Abstract
The putative ATPase chromatin-remodeling machine SRCAP was identified in a yeast two-hybrid protein screen by interaction with the histone acetylase CBP. SRCAP is implicated in the transcriptional coactivation of cyclic AMP- and steroid-dependent promoters, but no natural chromosomal targets for SRCAP regulation have been identified. DOM is the unique SRCAP homolog in Drosophila melanogaster. The goal of this study was to test whether SRCAP is a functional homolog of DOM and to identify potential activities and targets of SRCAP in vivo. We show that human SRCAP complements recessive domino mutant phenotypes. This rescue depends on an intact ATPase homology domain. SRCAP colocalizes extensively with DOM on Drosophila polytene chromosomes and is recruited to sites of active transcription, such as steroid-regulated loci, but not to activated heat shock loci. We show that SRCAP recruits Drosophila CBP to ectopic chromosomal sites, providing the first evidence to suggest that SRCAP and CBP interact directly or indirectly on chromosomes. We show that DOM is a Notch pathway activator in Drosophila and that wild-type SRCAP—but not an ATPase domain mutant—can substitute for DOM in Notch-dependent wing development. We show that SRCAP potentiates Notch-dependent gene activation in HeLa cells. Taken together, these data implicate SRCAP and DOM in developmental gene activation.
Most regulated gene expression in eukaryotes is the result of transcriptional regulation. Current models of transcriptional regulation invoke the targeted recruitment of enzymes that make covalent changes in histone and/or DNA structure, in ATPases that remodel histone-DNA interactions, and, in cases of gene activation, in RNA polymerase itself. A major challenge is to identify the diverse roles of chromatin modifiers in transcriptional regulation.
SRCAP (SNF2-related CBP-activating protein) shares homology with members of the SNF2/SWI2 class of chromatin-remodeling enzymes (20). SRCAP was originally identified in a yeast two-hybrid protein screen for proteins that interact with the histone acetyltransferase CBP (CREB-binding protein). Subsequent studies using transiently transfected reporters have implicated SRCAP as a coactivator for CREB- and nuclear hormone receptor-mediated transcriptional activation (30, 31). The mechanism by which SRCAP activates transcription has not been completely elucidated. Results from a number of studies, including mammalian two-hybrid and coimmunoprecipitation experiments, indicate SRCAP may function in part by recruitment of CBP or other coactivators, such as the arginine methyltransferase CARM-1 and the nuclear hormone receptor coactivator GRIP-1 (20, 31). A role for the conserved ATPase/helicase homology domain in regulation of transcription has not been established, since this activity is dispensable for SRCAP activity in transient transfection assays, nor have the natural chromosomal promoters that utilize SRCAP to regulate transcription been identified.
In mammals, the protein most homologous to SRCAP is p400. p400 was identified through its interactions with the adenoviral protein E1A and appears to play a critical role in E1A-mediated transformation (16). A role of p400 in the activation of transcription mediated by myeloid zinc finger protein (MZF-2A) has also been reported (34). Both SRCAP and p400 have been isolated as parts of large multiprotein complexes implicated in the remodeling of chromatin, including the DMAP1 complex (11), the TRRAP/TIP60 histone acetylase complex (4), and the NCoR-1 histone deacetylase complex (46). The yeast SWR1 complex (29), which catalyzes histone variant exchange in vitro, contains several subunits homologous to subunits of a recently identified SRCAP complex (5).
SRCAP-related proteins have been described for humans (20), rats (accession number XP_341933), fish (accession number CAG00637), Drosophila melanogaster (39), worms (8), the protozoan Toxoplasma gondii (43), and yeast (21), suggesting that SRCAP represents an ancient member of the conserved SWI2/SNF2 family of ATPase/helicase proteins. In Drosophila, the gene domino encodes the sole homolog of SRCAP and p400, which is termed DOM. Similar to SRCAP and p400, DOM can be isolated as part of a TRRAP/TIP60 histone acetylase complex (26). domino is essential for organismal viability and has been implicated in several aspects of fly development, including hematopoeisis, wing development, and female fertility (39). Clones of domino mutant cells cannot be recovered in the adult wing, suggesting that domino is essential for cell viability. Genetic interactions with mutations in several Polycomb group genes suggest that domino may play a role in homeotic gene silencing. domino alleles also suppress heterochromatic position effect variegation, suggesting a role in heterochromatin-mediated silencing as well.
Here we identify SRCAP as a homolog of DOM and provide genetic evidence that SRCAP is at least partially functionally equivalent to DOM. We show that human SRCAP can complement the recessive female sterility of hypomorphic domino alleles and that this rescue depends on an intact ATPase homology domain. We find that SRCAP binds to chromosomes, colocalizes extensively with DOM, and is recruited to some sites of active transcription, such as steroid (ecdysone)-regulated loci, but not to activated heat shock loci. We show that SRCAP can recruit Drosophila CBP to ectopic chromosomal sites, providing the first in vivo evidence to suggest that SRCAP and CBP interact directly or indirectly on chromosomes. We also show that domino is required for proper Notch pathway activity, and that wild-type SRCAP—but not a SRCAP ATPase domain mutant—can substitute for DOM in Notch-dependent wing development. Thus, SRCAP is implicated in Notch signaling. We test this implication using a Notch-dependent reporter in mammalian cells and show that SRCAP is a potentiator of Notch-dependent gene activation. Taken together, these data implicate SRCAP and its Drosophila ortholog, DOM, in developmental gene activation pathways.
MATERIALS AND METHODS
Fly stocks.
domino mutant stocks were a gift of Marie Meister. The various alleles used are described in the work of Ruhf et al. (39). Stocks carrying both domino alleles and transgene inserts on the third chromosome were balanced using the compound balancer T(2;3)CyOGFP-TM3GFP, Cy Ser GFP described by Rudolf et al. (38). Transgenic flies were made essentially as described previously (41), using embryos from a Df(1)w, y1 w67c23 stock for injection.
Transgene synthesis.
The pSRCAP 1-2971 and the pSRCAP K649R plasmids previously described (20, 30) were restriction digested with NotI and BamHI and subcloned into pBlueBac 4.5 (Invitrogen). The resulting plasmid was digested with NotI, allowing the excision of the SRCAP cDNAs containing an in-frame C-terminal six-histidine tag. This cDNA fragment was subcloned into a NotI site of the Drosophila expression plasmid pCasper-Chip, which places SRCAP under the control of the promoter of the chip gene (32). The pCasper-Chip plasmid was a gift of Dale Dorsett at Saint Louis University Medical School.
Immunolocalization of proteins on fixed polytene chromosomes.
For heat-shocked chromosomes, larvae were placed on prewarmed moist paper towels at 37°C for 20 min, and then stored on ice until dissection. Dissection, fixation, and staining of larval polytene chromosome squashes were done essentially as described in the work of Lis et al. (28). Antibodies were used at the following dilutions: rat anti-DomB (39) (gift of M. Meister), 1:330; fluorescein isothiocyanate goat anti-rat (Santa Cruz), 1:200; rabbit anti-six-histidine (Santa Cruz), 1:200; Cy3 goat anti-rabbit (Santa Cruz), 1:200; mouse anti-Pol IIo (H14; Covance), 1:200; Alexa Fluor 488 goat anti-mouse (μ chain; Molecular Probes), 1:200; chicken anti-Drosophila-CBP (gift of S. Smolik), 1:500; Alexa Fluor 488 goat anti-chicken (Molecular Probes), 1:200.
Wing phenotype quantitation.
Crosses were done at 25°C in uncrowded vials. Adults used for wing quantitation were harvested within the first 5 days after the first adults eclosed. Wings were mounted in Permount (Sigma) on glass slides. Digital photographs were made using a compound microscope, and measurements were made using Northern Eclipse software. For each wing, the shortest line that could be drawn from margin to margin that bisected the fourth longitudinal vein at the midpoint between the anterior and posterior cross vein was used.
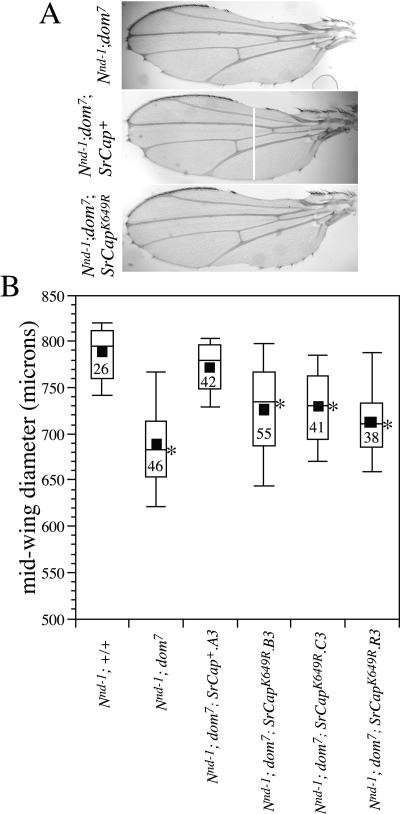
For statistical analysis of the different genotypes (see Fig. 5), we did one-way analysis of variance. The F value is 24.7, which is significant at a level of <0.001. Post hoc test results (Bonferroni) showed that the effects of the dom7 allele alone or in the presence of the ATPase mutant transgene are significantly different (at the 0.01 level) from those for flies carrying either a wild-type dom allele or dom7 together with a wild-type SRCAP transgene.
FIG. 5.
SRCAP functionally complements DOM in Notch signaling. (A) Examples of wings showing the extent of wing margin nicking observed in Nnd-1; dom7 flies (top panel), Nnd-1; dom7; SRCAP+ flies (middle panel), and Nnd-1; dom7; SRCAPK649R flies (bottom panel). The bar across the wing in the middle panel indicates the relative position of the midwing measurement line used in panel B (see Materials and Methods). (B) Box plot chart of wing measurements of Nnd-1; dom7, Nnd-1; dom7; SRCAP+, and Nnd-1; dom7; SRCAPK649R flies. The filled squares indicate means, the bars across the open rectangles indicate medians, the tops and bottoms of the open rectangles indicate the 75th- and 25th-percentile values, respectively, and the whiskers indicate the 10th- and 90th-percentile values. Numbers in the boxes indicate the numbers of wings scored. Asterisks indicate genotypes in which the mean difference from Nnd-1; +/+ and from Nnd-1; SRCAP+ is significant at the level of 0.01 by one-way analysis of variance (more statistical details are discussed in Materials and Methods).
Mammalian transfection assay.
HeLa cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were seeded at 1 × 105 cells/35 mm dish 18 h prior to transfection. Each transfection utilized 100 ng of the Hes-1 luciferase reporter plasmid and the indicated amounts of each additional plasmid. The pHes-1-luciferase, pNotch1-IC, and pSRCAP plasmids have previously been described (14, 19, 20). Each transfection was adjusted to contain equal molar amounts of CMV promoter by use of the pcDNA3.1 Myc/His plasmid. The Lipofectamine (Invitrogen) transfection method was used according to the manufacturer's directions. Following an overnight incubation, cells were harvested and assayed for luciferase activity. The relative luciferase activity reported was normalized for variation in transfection efficiencies between samples (the variation in the amounts of plasmid taken up by cells in each sample) using a statistical approach where each experimental point was performed in triplicate in at least three separate experiments. We used this approach since SRCAP regulates the transcription of common reporter genes used as internal controls, presumably because these reporters are also CBP responsive. Coexpression of SRCAP and pNotch1-IC did not result in the overexpression of pNotch1-IC (see Fig. S3 in the supplemental material).
RESULTS
DOM is the only Drosophila homolog of SRCAP.
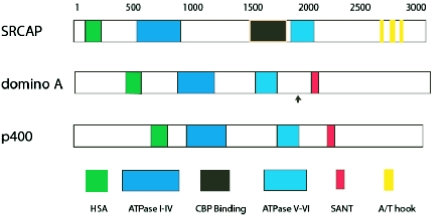
A BLASTP search of the National Center for Biotechnology Information database for SRCAP homologs revealed that Drosophila contains only one SRCAP-like protein, encoded by the domino gene. Two domino mRNA-encoded splice isoforms, termed domA and domB, have been identified; they are identical except for an alternate splice in the C-terminal end of the amino acid sequence of domB. The site of the alternative splice is indicated in Fig. 1. A Coffee-T alignment indicates that the domino gene product (DOM) shares extensive homology with SRCAP and p400 outside the conserved ATPase domain (see Fig. S1 and S2 in the supplemental material). In some regions, DOM exhibits a higher level of amino acid identity to SRCAP, whereas in other regions it exhibits greater identity to p400, suggesting that these proteins represent a family with overlapping but distinct functions. Comparison of the amino acid sequence of the helicase Sant-associated (HSA) domain in the SRCAP/p400/DOM family to that of the HSA domain within the members of the human SNF2/Drosophila brahma family indicates that while each family has the core sequences that define the HSA domain, they each also have additional sequences that distinguish these families from one another (see the supplemental material). For example, the sequence HWDY(L/C)EEEM(Q/V) is found in the SRCAP/p400/DOM family, whereas the sequence HQE(Y/F)LNSILQ is found in the human SNF2/Drosophila brahma family. In the HSA domain, SRCAP shares slightly more identity to DOM (53%) than does p400 (47%).
FIG. 1.
Comparison of the amino acid sequences of SRCAP, p400, and DOM. Shown is a schematic highlighting the positions of the HSA, the ATPase domain, the putative SANT domain, the CBP binding domain, and the A/T hook motif in SRCAP, DOM, and p400. The arrow marks the position of the alternate splice in which the C-terminal amino acids (2209 to 3203) of domA are replaced with a nonhomologous 489-amino-acid sequence to give the shorter (2,697 amino acids) domB isoform. Contained in the supplemental material data is a complete alignment of the amino acid sequences of human SRCAP, human p400, and Drosophila DOM proteins.
Additional conserved sequences that distinguish the SRCAP/p400/DOM family from the human SNF2/Drosophila brahma family occur in the conserved ATPase domain. Five of the seven motifs (I and Ia to IV) that make up the ATPase domain are located in the amino terminal end of SRCAP/p400/DOM. In the sequences surrounding these motifs, SRCAP shares 71% identity with DOM, while p400 has 44% identity. The remaining two motifs (V and VI) that complete the ATPase domain are located in the middles of these proteins. In the sequences surrounding these motifs, SRCAP and DOM share 84% identity, while p400 and DOM share 49% identity. A closer examination of the sequences adjacent to the various ATPase motifs in the SRCAP/p400/DOM family reveals that they have differences in sequencing and in spacing that distinguish them from the human SNF2/Drosophila brahma family. These differences for the most part appear to be conserved across species. For example, adjacent to motif Ia in SRCAP/p400/DOM is the sequence (L/F)AHLACE, whereas the sequence (I/V)TYLM(E/D) is found in the human SNF2/Drosophila brahma family. A characteristic six-amino-acid deletion that is found in the SRCAP/p400/DOM family adjacent to motif IV is not found in the human SNF2/Drosophila brahma family (see the supplemental material).
Although SRCAP and DOM have extensive homology in the HSA and ATPase domains, they diverge in their C-terminal ends. The C-terminal end of SRCAP lacks the polyglutamines and a putative SANT domain present in p400 and domA. In their places, SRCAP contains three A/T hook motifs (Fig. 1). The alternately spliced domB has a C-terminal end that also lacks the putative SANT domain and does not share homology with SRCAP, p400, or domA (data not shown). In addition to these differences, SRCAP contains a uniquely large spacer between ATPase motifs IV and V that is not present in domA/B or p400 (Fig. 1).
The existence of two closely related human proteins, SRCAP and p400, that have a single corresponding Drosophila homolog, DOM, is similar to the case of Drosophila brahma, which has two closely related human homologs, SNF2-alpha and -beta (also denoted BRM and BRG-1), which have similar (36, 42) but distinct (3) functions. To test the hypothesis that SRCAP has functions overlapping with those of DOM and to establish whether Drosophila can provide a model system for understanding the function of the SRCAP/p400 proteins, we tested whether SRCAP can substitute for DOM in vivo.
SRCAP can complement the semilethality and recessive female sterility of hypomorphic domino alleles.
To test whether the extensive sequence similarity between SRCAP and DOM reflects their functional equivalence, we generated transgenic flies that express SRCAP under the control of the constitutive Chip promoter (32). On a wild-type genetic background, SRCAP expression from this transgene has no obvious phenotypic effect. To test whether SRCAP can complement domino mutant phenotypes, we generated stocks that carry one of three dom hypomorphic alleles (dom7, dom9, and dom10). Each of these alleles results in semilethality and female sterility (39), so complementation by SRCAP should result in increased survival and female fertility.
Shown in the upper line of Table 1 are the results of crossing fly stocks with the dom7 and dom10 mutations. In this cross, the progeny with the genotype of dom/Cy would be expected with a 66% frequency and those with the genotype of dom7/dom10 would be expected with a 33% frequency if heteroallelic dom7/dom10 flies showed no recessive lethality (the genotype Cy/Cy is lethal and not observed). Since a total of 360 dom/Cy flies were observed, the smaller number of progeny with the dom7/dom10 genotype we observed (n = 72) relative to that we expected (n = 180) demonstrates the semilethal effect of these weak domino alleles. Flies that escape the recessive lethality are characterized by wing defects and female sterility, as previously described for homozygotes of these alleles (39). The total of observed escapers divided by the number of expected flies (72/180) is defined as the percentage of rescue of expected progeny, which, in the absence of a complementing transgene, is 40%. Crosses of fly stocks with the dom7 and dom9 genotypes (line 6) also had lower than the expected number of progeny with the dom7/dom9 genotype (22 observed/82 expected = 27% rescue), and the female progeny were sterile.
TABLE 1.
Complementation analysis of transgenic SRCAP and domino hypomorphic alleles
| Cross | Cy progeny | Cy+ progeny | Female fertility | Rescue (% of expected) |
|---|---|---|---|---|
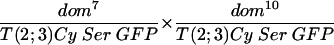 |
360 | 72 | No | 40 |
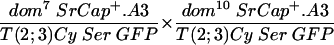 |
575 | 168 | Yes | 58 |
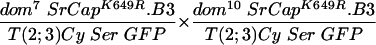 |
566 | 119 | No | 42 |
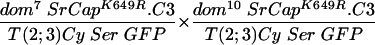 |
365 | 16 | No | 9 |
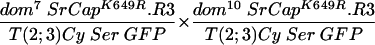 |
223 | 52 | No | 47 |
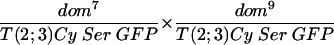 |
163 | 22 | No | 27 |
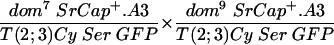 |
419 | 138 | Yes | 66 |
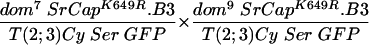 |
485 | 68 | No | 28 |
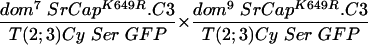 |
366 | 45 | No | 25 |
In contrast, in crosses of fly stocks carrying either dom7 or dom10 alleles together with the wild-type SRCAP transgene (line 2), the rescue of the expected number of progeny increased from 40% (line 1) to 58% (line 2). In crosses of fly stocks carrying either dom7 or dom9 alleles along with the SRCAP transgene (line 7), the rescue of the expected number of progeny also increased from 27% (line 6) to 66% (line 7). In both cases, the SRCAP transgene caused a restoration of female fertility. Overall, it appeared that wing defects were fewer and milder in the transgenic escapers, although this was not rigorously quantitated.
To test the functional significance of the ATPase homology domain of SRCAP, we generated transgenic flies expressing a mutant form of SRCAP in which a single missense mutation (K649R) was introduced in the putative ATPase active site lysine (Fig. 1 and the supplemental material). Lysine-to-arginine substitutions at the homologous positions extinguish the in vitro ATPase activities of the ATPases RAD3 (44), SNF2 (27), RecA (35), RAD51 (45), UvsW (7), hMSH6 (18), CSB (9), SRS2 (23), and BACH1 (6), as well as the in vivo functions of these proteins and of Drosophila BRM (1, 13). The mutant SRCAPK649R was unable to complement the recessive female sterility in any of the domino crosses tested (in Table 1, compare line 2 to lines 3 to 5 and line 7 to lines 8 and 9), indicating that the ATPase activity of SRCAP is required for its in vivo activities.
SRCAP binds to sites of active transcription on Drosophila chromosomes.
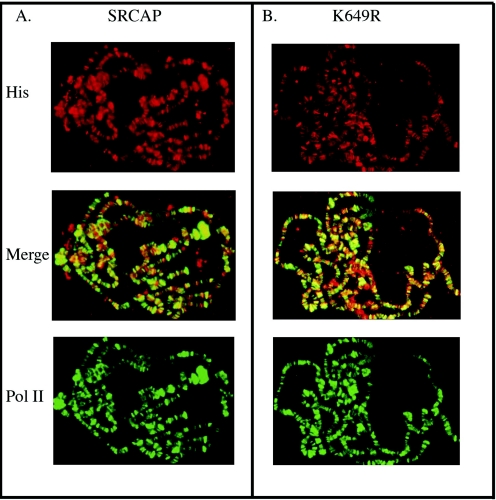
The in vivo binding sites of SRCAP in mammalian cells are unknown. To test whether SRCAP binds chromosomes in vivo and whether its binding sites might reveal something about its targets, transgenic SRCAP was immunolocalized on the salivary gland polytene chromosomes of transgenic third-instar larvae. As shown in Fig. 2A, SRCAP binds to a large number of sites on all Drosophila chromosomes. In many cases, SRCAP colocalizes with RNA polymerase II (Pol II) phosphorylated at serine 2 of the C-terminal-domain heptad repeats (Fig. 2A), which is thought to be the elongating form of Pol II (22).
FIG. 2.
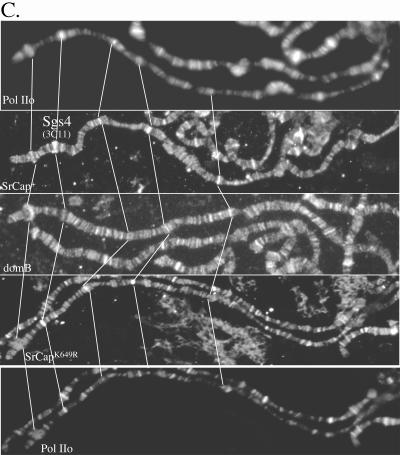
Transgenic SRCAP binds to diverse sites on polytene chromosomes. (A) Six-histidine-tagged SRCAP protein (His) and phosphorylated Pol II were immunolocalized to many chromosomal sites using antibody (Santa Cruz) to the six-histidine epitope tag and antibody to phosphorylated Pol II (H14; Covance). In the middle panel is the merged view; yellow color indicates sites of SRCAP and Pol II colocalization. (B) A SRCAP ATPase homology domain mutant protein is still recruited to polytene chromosomes. Chromosomes from a transgenic line expressing the six-histidine-tagged SRCAPK649R mutant protein were immunostained with antibody to the six-histidine epitope tag and antibody to phosphorylated Pol II. The middle panel shows the merged view. (C) Colocalization of phosphorylated Pol II (top and bottom panels), SRCAP (panel second from top), domB (middle panel), and SRCAPK649R (panel second from bottom) on the X chromosome. Images were obtained from separate, singly stained wild-type chromosomes (domB) or from doubly stained SRCAP transgenic chromosomes (Pol II and SRCAP).
To test whether the ATPase homology domain plays a role in SRCAP chromosome binding, we examined the distribution of the SRCAPK649R mutant protein on polytene chromosomes. As shown in Fig. 2B, the ATPase mutant protein has a widespread distribution and colocalizes with phosphorylated Pol II in a pattern apparently identical to that of wild-type SRCAP protein. Thus, the failure of the SRCAPK649R protein to complement dom7 is not due to its instability, its failure to bind chromosomes, or its gross mislocalization.
Detailed analysis of SRCAP distribution on the X chromosome (Fig. 2C) reveals significant overlap with the DOM-B protein product of domino, as well as with phosphorylated Pol II. Interestingly, the Sgs-4 locus at 3C is a site of staining for SRCAP, DOM-B, and phosphorylated Pol II. Sgs-4 is an ecdysone-regulated gene. SRCAP has previously been implicated as a coactivator in steroid-mediated gene activation using transient expression assays. This result showing SRCAP recruitment to a steroid-activated gene in Drosophila provides the first evidence for SRCAP binding to an endogenous steroid-regulated gene.
SRCAP is not recruited to active heat shock genes.
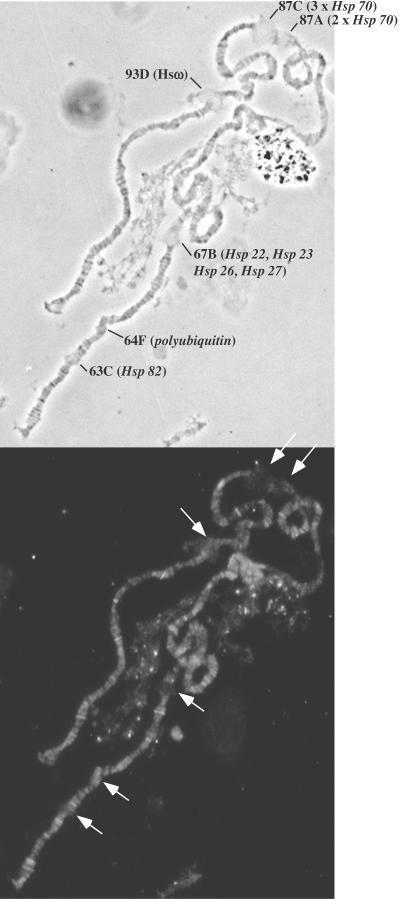
The observation that SRCAP is associated with developmentally regulated puffs and colocalizes with phosphorylated Pol II suggested that SRCAP could be a general transcriptional coactivator. To test this possibility, we examined the distribution of transgenic SRCAP protein after heat shock. Heat shock in Drosophila results in a disappearance of most mRNAs and corresponding protein synthesis and the concomitant appearance of high levels of a few heat shock mRNAs and their corresponding heat shock protein products. This dramatic change in gene expression is accompanied by a rapid mobilization of Pol II and associated factors to a small number of heat shock loci, which can be visualized by immunofluorescent localization (17). If SRCAP is a general transcription coactivator for Pol II, we would expect to find it similarly concentrated at heat shock puff sites. As shown in Fig. 3, there is a marked absence of SRCAP at all of the major heat shock loci, demonstrating that SRCAP is specifically recruited to only a subset of Pol II-transcribed genes.
FIG. 3.
Transgenic SRCAP protein is not recruited to sites of active heat shock genes. Transgenic larvae were subjected to a 20-min heat shock at 37°C, and fixed polytene chromosomes were immunostained with antibody to the six-histidine epitope tag. Top panel: phase contrast; bottom panel: immunofluorescence. Arrows indicate the positions of the major heat shock loci on the third chromosome.
SRCAP recruits CBP to ectopic chromosomal sites.
SRCAP was identified as an interactor with the CBP transactivation domain in a yeast two-hybrid protein assay (20), and a monoclonal antibody to SRCAP can pull down CBP from whole-cell lysates (48). Recently, SRCAP was found to be part of a protein complex that shares some subunits with the human NuA4 complex (11), but no CBP was detected in this complex. Similarly, domino-encoded protein was found as part of the Drosophila Tip60 complex (26), but no CBP was reported.
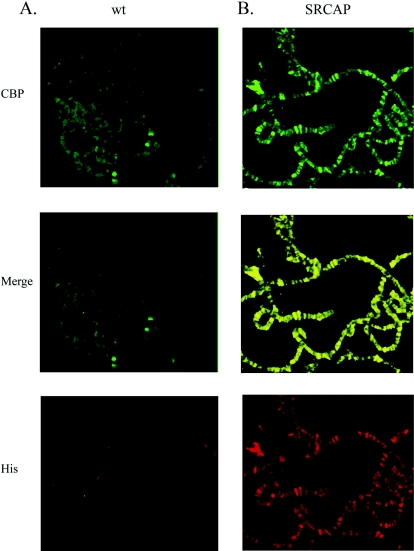
Antibodies to Drosophila CBP reveal a weak general distribution on wild-type Drosophila polytene chromosomes, with prominent concentrations at only a few sites (2) (Fig. 4A). In marked contrast, polytene chromosomes from transgenic larvae expressing SRCAP show a large number of CBP staining sites (Fig. 4B; compare the top panels in Fig. 5A and B). Furthermore, these novel sites of binding colocalize with sites of SRCAP binding. The simplest interpretation of this experiment is that CBP is recruited to the site of SRCAP binding.
FIG. 4.
CBP is recruited to chromatin by SRCAP. (A) Wild-type polytene chromosomes were immunostained with antibodies to Drosophila CBP (top panel) and to the six-histidine epitope tag (bottom panel). Note that no immunostaining is expected using the six-histidine antibody with wild-type chromosomes. (B) Chromosomes from a transgenic line expressing wild-type SRCAP and chromosomes from a transgenic line expressing the SRCAPK649R mutant were immunostained with antibody to Drosophila CBP (top panel) and to the six-histidine epitope tag (bottom panel).
To test whether CBP recruitment to SRCAP sites depends on an intact ATPase homology domain, we examined the distribution of CBP in transgenic larvae expressing the SRCAPK649R mutant protein. Again, CBP shows extensive binding to numerous chromosomal loci that correspond to SRCAP binding sites, suggesting that SRCAP can target CBP directly or indirectly to chromatin in vivo and that this targeting does not depend on the ATPase activity of SRCAP (data not shown).
SRCAP can complement the effect of DOM mutation on Notch signaling.
An important goal of this study was to use the DOM/SRCAP functional homology to identify new targets for SRCAP regulation. SRCAP and DOM have been purified as subunits of complexes that also contain the Tra1/TRRAP/Nipped A protein (11, 26). Mutations in the Drosophila Nipped A gene are modifiers of Notch pathway signaling in Drosophila (37). This suggests that Nipped A may act through the Tip60 coactivator complex to regulate the Notch pathway and leads to the hypothesis that SRCAP and DOM may also be regulators of Notch signaling.
To test this hypothesis, we tested whether a domino mutation, like Nipped A mutations, enhances the wing margin nicking caused by the hypomorphic Notch allele notchoid-1 (Nnd-1). Enhancement of wing nicking causes small nicks to fuse into large scallops; thus, the phenotype is best quantitated by measuring the distance across the wing using the shortest line that runs from anterior to posterior margin bisecting the midvein at a point equidistant from the two cross veins (Fig. 5A). As shown in Fig. 5B, a domino mutation dominantly enhances the Nnd-1 phenotype. This enhancement is due to a mutation in domino, since the reversion of the mutation by a precise excision of the P element that causes the domino mutation also leads to the loss of the enhancement activity (data not shown). SRCAP can functionally complement DOM in Notch signaling, since the enhanced wing nicking caused by the dom7 allele can be suppressed by transgenic SRCAP (Fig. 5B). SRCAP transgenes expressing the ATPase mutant form fail to complement in this assay, implicating the putative ATPase activity of SRCAP in the mechanism of SRCAP-dependent Notch coactivation.
SRCAP is a potentiator of Notch-dependent gene activation in mammalian cells.
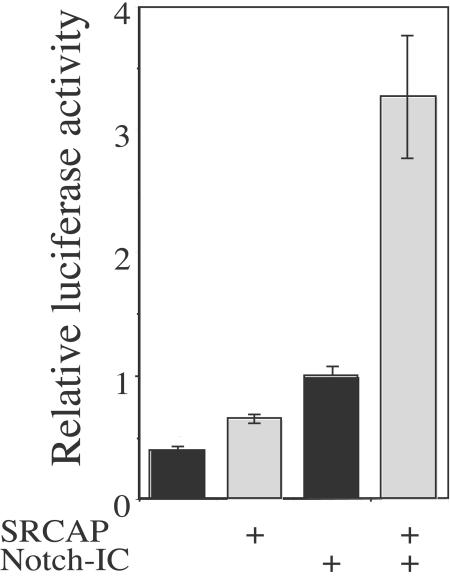
In mammalian cells. the binding of the Notch-1 intracellular domain to the DNA binding transcription factor CSL/CBF1 has been proposed to trigger the activation of transcription by the recruitment of a number of coactivators, including CBP (15, 33, 47). The ability of SRCAP to enhance CBP function (20) suggested that SRCAP also serves as a coactivator of Notch-mediated transcription. To test this hypothesis, HeLa cells were cotransfected with the Notch-1-dependent reporter gene expressing Hes-1 luciferase and plasmids expressing the Notch1 intracellular domain (Notch1-IC) and SRCAP (Fig. 6). Transcription of the Hes-1 luciferase promoter was enhanced to a modest extent by the expression of SRCAP alone (1.6-fold) or by the expression of Notch1-IC alone (2.5-fold). This is consistent with previous reports that found that Notch1-IC enhanced transcription of the Hes-1 promoter about threefold in HeLa cells (19). In contrast, the expression of both SRCAP and Notch1-IC resulted in a synergistic activation of transcription (eightfold). Since SRCAP actually results in a reduction in Notch1-IC mRNA (see Fig. S3 in the supplemental material), the synergism between SRCAP and Notch1-IC is, if anything, underestimated by this assay. Thus, SRCAP is a coactivator of Notch-dependent gene expression.
FIG. 6.
SRCAP regulates Notch1-mediated transcription in HeLa cells. The ability of SRCAP to enhance Notch1-mediated transcription in HeLa cells was assessed by transient transfection. Cells were transfected with 100 ng of the Notch-responsive reporter gene expressing Hes-1 luciferase and, where indicated, with 500 ng of the pNotch1-IC plasmid and 2,000 ng of the pSRCAP plasmid. Eighteen hours after transfection, the cells were harvested, and luciferase activity was measured and reported relative to the transcriptional activity induced by the pNotch1-IC plasmid. Values are the means and standard errors of three separate experiments in which each point is performed in triplicate.
DISCUSSION
The goals of this work were to exploit the homology between the human SRCAP and Drosophila DOM proteins to test previous models of SRCAP activities in vivo and to identify targets of SRCAP regulation. As a precondition to such experiments, we first demonstrated that SRCAP is a functional homolog of DOM. This functional complementation implies that at least some of the activities and molecular targets of SRCAP and DOM are evolutionarily conserved.
The distribution of SRCAP on Drosophila chromosomes is consistent with a role for SRCAP in gene activation. The significant overlap of SRCAP, phosphorylated Pol II, and DOM suggests that DOM is also recruited to transcriptionally active chromatin. This was not anticipated, as DOM had previously been proposed to be a transcriptional repressor (39). The colocalization of DOM with Pol II does not exclude a role for DOM in gene repression as well but suggests that DOM activity could have different consequences at different loci. Thus, at some loci, DOM may function analogously to the chromatin ATPase brahma, which is required for normal gene activity and broadly colocalized with Pol II (1), while at other loci, it may function like ISWI, which is also required for gene expression but shows little overlap with RNA Pol II (10). An alternative possibility is that the two splice isoforms of DOM, domA and domB, play distinct roles in gene expression. DOM-B is widely expressed throughout development, while DOM-A expression is restricted to the developing nervous system (39). DOM-B is the only isoform expressed in salivary glands (39) and is thus the only isoform whose chromosomal distribution was determined by immunostaining salivary gland polytene chromosomes.
The ATPase homology domain of SRCAP is required for function in vivo.
Several studies have implicated SRCAP as a transcriptional coactivator (20, 30, 31). However, the mechanism of this coactivation and its connection to ATP-dependent chromatin remodeling are uncertain. To date, assays for SRCAP coactivation have relied upon artificial reporter constructs and transient transfection assays. While transfected DNA can assemble into nucleosomes, transfected templates do not form the same nucleosomal complexes as chromosomal templates (40). Furthermore, a truncated SRCAP in which most of the ATPase homology domain was deleted had activity similar to that of full-length SRCAP in a transient transfection assay (30), suggesting that SRCAP has an ATP-independent coactivator activity and that the ATPase activity is dispensable in transient expression assays.
Here we show that an ATPase mutation in SRCAP abolishes its ability to complement domino mutations in transgenic flies. While we have not tested directly whether the K649R mutation abolishes ATPase activity in SRCAP, we note that the homologous substitution in other ATPases results in a loss of biochemical and/or genetic activity. The simplest interpretation of this result is that the ATPase activity (and by extension, the putative helicase activity) of SRCAP is required for its function in chromatin.
CBP is recruited to SRCAP binding sites on chromosomes.
Several lines of evidence support the view that SRCAP interacts with CBP (20, 48). Here, we show evidence for a SRCAP-CBP interaction on chromosomes, although this interaction could be either direct or indirect. Given the functional homology between SRCAP and DOM, then, it is puzzling that the normal distribution of CBP in wild-type chromosomes is strikingly different from the distribution of DOM. One possible explanation is that CBP interacts with greater affinity to SRCAP than to DOM. Consistent with this hypothesis, only a portion of the domain which directs SRCAP interaction with CBP (amino acids 1577 to 1925) is found within DOM or p400 (20). Deletion of the conserved region shared in SRCAP/p400/DOM (amino acids 1867 to 1925) reduces but does not abolish SRCAP-CBP interaction in a mammalian two-hybrid assay, thus indicating that additional contacts between CBP and SRCAP occur in the nonconserved domain (amino acids 1577 to 1866) (J. C. Chrivia, unpublished observation). Interestingly, this region accounts for part of the unique large spacer located between the ATPase IV and V motifs found in SRCAP but not in DOM or p400. Thus, in mammals, the insertion of this unique spacer may have evolved to allow enhanced CBP binding by SRCAP.
Endogenous targets of SRCAP coactivation.
Previous studies implicated SRCAP in cyclic AMP- and steroid-mediated gene activation using transfection assays. However, this is the first demonstration of specific endogenous gene targets for SRCAP. By immunolocalizing SRCAP on polytene chromosomes, we confirm that SRCAP is recruited to several loci known to contain ecdysone-induced genes. A detailed comparison reveals significant overlap between DOM binding sites, SRCAP binding sites, and sites of binding by phosphorylated Pol II on polytene chromosomes, suggesting that both SRCAP and its Drosophila homolog DOM are transcriptional coactivators in vivo.
A novel insight into the normal physiological targets for SRCAP activation that emerged from this study was the evolutionarily conserved role of SRCAP and DOM in Notch pathway activation. The activation of this pathway might occur by SRCAP interaction with CBP, which is recruited to chromatin by mastermind, a component of the Notch ICD-CBF1 enhancer complex (15). However, the Notch intracellular domain was not identified among the proteins associated with SRCAP/DOM and Tra1/TRRAP/Nipped A either in the human NuA4 or Drosophila Tip60 complexes or in the recently identified SRCAP complex that contains the histone variant H2AZ (5). Similarly, CBP was not found as part of these complexes. It seems likely, then, that SRCAP and DOM participate in one or more protein complexes distinct from the currently characterized complexes.
Finally, an important question is the degree to which SRCAP is serving a function in flies that resembles its activity in mammalian cells. The partial complementation of domino mutations by mammalian SRCAP we observed could be explained by the inadequate expression of the mammalian protein in transgenic flies or, alternatively, by the possibility that some functions of DOM are not being complemented by SRCAP because these functions are performed uniquely by p400 in mammals. In yeast, the p400 homolog Eaf1 and the SRCAP homolog Swr1 are found in distinct complexes (11, 12, 21, 24, 29), and data suggest that they perform distinct functions (25). Similarly, in mammals, the NuA4 complex contains p400 but not SRCAP, while a distinct SRCAP-containing complex has been described previously (5). The respective functions of these mammalian complexes and their targets in vivo are unknown. The two splice isoforms of DOM may represent distinct functional homologs of p400 (DOM-A) and SRCAP (DOM-B). The homology within the HSA and ATPase domains suggests that both DOM isoforms are ancestrally more related to SRCAP; however, the C-terminal SANT domain, which is found only in the DOM-A splice isoform, makes it somewhat more homologous to p400 than is DOM-B.
It remains an open question whether the DOM functions complemented by SRCAP are those that represent SRCAP-dependent pathways in mammals or whether some of these DOM functions are normally p400 dependent in mammals. This question can be answered only when direct chromosomal targets of DOM, SRCAP, and p400 are identified and the roles of DOM/SRCAP/p400 in chromatin modification at these loci are determined by biochemical assays. The cytological and genetic data from this study suggest that steroid- and Notch-regulated genes are likely among such targets and should be the focus of future study.
Supplementary Material
Acknowledgments
We thank M. Meister and M. Ruhf for domino mutant stocks, DomB antibody, and useful advice; A. Bigas for the HES-1 luciferase reporter gene and for critical reading of the manuscript; S. Smolik for anti-Drosophila CBP serum; M. Gause for help in scoring wing phenotypes; A. Monroy for performing preliminary immunostaining experiments; D. Dorsett for the Notchnd-1 mutant stock, the Chip expression vector, and much helpful advice and encouragement; M. Donlin for help with statistics; and P. Harte for acutely critical reading of the manuscript.
This work was supported by NSF grant MCB 0131414 (J.C.E.) and NIH grant DK58262 (J.C.C.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armstrong, J. A., O. Papoulas, G. Daubresse, A. S. Sperling, J. T. Lis, M. P. Scott, and J. W. Tamkun. 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantignies, F., R. H. Goodman, and S. M. Smolik. 2000. Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol. Cell. Biol. 20:9317-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, and G. Crabtree. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 4.Cai, Y., J. Jin, C. Tomomori-Sato, S. Sato, I. Sorokina, T. J. Parmely, R. C. Conaway, and J. W. Conaway. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278:42733-42736. [DOI] [PubMed] [Google Scholar]
- 5.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP-TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665-13670. [DOI] [PubMed] [Google Scholar]
- 6.Cantor, S., R. Drapkin, F. Zhang, Y. Lin, J. Han, S. Pamidi, and D. M. Livingston. 2004. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. USA 101:2357-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carles-Kinch, K., J. W. George, and K. N. Kreuzer. 1997. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 16:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceol, C. J., and H. R. Horvitz. 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6:563-576. [DOI] [PubMed] [Google Scholar]
- 9.Citterio, E., S. Rademakers, G. T. J. van der Horst, A. J. van Gool, J. H. J. Hoeijmakers, and W. Vermeulen. 1998. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 273:11844-11851. [DOI] [PubMed] [Google Scholar]
- 10.Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas, M. Prestel, G. Daubresse, M. Verardo, S. L. Moseley, M. Berloco, T. Tsukiyama, C. Wu, S. Pimpinelli, and J. W. Tamkun. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5:355-365. [DOI] [PubMed] [Google Scholar]
- 11.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Côté. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Côté. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276:3484-3491. [DOI] [PubMed] [Google Scholar]
- 13.Elfring, L. K., C. Daniel, O. Papoulas, R. Deuring, M. Sarte, S. Mosely, S. J. Beek, W. R. Waldrip, G. Daubresse, A. DePace, J. A. Kennison, and J. W. Tamkun. 1998. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa, L., J. Ingles-Esteve, A. Robert-Moreno, and A. Bigas. 2003. IκBα and p65 regulate the cytoplasmic shuttling of nuclear corepressors: cross-talk between Notch and NFκB pathways. Mol. Biol. Cell 14:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, M., J. Gerber, T. Ikura, S. Sif, R. Drapkin, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 17.Greenleaf, A. L., U. Plagens, M. Jamrich, and E. K. F. Bautz. 1978. RNA polymerase B (or II) in heat induced puffs of Drosophila polytene chromosomes. Chromosoma 65:127-136. [DOI] [PubMed] [Google Scholar]
- 18.Iaccarino, I., G. Marra, F. Palombo, and J. Jiricny. 1998. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 17:2677-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarriault, S., O. Le Bail, E. Hirsinger, O. Pourquié, F. Logeat, C. F. Strong, C. Brou, N. G. Seidah, and A. Israël. 1998. Delta-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18:7423-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, H., J. Kneer, I. Chackalaparampil, P. Yaciuk, and J. Chrivia. 1999. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 274:16370-16376. [DOI] [PubMed] [Google Scholar]
- 21.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 23March2004, posting date. [Online]. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:e131. 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krejci, L., M. Macris, Y. Li, S. Van Komen, J. Villemain, T. Ellenberger, H. Klein, and P. Sung. 2004. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279:23193-23199. [DOI] [PubMed] [Google Scholar]
- 24.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 25.Krogan, N. J., K. Baetz, M.-C. Keogh, N. Datta, C. Sawa, T. C. Y. Kwok, N. J. Thompson, M. G. Davey, J. Pootoolal, T. R. Hughes, A. Emili, S. Buratowski, P. Hieter, and J. F. Greenblatt. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101:13513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusch, T., L. Florens, W. H. MacDonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 27.Laurent, B. C., S. Yang, and M. Carlson. 1992. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 12:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuguchi, G., X. Shen, J. Landry, W.-H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 30.Monroy, M. A., D. D. Ruhl, X. Yu, D. K. Granner, P. Yaciuk, and J. C. Chrivia. 2001. Regulation of cAMP-responsive element-binding protein-mediated transcription by the SNF2/SWI-related protein, SRCAP. J. Biol. Chem. 276:40721-40726. [DOI] [PubMed] [Google Scholar]
- 31.Monroy, M. A., N. M. Schott, L. Cox, J. D. Chen, M. Ruh, and J. C. Chrivia. 2003. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor-mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 17:2519-2528. [DOI] [PubMed] [Google Scholar]
- 32.Morcillo, P., C. Rosen, M. K. Baylies, and D. Dorsett. 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam, Y., A. P. Weng, J. C. Aster, and S. C. Blacklow. 2003. Structural requirements for assembly of the CSL/intracellular Notch1/Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278:21232-21239. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, H., T. Ueda, T. Aoyama, A. Aronheim, S. Nagata, and R. Fukunaga. 2003. A SW12/SNF2-type ATPase/helicase protein, mDomino, interacts with myeloid zinc finger protein 2A (MZF-2A) to regulate its transcriptional activity. Genes Cells 8:325-339. [DOI] [PubMed] [Google Scholar]
- 35.Rehrauer, W. M., and S. C. Kowalczykowski. 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268:1292-1297. [PubMed] [Google Scholar]
- 36.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollins, R. A., P. Morcillo, and D. Dorsett. 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudolph, T., B. Lu, T. Westphal, J. Szidonya, J. Eissenberg, and G. Reuter. 1999. New type of CyO and TM3 green balancers. Drosophila Inf. Serv. 82:99-100. [Google Scholar]
- 39.Ruhf, M.-L., A. Braun, O. Papoulas, J. W. Tamkun, N. Randsholt, and M. Meister. 2001. The domino gene of Drosophila encodes novel members of the SWI/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128:1429-1441. [DOI] [PubMed] [Google Scholar]
- 40.Smith, C. L., and G. L. Hager. 1997. Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J. Biol. Chem. 272:27493-27496. [DOI] [PubMed] [Google Scholar]
- 41.Spradling, A. C. 1986. P element mediated transformation, p. 175-197. In D. B. Roberts (ed.), Drosophila, a practical approach. IRL Press, Oxford, United Kingdom.
- 42.Strobeck, M. W., D. N. Reisman, R. W. Gunawardena, B. L. Betz, S. P. Angus, K. E. Knudsen, T. F. Kowalik, B. E. Weissman, and E. S. Knudsen. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782-4789. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, W. J., Jr., M. A. Monroy, W. Bohne, J. Chrivia, P. Yaciuk, K. C. Nallani, C. K. Smith II, and S. F. Queener. 2003. Molecular cloning and characterization of an SRCAP chromatin remodeling homologue in Toxoplasma gondii. Parasitol. Res. 90:1-8. [DOI] [PubMed] [Google Scholar]
- 44.Sung, P., D. Higgins, L. Prakash, and S. Prakash. 1988. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 7:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 46.Underhill, C., M. S. Qutob, S.-P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 47.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by Notch intracellular domains in vitro. Mol. Cell. Biol. 22:7812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, X., I. Chackalaparampil, M. A. Monroy, M. T. Cannella, E. Pesek, J. Chrivia, and P. Yaciuk. 2001. Adenovirus DNA binding protein interacts with the SNF2-related CBP activator protein (SrCAp) and inhibits SrCap-mediated transcription. J. Virol. 75:10033-10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.