjugating enzymes. The UBC4-testis isoform is testis specific, is induced in round spermatids, and demonstrates biochemical functions distinct from a ubiquitously expressed isoform UBC4-1. To explore further the function of UBC4-testis, mice bearing inactivation of this gene were produced. Homozygous (−/−) mice showed normal body growth and fertility. Although testis weight and morphology were normal in testes from adult mice, examination of young mice during the first wave of spermatogenesis revealed that testes were ∼10% smaller in weight at 40 and 45 days of age but had become normal at 65 days of age. Overall protein content, levels of ubiquitinated proteins, and ubiquitin-conjugating activity did not differ between wild-type and homozygous (−/−) mice. Spermatid number, as well as the motility of spermatozoa isolated from the epididymis, was also normal in homozygous (−/−) mice. To determine whether the germ cells lacking UBC4-testis might be more sensitive to stress, testes from wild-type and knockout mice were exposed to heat stress by implantation in the abdominal cavity. Testes from both strains of mice showed similar rates of degeneration in response to heat. The lack of an obvious phenotype did not appear to be due to induction of other UBC4 isoforms, as shown by two-dimensional gel immunoblotting. Our data indicate that UBC4-testis plays a role in early maturation of the testis and suggest that the many UBC4 isoforms have mixed redundant and specific functions.
Spermatogenesis is a complex developmental process in which undifferentiated stem cells become committed to the spermatid lineage and then pass through the spermatocyte stages, during which DNA replication and genetic recombination occur. The subsequent two divisions of meiosis generate haploid round spermatids, which then finally differentiate into mature elongated spermatids (reviewed in reference 5). This last process involves condensation and removal of much of the cytoplasm, including structures such as the endoplasmic reticulum. New structures such as the acrosome, manchette, and flagellum are assembled to confer some of the distinct and essential functions of spermatozoa. In addition, there is a major reorganization of the chromatin into a tighter structure by replacement of histones initially by transition proteins and ultimately by protamines (reviewed in references 11 and 13). The molecular mechanisms regulating these dramatic transformations remain unclear. Elements of the apoptotic pathway may be involved (1). Removal of the residual body from the spermatid occurs as a result of phagocytosis by Sertoli cells (16, 23). However, the mechanisms by which the reduction of cytoplasmic volume occurs to generate the residual body remain unclear.
Recent evidence indicates that the ubiquitin system plays a role in this developmental process. The posttranslational conjugation of ubiquitin to proteins is now recognized to play an important role in many cellular processes. The attachment of polyubiquitin chains to proteins targets them for recognition and degradation by the proteasome (reviewed in reference 7). In addition, monoubiquitination of proteins mediates nonproteolytic functions of ubiquitin, such as intracellular trafficking to other organelles, regulation of transcription, or modification of chromatin (reviewed in references 6, 17, and 24). We have previously shown that ubiquitin conjugation is activated during spermatogenesis and that this activation of conjugation is dependent on the UBC4/UBC5 family of ubiquitin-conjugating enzymes (E2s) (20). In yeast, the UBC4/UBC5 enzymes are essential for the degradation of short-lived and abnormal proteins and are responsible for the bulk of steady-state ubiquitination in the yeast cell (25). Thus, these enzymes are good candidates for the involvement in the large-scale degradation of proteins occurring during sperm maturation. We therefore cloned a family of rat E2 isoforms that are homologous to the Saccharomyces cerevisiae E2s UBC4/UBC5 (27, 28). Although these rat isoforms are >90% identical in their amino acid sequences, one of the isoforms has several distinct properties. This UBC4-testis isoform (27) (previously referred to as the 8A isoform of E217k) is unique in being testis and spermatid specific. Indeed, its expression is only apparent at the mRNA level in round spermatids, although the protein product persists into elongating spermatids. Structurally, unlike the other isoforms, which have a basic pI, UBC4-testis has an acidic pI. Although UBC4-testis and the constitutive isoform UBC4-1 can both support in vitro ubiquitination by some ubiquitin protein ligases (E3s) (18), for other E3s the two isoforms are quite distinct (3; S. S. Wing, unpublished observations). Since E3s are the substrate recognition components of the pathway, such differential interactions with E3s should lead to different patterns of conjugation of ubiquitin to proteins. Indeed, consistent with this partially overlapping panel of interacting E3s, UBC4-testis can support conjugation to proteins found in some fractions of testis proteins but not all those supported by the constitutive UBC4-1 isoform (27). Since the specific expression and induction in the testis and the distinct biochemical properties of UBC4-testis suggested a unique role during spermatogenesis, we explored this possibility by inactivating the gene in the mouse by homologous recombination.
MATERIALS AND METHODS
Cloning of the UBC4-testis gene.
The UBC4-testis gene was isolated from a mouse sv129 genomic library in lambda phage kindly supplied by J. Rossant. The library was screened with a probe derived from the 3 prime noncoding region of the UBC4-testis cDNA because a coding region probe would also detect other UBC4 isoforms. DNA was isolated from positive clones and subcloned into pBluescript and analyzed by sequencing and restriction enzyme analysis. This determined that the UBC4-testis gene is intronless (Fig. 1A). Since there are intron-containing paralogs, this suggests that the UBC4-testis gene may be a retrogene. Examination of various mammalian genome sequences as they became available indicated that UBC4-testis is found in mouse and rat genomes but appears not to be in human, chimpanzee, dog, frog, and fish genomes. This does not necessarily indicate that UBC4-testis lacks functional importance since it is expressed in spermatids during a period of extensive remodeling. Since the ultimate shapes of spermatids differ in various mammalian species, UBC4-testis could play an important role in such species specific remodeling.
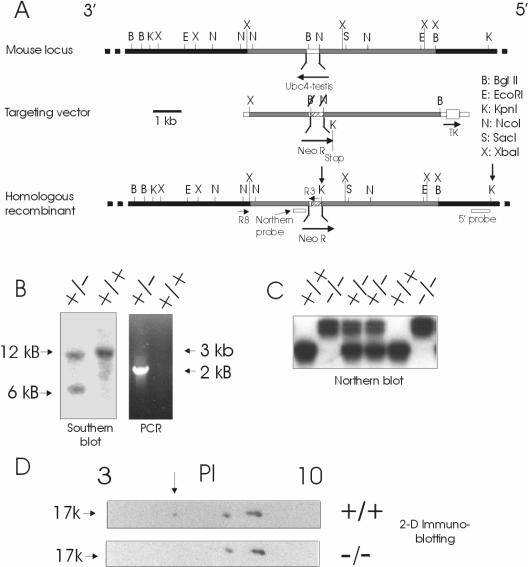
FIG. 1.
Inactivation of the mouse UBC4-testis gene. (A) Design of targeting vector for inactivation of the mouse UBC4-testis gene. The UBC4-testis gene is intronless. Shaded flanking regions were subcloned around a neomycin resistance gene in the pNT vector, which also contains a thymidylate kinase gene (TK) for positive selection. The linearized targeting vector was transfected into ES cells and homologous recombination selected for by using G418 and FIAU. Shown on the structure of the homologous recombinant are the positions of the 5′ probe for Southern analysis, the R3, R8 primers used for PCR screening and the 3′ noncoding probe used for Northern analysis. Arrows indicate directions of gene transcription. (B) Screening of resistant clones by Southern analysis and by PCR. Homologous recombination with the targeting vector results in a new 6-kb fragment when genomic DNA is digested by KpnI (arrows on homologous recombinant in panel A) and probed with a 5′ probe and in amplification of a 2-kb DNA fragment with primer R8 from the 3′ flank and primer R3 from the neomycin resistance gene. (C) Northern analysis of testis RNA from wild type, heterozygous, and homozygous (−/−) mice using a probe derived from the 3′ noncoding region of the UBC4-testis gene. Homozygous (−/−) mice lack the ∼1.0-kb UBC4-testis transcript but reveal instead a larger transcript that is due to transcription through the larger neomycin resistance cassette. (D) Two-dimensional gel electrophoresis of testis proteins from wild-type and knockout mice, followed by immunoblotting with anti-UBC4 antibodies reveals the presence of the acidic UBC4-testis isoform (arrow) in the wild-type mice but absent from the homozygous (−/−) mice.
Generation of mice lacking the UBC4-testis gene.
An ∼4-kb BglII/NcoI fragment of 5′ flanking genomic sequence and an ∼2.2-kb BglII/XbaI fragment of 3′ flanking genomic sequence were subcloned downstream and upstream, respectively, of the neomycin resistance gene in the pNT vector. In addition, the thymidylate kinase gene was placed on the opposite side of the 5′ flank to also permit negative selection (Fig. 1A). After linearization of the plasmid, the DNA was electroporated into RS1 embryonic stem (ES) cells (kindly supplied by J. Rossant). Growth of the cells in the presence of G418 and FIAU selected for the presence of the neomycin resistance gene and the absence of the thymidylate kinase gene (15), events that would occur as a result of homologous recombination across the two flanks. This homologous recombination results in replacement of the segment of the UBC4-testis gene encoding amino acid residues 2 to 126 with the neomycin resistance cassette. In the unlikely event that any of the UBC4-testis transcript could still be translated, this deletion in the UBC4-testis gene product would result in an inactive enzyme since it removes 124 of the 147 residues, including all conserved elements making up the active site. Approximately 16% of the resistant clones screened were found to have undergone the appropriate homologous recombination. Cells from two different clones were then injected into mouse blastocysts and reimplanted in pseudopregnant female BALB/c mice. Chimeric male mice were backcrossed with BALB/c females to identify mice in whom germ line transmission of the mutant allele had taken place. These heterozygotes were backcrossed with BALB/c mice for a minimum of seven generations before they were used in studies. For these studies, heterozygotes were mated to obtain homozygous knockout males and wild-type littermates.
Protein analyses and assays.
For Western blot analyses of testis proteins with anti-ubiquitin or anti-UBC4 antibodies, testes were sliced and homogenized in 10 ml of 0.25 M sucrose, 50 mM Tris (pH 7.5), 5 mM N-ethylmaleimide, and 1 mM EDTA/g of tissue by using a Polytron tissue disruptor. N-Ethylmaleimide was included to inactivate deubiquitinating enzymes and thereby preserve endogenous ubiquitinated proteins. The extracts were centrifuged sequentially at 10,000 × g for 10 min and 100,000 × g for 1 h. The pellets from these centrifugations were solubilized in 25 mM Tris (pH 7.5)-2% sodium dodecyl sulfate (SDS). Ubiquitinated proteins were detected in the final supernatant, as well as the pellet fractions with anti-ubiquitin antibodies (Sigma). To identify various isoforms of UBC4, two-dimensional gel electrophoresis was performed, followed by transfer to nitrocellulose membranes and standard Western blotting. First-dimension isoelectric focusing was done by using 7-cm pH 3 to 10 IEF gel strips (Amersham Pharmacia) on an IPGphor isoelectric focusing apparatus (Amersham Pharmacia).
For enzymatic assays, testes were sliced and homogenized in 5 ml of 25 mM Tris (pH 7.5), 1 mM dithiothreitol, 0.25 M sucrose, 1 mM EDTA, and protease inhibitors (leupeptin at 10 μg/ml, pepstatin A at 10 μg/ml, and 1 mM phenylmethylsulfonyl fluoride)/g of tissue in a Potter-Elvehjem homogenizer. Protein content was determined by either the Bradford method or the bicinchoninic acid method. The lysates were clarified by centrifugations at 7,000 × g and 100,000 × g, and the remaining supernatants were frozen at −80°C until used. Rates of ubiquitination in these extracts were measured by incubating 25 μg of testis proteins in 20 μl of 50 mM Tris (pH 7.5), 1 mM dithiothreitol, 2 mM MgCl2, 2 mM AMPPNP, 5 μM ubiquitin aldehyde, and 5 μM 125I-ubiquitin (∼3,000 cpm/pmol [radiolabeled by chloramine T]). AMPPNP will support ubiquitination but not proteasome-mediated degradation of ubiquitinated proteins since ubiquitin-activating enzyme converts ATP to AMP (8), but the proteasome converts ATP to ADP. Ubiquitin-aldehyde will prevent loss of the ubiquitinated protein products by deubiquitinating enzymes (12, 19). Reactions were incubated at 37°C for 10 min (conditions under which the assay is known to be linear) (19) and quenched with Laemmli sample buffer. The reaction products were resolved from free ubiquitin by electrophoresis on SDS-10% polyacrylamide gels, stained with Coomassie blue, and then dried. The gels were exposed to film, and then the lanes were cut and counted in a gamma counter to quantitate the incorporated 125I-labeled ubiquitin.
RNA analysis.
Total RNA was prepared from testes either by guanidinium isothiocyanate solubilization followed by centrifugation on a cushion of CsCl (2) or by the phenol-guanidinium isothiocyanate extraction method (4). Both methods yielded similar amounts of RNA from this tissue. Northern blots were prepared and probed with 32P-labeled probes as previously described (27).
Surgical cryptorchidism and histological analyses.
Under general anesthesia, mice of ∼100 days of age were subjected to laparotomy. After identification of both testes, one testis was sutured via its perigonadal fat pad to the lowest rib of the thoracic cage. The other was returned to the scrotum. The abdominal cavity was closed, and animals were sacrificed at various days up to 10 days postsurgery. The testes were quickly isolated, weighed, and immersed in Bouin's preservative. After dehydration with progressively increasing concentrations of ethanol, the testes were sectioned and analyzed by hematoxylin-eosin staining.
To identity apoptotic cells, sections were subjected to TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining by using the ApopTag peroxidase in situ apoptosis detection kit (Chemicon). Testes sections were processed in accordance with the manufacturer's protocol and then counterstained with Harris modified hematoxylin solution. All round tubules were identified in the sections, and the stained cells were counted. Sequential sections from each testis were completely analyzed until at least 40 tubules from each animal were examined (average, 60 tubules per animal).
Sonication-resistant spermatid number and sperm motility.
To count spermatids, each frozen testis was homogenized in 5 ml of 0.9% NaCl, 0.5% Triton X-100, and 0.01% thimerosal by using the Polytron tissue disruptor. The suspensions were filtered through a nylon mesh filter (Nitex 70). The spermatid heads in the filtrate were then counted by using a hemacytometer. For motility analyses, sperm were isolated from the cauda epididymis by cutting it while immersed in M199 medium containing 0.5% (wt/vol) bovine serum albumin and adjusted to pH 7.3-7.4. Once the sperm cells were released, the epididymis was removed, and the sperm cells were incubated at 37°C for at least 5 min to permit their dispersal. Aliquots were then diluted 10-fold in the same warmed media and analyzed by using a Hamilton Thorn sperm analyzer.
RESULTS
Generation of mice lacking the UBC4-testis gene.
Transfection of the ES cells with the targeting vector resulted in correct homologous recombination in 16% of clones that grew in the presence of G418 and FIAU. Correct homologous recombination was confirmed by both PCR and Southern analysis (Fig. 1A and B). In addition, disruption of the UBC4-testis gene was confirmed by Northern blotting (Fig. 1C). To avoid cross binding to other UBC4 isoforms which have very similar coding region sequences, a probe specific to the UBC4-testis gene was obtained by using 3′ noncoding sequence (27). In RNA from the homozygous (−/−) mice, the usual ∼1.4-kb UBC4-testis transcript was now lost compared to samples from wild-type and heterozygous mice. Heterozygous and homozygous (−/−) mice, however, demonstrated the presence of a larger transcript. This represents RNA transcripts arising from transcription from the endogenous UBC4 promoter through the neomycin resistance gene (in reverse orientation) and back into the region encoding the 3′ noncoding sequence, which was not deleted by the homologous recombination. The homologous recombination resulted in deletion of codons 2 to 126 of UBC4-testis, removing 85% of the protein coding potential of the wild-type gene. This deletion includes the loops containing the highly conserved residues 76 to 96 that contribute critical parts of the active site, including the essential cysteine that forms the thiol ester linkage with ubiquitin. Two-dimensional gel electrophoresis, followed by immunoblot analysis, was performed to confirm the absence of UBC4-testis protein in homozygous (−/−) mice (Fig. 1D). Analysis of testis protein from wild-type animals revealed the presence of four immunoreactive spots at ∼17 kDa. The spot migrating most acidicly did correspond to the predicted pI of 5.4 for UBC4-testis. It was a quantitatively minor isoform, representing ∼10% of the total UBC4 immunoreactivity. This spot was, as expected, absent in samples from homozygous (−/−) mice.
Effects of loss of UBC4-testis on fertility, testis development, and morphology.
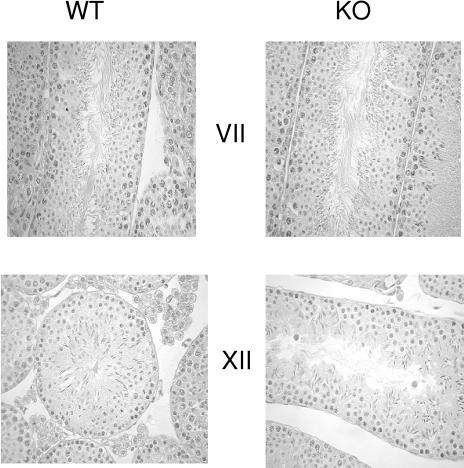
Mice lacking UBC4-testis grew normally (data not shown). Since the gene appears to be expressed only in the testis, we focused our search for a phenotype on this tissue and its functions. The fertility of the male knockout mice appeared normal (Table 1). Therefore, testes were analyzed for more specific defects. Overall, masses of the testes were similar between adult wild-type and homozygous (−/−) mice. The testes were subjected to histological analyses. Both spermatogenesis and spermiogenesis appeared to be normal in the mice lacking UBC4-testis (Fig. 2).
TABLE 1.
Normal fertility, testis size, and protein content in UBC4 wild-type and knockout micea
| Parameter | Mean ± SEM (n)
|
|
|---|---|---|
| UBC4 (+/+) | UBC4 (−/−) | |
| Litter size (no. of pups) | 7.7 ± 0.6 (9) | 8.1 ± 0.9 (11) |
| Testis wt (mg) | 102 ± 2 (10) | 96 ± 2 (11) |
| Testis protein (mg/testis) | 9.3 ± 1 (5) | 10.8 ± 0.4 (4) |
| Soluble testis protein (mg/testis) | 2.60 ± 0.24 (7) | 2.95 ± 0.12 (8) |
Wild-type or knockout male mice were mated with wild-type females, and the sizes of the litters were determined. Testis weights and soluble testis proteins (measured in supernatants of the extracts centrifuged at 100,000 × g) were determined for 15-week-old mice. Total testis proteins were determined by homogenizing testes from 11-week-old mice and determining the protein content in the crude extracts. n, number of animals studied.
FIG. 2.
Inactivation of the UBC4-testis gene does not affect spermatogenesis in adult testis. Shown are representative sections of the testis stained with hematoxylin-eosin at stages VII and XII from wild-type (WT) and homozygous (KO) mice.
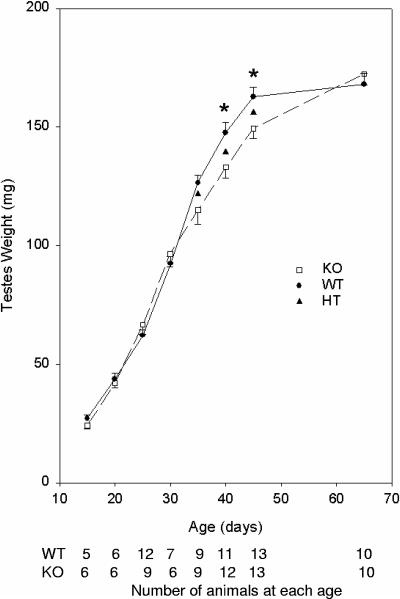
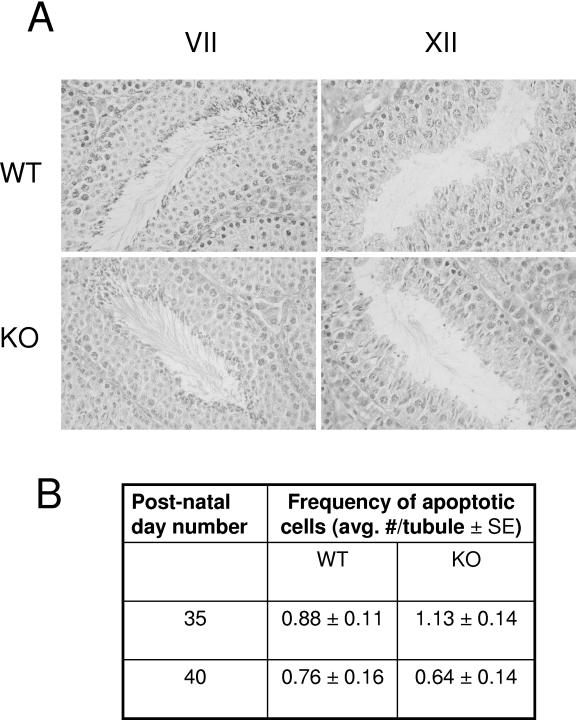
To evaluate this further and to detect possible defects in testis development, testes were analyzed at early time points after birth during the first wave of spermatogenesis (Fig. 3). The weights of the testes increased normally from day 15 to day 30. However, at days 35, 40, and 45, the mean combined testis weights from knockout mice were 9, 10, and 8% less, respectively, than the testis weights from wild-type animals, with the difference being of borderline statistical significance at 35 days (P = 0.06) and clearly significant at days 40 and 45 (P < 0.01). Subsequently, the differences disappeared as testes from 65-day-old mature mice (and 105-day-old mice [data not shown]) were similar in weight between wild-type and knockout mice. To explore whether the differences in testis weights were due to a delay in spermatid maturation in the knockout mice, histologic sections of the testis during development were analyzed. No reproducible differences were observed in sections from wild-type or knockout mice at 40 days of age (Fig. 4A) or at 35 and 45 days of age (data not shown). To explore whether the differences might be due to increased apoptosis during spermatogenesis in the knockout mice, testis sections were subjected to analysis by TUNEL staining. The numbers of apoptotic cells were similar in knockout and wild-type mice at both 35 and 40 days of age (Fig. 4B).
FIG. 3.
Inactivation of the UBC4-testis gene results in a delay in the postnatal development of the testis. Testes weight is significantly decreased at days 40 and 45 in homozygous mice compared to wild-type mice. At the indicated days after birth, testes were removed and weighed. The means ± the standard error (SE) of 5 to 13 mice are given. Significant differences were identified by two-way analysis of variance. ✽, P < 0.01. KO, homozygous; WT, wild type.
FIG. 4.
(A) Morphology of spermatogenesis appears normal in the testis of wild-type and UBC4-testis homozygous mice at day 40 of life. Shown are representative sections of the testis stained with hematoxylin-eosin at stages VII and XII. (B) Rates of apoptosis in the testis are similar in wild-type and UBC4-testis homozygous mice at days 35 and 40. Testis sections were subjected to TUNEL staining, and the numbers of apoptotic cells per tubule were determined (n = 5 for each group at day 40, n = 9 for each group at day 35.). KO, homozygous; WT, wild type.
Effects of loss of UBC4-testis on ubiquitination in the testis.
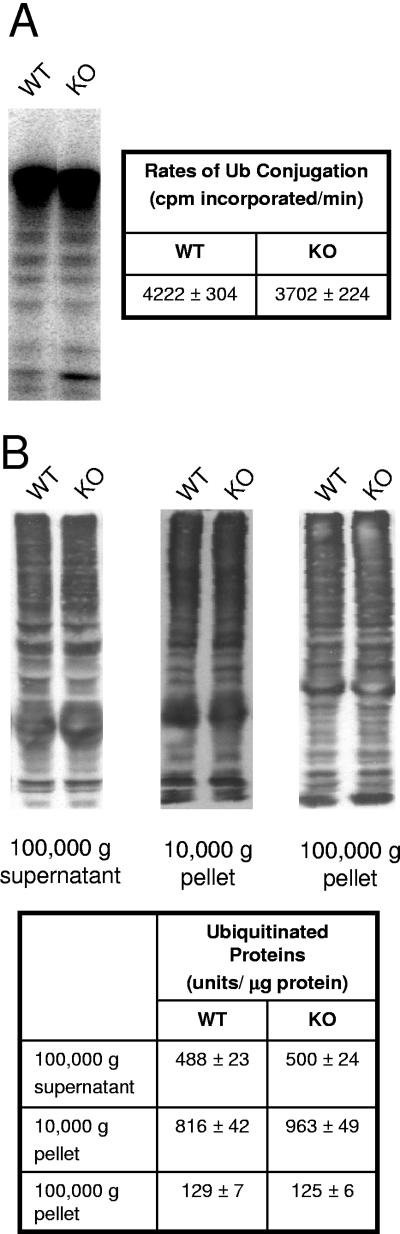
Since a major function of ubiquitination is the targeting of proteins for degradation, we assessed protein content in the testis as well as overall rates of ubiquitination. Protein content was similar in testes from wild-type and knockout mice (Table 1). Previously, we had observed that UBC4 isoforms together are responsible for the major part of the ubiquitination activity in testis extracts (20). Therefore, to evaluate whether UBC4-testis contributes a significant part to this ubiquitination activity, we measured rates of ubiquitination in extracts prepared from testes of wild-type and homozygous (−/−) mice. Rates were similar in both strains (Fig. 5A). Since proteins susceptible to ubiquitination in these extracts may not reflect exactly the ubiquitinated substrates found in the steady state in vivo, we measured levels of endogenous ubiquitinated proteins by using anti-ubiquitin antibodies. Since ubiquitination of proteins is now recognized to occur in membrane compartments, we analyzed samples from both soluble proteins and from solubilized pellets from centrifugations of the extracts at 10,000 × g and 100,000 × g. The levels of ubiquitinated proteins were similar between the two strains (Fig. 5b). We therefore measured UBC4 protein levels in the testis. Because of the identical size and high degree of sequence similarity between the many isoforms, only the total levels of the UBC4 isoforms together can be obtained from one-dimensional immunoblotting. Such analysis revealed that the total UBC4 levels were similar between wild-type and knockout mice (data not shown). This is not surprising since UBC4-testis only accounts for ∼10% of the total UBC4 immunoreactivity (Fig. 1D), and such a difference is within the error of such measurements. Nevertheless, to evaluate for possible induction of other isoforms of UBC4, samples were analyzed by two-dimensional gel electrophoresis, followed by immunoblotting to resolve the various UBC4 isoforms. As expected UBC4-testis expression was absent from knockout animals (Fig. 1D). Levels of the remaining isoforms appeared similar (Fig. 1D), and their quantitation did not reveal any significant differences between wild-type and knockout animals (data not shown). Similarly, Northern blot analyses with probes specific for several other UBC4 gene transcripts (27, 28) did not reveal any induction in testis of knockout mice (data not shown).
FIG. 5.
Overall ubiquitination of testis proteins is similar in wild-type (WT) and UBC4-testis homozygous (KO) mice. (A) Rates of in vitro ubiquitination were measured by incubation of testis extracts with 125I-labeled ubiquitin. Reaction products were resolved of free ubiquitin by SDS-polyacrylamide gel electrophoresis. Coomassie blue-stained gels were dried and exposed to film, and then the lanes of the gels were cut and counted in a gamma counter to quantitate theradioactivity incorporated into proteins. (B) Endogenous ubiquitinated proteins were detected by immunoblotting with anti-ubiquitin antibodies. Detection was by use of 125I-labeled secondary antibody, and quantitation was done by phosphorimager analysis.
Sperm number and motility.
Although the size and developmental morphology in the adult testis appeared normal, it is possible that defects were present at a physiological level that would be obvious only by examining closely more mature germ cells, including those from the epididymis. First, we tested for any differences in the number of spermatids as determined by counting the number of sonication-resistant spermatids in testis (Table 2). The numbers of spermatids were similar between the two groups. We then assessed for possible defects in motility. The percentages of motile sperm, as well as their movement characteristics—path velocity, progressive velocity, or curvilinear velocity, and lateral amplitude movements-were similar in sperm from wild-type or UBC4-testis knockout mice.
TABLE 2.
Comparison of UBC4 +/+ and −/− animalsa
| Parameter | Mean ± SEM
|
|
|---|---|---|
| UBC4 (+/+) | UBC4 (−/−) | |
| Testis sperm count (106/testis) | 27.8 ± 2.1 | 27.7 ± 1.3 |
| % Motility | 88 ± 4 | 87 ± 2 |
| Motility (path velocity [mm/s]) | 176 ± 16 | 169 ± 9 |
| Motility (progressive velocity or straight-line velocity [mm/s]) | 146 ± 14 | 138 ± 8 |
| Motility (track speed or curvilinear velocity [mm/s]) | 284 ± 24 | 263 ± 14 |
| Motility (lateral amplitude [mm]) | 7.7 ± 0.9 | 7.1 ± 0.5 |
Testis sperm counts were determined as the number of sonication-resistant spermatids. For motility studies, sperm were isolated from the cauda epididymus and analyzed in a Hamilton-Thorn analyzer. Values are the means of four +/+ and five −/− animals.
Response of the testis to heat stress of cryptorchidism.
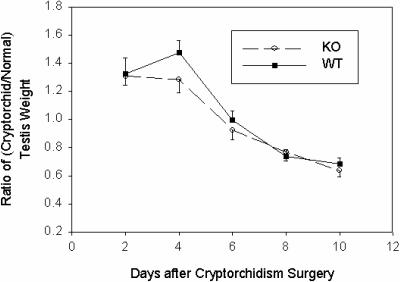
Our analyses to this point of mice lacking UBC4-testis only identified a developmental delay and did not reveal any defects in the adult testis under normal healthy conditions. However, it remains possible that the response to a stress in the testis may be defective. A frequently used model of stress of the testis is that of cryptorchidism. When placed in the higher-temperature environment of the abdominal cavity, all germ cell types except spermatogonia become depleted in the testis largely through death by apoptosis. Since different cell types have different sensitivity to heat stress, a certain temporal profile for depletion occurs. We therefore implanted one of the two testes in the abdominal cavity in both wild-type and mutant mice. Animals were killed at various time points, and the size and morphology of the cryptorchid testis were compared to that of the testis in the scrotum. Initially, the cryptorchid testis gains weight relative to the normal testis, and this is likely due to the acute inflammation that takes place in the heat-stressed testis. The testis mass then diminishes steadily in the cryptorchid testis as the germ cells disintegrate. Both wild-type and mutant mice displayed similar time courses for the disappearance of testis mass (Fig. 6).
FIG. 6.
Inactivation of UBC4-testis does not affect the rate of testicular involution in response to cryptorchidism. Wild-type (WT) or knockout (KO) mice were subjected to unilateral surgical cryptorchidism in which one of the testes was sutured to a lower rib. At the indicated times, mice were killed, and the cryptorchid and normal testis were isolated and weighed. The ratios of cryptorchid to normal testis weights ± the SE were plotted. There were no significant differences between the wild-type and knockout animals.
DISCUSSION
We have reported here the creation and extensive morphological, biochemical, and functional characterization of mice lacking a spermatid-specific ubiquitin-conjugating enzyme. Adult knockout mice were fertile and showed normal testes weights, spermatid number, protein content, and rates of ubiquitination. In vitro analyses of sperm showed no evident defects in motility compared to sperm from wild-type mice. Even when subjected to the heat stress of cryptorchidism, the temporal profile of loss of germ cells was not significantly different from wild-type mice.
However, the knockout mice did manifest a subtle delay in postnatal development during the first wave of spermatogenesis. This developmental defect was apparent at days 40 and 45 of life but resolved by day 65. Although this defect was subtle, it appears to be a real finding. Large numbers of mice derived from crosses of different heterozygous parents over a period of 2 years were analyzed at these time points to render it very unlikely that these findings were spurious. Furthermore, heterozygous mice showed a defect intermediate between wild-type and mutant mice, a finding consistent with a gene dosage effect. Although differences were statistically significant only at days 40 and 45, it should be noted that at day 35 the mean of the knockout group was also less than the mean of the wild-type group, but the P value of the difference was borderline (0.06). The likelihood that differences would occur by chance in three consecutive time points when UBC4-testis is normally expressed in wild-type mice would be very remote.
These findings indicate a role for UBC4-testis in regulating testis maturation. This is consistent with our previous finding that UBC4-testis is induced in round spermatids beginning at day 25 of life (27). However, in the adult testis, there was no evidence of abnormal distribution of germ cell types and in particular no prolongation of duration in the evolution of round into elongating spermatids, as might be revealed by a loss of the normal pattern of cells seen at each stage of spermatogenesis (Fig. 4). This shows that UBC4-testis is not required per se for spermatogenesis. Instead, it may have a specific role in promoting the evolution of the first wave of spermatogenesis. Due to the small differences between the testes of wild-type and knockout mice, it has been very difficult to explore further the cellular and biochemical bases for the differences.
Alternatively, UBC4-testis may play a role in regulating the postnatal growth of the testis. Possibly, during the first wave of spermatogenesis, UBC4-testis positively regulates the function of a specific factor that promotes the growth of the testis or it negatively regulates an inhibitor of such evolution. Presumably, UBC4-testis does so by interacting with specific ubiquitin protein ligases that preferably interacts with it as opposed to other UBC4 isoforms. The catch up to the normal size with time suggests that other UBC4 isoforms may interact weakly with this ubiquitin protein ligase(s) and therefore be less effective at mediating ubiquitination with these putative ligase(s). However, over time, an equivalent amount of ubiquitination is completed and a catch up in maturation occurs. Up to now we have not been able to identify any such UBC4-testis specific ligases. We have only identified some ligases, such as ARNIP/Pirh2 (3) and SCFβ-TRCP (Y. Ben-Neriah and S. S. Wing, unpublished observations), which function with the ubiquitous UBC4-1 but not UBC4-testis. Therefore, we cannot rule out at this time that these differences are due to a gene dosage effect occurring in isoforms with redundant functions. Overall, UBC4 levels do increase during the elongation of spermatids (20) and so by mass action could increase ability to bind and support weaker interacting ligases or simply replace the loss of the UBC4-testis isoform. These other UBC4 isoforms are highly similar to UBC4-testis with 91 to 93% amino acid identity so such overlapping or replacement function would not be surprising.
Taken together, our biochemical and genetic data support the concept that highly similar isoforms are not necessarily redundant. For example, markedly reduced expression of UBCM4 (the mouse ortholog of human UBCH7), an isoform more distantly related to UBC4/UBC5, leads to a defect in implantation of the embryo (9). Similarly, there are two isoforms of UBC2 (HHR6A and HHR6B) that are 96% amino acid identical (14). They show ubiquitous expression in all cells studied to date, and there is no evidence of any differences in biochemical function between these two isoforms. However, when HHR6B is inactivated in the mouse, it leads to male infertility due to incomplete spermatogenesis (22). The mutant mice manifest a heterogeneous population of abnormal spermatids. Interestingly, inactivation of the HHR6A isoform in the mouse does not lead to male infertility but female infertility due to an apparent defect in progression of embryos past the two cell stage (21). Although these results could be due to gene dosage effects due to relatively high expression of HHR6A in oocytes and HHR6B in spermatids, these findings could also be due to specific functions of the two isoforms. In support of this latter possibility, the putative ligase cyclophilin CYC4/hCyP-60 requires HHR6B but not HHR6A (10), whereas another RING ligase, Rfpl4, interacts with HHR6A but apparently not HHR6B in a yeast two-hybrid screen (26).
With the availability of sequences of various genomes, it is clear now that for several of the yeast ubiquitin-conjugating enzymes such as UBC2 and UBC4/UBC5, there are multiple orthologous genes in the mammalian genomes. Our data suggest that these isoforms can serve redundant complementary functions but, in addition, may also have specific roles that help meet the more complex functional requirements of higher organisms.
Acknowledgments
P.H. was the recipient of a fellowship from the Medical Research Council of Canada. Z.P. is the holder of a studentship from the McGill University Health Centre Research Institute. S.S.W. and J.T. are recipients of Chercheur National salary awards from the Fonds de la Recherche en Santé du Québec. This research was supported by operating grant MT12121 from the Canadian Institutes of Health Research.
REFERENCES
- 1.Arama, E., J. Agapite, and H. Steller. 2003. Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev. Cell 4:687-697. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, D. D. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Beitel, L. K., Y. A. Elhaji, R. Lumbroso, S. S. Wing, V. Panet-Raymond, B. Gottlieb, L. Pinsky, and M. A. Trifiro. 2002. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29:41-60. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.de Kretser, D. M., K. L. Loveland, A. Meinhardt, D. Simorangkir, and N. Wreford. 1998. Spermatogenesis. Hum. Reprod. 13:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Di Fiore, P. P., S. Polo, and K. Hofmann. 2003. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell. Biol. 4:491-497. [DOI] [PubMed] [Google Scholar]
- 7.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 8.Haas, A. L., and I. A. Rose. 1982. The mechanism of ubiquitin activating enzyme. J. Biol. Chem. 257:10329-10337. [PubMed] [Google Scholar]
- 9.Harbers, K., U. Muller, A. Grams, E. Li, R. Jaenisch, and T. Franz. 1996. Provirus integration into a gene encoding a ubiquitin-conjugating enzyme results in a placental defect and embryonic lethality. Proc. Natl. Acad. Sci. USA 93:12412-12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 11.Hecht, N. B. 1988. Post-meiotic gene expression during spermatogenesis, p. 291-313. In F. P. Haseltine and N. L. First (ed.), Meiotic inhibition: molecular control of meiosis. Alan R. Liss, Inc., New York, N.Y.
- 12.Hershko, A., and I. A. Rose. 1987. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc. Natl. Acad. Sci. USA 84:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kistler, W. S. 1989. Histones and other basic nuclear proteins. CRC Press, Inc., Boca Raton, Fla.
- 14.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansour, S. L., K. R. Thomas, and M. R. Capecchi. 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336:348-352. [DOI] [PubMed] [Google Scholar]
- 16.Morales, C., Y. Clermont, and L. Hermo. 1985. Nature and function of endocytosis in Sertoli cells of the rat. Am. J. Anat. 173:203-217. [DOI] [PubMed] [Google Scholar]
- 17.Osley, M. A. 2004. H2B ubiquitylation: the end is in sight. Biochim. Biophys. Acta 1677:74-78. [DOI] [PubMed] [Google Scholar]
- 18.Oughtred, R., N. Bédard, A. Vrielink, and S. S. Wing. 1998. Identification of amino acid residues in a class I ubiquitin conjugating enzyme involved in determining specificity of conjugation of ubiquitin to proteins. J. Biol. Chem. 273:18435-18442. [DOI] [PubMed] [Google Scholar]
- 19.Rajapurohitam, V., N. Bedard, and S. S. Wing. 2002. Control of ubiquitination of proteins in rat tissues by ubiquitin conjugating enzymes and isopeptidases. Am. J. Physiol. 282:E739-E745. [DOI] [PubMed] [Google Scholar]
- 20.Rajapurohitam, V., C. R. Morales, M. El-Alfy, S. Lefrancois, N. Bédard, and S. S. Wing. 1999. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev. Biol. 212:217-228. [DOI] [PubMed] [Google Scholar]
- 21.Roest, H. P., W. M. Baarends, J. de Wit, J. W. van Klaveren, E. Wassenaar, J. W. Hoogerbrugge, W. A. van Cappellen, J. H. Hoeijmakers, and J. A. Grootegoed. 2004. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24:5485-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. M. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. M. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. J. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 23.Russell, L. C., and Y. Clermont. 1976. Anchoring device between Sertoli cells and late spermatids in rat seminiferous tubules. Anat. Rec. 185:259-278. [DOI] [PubMed] [Google Scholar]
- 24.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 25.Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzumori, N., K. H. Burns, W. Yan, and M. M. Matzuk. 2003. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc. Natl. Acad. Sci. USA 100:550-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wing, S. S., N. Bedard, C. Morales, P. Hingamp, and J. Trasler. 1996. A novel rat homologue of the Saccharomyces cerevisiae ubiquitin conjugating enzyme UBC4 with distinct biochemical features is induced during spermatogenesis. Mol. Cell. Biol. 16:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wing, S. S., and P. Jain. 1995. Molecular cloning, expression, and characterization of a ubiquitin conjugation enzyme (E2 17KB) highly expressed in rat testis. Biochem. J. 305:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]