Abstract
We report the identification and characterization of a new ischemia/reperfusion-inducible protein (IRIP), which belongs to the SUA5/YrdC/YciO protein family. IRIP cDNA was isolated in a differential display analysis of an ischemia/reperfusion-treated kidney RNA sample. Mouse IRIP mRNA was expressed in all tissues tested, the highest level being in the testis, secretory, and endocrine organs. Besides ischemia/reperfusion, endotoxemia also activated the expression of IRIP in the liver, lung, and spleen. The transporter regulator RS1 was identified as an IRIP-interacting protein in yeast two-hybrid screening. The interaction between IRIP and RS1 was further confirmed in coimmunoprecipitation assays. A possible role of IRIP in regulating transporter activity was subsequently investigated. IRIP overexpression inhibited endogenous 1-methyl-4-phenylpyridinium (MPP+) uptake activity in HeLa cells. The activities of exogenous organic cation transporters (OCT2 and OCT3), organic anion transporter (OAT1), and monoamine transporters were also inhibited by IRIP. Conversely, inhibition of IRIP expression by small interfering RNA or antisense RNA increased MPP+ uptake. We measured transport kinetics of OCT2-mediated uptake and demonstrated that IRIP overexpression significantly decreased Vmax but did not affect Km. On the basis of these results, we propose that IRIP regulates the activity of a variety of transporters under normal and pathological conditions.
Ischemia/reperfusion (I/R) is the major cause of tissue injury under many pathophysiological conditions such as stroke, myocardial infarction, and acute renal failure (1, 11). Similar to other stress conditions, I/R causes dramatic changes in cell physiology and metabolism. Ischemic tissue is deprived of oxygen and essential substrates for energy metabolism. As the cellular oxygen is depleted, mitochondrial oxidative phosphorylation and ATP production rates decrease. There are several important consequences of ATP depletion. First, the activities of energy-dependent Na+/K+-ATPases are diminished. As a result, cells accumulate sodium and other solutes while losing potassium, causing the cell and cellular organelles (mitochondria and endoplasmic reticulum [ER]) to swell due to water gained from osmotic balance. Second, cytosolic free Ca2+ concentration increases because the activities of membrane-associated ATP-dependent Ca2+ pumps are reduced, resulting in a net influx of Ca2+ from extracellular space, mitochondria, and ER lumens. This has profound effects on cellular functions due to the activation of a wide range of proteins by Ca2+, including proteases, endonucleases, phospholipases, protein kinases/phosphatases, and transcription factors. Many of these proteins mediate degradation of cellular components (protein, nucleic acid, lipid) in necrosis and apoptosis. Third, severe mitochondrial damage occurs, including breakdown of membrane lipid, modification of electron transfer chain proteins, and most importantly, mitochondrial permeability transition (15). If ischemia is transient and blood flow restored within a short time, most ischemic damage can be reversed. However, reperfusion causes additional damage (reperfusion injury). It is believed that reactive oxygen species produced in reperfused cells and inflammatory responses are the key mediators of reperfusion injury (29).
The transport of small molecules across biological membranes is essential for maintaining cellular homeostasis and adaptation to environmental changes. An important aspect of cellular function during I/R is the modulation of metabolite transport across the plasma membrane, which may be involved in cell injury or repair mechanisms. For example, the expression levels of Na+-glucose, Na+-phosphate, and Na+-succinate cotransporters are significantly reduced at 24 h postreperfusion in a rabbit kidney I/R model, which leads to the inhibition of Na+-dependent uptake of glucose and impairment in Na+-dependent reabsorption of solutes (12). I/R causes extracellular accumulation of glutamate (in the central nervous system) and monoamines (in the sympathetic nervous system) through reverse uptake by respective transporters, which may cause additional cell damage such as excitotoxicity (21, 22). Therefore, studies on the regulation of plasma membrane transporters under normal and I/R stress conditions will contribute to a better understanding of the injury mechanisms and to the development of novel therapeutics.
In the past decade a variety of genetic, molecular and pharmacological approaches were undertaken to establish the mechanism of regulation for these transporters. It is becoming clear that transporter activities can be regulated at multiple levels: transporter protein expression, posttranslational modification, oligomerization, and protein trafficking (13). The extracellular loops of many transporters contain potential glycosylation sites that have been shown to play an important role in transporter trafficking and stability (17, 19). There are also potential phosphorylation sites for serine/threonine and tyrosine kinase in the amino and carboxy termini and cytoplasmic loops (26). A variety of transporter-interacting proteins that are capable of regulating transporter activity and trafficking have been reported (18). The physiological significance of these interactions, in most cases, is not clear at this stage and attracts intense investigation, especially for regulators of plasma membrane monoamine transporters (24).
In this study, we identified and cloned a new I/R-inducible protein (IRIP) using an AU-motif targeted differential display approach. IRIP can negatively regulate the uptake activity of several organic cation transporters (OCT2 and OCT3), organic anion transporter (OAT1), and three monoamine transporters: dopamine transporter (DAT), norepinephrine transporter (NET), and 5-hydroxytryptamine (serotonin) transporter (SERT). We hypothesize that IRIP, together with RS1, represents a novel regulatory pathway that controls the activity of several transporter families.
MATERIALS AND METHODS
Kidney I/R operation.
The I/R surgical procedure was described previously (30). Briefly, mice were anesthetized with sodium pentobarbital (25 mg/kg) and xylazine (10 mg/kg), followed by subcutaneous heparin administration (300 USP units/kg) before the surgery. Unilateral renal ischemia was induced by occluding the left side renal vein and artery with a microaneurysm clamp. After 32 min, the microaneurysm clamp was removed to allow reperfusion and right nephrectomy was performed at the same time. Body temperature was maintained at 37°C during the entire procedure. Sham surgery consisted of a surgical procedure that was identical, except that the microaneurysm clamp was not applied. Mice were sacrificed at 6 or 24 h after reperfusion. Kidneys were perfused with cold phosphate-buffered saline (PBS; 150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, 5 mM β-mercaptoethanol, pH 7.3), immediately removed, and frozen in liquid nitrogen.
Differential display.
The experimental design was adapted from a previously published method (7). Four sets of PCRs were carried out for each reverse transcription (RT) reaction, with primers that differ in the anchoring base residue (common AT-rich sequences are underlined) (RT1, 5′-GGGGGGGTATTTATTTANTTTTTTTTTTTTTTT(A/C/G)-3′; PCR1, 5′-GGGGGGGTATTTATTTAA-3′, 5′-GGGGGGGTATTTATTTAC-3′, 5′-GGGGGGGTATTTATTTAG-3′, and 5′-GGGGGGGTATTTATTTAT-3′; RT2, 5′-GGTGGGTGGTATTTATTTANTTTTTTTTTTTTTTT(A/C/G)-3′; and PCR2, 5′-GGTGGGTGGTATTTATTTAA-3′, 5′-GGTGGGTGGTATTTATTTAC-3′, 5′-GGTGGGTGGTATTTATTTAG-3′, and 5′-GGTGGGTGGTATTTATTTAT-3′). A single primer was used in differential display PCR (DD-PCR) that would anneal with the 5′ end of the first-strand cDNA as well as the AUUUA motif at the 3′-untranslated region (UTR) of stress-responsive genes. PCR master mix was prepared as follows: (for 40 μl master mix) 5 μl 10× Mg2+-free buffer, 1.5 μl MgCl2 (50 mM), 1 μl deoxynucleoside triphosphate (dNTP; 10 mM), 3 μl primer (50 pmol/μl), and 28.5 μl double-distilled H2O were mixed and 1 μl DyNAZyme polymerase (1 unit; Finnzymes) was then added to make the master mix. For each DD-PCR, 8 μl of the master mix was aliquoted into a 500-μl Eppendorf tube, covered with mineral oil, and heated at 96°C for 2 min. cDNA template (2 μl) from previous RT reaction and 0.25 μl [α-35S]dATP (10 μCi/μl; NEN) was added underneath the mineral oil and the PCR cycle (94°C for 30 s, 42°C for 1 min, 70°C for 1 min, 40 cycles) was started immediately. PCR product (3.5 μl) was mixed with 2 μl loading solution (95% formamide, 0.09% bromophenol blue, and 0.09% xylene cyanole FF), heated at 95°C for 3 min, and loaded on a 6% polyacrylamide gel containing 2× Tris-borate-EDTA (TBE) buffer and 6 M urea. Electrophoresis was run at constant power of 40 W until the xylene cyanole FF dye reached the gel bottom. Gels were dried and visualized by autoradiography. Selected differential bands were excised from the dried gel after aligning the gels with the autoradiography films. Gel slices were rehydrated and reamplified by PCR using the same primer as in the original DD-PCR. The amplified DNAs were then TA cloned into pCR2.1-TOPO (Invitrogen).
RACE.
Total RNA was isolated as described previously. V(10) poly(A)+ RNA was then isolated with RNeasy mRNA purification kit (QIAGEN). Rapid amplification of cDNA ends (RACE) reactions were performed with the Marathon cDNA amplification kit (Clontech) according to the manufacturer's instruction. RACE products were cloned into TA cloning vector pCR2.1-TOPO (Invitrogen).
Northern blotting.
RNA samples (20 μg) were loaded on 1.2% agarose gels containing 2.2 M formaldehyde. After electrophoresis, RNA was transferred to a GeneScreen nylon membrane (New England Nuclear, Boston, MA) using a PosiBlot pressure blotter (Stratagene, La Jolla, CA). The membranes were then fixed and hybridized with α-32P-labeled DNA probe made with a random-priming DNA labeling kit (Roche Biochemicals) or RNA probe labeled with the Riboprobe Combination system (T3/T7). The DNA probe was hybridized at 42°C as previously described (10), whereas the RNA probe was hybridized at 65°C overnight in a hybridization solution (50% formamide, 1× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 4% dextran sulfate, 5× Deinhardt's, 1% sodium dodecyl sulfate [SDS], 0.25 mg/ml tRNA, and 0.1 mg/ml single-stranded DNA). Labeled β-actin or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) DNA probes were used for hybridization as an internal control. To quantify levels of expression, bands on the films with linear exposure range were scanned on a model 300A densitometer using ImageQuant software (Amersham Biosciences).
Primer extension.
Oligonucleotide primers 5′-GGCCAGAACTAGCCGGCCCGTCGCTCAA-3′ (mouse PE1) and 5′-GTCACAGGGCCTGGGAACAGGTCTTCCA-3′ (mouse PE2) were used. Primers were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (PNK). For a 10-μl reaction, 1 μl 10× PNK buffer, 1 μl primer (20 μM), 3 μl [γ-32P]ATP (10 μCi/μl), 4 μl RNase-free water, and 1 μl T4 PNK (New England Biolabs [NEB]) were mixed and incubated at 37°C for 1 h followed by heat inactivation at 90°C for 5 min. For a 20-μl primer extension reaction, 10 μg total RNA was mixed with 2 μl 5× avian myeloblastosis virus (AMV)-RT buffer, 0.5 μl dNTP (10 mM), 1 μl labeled primer, and RNase-free water to make a volume of 9.5 μl. The mixture was incubated at 65°C for 5 min and quickly chilled on ice. RNase inhibitor (0.25 μl) and AMV-RT enzyme (0.25 μl) (Roche Applied Science) were added and mixed. The reaction mixture was incubated at 25°C for 10 min and then at 42°C for 1 h. The reaction was stopped by addition of 6 μl stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanole FF). The samples were loaded on a 6% polyacrylamide gel containing 6 M urea and 2× TBE buffer. Gel was dried after electrophoresis and exposed to X-ray film (Kodak).
Real-time PCR.
Real-time PCRs were performed using a SYBR Green PCR Core Reagents kit (Applied Biosystems) in a DNA engine Opticon continuous fluorescence detection system (MJ Research) with the following program: initial incubation at 95°C for 10 min, followed by 40 cycles consisting of two steps, 95°C for 15 s and 60°C for 1 min. The following primers were used for amplification of mIRIP, 5′-GCTCAACAAAGACCTGAACCCC-3′ (forward) and 5′-GCAAGTGGTCCCCCAAACAT-3′ (reverse); mGADH, 5′-GCAAAGTGGAGATTGTTGCCAT-3′ (forward) and 5′-CCTTGACTGTGCCGTTGAATTT-3′ (reverse); hIRIP, 5′-GCAAGCGGACCCTCAAACAT-3′ (forward) and 5′-GCTCAACAAGGACCTAAACCCT-3′ (reverse); hGAPDH, 5′-AGATCATCAGCAATGCCTCCTG-3′ (forward) and 5′-TGGCATGGACTGTGGTCATG-3′ (reverse).
Expression of GST-mIRIP fusion protein and raising specific antibody.
Glutathione S-transferase (GST)-mIRIP fusion protein expression plasmid was constructed as follows. The coding sequence of mIRIP was amplified from pUC19-mIRIP with the following primers: 5′-CATATGTCTACGGCGCGTCCG-3′ and 5′-GGATCCTCATGAACAGGACCCCTG-3′. The PCR product was first cloned into the NdeI/BamHI sites of pET-GST-Km (Novagen). The resulting plasmid clone was designated as pET-GST-mIRIP. The overnight culture of Escherichia coli cells BL21-DE3 (ndk:cam [F− ompT gal [dcm] [lon] hsdSB with DE3, a λ prophage carrying the T7 RNA polymerase gene]) transformed with pET-GST-mIRIP was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) solution (1 mM final concentration) and the culture was grown for an additional 2 h. Cells were harvested, and the cell pellet was suspended in 10 ml PBS. Cells were broken open with a French press instrument (Aminco) at 1,000 lb/in2. Cell debris and membrane fragments were removed by serial centrifugation at 8,000 × g for 10 min and then at 134,000 × g for 1 h. The supernatant was applied on a prepacked glutathione Sepharose 4B column. The column was washed with 2× 10 ml PBS, and the bound protein was eluted with 10 ml elution buffer (50 mM Tris-HCl, 5 mM reduced glutathione, pH 8.0) and collected in 1.5-ml fractions. Purified GST-mIRIP was digested with thrombin. The digestion mixture was separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. The gel was stained with Coomassie brilliant blue. The band that corresponded to mIRIP was excised, and the gel slice was used as an antigen for immunization. Polyclonal antibody against mIRIP was raised in rabbit (service provided by Pocono Rabbit Farm, Inc). The specificity of the antibody was verified by running a Western blot with GST-mIRIP as well as myc-mIRIP expressed in NIH 3T3 cells.
Metabolic labeling of NIH 3T3 cells.
[35S]methionine labeling was performed as previously described (6). Equal amounts of [35S]methionine-labeled proteins were analyzed by SDS-PAGE followed by fluorography. Immunoprecipitation reactions with polyclonal rabbit antibodies were carried out overnight at 4°C.
Cell cultures, expression plasmids, and transfection.
Mammalian expression plasmids pEF/myc/cyto-mIRIP and pEF/myc/cyto-hIRIP were constructed by subcloning mIRIP and hIRIP coding sequences into the NcoI/NotI sites of pEF/myc/cyto vector (Invitrogen). To obtain pcDNA-FLAG-mRS1 plasmid for transient expression of RS1, the coding sequence of RS1 was PCR amplified from mouse liver cDNA using a 5′-primer with FLAG tag sequence fused in frame with RS1 sequence. PCR product was then cloned into pCDNA 3.1 for expression. NIH 3T3, HeLa, and COS-7 were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin, in a humidified incubator with 5% CO2. Cell transfection was performed with Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Stably transfected cell lines were established by selecting transfected HeLa cells in the presence of Gx418 (450 μg/ml). After transfection with pEF/myc/cyto-hIRIP and selection, we obtained several cell lines in which expression of IRIP was confirmed by Western blotting. Two cell lines, obtained with the pEF/myc/cyto vector and pEF/myc/cyto-hIRIP (HeLa-2 and HeLa-4, respectively) were used in most of the experiments. HeLa-dm was a stable cell line that overexpressed the T87L-K110I double-mutant myc-hIRIP. Mutations were introduced using QuikChange site-directed mutagenesis kit (Stratagene). For obtaining stable cell lines with inducible expression of sense and antisense hIRIP RNA, IRIP cDNA fragment (position +20 to +820 bp) was inserted in both orientations in plasmid pUHD10-3-hygro (pUHD10-3-hygro was a gift from H. Bujard, University of Heidelberg, Heidelberg, Germany): plasmid As with an antisense orientation of RNA and plasmid S with a sense orientation. In this vector, RNA was expressed in a Tet-OFF system, under the control of a minimal cytomegalovirus promoter and seven tetracycline operator sites. Upon transfection of HeLa-tTA cells (8) with S and As plasmids, stable clones were selected in the presence of hygromycin (200 μg/ml). Expression of sense and antisense RNA was induced in selected cell lines upon withdrawal of tetracycline according to Northern blot analysis using labeled sense and antisense 32P-labeled RNA probes.
Subcellular fractionation.
Kidney tissue was homogenized in 10 volumes (wt/vol) of 0.25 M sucrose/buffer A (10 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]), supplemented with Complete protease inhibitor cocktail tablets (Boehringer Mannheim) and centrifuged at 1,300 × g for 15 min at 4°C. The supernatants were centrifuged at 16,000 × g for 15 min at 4°C, and pellets were used for plasma membrane isolations by centrifugation through 1.12 M sucrose cushion as described before (3). Supernatants were centrifuged at 200,000 × g for 1 h at 4°C, and the resulting supernatants were taken as the cytosolic fractions. The pellets from the initial centrifugation step were resuspended in 0.25 M sucrose-buffer A and then layered on top of 1.8 M sucrose-buffer A cushion. Nuclei were pelleted by centrifugation at 30,000 × g for 45 min at 4°C. The pellet was resuspended and washed by low-speed centrifugation. The final pellet was designated as the nuclear fraction.
Western blotting.
Proteins were separated on SDS-polyacrylamide gels (10% for the separation of RS1 and 15% for the testing of IRIP). After electrophoresis, the gel was soaked for 20 min in transfer buffer (24.8 mM Tris, 192 mM glycine, 20% methanol, pH 8.3). Proteins were transferred to polyvinylidene difluoride membrane in the presence of transfer buffer in a Trans-Blot semidry transfer cell (Bio-Rad). The membranes were incubated in blocking solution containing 5% milk for 1 h, and then incubation continued in the presence of appropriate primary antibodies overnight at 4°C. The membranes were washed and incubated in blocking solution with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Inc.). Finally, the proteins were detected using the enhanced chemiluminescence reagent (NEB). Anti-myc monoclonal antibody (MAb) (Invitrogen) and anti-FLAG MAb (Sigma) were used at a 1:5,000 dilution. Anti-IRIP polyclonal antibody was routinely used at a 1:1,000 dilution. Anti-rabbit and anti-mouse immunoglobulin G (IgG) HRP-conjugated secondary antibodies (NEB) were used at a 1:5,000 dilution. For demonstration of the antibody specificity, immunoneutralization with GST-IRIP was conducted. One to two microliters of antibodies was incubated with 5 μg of purified GST-IRIP or without the antigen in 100 μl of PBS for 1 h at 37°C and for 4 h at 4°C. After centrifugation, antibodies were used at 1:2,500 dilutions for Western blotting.
Construction of yeast two-hybrid cDNA library.
HybriZAP-2.1 XR cDNA library construction kit (Stratagene) was used to generate the library, according to the manufacturer's protocol. Briefly, 5 μg poly(A)+ RNA isolated from liver tissue of C57BL/6 mice at 18 h after injection of lipopolysaccharide (LPS; 16 mg/kg of body weight) were transcribed into first-strand cDNA with primer 5′-GAGAGAGAGAGAGAGAGAGAACTAGTCTCGAGTTTTTTTTTTTTTTTTTT-3′ and Stratascript reverse transcriptase. Second-strand cDNAs were then synthesized with DNA polymerase I and size fractionated through a Sepharose CL-2B gel filtration column. cDNA fractions more than 400 bp in size were subsequently cloned into HybriZAP-2.1 vector digested with EcoRI and XhoI. The primary library contained about 1.2 × 106 PFU. The cDNA inserts of individual phage clones contained inserts with average size about 1 kb. The primary library was then amplified in XL1-Blue MRF′ cells. The amplified library was subjected to mass in vivo excision in XL1-Blue MRF′ cells in the presence of ExAssist helper phage. The resulting phagemid library was amplified in XLOLR cells, and finally the phagemid DNA was isolated for use in yeast transformation.
Yeast two-hybrid screen.
For construction of the bait plasmid, coding sequence of mIRIP was amplified by PCR using the following primers: 5′-TAGAATTCCCGGAGAGCGAGCCC-3′ and 5′-ACTGTCGACTCATGAACAGGACCC-3′. The EcoRI-SalI fragment was cloned into EcoRI and SalI sites of pBD-GAL4-Cam (Stratagene). For library-scale transformation, 40 μg library DNA was used to transform AH109 cells from 1 liter of culture. All other reagents were scaled up accordingly. Library transformation mixture was spread on 60 SD− Trp− Leu− His plates (150 mm) and incubated at 30°C for 5 to 7 days until colonies grew to >2 mm in diameter. Trp+ Leu+ His+ colonies were transferred to SD− Trp− Leu− His− Ade plates in a grid format to select for both HIS and ADE reporter. Trp+ Leu+ His+ Ade+ clones were grown on SD− Trp− Leu− His− Ade plates in 1-cm2 patches. Colonies growing on selection media, but not on SD plates, were further cultivated and analyzed for β-galactosidase activity. Cells from selected colonies were harvested for plasmid DNA isolation, which was used to retransform the reporter strain along with the bait plasmid in order to confirm the detected protein-protein interactions. Finally, plasmid DNA was transformed in E. coli DH5α cells and DNA sequences of positive clones were then determined by manual sequencing.
Coimmunoprecipitation analysis.
To test whether full-length RS1 and IRIP can interact upon expression in mammalian cells, we transfected COS-7 cells with expression plasmids pCDNA-FLAG-mRS1 and pEF/myc-mIRIP, individually or in combination using Lipofectin (Invitrogen) according to the manufacturer's recommendations. The cultures were harvested 48 h after transfection and lysed in radioimmunoprecipitation assay buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors (1 mM PMSF, 50 μg/ml leupeptin, 50 μg/ml antipain, 0.1 mg/ml aprotinin, 10 mM NaF, 1 mM NaVO4, 1.5 mM Na2MoO4, 1 mM benzamidine, 20 mM glycerophosphate, and 20 mM p-nitrophenyl phosphate). Cellular extracts were immunoprecipitated with anti-myc (Invitrogen) or anti-FLAG (Sigma) antibodies immobilized on the beads overnight at 4°C. Beads were washed four times in radioimmunoprecipitation assay buffer; bead-bound proteins were denatured in sample buffer, separated by SDS-PAGE, and detected by immunoblotting with anti-myc or anti-FLAG HRP-conjugated antibodies and Western Lighting chemiluminescence reagent (Perkin-Elmer Life Sciences, Inc.).
Transport studies.
HeLa stable cell lines described above (HeLa-2, HeLa-4, and HeLa-dm, S and As) as well as transiently transfected HEK-293T cells were used for transport studies. Human OCT2 and OCT3 cDNA expression plasmids were provided by D. Grundemann (University of Cologne, Cologne, Germany), human DAT and NET plasmids by M. G. Caron (Duke University Medical Center, NC), human SERT (hSERT) plasmid by R. Blakely (Vanderbilt University, TN), and mouse OAT1 by G. You (Rutgers University, Piscataway, NJ). Cells were seeded on 12-well plates at 1 × 105 to 2 × 105 cells/well about 18 h before the assay. At the beginning of the assay, cells were washed with Krebs-Ringer-HEPES (KRH) buffer (25 mM HEPES, 125 mM NaCl, 4.8 mM KCl, 5.6 mM d-glucose, 1.2 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, pH 7.4) and then incubated at 37°C in KRH buffer containing radiolabeled substrates (10 nM [3H]MPP+, 80 Ci/mmol, adjusted with cold MPP up to 100 nM, 50 nM [3H]norepinephrine hydrochloride, 7.94 Ci/mmol; 40 nM [3H]dihydroxyphenylethylenamine, 20 Ci/mmol; all isotopes from Perkin-Elmer Life Sciences, Boston, MA; or 75 nM [3H]-5-hydroxytryptamine creatinine sulfate, 11 Ci/mmol; Amersham, Piscataway, NJ). Uptake was stopped by washing cells with 3× 1 ml ice-cold KRH buffer. Cells were solubilized with 0.5 ml RIPA buffer and the radioactivity of the lysates was measured with a liquid β-scintillation counter. Aliquots were removed from the lysates for protein measurement. Uptake rates were expressed in pmol per mg of protein. For saturation studies of OCT2-mediated uptake, transport was performed for 8 min in the presence of 10 nM [3H]MPP+ and various concentrations of nonlabeled MPP+ (0 to 200 μM). For the anion transport assay, HEK-293T cells were cultured in Transwell chambers (0.4-μm pores) in 12-well plates; 5 μM [14C]para-aminohippuric acid (Perkin-Elmer Life Sciences, Boston, MA) and 15 μM of cold para-aminohippuric acid PAH were added to the uptake solution, and transport was assayed as previously reported (32).
Inhibition of IRIP expression by siRNA.
IRIP-specific siRNA samples were produced with the Silencer siRNA construction kit (Ambion) following the manufacturer's protocol. The following DNA template oligonucleotides were used: forward, 5′-AACTGTGGTTGATTTGTCTGTCCTGTCTC-3′; and reverse, 5′-AAACAGACAAATCAACCACAGCCTGTCTC-3′. Scrambled siRNA was used as a negative control. Transfection of siRNA was performed with the Silencer siRNA transfection kit (Ambion). Transfected cells were incubated for 24 h, and transporter assays were performed as described before. In a similar experiment, RNA was purified from transfected cells and the level of IRIP mRNA was measured using real-time PCR as described above.
RESULTS
Cloning of a new ischemia/reperfusion-inducible gene.
We employed a “targeted differential display” approach in an attempt to identify new genes activated during early ischemia/reperfusion injury. According to our previous observations, a variety of stress-responsive genes important for the development of tissue injury are induced at 2 to 6 h following reperfusion (10). One common feature shared by many of these genes is the presence of an AU-rich element in the 3′-untranslated region of their mRNA (4). A conserved pentanucleotide sequence motif (AUUUA) is present in most AU-rich elements, often in tandem repeats (4). Our differential display protocol was adapted from a previously reported procedure, which was designed to specifically amplify target genes that contain the AUUUA motif (7). The differential display PCR products would correspond to the 3′-UTR of the target genes, from the AUUUA motif to the poly(A) tail (7). RNA samples from sham-operated and I/R mouse kidneys (taken at 6 h postreperfusion) were subjected to differential display analysis. Candidate clones from this analysis were screened using a reverse Northern approach, and positive clones were chosen for further sequence analysis. Among the positive clones were genes for chemokine KC and leukemia inhibitory factor. This result was consistent with our previous finding that the expression of chemokine KC was activated in I/R tissue (10) and confirmed the validity of this approach. Another positive clone contained a fragment of an unknown gene. The reverse Northern analysis result showed that mRNA expression of this unknown gene was robustly activated in the I/R sample.
Full-length cDNA of this gene was cloned using RACE. We named the gene (and its encoded protein) IRIP. IRIP cDNA and predicted protein sequences are shown in Fig. 1. It is noted that IRIP cDNA contains canonical AU-rich elements in the 3′-UTR.
FIG. 1.
Mouse IRIP cDNA and encoded amino acid sequences. The sequence of 1,368-bp IRIP mRNA and the 263-residue amino acid sequence of IRIP are shown. The predicted initiation and termination codons as well as two AUUUA sequence motifs are marked.
cDNA and amino acid sequence analysis of IRIP.
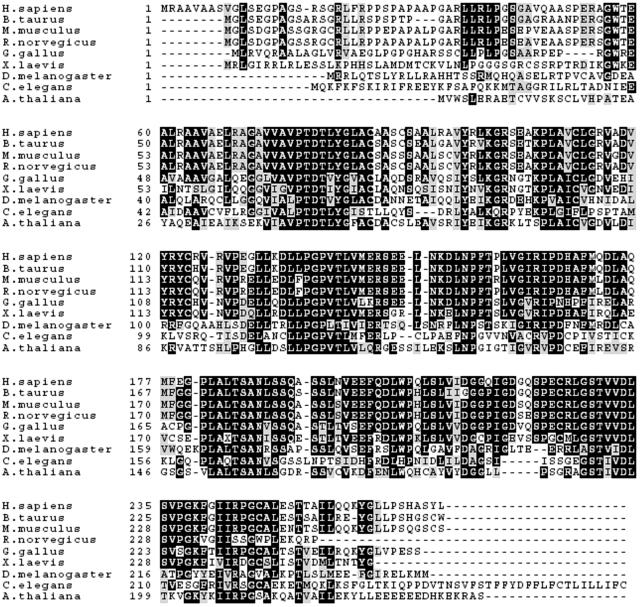
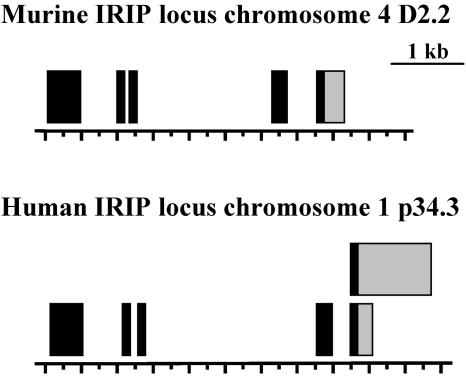
Upon search of the corresponding databases, IRIP homologues have been found in almost every multicellular eukaryotic organism for which genome or expressed sequence tag sequences are available. Functions of any of the homologue proteins are not known. Therefore, IRIP represents an evolutionary conserved, yet unique protein family. As demonstrated in a multiple sequence alignment (Fig. 2), the amino acid sequences of IRIP homologues are highly conserved across species and several such sequence segments are identified, suggesting that IRIP homologues possibly have very similar functions and three-dimensional structures. The structures of mouse and human IRIP genes were analyzed by using gene prediction programs and by comparing cDNA sequences with corresponding genomic sequences. The chromosomal regions of both genes span about 5 kb (Fig. 3). Two cDNA sequences with alternative polyadenylation sites were identified upon analysis of hIRIP transcripts in human cell lines (see next section). The human IRIP gene is located on chromosome 1 and the mouse IRIP gene is located on chromosome 4. The GenBank accession numbers of IRIP cDNA sequences are AY283537 (mouse) and AY286019/AY286020 (human).
FIG. 2.
Multiple sequence alignments of IRIP homologues. The sequences were aligned with the ClustalW program, and the graphical presentation was created with Boxshade. Identical amino acid residues are shaded in black, and similar amino acid residues are in shaded in gray.
FIG. 3.
Schematic representation of intron/exon structures of mouse and human IRIP genes. Exons are shown as black bars, whereas 3′-untranslated regions are shown as gray bars. The human IRIP gene has two transcripts that differ in the length of the 3′-untranslated regions.
Human IRIP has a predicted molecular mass of 28.3 kDa, and mouse IRIP has a molecular mass of 27.8 kDa. IRIP does not contain transmembrane domains and appears to be a compact globular protein (The PredictProtein server; http://cubic.bioc.columbia.edu). Human IRIP sequence was scanned against protein motif databases PROSITE, Pfam, and COG using several web-based programs. Two sequence motifs were identified with a significant P value: a protein kinase ATP-binding signature (PROSITE accession no. PS00107) and SUA5/yciO/yrdC family signature (PROSITE accession no. PDOC00883; COG designation no. COG0009; Pfam designation, pfam01300). The SUA5/yciO/yrdC domain is also found in many prokaryotic proteins of unknown functions. It contains an average of about 180 amino acid residues. The founding members of this protein family include SUA5 (Saccharomyces cerevisiae) and YrdC (E. coli) proteins.
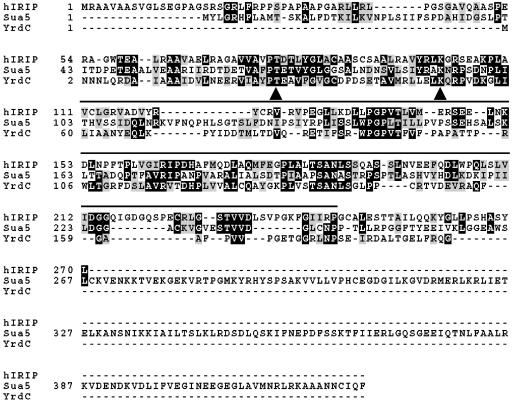
A multiple sequence alignment of human IRIP, SUA5, and YrdC is shown in Fig. 4. The overall sequence similarities between the three proteins are quite modest (approximately 25 to 30% identity with many mismatches and large gaps), which is probably the reason why SUA5 and YrdC were not identified in the initial BLAST database search. However, several of the highly conserved sequence blocks in IRIP homologues are also conserved in SUA5 and YrdC, suggesting that these proteins probably have a common ancestry and may have diverged during evolution. A number of amino acid residues are identical in all these three proteins (Fig. 4).
FIG. 4.
Multiple sequence alignments of hIRIP, SUA5, and YrdC. The alignment file was generated with the ClustalW program, and the graphical presentation was created with Boxshade. Identical amino acid residues are shaded in black, and similar amino acid residues are shaded in gray. Indicated by arrows are two residues (T87 and K110 in hIRIP) that are conserved in all three proteins and located on the surface of YrdC protein in the crystal structure. These two residues were mutated to obtain an IRIP double mutant, T87L/K110I. The SUA5/YrdC/YciO family domain is underlined.
Expression of IRIP mRNA.
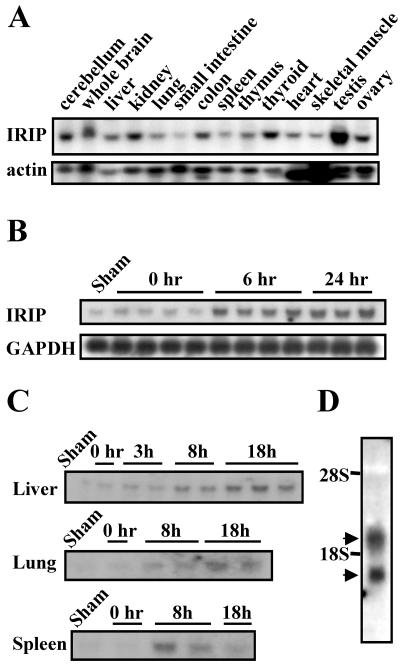
The expression of IRIP mRNA in mouse tissues was examined by Northern blot analysis. A single IRIP transcript was detected with an estimated size of 1.4 kb, which matched the size of cloned cDNA (Fig. 5A). IRIP mRNA was expressed at a low level in the spleen, muscle, heart, and small intestine and at a relatively high level in testis, thyroid, ovary, colon, kidney, and brain. This result is in excellent agreement with microarray data from SymAtlas database (Genomics Institute of the Novartis Research Foundation; http://expression.gnf.org). Testis and thyroid were also the tissues with the highest expression level of human IRIP, according to the SymAtlas database. We identified the IRIP gene as an I/R-inducible gene in a differential display analysis. Therefore, we examined the expression of IRIP mRNA in mouse kidney at different time points after I/R. In normal kidney, IRIP was expressed at a basal level and it was activated 1.7-, 4.2-, and 5.1-fold after ischemia and at 6 and 24 h of reperfusion, respectively (Fig. 5B).
FIG. 5.
Mouse IRIP mRNA expression. Twenty micrograms of total RNA was applied in each lane for the Northern blot analysis. (A) mRNA expression in different mouse tissues. β-Actin mRNA was probed as a loading control. (B) IRIP expression in kidney samples after 32 min of ischemia and 0, 6, or 24 h of reperfusion. GAPDH mRNA was used as a loading control. (C) Effect of endotoxic shock induced by intraperitoneal injection of LPS (16 mg/kg of body weight) on IRIP mRNA expression in liver, lung, and spleen. RNA was purified from tissues at 0, 3, 8, and 18 h after LPS injection. Sham-control animals received only PBS. (D) Two mRNA IRIP species observed in human Wish cells (∼1,300 and 1,900 bases). The Northern blots were hybridized with 32P-labeled RNA (A) or DNA (B to D) probes. Each sample on the blots represents an independently tested animal.
Endotoxemia can induce multiple organ ischemia due to microvascular constriction (9). We decided to characterize expression of IRIP in the mouse model of LPS-induced endotoxemia. After LPS administration (16 mg/kg of body weight), the IRIP mRNA level significantly increased in all tested organs, such as the liver, lungs, and spleen (Fig. 5C). Sustained activation was observed in the liver and lungs up to 18 h and in the spleen up to 8 h. At this moment, the molecular mechanism of IRIP activation in I/R and endotoxemia remains unknown. IRIP was also expressed in mouse fibroblasts (NIH 3T3), proximal tubular and mesangial cells, brain cortical neurons (data not shown), and human HeLa and Wish cells. Interestingly, two IRIP transcripts were observed in human cells, with apparent sizes of 1.9 and 1.2 kb, respectively (Fig. 5D; only Wish cells are shown). Both hIRIP transcripts were cloned and sequenced. They have identical 5′-end sequences, the same open reading frame, and encode the same protein. The longer transcript contain an additional ∼600 bp in the 3′-untranslated region (Fig. 3). Further examination of hIRIP sequence revealed two potential polyadenylation signal sites that correspond to the two transcripts, suggesting that they could be the product of alternative polyadenylation (2).
Determination of IRIP transcriptional start site.
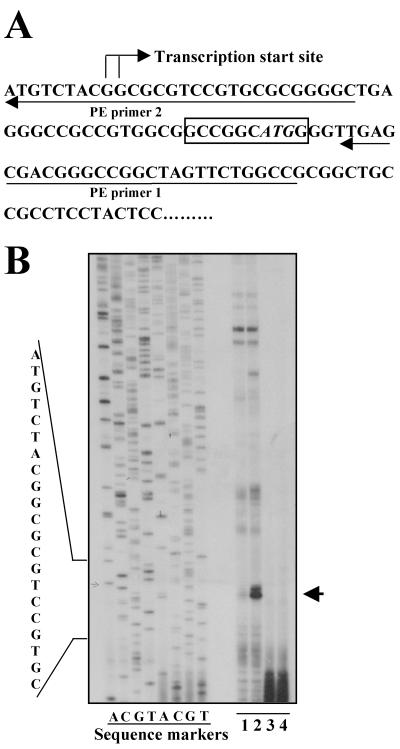
Cloning full-length cDNA with RACE can sometimes be problematic. In the cloned human and mouse IRIP cDNA sequences, the predicted 5′-UTRs form continuous reading frames with the predicted coding regions, which raised the possibility that the predicted 5′-UTR is actually part of the coding region and there is an additional upstream sequence missing in the cloned cDNA. To address this issue, primer extension experiments were performed to determine the transcriptional start site of the mouse IRIP gene. Two oligonucleotide primers were used: one upstream and one downstream of the predicted AUG start codon (Fig. 6). A predominant primer extension product was observed in kidney RNA sample. This primer extension product was inducible by I/R and corresponded to a transcription start site at two adjacent guanine residues upstream of the predicted start codon (Fig. 6). Accordingly, we confirmed that the cloned mouse IRIP cDNA is full length.
FIG. 6.
Identification of the transcription start site of the mouse IRIP gene by primer extension. Primer extension experiments and probe preparation are described in Materials and Methods. Total RNA was isolated from normal or 6-h postreperfusion kidney tissues. (A) DNA sequence around IRIP transcriptional start site. The start site and two primers used for primer extension experiment are indicated. The predicted AUG start codon is shown in italics. (B) A representative primer extension gel. Manual sequencing was performed with an mIRIP cDNA-containing plasmid, and reaction mixtures were loaded as sequence markers. Lane 1, primer 1, normal kidney; lane 2, primer 1, I/R kidney; lane 3, primer 2, normal kidney; lane 4, primer 2, I/R kidney.
Expression of IRIP.
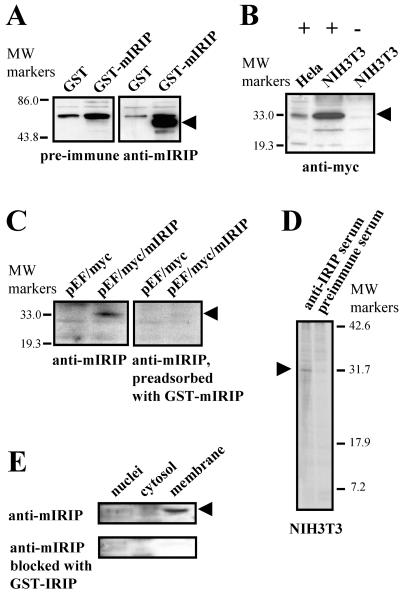
To generate polyclonal anti-IRIP antibodies, we overexpressed and purified GST-mIRIP fusion protein from E. coli. The GST moiety was removed by thrombin digestion, and the digestion mixture was separated on SDS-PAGE. The band corresponding to IRIP was excised from the gel and used as an immunizing antigen. The specificity of anti-IRIP serum was tested by Western blotting. The original GST-mIRIP protein as well as human and mouse myc-tagged proteins in transient and stable transfected HeLa and NIH 3T3 cells (Fig. 7A and B, only transient transfected cells are shown) can be readily detected. Specificity of detection was confirmed by blocking antibodies with GST-mIRIP (Fig. 7C).
FIG. 7.
Generation of anti-IRIP antibiotics and expression of IRIP in cell lines and kidney subcellular fractions. (A) Purified mIRIP protein expressed in E. coli was used for generation of rabbit polyclonal antibodies. The purified GST-mIRIP was detected by antibodies in Western blots. (B) Cellular extracts were prepared from HeLa and NIH 3T3 cells transiently transfected with myc-tagged mIRIP construct (+) or cells transfected with expression vector (−). The recombinant protein was detected only in cells transfected with mIRIP. (C) Specificity of antibodies was also confirmed by testing cellular extracts from myc-tagged mIRIP (pEF/myc/mIRIP)-transfected cells with antibodies which were preadsorbed with GST-mIRIP. After blocking, mIRIP was not detected; its position is shown by an arrowhead. (D) Immunoprecipitation of endogenous IRIP with polyclonal rabbit antibodies. Mouse NIH 3T3 cells were metabolically labeled with [35S]methionine and [35S]cysteine. Total cell extracts were prepared and immunoprecipitated with either preimmune or anti-mIRIP antibodies. The precipitates were resolved on 15% SDS-PAGE gel. The ∼30-kDa bands, visible only in samples immunoprecipitated with the antiserum, are indicated with arrows. (E) Intracellular distribution was tested in subcellular fractions from mouse kidney. Cytosolic, nuclear, and plasma membrane fraction extracts were used for Western blot analysis with anti-mIRIP antibodies. The lowest blot shows that there was no labeling in any of the samples after blocking antibodies with the purified GST-IRIP fusion protein.
To detect endogenous IRIP, NIH 3T3 cells were metabolically labeled with [35S]methionine and [35S]cysteine. Total cell extracts were immunoprecipiated with either preimmune serum or anti-IRIP serum, and proteins bound to beads were resolved by SDS-PAGE (Fig. 7D). The anti-IRIP serum was able to precipitate an ∼31-kDa protein that was absent in the preimmune serum precipitate. The apparent size of this band was slightly higher than the theoretical molecular mass of mIRIP (27.8 kDa), suggesting possible posttranslational modifications. A similar result was observed in a human Wish cell line (data not shown).
The subcellular localization of IRIP was investigated. Mouse kidney tissue was homogenized and fractionated into nuclear, cytosolic, and plasma membrane enriched fractions. The presence of IRIP in each fraction was detected by Western blotting. In kidney, IRIP is predominantly located in the plasma membrane enriched fraction (Fig. 7E). When anti-IRIP serum was preadsorbed with GST-IRIP protein before Western blot analysis, the ∼31-kDa IRIP band disappeared, confirming the specificity of the detection (Fig. 7E, lower panel).
IRIP interacts with RS1 protein in yeast two-hybrid system.
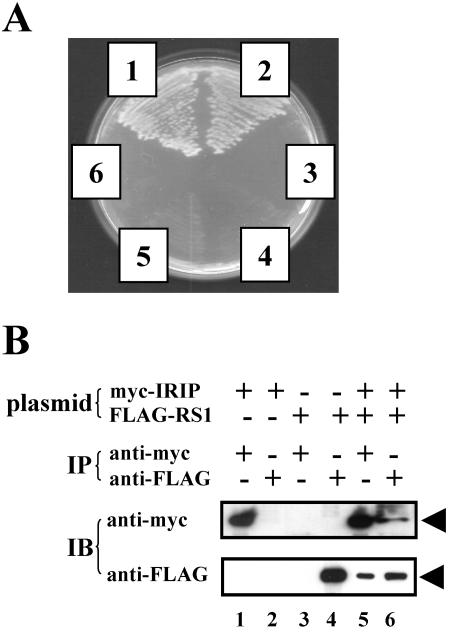
To elucidate the cellular function of IRIP, we performed a yeast two-hybrid screening to identify proteins that interacted with IRIP. After an unsuccessful attempt using a commercially available HeLa cDNA library, we constructed our own library from a source in which the level of IRIP expression is relatively high. Liver tissue of endotoxemic mice was chosen because IRIP expression was significantly induced in this tissue. To eliminate possible adverse effects of the highly positively charged N-terminal sequence of IRIP, a bait plasmid carrying a truncated IRIP in which 34 amino acid residues are deleted from the N terminus, was constructed. Using the truncated bait and the new cDNA library, we obtained positive clones with the His+ Ade+ phenotype and enhanced β-galactosidase activity. One of the positive clones (clone 81) encoded the C-terminal part (Leu347-Thr582) of the RS1 protein, a protein that regulates the sodium-glucose cotransporter (Fig. 8A).
FIG. 8.
Interaction between mouse IRIP and mouse RS1. (A) Interaction of mIRIP and mRS1 in the yeast two-hybrid system. AH109 yeast cells were transformed with the following plasmids: 1, pAD-λcI plus pBD-λcI (positive control); 2, RS1 containing clone plus truncated IRIP bait; 3, RS1 containing clone plus pBD-lamin C (negative control); 4, RS1 containing clone plus empty pBD vector (negative control); 5, RS1 containing clone plus pBD-λcI (negative control); and 6, pAD-λcI plus truncated IRIP bait (negative control). Individual transformant colonies were streaked on an SD− Trp− Leu− His− Ade plate and incubated at 30°C for 4 days. The specificity of protein-protein interactions was also confirmed using the β-galactosidase reporter and X-Gal indicator plates (data not shown). (B) mIRIP interacts with mRS1 in vivo. COS-7 cells were transfected with pEF/myc-mIRIP (myc-IRIP) or pCDNA-FLAG-mRS1 (FLAG-RS1) or both plasmids. Whole-cell extracts were immunoprecipitated (IP) with either anti-myc or anti-FLAG antibody. The presence of IRIP or RS1 protein was detected by Western blot analysis (immunoblotting [IB]) using anti-myc or anti-FLAG antibody, respectively. The arrows indicate the positions of both proteins.
The RS1 gene was originally isolated from an expression cDNA library of porcine kidney cortex by screening the library with a monoclonal antibody that stimulated high-affinity phloridzin (a specific Na+-d-glucose cotransporter 1 [SGLT] inhibitor) binding but did not react with the plasma membrane Na+-glucose cotransporter SGLT1 (27). When expressed in Xenopus oocytes, RS1 changed the activity of human SGLT1 (hSGLT1), Na+-myo-inositol cotransporter, and hOCT2 but not the activity of Na+-γ-aminobutyric acid, Na+-l-glutamate cotransporters, Na+-K+-ATPase, and Na+-independent glucose transporter GLUT1 (20, 27). Porcine RS1 is a hydrophilic 67-kDa protein with a hydrophobic C terminus that is encoded by an intronless gene. Homologous proteins were cloned from rabbit and human (16, 20, 27). RS1 is associated with the inner side of plasma membrane (25); however, other subcellular localizations were also proposed. RS1 is currently assumed to be a modulator of membrane transport. The molecular mechanism of its actions is not clear, and according to the latest data, it might be involved in transporter trafficking (28).
IRIP interacts with RS1 in vivo.
To investigate the interaction between RS1 and IRIP in mammalian cells, we constructed recombinant plasmids for expression of both full-length proteins with N-terminal tags (pcDNA3.1-FLAG-mRS1 and pEF/myc/cyto-IRIP, which expressed FLAG-tagged RS1 and myc-tagged IRIP, respectively). COS-7 cells were transfected with individual or both plasmids. Whole-cell extracts were immunoprecipitated with either anti-myc or anti-FLAG antibody. The presence of myc-IRIP or FLAG-RS1 in the precipitates was detected in Western blotting with corresponding anti-Tag antibodies. As shown in Fig. 8B, myc-IRIP protein can be detected in anti-FLAG precipitate from cells cotransfected with pEF/myc-mIRIP and pcDNA-FLAG-mRS1 plasmids (upper panel, lane 6). Similarly, FLAG-RS1 protein is present in anti-myc precipitate from the same extract (lower panel, lane 5). As a control, anti-myc antibody did not pull down FLAG-RS1 protein in the absence of myc-IRIP expression (lower panel, compare lane 3 with lane 5) and anti-FLAG antibody did not pull down myc-IRIP protein in the absence of FLAG-RS1 expression (upper panel, compare lane 2 with lane 6). These results indicate that full-length IRIP and RS1 associate with each other when expressed in mammalian cells.
IRIP negatively modulates transporter activities.
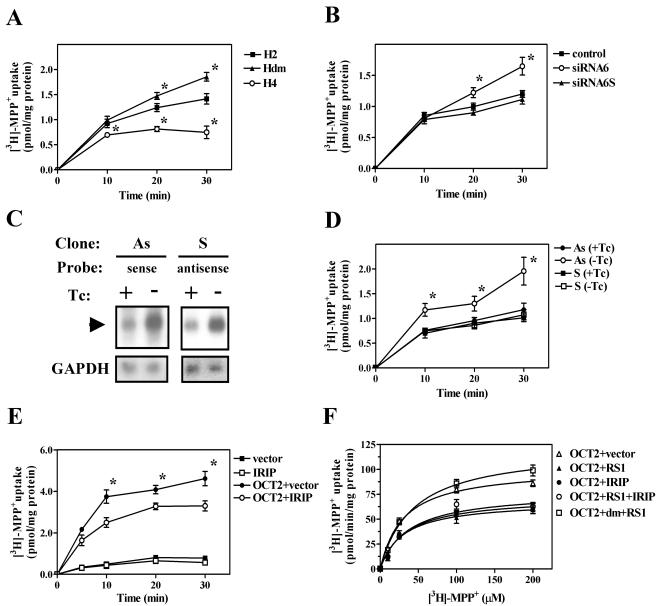
Yeast two-hybrid and coimmunoprecipitation experiments showed that IRIP was able to associate with RS1. RS1 is a protein that regulates the activity of a number of plasma membrane transporters (27, 28). Because of the important physiological function and medical implication of these transporters, we decided to test whether changes in the level of IRIP expression can influence transporter activity. First, we compared endogenous transport activity of MPP+ (a known substrate for organic cation and monoamine transporters) in HeLa cell lines that were stably transfected with hIRIP expression plasmid (HeLa-4) or corresponding vector alone (HeLa-2), respectively. Normal HeLa cells have an efficient MPP+ uptake activity, mostly mediated by organic cation transporters (31). Cells overexpressing hIRIP displayed decreased ability for MPP+ transport (Fig. 9A). Similar results were obtained for other stably transfected cell lines overexpressing hIRIP (data not shown). Further experiments were performed in HeLa-2 and HeLa-4 cell lines. Moreover, cell line HeLa-dm that expresses a mutant hIRIP (double point mutation T87L and K110I) displayed enhanced MPP+ transport. HeLa-4 and HeLa-dm cell lines expressed hIRIP at similar levels, suggesting the dominant-negative effect of mutant hIRIP was due to the introduced mutations (data not shown). T87 and K110 were mutated because these residues are highly conserved among IRIP homologues. In the crystal structure of YrdC protein, the corresponding threonine and lysine residues are located in a conserved patch on the protein surface (23). We hypothesize that T87 and K110 are also located on the surface of IRIP and are essential for IRIP function.
FIG. 9.
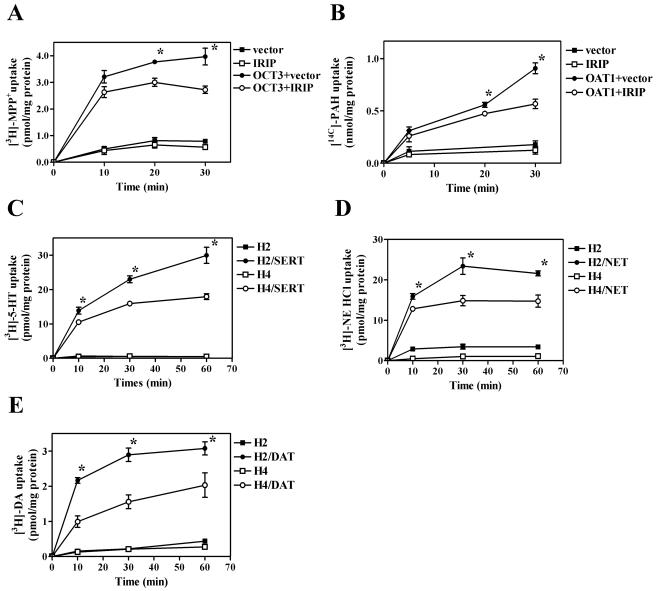
Effect of IRIP expression on endogenous and OCT2-mediated uptake activity. (A) Time course of [3H]MPP+ uptake in HeLa cell lines, stably transfected with vector (H2) or pEF/myc-hIRIP (H4). The Hdm HeLa cell line expressed a double mutant of hIRIP (T87L/K110I). (B) Time course of [3H]MPP+ uptake in HeLa cells, transfected with anti-IRIP siRNA (siRNA6) or the scrambled siRNA6s or treated with transfection reagent alone (control). (C) Effect of tetracycline (Tc) withdrawal on the expression of the sense (S) and antisense (As) hIRIP RNA in stable transfected cell lines S and As. Some background levels of corresponding S and As RNA expression was observed in both cell lines in the presence of Tc, likely due to leaky promoter activity, whereas the levels of expression of these RNAs were induced four to five times in the absence of the antibiotic in the culture medium. (D) Time course of [3H]MPP+ uptake in stable HeLa cell lines expressing sense or antisense hIRIP RNA. For induction of expression, cells were transferred from the medium with Tc (+Tc) into the medium without antibiotic (−Tc). (E) Time course of [3H]MPP+ uptake in 293T cells transfected with hOCT2 along with vector or pEF/myc-hIRIP. Background uptake was also measured in the absence of exogenous OCT2. (F) Concentration dependence of hOCT2-mediated uptake of [3H]MPP+ in 293T cells. Uptake rates were determined for a period of 8 min in the presence of overexpressed hRS1, hIRIP, hRS1 plus hIRIP, and RS1 plus double-mutant hIRIP (dm). OCT2-mediated uptake was calculated as the difference between the uptake of OCT2-transfected cells and the uptake of cells transfected with the vector alone. The curves were obtained by fitting the Michaelis-Menten equation to the data using GraphPad Prizm software (GraphPad Sotware, Inc.). Values of Km and Vmax are presented in Table 1. For all the curves in Fig. 9, mean ± standard error values are presented. Three to four independent experiments are presented in each graph. *, P < 0.05 in comparison to cell lines transfected with vector (A and E), transfection reagent alone (B), or cells incubated in the presence of Tc (D).
Next, we used siRNA approach to inhibit IRIP expression in HeLa cells. We were able to achieve about 60% inhibition of IRIP mRNA expression, as measured by quantitative real-time RT-PCR. In HeLa cells transfected with the hIRIP siRNA6, MPP+ transport was not different from mock-transfected cells during the first 10 min of the assay, but was significantly enhanced after 20 min. The scrambled siRNA6s had no effect on MPP+ transport (Fig. 9B). We also used stable transfected NIH 3T3 cell lines expressing an antisense IRIP RNA or the same fragment in a sense orientation in a tetracycline-dependent manner. In both cell lines (As and S) RNA transcripts were inducible by the withdrawal of tetracycline from the culture medium (Fig. 9C). Similar to the effect of siRNA, cells expressing antisense transcript demonstrated increased transport of MPP+ (Fig. 9D). These results indicate that inhibition of endogenous IRIP expression leads to enhanced MPP+ uptake activity.
We then examined the effect of IRIP overexpression on transport mediated by individual transporters. HEK-293T cells were cotransfected with hOCT2 expression plasmid and either pEF/myc or pEF/myc-hIRIP. When IRIP was coexpressed, hOCT2-mediated MPP+ transport was significantly decreased (Fig. 9E). To test the effect of IRIP overexpression on the kinetic properties of OCT2 transporter, we compared the substrate concentration dependence of the hOCT2-mediated uptake. HEK-293T cells were cotransfected with hOCT2 plasmid and either vector, hRS1, or hIRIP plasmid. The results of this experiment are shown in Fig. 9F and Table 1. The expression of either RS1 or IRIP significantly decreased the maximal velocity of substrate transport (Vmax) but had no effect on the substrate affinity of OCT2 transporter (Km). No additive or synergistic interaction between effects of IRIP and RS1 overexpression on OCT2 transporter activity was observed upon cotransfection of both proteins into 293T cells (see Fig. 9F and Table 1). This result suggests that both proteins may regulate transporter activity through a similar pathway. To test if presence of functional IRIP is necessary for the RS1 inhibitory effect, 293T cells were transfected with hRS1 and inactive T87L-K110I double-mutant myc-hIRIP (Fig. 9F). Interference with the activity of endogenous hIRIP reversed inhibition of OCT2-mediated transport by hRS1. These data support the biological significance of functional interaction between RS1 and IRIP in regulation of the OCT2 transporter.
TABLE 1.
Kinetics of [3H]MPP+ uptake in 293T cells following transfection with expression plasmidsa
| Transfected plasmid(s) | Km (μM) | Vmax (pmol/min/mg of protein)b |
|---|---|---|
| OCT2 + vector | 30.43 ± 4.11 | 101.40 ± 3.73 |
| OCT2 + RS1 | 29.76 ± 7.50 | 68.15 ± 4.90* |
| OCT2 + IRIP | 32.88 ± 7.18 | 72.67 ± 4.92* |
| OCT2 + RS1 + IRIP | 36.54 ± 6.21 | 77.60 ± 4.35* |
Cells were transfected with plasmids expressing corresponding proteins and assayed for [3H]MPP+ transport as described in Materials and Methods. Km and Vmax values for [3H]MPP+ uptake are the mean ± standard error of the mean of three independent experiments, each performed in triplicate. Km and Vmax values were obtained by nonlinear least-squares regression analysis.
*, P < 0.05, two-tailed Student's t test.
Similar inhibitory effects of IRIP on transporter activity were also observed for transfected OCT3 and OAT1 transporters, though the effect on OAT1-mediated transport was less dramatic at early time points (Fig. 10 A and B). One of the physiological functions of OCT transporters is to mediate monoamine transport in extraneuronal cells, whereas specific monoamine transporters functionally related to OCTs mediated monoamine transport in neuronal cells. Therefore, we examined whether hIRIP would also affect the activity of specific monoamine transporters. In this context, we tested closely related transporters hDAT, hNET, and hSERT. We used stable cell lines HeLa-2 and HeLa-4 that were transiently transfected with empty vector or plasmids expressing individual transporters; transport was measured using individual substrates. In HeLa-4 cells that overexpressed IRIP, transport of all three monoamine substrates was significantly inhibited in comparison to the transport in the control HeLa-2 cell line (Fig. 10C, D, and E). In summary, IRIP can negatively modulate the activities of organic cation transporters (OCT2 and OCT3), organic anion transporter OAT1, and monoamine transporters (DAT, NET, and SERT).
FIG. 10.
Time course of MPP+ and monoamine uptake in HeLa and 293 T cells expressing IRIP. 293T cells were transfected with hOCT3 (A) or mOAT1 (B) transporters in the presence of either vector pEF/myc-hIRIP or pEF/myc-mIRIP. To evaluate the effect of IRIP overexpression on the uptake activity of neuronal monoamine transporters, HeLa-2 (H2) and HeLa-4 (H4) cells were transfected with hSERT (C), hNET (D), or hDAT (E). Transport activity was measured after adding labeled substrates for different time periods. For all curves, means ± standard errors are presented. Three to four independent experiments are presented in each graph. *, P < 0.05 in comparison to cell lines transfected with transporter and vector plasmid.
DISCUSSION
We report here the cloning and characterization of a new ischemia/reperfusion-inducible gene, coding for IRIP. Homologues of IRIP are found in almost all higher eukaryotic organisms, including vertebrates, insects, nematodes, and plants. IRIP homologues are highly conserved in amino acid sequences, suggesting that they share a common function. As evident from the multiple amino acid sequence alignment, several internal regions are almost identical in these proteins, whereas the N-terminal regions are less conserved (Fig. 2). Notably, IRIP shows moderate sequence similarity with members of the SUA5/YrdC/YciO protein family and most likely represents the mammalian member of this family.
The crystal structures of several prokaryotic members of this protein family have been determined, including YrdC, Tf1, and YciO proteins from E. coli and Mth1692 from Methanobacterium thermoautotrophicum. However, their functions are still unknown. YrdC protein adopts a novel fold that can be classified as an α/β twisted open-sheet structure (23). One prominent feature of this structure is a large concave surface on one side of the protein with a strong positive electrostatic potential. Based on the overall shape and the positive electrostatic potential of the depression, the authors proposed that YrdC may be an RNA-binding protein and purified YrdC was shown to bind synthetic double-stranded RNA and tRNA. The predicted secondary structure topology of human and mouse IRIP is very similar to that of YrdC, suggesting that IRIP may adopt a similar three-dimensional fold. Most of the amino acid residues conserved among IRIP, SUA5, and YrdC are located on the surface of the YrdC structure. Two of these residues (T27 and K50) are highlighted with arrows in Fig. 4, and based on the degree of conservation and the positions on the protein surface, they are considered to be important for YrdC function. The corresponding residues in hIRIP (T87 and K110) were mutated to leucine (T87L) and isoleucine (K110I), respectively, by site-directed mutagenesis. This double-mutant of hIRIP showed a loss-of-function or even a dominant-negative phenotype in modulating the MPP+ uptake activity.
Mouse IRIP mRNA is expressed in all tested tissues, with the highest expression in testis. A very similar profile of mRNA expression is reported for human tissues by the SymAtlas database (http://expression.gnf.org). Secretory and endocrine organs (adrenal gland, thyroid, salivary gland) and epithelia (medial olfactory epithelium in mice and bronchioepithelial cells in humans) express significantly higher levels of IRIP mRNA than other tissues. IRIP mRNA expression is induced by ischemia/reperfusion and endotoxemia, and the biological significance of this response is currently under investigation. While this study was under way, human IRIP was reported to be an interactive partner of the retinoblastoma binding protein RBBP10 in a yeast two-hybrid screening. In this report, however, the interaction between IRIP and RBBP10 was not confirmed by an alternative approach, nor were any experimental data presented to indicate the biological activity of IRIP (5).
Our yeast two-hybrid screening identified RS1 as an interactive partner of IRIP. The interaction between exogenously expressed RS1 and IRIP was confirmed using a coimmunoprecipitation assay in COS-7 cells. RS1 was originally discovered as a protein that is able to modulate the activity of Na+-glucose cotransporter SGLT1 in Xenopus oocytes. Based on these observations, the authors initially proposed that RS1 was a regulatory subunit of SGLT1. Later, it was found that coexpression of human RS1 in Xenopus laevis oocytes inhibited the activity of not only human SGLT1 but also that of human organic cation transporter OCT2 (20). Therefore, it appeared that the regulatory function of RS1 was not specific for SGLT1, but probably for a number of related transporters. The effects of RS1 were later confirmed by cell culture studies. It was also reported that the mRNA level of OCT2 was decreased in cells overexpressing RS1 (14), which can explain, at least in part, the inhibitory effect of RS1 on transporter-mediated substrate uptake. It is not clear how the plasma membrane-associated RS1 protein could regulate transcription, though it was recently reported that RS1 protein can migrate into the nucleus (28).
Like the RS1 protein, IRIP also plays an inhibitory role in regulating transporter activities. Overexpression of IRIP leads to the inhibition of endogenous as well as exogenously expressed transporter uptake activity. Among these affected transporters are hOCT2, hOCT3, mOAT1, hDAT, hNET, and hSERT. We observed some species specificity of this effect. For example, hIRIP was not able to modulate rat SERT uptake activity (data not shown). The basis for such specificity is presently not known. In addition, downregulation of endogenous IRIP expression leads to increased transporter uptake activity, further supporting the inhibitory function of IRIP. Interestingly, the level of RS1 mRNA was also upregulated in mouse kidney after ischemia/reperfusion, whereas mRNA levels of SGLT1, OCT1, and OCT3 transporters were decreased (data not shown). At this moment, we do not know the molecular mechanism of how IRIP or RS1 inhibits transporter activity. Both IRIP and RS1 decrease the Vmax of OCT2-mediated MPP+ transport, with no effect on Km, suggesting that IRIP or RS1 does not affect the intrinsic catalytic activity of transporters (28). Consistent with this notion, it was recently reported that RS1 can influence dynamin-dependent trafficking of intracellular vesicles containing hSGLT1 and protein kinase C-dependent regulation of the transporter activity (28).
It becomes evident that transporters undergo very complex regulation in normal and disease conditions, which include control of protein synthesis, posttranslational modifications, assembly, oligomerization, cell surface delivery, and endocytic retrieval/recycling. Our understanding of these regulatory mechanisms is still very limited. Based on the evidence that IRIP and RS1 physically interact with each other and they both negatively regulate the activity of a variety of transporters, we propose that IRIP and RS1 are part of a common regulatory pathway controlling transporter activities. The regulatory mechanism and other components of this pathway clearly warrant further investigation. It also remains to be determined what other classes of transporters are subjected to such regulation.
During the preparation of this article, RS1-knockout mice were reported (26). RS1-null mice were obese, with 30% more body weight, 80% more body fat, and 30% more serum cholesterol than wild-type mice. The protein level of Na+-glucose cotransporter SGLT1 was upregulated sevenfold in the small intestine of the RS1-null mice, but was unchanged in kidney. Given that IRIP is expressed in a wide variety of tissues, such as thyroid, testis, ovary, colon, kidney, and cerebellum, it is tempting to speculate that ablation of the IRIP gene in vivo will result in changes in key transporter activities and profound physiological phenotypes.
Acknowledgments
We thank You G. for advice. We thank K. Reuhl and S. Phadtare for critical reading of the manuscript.
REFERENCES
- 1.Anaya-Prado, R., L. H. Toledo-Pereyra, A. B. Lentsch, and P. A. Ward. 2002. Ischemia/reperfusion injury. J. Surg. Res. 105:248-258. [DOI] [PubMed] [Google Scholar]
- 2.Beaudoing, E., S. Freier, J. R. Wyatt, J. M. Claverie, and D. Gautheret. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, E., S. E. Schellhorn, J. M. Zabolotny, S. Martin, E. Tozzo, O. D. Peroni, K. L. Houseknecht, A. Mundt, D. E. James, and B. B. Kahn. 2004. GLUT4 overexpression or deficiency in adipocytes of transgenic mice alters the composition of GLUT4 vesicles and the subcellular localization of GLUT4 and insulin-responsive aminopeptidase. J. Biol. Chem. 279:21598-21605. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., C. Ji, S. Gu, E. Zhao, J. Dai, L. Huang, J. Qian, K. Ying, Y. Xie, and Y. Mao. 2003. Isolation and identification of a novel cDNA that encodes human yrdC protein. J. Hum. Genet. 48:164-169. [DOI] [PubMed] [Google Scholar]
- 6.Datta, R., P. Choudhury, M. Bhattacharya, F. Soto Leon, Y. Zhou, and B. Datta. 2001. Protection of translation initiation factor eIF2 phosphorylation correlates with eIF2-associated glycoprotein p67 levels and requires the lysine-rich domain I of p67. Biochimie 83:919-931. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez, O., Y. Ashhab, L. Sabater, E. Belloso, P. Caro, and R. Pujol-Borrell. 1998. Cloning of ARE-containing genes by AU-motif-directed display. Genomics 54:278-286. [DOI] [PubMed] [Google Scholar]
- 8.Gossen, M., S. Freundlieb, G. Bender, G. Muller, W. Hillen, and H. Bujard. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766-1769. [DOI] [PubMed] [Google Scholar]
- 9.Holmes, C. L., and K. R. Walley. 2003. The evaluation and management of shock. Clin. Chest Med. 24:775-789. [DOI] [PubMed] [Google Scholar]
- 10.Ishibashi, N., M. Weisbrot-Lefkowitz, K. Reuhl, M. Inouye, and O. Mirochnitchenko. 1999. Modulation of chemokine expression during ischemia/reperfusion in transgenic mice overproducing human glutathione peroxidases. J. Immunol. 163:5666-5677. [PubMed] [Google Scholar]
- 11.Jennings, R. B., and K. A. Reimer. 1991. The cell biology of acute myocardial ischemia. Annu. Rev. Med. 42:225-246. [DOI] [PubMed] [Google Scholar]
- 12.Jung, J. S., R. H. Lee, S. H. Koh, and Y. K. Kim. 2000. Changes in expression of sodium cotransporters and aquaporin-2 during ischemia-reperfusion injury in rabbit kidney. Renal Fail. 22:407-421. [DOI] [PubMed] [Google Scholar]
- 13.Koepsell, H., B. M. Schmitt, and V. Gorboulev. 2003. Organic cation transporters. Rev. Physiol. Biochem. Pharmacol. 150:36-90. [DOI] [PubMed] [Google Scholar]
- 14.Korn, T., T. Kuhlkamp, C. Track, I. Schatz, K. Baumgarten, V. Gorboulev, and H. Koepsell. 2001. The plasma membrane-associated protein RS1 decreases transcription of the transporter SGLT1 in confluent LLC-PK1 cells. J. Biol. Chem. 276:45330-45340. [DOI] [PubMed] [Google Scholar]
- 15.Kowaltowski, A. J., and A. E. Vercesi. 1999. Mitochondrial damage induced by conditions of oxidative stress. Free Radic. Biol. Med. 26:463-471. [DOI] [PubMed] [Google Scholar]
- 16.Lambotte, S., M. Veyhl, M. Kohler, A. I. Morrison-Shetlar, R. K. Kinne, M. Schmid, and H. Koepsell. 1996. The human gene of a protein that modifies Na(+)-D-glucose co-transport. DNA Cell Biol. 15:769-777. [DOI] [PubMed] [Google Scholar]
- 17.Li, L. B., N. Chen, S. Ramamoorthy, L. Chi, X. N. Cui, L. C. Wang, and M. E. Reith. 2004. The role of N-glycosylation in function and surface trafficking of the human dopamine transporter. J. Biol. Chem. 279:21012-21020. [DOI] [PubMed] [Google Scholar]
- 18.Melikian, H. E. 2004. Neurotransmitter transporter trafficking: endocytosis, recycling, and regulation. Pharmacol. Ther. 104:17-27. [DOI] [PubMed] [Google Scholar]
- 19.Ozaslan, D., S. Wang, B. A. Ahmed, A. M. Kocabas, J. C. McCastlain, A. Bene, and F. Kilic. 2003. Glycosyl modification facilitates homo- and hetero-oligomerization of the serotonin transporter. A specific role for sialic acid residues. J. Biol. Chem. 278:43991-44000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Reinhardt, J., M. Veyhl, K. Wagner, S. Gambaryan, C. Dekel, A. Akhoundova, T. Korn, and H. Koepsell. 1999. Cloning and characterization of the transport modifier RS1 from rabbit which was previously assumed to be specific for Na+-D-glucose cotransport. Biochim. Biophys. Acta 1417:131-143. [DOI] [PubMed] [Google Scholar]
- 21.Schomig, A., and G. Richardt. 1990. The role of catecholamines in ischemia. J. Cardiovasc. Pharmacol. 16(Suppl. 5):S105-S112. [PubMed] [Google Scholar]
- 22.Seal, R. P., and S. G. Amara. 1999. Excitatory amino acid transporters: a family in flux. Annu. Rev. Pharmacol. Toxicol. 39:431-456. [DOI] [PubMed] [Google Scholar]
- 23.Teplova, M., V. Tereshko, R. Sanishvili, A. Joachimiak, T. Bushueva, W. F. Anderson, and M. Egli. 2000. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci. 9:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres, G. E., R. R. Gainetdinov, and M. G. Caron. 2003. Plasma membrane monoamine transporters: structure, regulation and function. Nat. Rev. Neurosci. 4:13-25. [DOI] [PubMed] [Google Scholar]
- 25.Valentin, M., T. Kuhlkamp, K. Wagner, G. Krohne, P. Arndt, K. Baumgarten, W. Weber, A. Segal, M. Veyhl, and H. Koepsell. 2000. The transport modifier RS1 is localized at the inner side of the plasma membrane and changes membrane capacitance. Biochim. Biophys. Acta 1468:367-380. [DOI] [PubMed] [Google Scholar]
- 26.Vaughan, R. A. 2004. Phosphorylation and regulation of psychostimulant-sensitive neurotransmitter transporters. J. Pharmacol. Exp. Ther. 310:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Veyhl, M., J. Spangenberg, B. Puschel, R. Poppe, C. Dekel, G. Fritzsch, W. Haase, and H. Koepsell. 1993. Cloning of a membrane-associated protein which modifies activity and properties of the Na(+)-D-glucose cotransporter. J. Biol. Chem. 268:25041-25053. [PubMed] [Google Scholar]
- 28.Veyhl, M., C. A. Wagner, V. Gorboulev, B. M. Schmitt, F. Lang, and H. Koepsell. 2003. Downregulation of the Na(+)-D-glucose cotransporter SGLT1 by protein RS1 (RSC1A1) is dependent on dynamin and protein kinase C. J. Membr. Biol 196:71-81. [DOI] [PubMed] [Google Scholar]
- 29.Warner, D. S., H. Sheng, and I. Batinic-Haberle. 2004. Oxidants, antioxidants and the ischemic brain. J. Exp. Biol. 207:3221-3231. [DOI] [PubMed] [Google Scholar]
- 30.Weisbrot-Lefkowitz, M. 1997. The use of transgenic mice to evaluate the role of overexpression of human antioxidants in chemically and physiologically induced models of oxidative stress. Ph.D. thesis. Graduate School of Biomedical Sciences, University of Medicine and Dentistry of New Jersey, Piscataway.
- 31.Wu, X., R. Kekuda, W. Huang, Y. J. Fei, F. H. Leibach, J. Chen, S. J. Conway, and V. Ganapathy. 1998. Identity of the organic cation transporter OCT3 as the extraneuronal monoamine transporter (uptake2) and evidence for the expression of the transporter in the brain. J. Biol. Chem. 273:32776-32786. [DOI] [PubMed] [Google Scholar]
- 32.You, G., K. Kuze, R. A. Kohanski, K. Amsler, and S. Henderson. 2000. Regulation of mOAT-mediated organic anion transport by okadaic acid and protein kinase C in LLC-PK(1) cells. J. Biol. Chem. 275:10278-10284. [DOI] [PubMed] [Google Scholar]