Abstract
Oxygen-dependent proteolysis is the primary means of regulating the hypoxia-inducible factor (HIF) family of transcription factors. The alpha-subunit of HIF factor 1 (HIF-1) contains two highly conserved oxygen-dependent degradation domains (402 ODD and 564 ODD), each of which includes a proline that is hydroxylated in the presence of oxygen, allowing the von Hippel-Lindau (VHL) E3 ubiquitin ligase to interact and target HIF-1α to the proteasome for degradation. Mutation of either proline is sufficient to partially stabilize HIF-1α under conditions of normoxia, but the specific contributions of each hydroxylation event to the regulation of HIF-1α are unknown. Here we show that the two ODDs of HIF-1α have independent yet interactive roles in the regulation of HIF-1α protein turnover, with the relative involvement of each ODD depending on the levels of oxygen. Using hydroxylation-specific antibodies, we found that under conditions of normoxia proline 564 is hydroxylated prior to proline 402, and mutation of proline 564 results in a significant reduction in the hydroxylation of proline 402. Mutation of proline 402, however, has little effect on the hydroxylation of proline 564. To determine whether the more rapid hydroxylation of the proline 564 under conditions of normoxia is due to a preference for the particular sequence surrounding proline 564 or for that site within the protein, we exchanged the degradation domains within the full-length HIF-1α protein. In these domain-swapping experiments, prolyl hydroxylase domain 1 (PHD1) and PHD2 preferentially hydroxylated the proline located in the site of the original 564 ODD, while PHD3 preferred the proline 564 sequence, regardless of its location. At limiting oxygen tensions, we found that proline 402 exhibits an oxygen-dependent decrease in hydroxylation at higher oxygen tensions relative to proline 564 hydroxylation. These results indicate that hydroxylation of proline 402 is highly responsive to physiologic changes in oxygen and, therefore, plays a more important role in HIF-1α regulation under conditions of hypoxia than under conditions of normoxia. Together, these findings demonstrate that each hydroxylated proline of HIF-1α has a distinct activity in controlling HIF-1α stability in response to different levels of oxygenation.
A central component to the molecular and cellular response to low-oxygen tensions is the transcription factor hypoxia-inducible factor 1 (HIF-1). HIF-1 is a heterodimer composed of an oxygen-sensitive alpha subunit and a constitutive beta subunit (52, 55, 56). Stringent regulation of HIF-1 is necessary for the appropriate and coordinated expression of a variety of target genes involved in oxygen homeostasis, angiogenesis, metabolism, cell proliferation and viability, tissue remodeling, and erythropoiesis (34, 43, 50, 51). The predominant means of HIF-1 regulation under conditions of low-oxygen tensions is through protein stabilization of the oxygen-labile alpha subunit (17, 48, 49, 55).
HIF-1α contains two oxygen-degradation domains, the 564 oxygen-dependent degradation domain (564 ODD) and the 402 oxygen-dependent degradation domain (402 ODD). Each oxygen-dependent degradation domain contains a highly conserved proline residue, proline 564 and proline 402, respectively, which is modified by a family of 4-prolyl hydroxylases. These enzymes catalyze a hydroxylation reaction, which requires oxygen and 2-oxoglutarate as substrates and iron and ascorbate as cofactors. These posttranslational modifications allow the von Hippel-Lindau (VHL) tumor suppressor protein product to recognize HIF-1α. VHL in conjunction with elongins B and C functions as an E3 ubiquitin ligase, covalently attaching ubiquitin modifiers to substrate proteins such as HIF-1α (19, 27). Ubiquitin ladders mark the respective client protein for degradation by the proteasome (11, 13, 30, 41). Thus, under normal oxygenated conditions, HIF-1α protein is constantly turned over due to hydroxylation, recognition by VHL, and subsequent destruction by the proteasome. In contrast, the 4-prolyl hydroxylases have reduced activity under conditions of hypoxia, as oxygen is a requisite substrate of the hydroxylation reaction. In the absence of hydroxylation, HIF-1α escapes recognition by VHL, resulting in HIF-1α protein stabilization. Once stabilized, HIF-1α dimerizes with its binding partner, HIF-1β. Together HIF-1α and HIF-1β bind to a core sequence of 5′-RCGTG-3′ in the enhancer elements of target genes to initiate gene transcription.
HIF-1α protein is heavily posttranslationally modified (4, 6, 14, 22-24, 26, 28, 29, 31, 32, 36, 42, 44, 53, 57). These modifications affect both HIF-1α protein stability and its ability to transactive. The best-characterized modifications of HIF-1α protein are the hydroxylations of proline residues 402 and 564 and asparagine 803. Hydroxylation of asparagine 803 regulates HIF-1 transactivation (14, 28, 29, 32, 36). In contrast, both proline hydroxylation sites are involved in degradation as mutation of either site results in some protein stabilization (9, 29, 33). However, the relative contributions of each proline hydroxylation site in regulating the degradation of HIF-1α have not yet been fully elucidated. Here we employ two proline hydroxylation-specific antibodies to investigate the role of each hydroxylation site under conditions of normoxia and hypoxia, the interactions between the two sites, and their interactions with each of the prolyl hydroxylases. We found a nonreciprocal interaction in which the hydroxylation of proline 564 occurs first under conditions of normoxia and is necessary for the efficient hydroxylation of proline 402, leading to HIF-1α degradation. In contrast, under conditions of hypoxia, proline 402 is more responsive to changes in oxygen tension. These findings reveal the differential regulation and functionality of the two hydroxylation sites in directing HIF-1α degradation.
MATERIALS AND METHODS
Reagents.
MG132 was purchased from Calbiochem. Cobalt chloride (CoCl2) and desferrioaxmine (DFO) were purchased from Sigma. Dimethyloxalyglycine (DMOG) was a generous gift from Wayne Zundel (University of Louisville Brown Cancer Center).
Antibodies and immunoblotting.
Antisera to hydroxylated HIF-1α were produced in rabbits immunized with a modified peptide of amino acids 398 to 407 or amino acids 558 to 565 of HIF-1α conjugated to keyhole limpet hemocyanin (Custom Antibody Group; ZYMED) (9). Hydroxylated HIF-1α antibody was then affinity purified by binding to immobilized modified peptide (SulfoLink gel; Pierce). Hydroxylated HIF-1α P402 antibody was also purified against immobilized unmodified peptide. Dot blot analyses were performed by serially diluting both modified peptide and unmodified peptide onto nitrocellulose (Bio-Rad) or polyvinylidene difluoride (Hybond-P; Amersham) membranes. Quantitative analysis of relative antibody sensitivities was performed by ImageQuant (Molecular Dynamics). For immunoblotting, cells were lysed in urea lysis buffer (9 M urea, 75 mM Tris [pH 7.5], 150 mM β-mercaptoethanol). Cells were sonicated briefly (10 seconds). A total of 50 to 100 μg of protein (as determined by a Bradford assay; Bio-Rad) was electrophoresed on 8% or 12% sodium dodecyl sulfate-polyacrylamide gels and then transferred onto polyvinyl difluoride membranes. Intensity of exposure was adjusted for Fig. 1 and 2 (also see Fig. 6) to determine whether a small fraction of HIF-1α was hydroxylated. Furthermore, the preparations for Fig. 3 and 4 were carefully controlled for equivalent conditions and exposures. Total HIF-1α protein was detected with anti-HIF-1α antibody from Transduction Laboratories (clone 54). Anti-α-tubulin was from Research Diagnostics, Inc. (clone DM1A). Anti-VHL was from BD Pharmingen (clone Ig32). Anti-cytosine deaminase (anti-CD) was from mtm laboratories (clone 16D8F2).
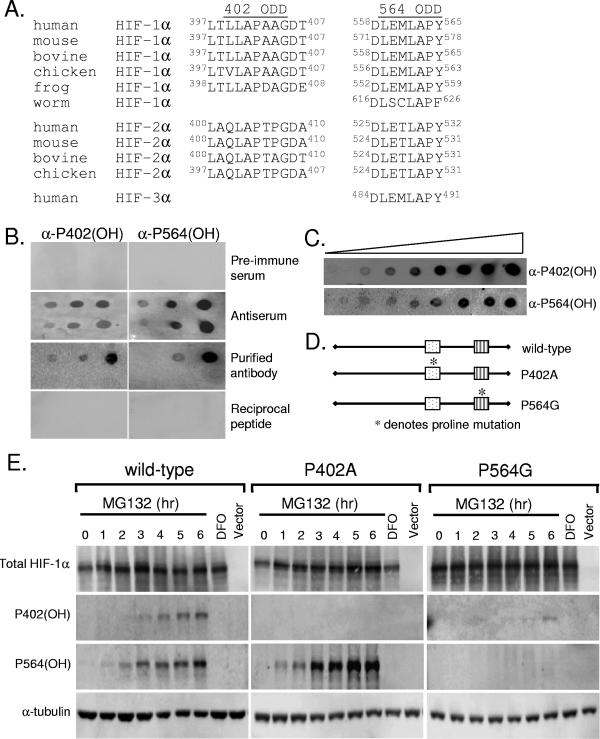
FIG. 1.
Proline 564 is hydroxylated prior to proline 402. (A) Protein sequence alignment of HIF-1α, HIF-2α, and HIF-3α subunits for 402 ODD and 564 ODD domains across species demonstrates high conservation. (B) Hydroxylation-specific antibodies recognize only hydroxylated prolines 402 and 564 of HIF-1α. Polyvinyl difluoride membranes were dotted with 10 ng, 100 ng, and 1 μg of hydroxylated (top) and unhydroxylated (bottom) proline 402 or proline 564 peptides and immunoblotted. (C) Nitrocellulose membranes were dotted with twofold serial dilutions of either hydroxylated proline 402 peptide (top) or hydroxylated proline 564 peptide (bottom) and immunoblotted. (D) Constructs transfected into HIF knockout (HKO) mouse embryo fibroblasts (MEFs). The asterisk indicates prolines that are mutated into residues that cannot be hydroxylated. (E) HKO cells were transfected with full-length HIF-1α, P402A HIF-1α, P564G HIF-1α, or empty vector and treated with MG132 (10 μM) for the indicated time or DFO (100 μM, 4 h). Immunoblot analyses were carried out to examine the levels of total HIF-1α, hydroxylated P402 HIF-1α, hydroxylated P564 HIF-1α, and α-tubulin.
FIG. 2.
Proline 564 hydroxylation influences the hydroxylation of proline 402. (A) Constructs transfected into HIF knockout (HKO) mouse embryo fibroblasts (MEFs). The asterisk indicates prolines that are mutated into residues that cannot be hydroxylated. (B and C) Hydroxylation of proline 402 is influenced by hydroxylation of proline 564. HKO MEFs were transfected with full-length HIF-1α or mutant HIF-1α along with the indicated prolyl hydroxylases and treated with MG132 (10 μM). Immunoblots were carried out to determine the levels of total HIF-1α, hydroxylated P402, hydroxylated P564, and α-tubulin. Intensity of exposure was adjusted to illustrate that a small fraction of proline 402 is still hydroxylated in the proline 564 mutant. (D) The effect of endogenous hydroxylases in HKO MEFs was examined. HKO MEFs were transfected with full-length HIF-1α or mutant HIF-1α with and without MG132. Immunoblot analyses were carried out to determine the levels of total HIF-1α, hydroxylated P402, hydroxylated P564, and α-tubulin.
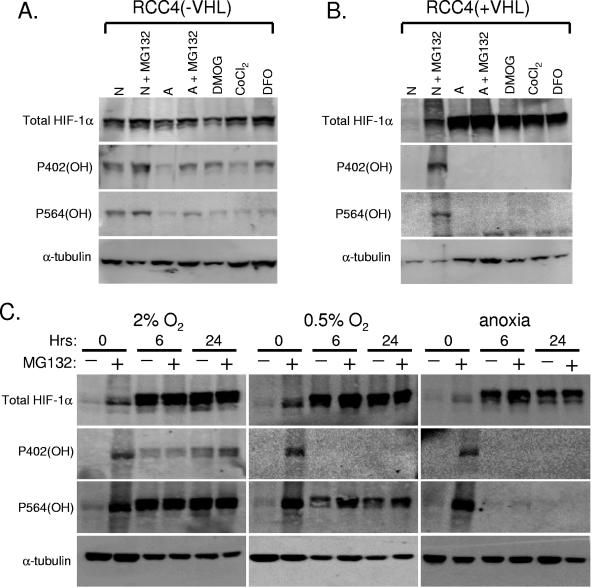
FIG. 6.
HIF-1α is hydroxylated under conditions of mild hypoxia but is still stable. (A) Total cell extracts were prepared from RCC4 without VHL. Cells were treated with the proteasome inhibitor MG132 (10 μM; 1 h), anoxia (>0.2%; 6 h), CoCl2 (100 μM; 6 h), and/or DFO (100 μM; 6 h). Immunoblot analyses were carried out to show the levels of total HIF-1α, hydroxylated proline 402 of HIF-1α, hydroxylated proline 564 of HIF-1α, and α-tubulin. (B) Total cell extracts were prepared from RCC4 with functional VHL. Cells were treated with the proteasome inhibitor MG132 (10 μM; 1 h), anoxia (>0.2%; 6 h), CoCl2 (100 μM; 6 h), and/or DFO (100 μM; 6 h). Immunoblot analyses were carried out to show the levels of total HIF-1α, hydroxylated proline 402 of HIF-1α, hydroxylated proline 564 of HIF-1α, and α-tubulin. (C) RCC4 with functional VHL were treated with mild hypoxia (2% or 0.5% O2) or anoxia for 6 to 24 h and harvested in a hypoxic environment. Cells were also treated with proteasome inhibition for the last hour of hypoxic/anoxic treatment. Total cell extracts were subjected to immunoblot analysis with antibodies to total HIF-1α, hydroxylated proline 402 of HIF-1α, hydroxylated proline 564 of HIF-1α, and α-tubulin.
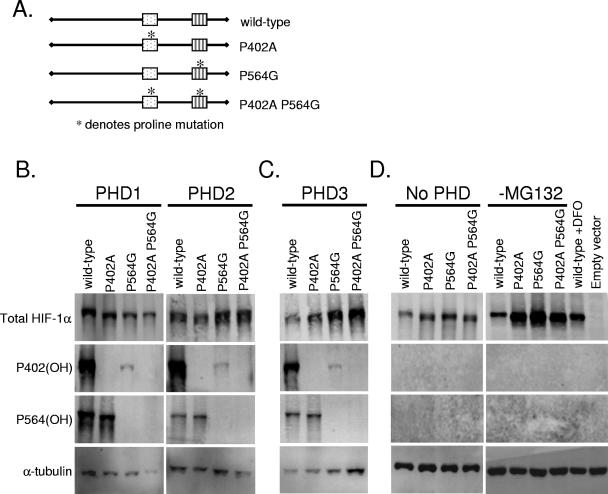
FIG. 3.
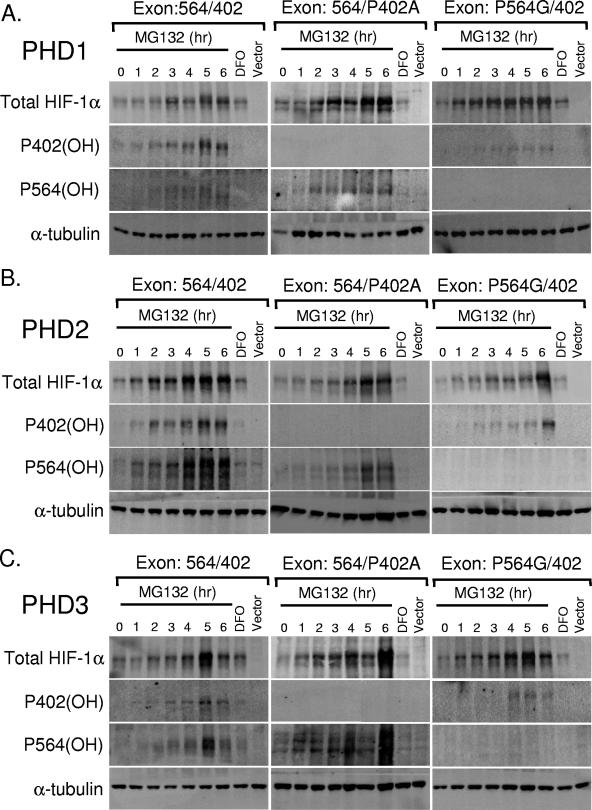
Difference in hydroxylation is due to structural determinants in full-length HIF-1α for PHD1 and PHD2, but PHD3 is sequence specific. (A) Oxygen-dependent degradation domain-swapped constructs transfected into HKO MEFs. The asterisk indicates prolines that are mutated to residues that cannot be hydroxylated. (B, C, and D) HKO cells were transfected with degradation domain-swapped mutant HIF-1α and the indicated prolyl hydroxylase. The day following transfection, the cells were treated with MG132 (10 μM) for the indicated time or DFO (100 μM, 4 h). Immunoblot analyses were carried out to examine the levels of total HIF-1α, hydroxylated P402 HIF-1α, hydroxylated P564 HIF-1α, and α-tubulin.
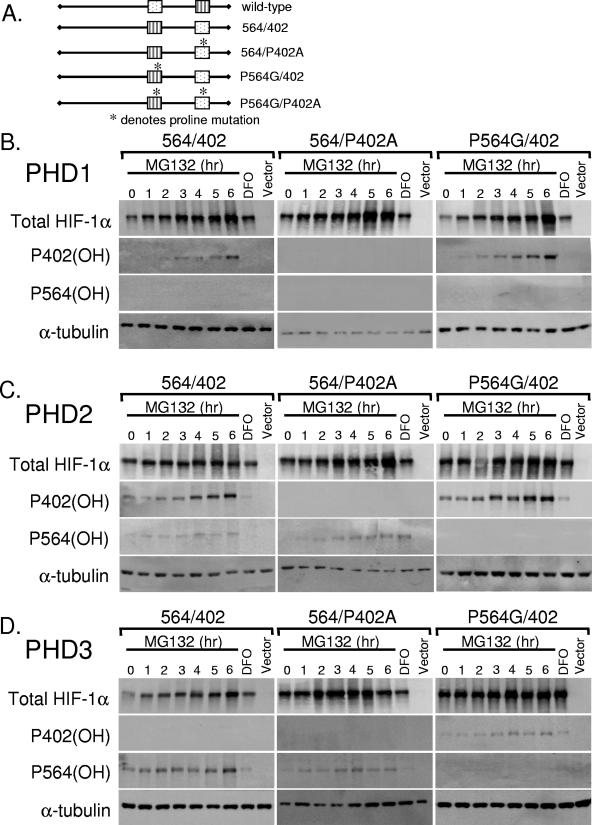
FIG. 4.
Kinetics of hydroxylation of exon-swapped mutant HIF-1α. (A, B, and C) HKO cells were transfected with exon-swapped mutant HIF-1α and the indicated prolyl hydroxylase. The day following transfection, the cells were treated with MG132 (10 μM) for the indicated time or DFO (100 μM, 4 h). Immunoblot analyses were carried out to examine the levels of total HIF-1α, hydroxylated P402 HIF-1α, hydroxylated P564 HIF-1α, and α-tubulin.
Cell lines and transient transfections.
RCC4(−VHL) cells and HIF-1α knockout mouse embryo fibroblasts and mouse embryo fibroblasts with HIF-1α alleles flanked by loxP sites were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (19, 35, 46). Cells of the RCC4(+VHL) renal carcinoma cell line with reintroduced VHL were generated by transfecting full-length VHL and selecting with G418. Transient transfections were performed using Lipofectamine-Plus reagent (Invitrogen) according to the manufacturer's directions.
Anoxia and hypoxia.
Anoxia (<0.2% O2) treatment was achieved using a Sheldon Labs anaerobic chamber. Hypoxia treatment was achieved using a variable hypoxia chamber (Biotrace) (0.5% O2 or 2% O2).
Plasmids.
HIF-1α mutant constructs were generated as described in reference 12. Briefly, plasmids were generated using a site-directed mutagenesis kit (QuikChange; Stratagene) and confirmed by DNA sequencing. Degradation domain-swapped mutants were created by two serial rounds of insertional mutagenesis PCR in which the 402 degradation domain and the 564 degradation domain were interchanged. We defined the 402 degradation domain as the 10-amino-acid peptide sequence used as an antigen for antibody production and the 564 degradation domain as the 8-amino-acid peptide sequence used in antibody production of the second antibody. Although these two regions are short and there may be other flanking sequences necessary for proper conformational folding, they both encompass the canonical LXXLAP motif for HIF-1α prolyl hydroxylation (15, 16, 33). For the first round, the 402 degradation domain, defined as the antigen peptide sequence, was inserted into the 564 degradation domain site to create a plasmid with two 402 degradation domain sequences. This mutant was then used as the template for the next round of fusion PCR in which the 564 degradation domain, again defined as the antigen peptide sequence, was inserted into the 402 degradation domain site. Similarly, degradation domain-swapped proline point mutants were also created in this fashion. Exon-swapped constructs were created in a similar fashion using megaprimer, fusion PCR. Exon 9 (74 amino acids), which contains proline 402, was switched into the site of exon 12 (146 amino acids). Likewise, exon 12, which contains proline 564, was switched into the site of exon 9. The corresponding proline point mutants were also constructed in this manner. Human VHL expression plasmid was a gift from the laboratory of Judith Frydman (Stanford University). ODD-CD plasmids were created by PCR amplification of the region encompassing amino acids 338 to 603 of HIF-1α and then subcloned into a vector containing Escherichia coli cytosine deaminase.
RESULTS
Proline 564 is hydroxylated prior to proline 402.
To examine the roles of each hydroxylation site in the full-length HIF-1α protein, we generated polyclonal antibodies against each of the hydroxylated proline residues of HIF-1α (9). The two peptide antigens chosen are highly conserved within HIF-α subunits as well as evolutionarily conserved across species from worms to humans, suggesting a critical function in protein degradation (Fig. 1A). Furthermore, the peptide antigens both encompass the canonical LXXLAP motif for HIF-1α prolyl hydroxylation (15, 16, 33). Initial verification of antibody specificities was performed by dot blot analysis. The unpurified antiserum recognizes both the modified and unmodified peptide antigens, whereas the preimmune serum recognized neither form. Affinity purification of the antiserum resulted in antibodies that specifically recognized their hydroxylated antigens with greater than 100-fold specificity over unmodified peptides (Fig. 1B). As a control, the reciprocal peptides were also probed to demonstrate that the antibodies are specific to their respective hydroxylated proline and are not pan-hydroxylation antibodies. Relative efficiency of antibody detection was examined by dot blot analysis using twofold serial dilutions. By quantitative analysis, α-proline 402(OH) recognizes its respective antigen with approximately eightfold more sensitivity than α-proline 564(OH) (Fig. 1C). These results indicate that we are able to monitor the hydroxylation of the proline residues individually with these antibodies.
Using these antibodies, we examined the kinetics of hydroxylation of the two proline residues to determine whether or not the two hydroxylation events are equivalent. Full-length human HIF-1α was transiently transfected into HIF-1α knockout mouse embryo fibroblasts (MEFs), allowing us to exclusively examine HIF-1α with specific point mutations without the confounding effects of endogenous HIF-1α protein (Fig. 1D). At 24 h posttransfection, these cells were treated with the proteasome inhibitor MG132 to prevent the rapid degradation of hydroxylated HIF-1α and harvested to make whole-cell protein extracts every hour. Despite the greater sensitivity of the α-P402(OH) antibody (Fig. 1B and C), hydroxylated proline 564 accumulated and was detected earlier than proline 402 (1 h versus 3 h), suggesting that proline 564 is hydroxylated prior to proline 402 (Fig. 1E). In parallel, we transiently transfected two point mutants, proline 402 mutated to an alanine (P402A) and proline 564 mutated to a glycine (P564G). Transfecting the proline residue into another amino acid prevents the residue from being hydroxylated but leaves the second hydroxylation site intact, allowing us to examine individual proline hydroxylation sites (Fig. 1D). Mutation of proline 564 altered the kinetics of hydroxylation of the remaining intact proline 402 compared to that of wild-type HIF-1α (Fig. 1E). Specifically, mutation of proline 564 delays and reduces the hydroxylation of proline 402, while mutation of proline 402 had relatively little influence on the hydroxylation of proline 564. These data suggest that hydroxylation of proline 564 occurs first, which in turns promotes the hydroxylation of proline 402. This influence, however, appears to be unidirectional in that hydroxylation of proline 564 remains unaffected by mutation of proline 402.
Proline 564 influences hydroxylation of proline 402.
To investigate whether the influence of proline 564 hydroxylation on proline 402 hydroxylation is restricted to the activity of specific prolyl hydroxylases, we examined hydroxylation of wild-type HIF-1α and the proline point mutants by individual prolyl hydroxylases. Full-length HIF-1α, the two proline point mutants, or a double proline mutant was cotransfected with one of three prolyl hydroxylases (prolyl hydroxylase domain 1 [PHD1] to -3) (8, 12, 40) and treated with proteasome inhibitor for 1 h to examine the protein levels by immunoblot analysis (Fig. 2A to C). Over this time course, we observed that there was no detectable hydroxylation by the endogenous PHDs, allowing us to examine only the hydroxylation by the transfected PHDs (Fig. 2D). We found that each prolyl hydroxylase is active on both proline hydroxylation sites (Fig. 2B and 2C) and that proline 564 is hydroxylated at equivalent levels on wild-type HIF-1α or when proline 402 is mutated (9). In contrast, proline 402 is hydroxylated much more efficiently by each of the three prolyl hydroxylases when both hydroxylation sites are intact compared to the results seen when proline 564 is mutated. These data indicate that the hydroxylation of proline 564 is the primary target of all three prolyl hydroxylases within the full-length HIF-1α protein, and it in turn affects the hydroxylation of proline 402.
Differential recognition of the hydroxylation sites by distinct hydroxylases.
We next examined whether the preferential recognition of the proline 564 over proline 402 by individual PHDs depends primarily on the sequence surrounding the prolines or on the conformational cues conferred by the location of the prolines within the full-length protein. To distinguish between these two models, we created a series of degradation domain-swapped constructs, replacing the 402 degradation domain with the 564 degradation domain and transferring the 564 degradation domain into the 402 site, along with the corresponding point mutants (Fig. 3A; see also Materials and Methods). These mutants allow us to determine whether either the 564 degradation domain sequence alone or its position within the full-length protein is sufficient to confer preferential activity of the individual PHDs. We transfected these constructs along with individual prolyl hydroxylases (PHD1 to -3) and treated cells for the indicated times with proteasome inhibitor (Fig. 3B to D). Hydroxylated proline 402 in the 564 site accumulated as early as h 1 to 2 when hydroxylated by PHD1 or PHD2 (Fig. 3B and C). Hydroxylated proline 564 in the 402 site was detectable at a low level when hydroxylated by PHD2, suggesting that PHD2 is responsible for hydroxylation of both proline residues regardless of location (Fig. 3C). PHD3 specifically hydroxylated proline 564, irrespective of location within the full-length protein (Fig. 3D). Furthermore, proline 402 was hydroxylated at a low level by PHD3 when proline 564 was mutated and HIF-1α stabilized. These results indicate that PHD1 and PHD2 recognize the prolyl hydroxylation sites on the basis of their spatial, conformational position within the full-length, folded protein. PHD3 recognizes proline 564 in a sequence-specific manner, regardless of its position within the protein, confirming that the previous observations of others using peptides extend to the full-length protein (12, 15).
Since the domains we swapped were short and did not include previously identified amino acids required for prolyl hydroxylase recognition (12, 15, 16, 18), we created a second series of domain-swapped constructs, switching exon 9 for exon 12. Exon 9 contains proline 402, whereas exon 12 contains proline 564. These exon-swapped constructs span larger regions, including several key residues, such as tyrosine 565 and leucine 574, which were determined to be essential for VHL binding to HIF-1α (16, 18, 25). Switching exons allowed us to examine the interactions between individual prolyl hydroxylases, the two conserved proline residues, and the surrounding sequences within the context of the full-length HIF-1α protein. Treatment with the hypoxia mimetic clearly demonstrates that DFO treatment of the transfected cells inhibited prolyl hydroxylation. Consistent with our above-described results, Western blot analysis of these constructs indicates that upon switching the sites of the two exons, PHD1 continues to recognize the 564 site and only weakly hydroxylating proline 564 in the 402 site (Fig. 4A). Using a more extensive region of swapping, we determined that PHD2 has a twofold requirement for both site and flanking sequences, since proline 564 now in the 402 site is more strongly hydroxylated when a larger region is swapped (Fig. 4B). Again, PHD3 prefers proline 564, regardless of its position within the full-length protein, but will weakly hydroxylate proline 402 (Fig. 4C). Consistent with the findings using shorter domains, these results indicate that PHD1 and PHD2 differentially recognize the two prolyl hydroxylation sites, primarily on the basis of location, although PHD2 appears to have a larger sequence requirement. PHD3, in contrast, is sequence specific in its recognition of its hydroxylation targets.
Proline mutants of HIF-1α can still be degraded.
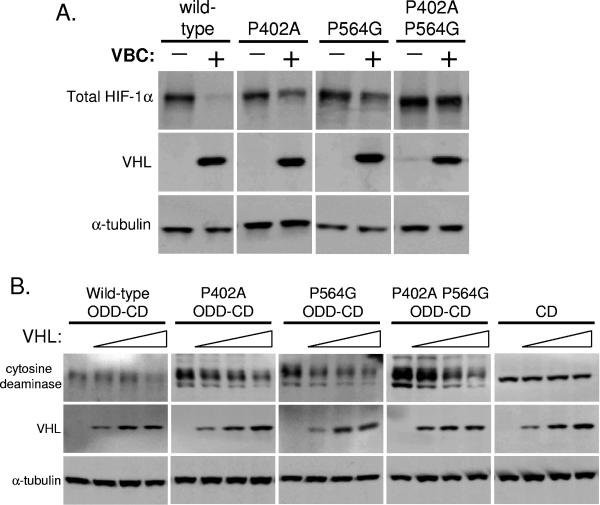
We next examined how differences in hydroxylation of the two prolines affected the degradation of HIF-1α. Previous studies have indicated that short peptides encompassing either the 402 ODD or the 564 ODD were able to interact with VHL and thus resulted in degradation, but these studies were unable to examine in the context of the full-length HIF-1α (20, 21, 33, 58). Given that our present results suggest a predominant role for proline 564 in regulating hydroxylation of proline 402, we wanted to determine whether having a single intact proline residue at 402 is sufficient for degradation in the context of the full-length protein. We transfected HIF-1α wild type, P402A, P564G, or the double proline mutant (P402A P564G) into the HIF knockout (HKO) MEFs. Cotransfection of VHL, elongin B, and elongin C (VBC complex) demonstrated that the single proline mutants could still be targeted for destruction in the presence of sufficiently high levels of the VBC complex, although notably to a lesser extent than wild-type HIF-1α (Fig. 5A). The double proline mutant, however, was not degraded by coexpression of the VBC complexes. These data indicate that despite stabilizing mutations (P402A and P564G), a single intact proline residue is sufficient for VHL-mediated degradation, but both sites are necessary for optimal targeting to the proteasome.
FIG. 5.
Single proline point mutants are still targeted for degradation by VHL. (A) HKO cells were transiently transfected with wild-type HIF-1α or mutant HIF-1α. In addition, cells were cotransfected with empty vector or VHL, elongin B, and elongin C. Immunoblot analyses were performed to examine the protein levels of total HIF-1α, VHL, and α-tubulin. (B) HKO cells were transiently transfected with CD fused with wild-type HIF-1α ODD (amino acids 338 to 603) or mutant HIF-1α along with human VHL in increasing amounts of plasmid. Immunoblot analyses were performed to examine the protein levels of cytosine deaminase, VHL, and α-tubulin.
Although an intact proline 402 is sufficient to mediate degradation of HIF-1α when proline 564 is mutated, we wanted to determine whether this mode of regulation of the degradation domains could be generalized in a chimeric protein setting. We constructed a fusion protein of E. coli cytosine deaminase (CD) and a region encompassing both degradation domains of HIF-1α (amino acids 338 to 603). This fusion, ODD-CD, or the corresponding proline point mutants were transfected into the HKO MEFs along with increasing amounts of VHL (Fig. 5B). ODD-CD with both proline sites intact (wild type) was degraded efficiently by low levels of VHL. It is noteworthy that in the chimeric setting, elimination of both proline residues does not completely abolish VHL-dependent degradation, suggesting that there are potentially additional, unknown mechanisms of regulation. This experiment suggests that targeting of heterologous proteins is possible on the basis of the interaction between HIF-1α and VHL and, potentially, oxygen tension.
Proline 402 is more responsive to changing oxygen tensions.
The predominant model for the regulation of HIF-1α stability, developed primarily from biochemical analyses, predicts that stabilizing stresses (e.g., hypoxia and prolyl hydroxylase inhibitors) function to increase HIF-1α protein levels by preventing the hydroxylation of proline 564 and proline 402 (9). We have previously shown that at least one oncogenic stabilizing stress does not reduce hydroxylation of proline 564, and our current data demonstrate that preventing the hydroxylation of one site is sufficient to significantly stabilize HIF-1α (Fig. 5A). This raises the question of whether stabilizing stresses work coordinately or independently to inhibit hydroxylation at each proline site. To examine endogenous HIF-1α hydroxylation of both proline 564 and proline 402 in response to various stabilizing stresses, RCC4 cells with and without functional VHL were subjected to hypoxia and the prolyl hydroxylase inhibitor dimethyloxalglycine (DMOG), cobalt chloride (CoCl2), or desferrioxamine (DFO). RCC4 cells are a renal clear-cell carcinoma cell line defective in VHL, which results in constitutive, normoxic HIF-1α protein. Introduction of wild-type VHL into these cells restores HIF-1α protein regulation. Immunoblot analysis of total cell extracts shows that RCC4 cells lacking functional VHL had detectable levels of hydroxylated 402 and hydroxylated 564 under normoxic conditions (Fig. 6A). Inhibition of the proteasome with MG132 resulted in stabilization of hydroxylated HIF-1α, allowing detection of both hydroxylated proline 402 and hydroxylated proline 564. Hydroxylation of HIF-1 at both sites decreases under conditions of severe oxygen deprivation and in the presence of any of the three prolyl hydroxylase inhibitors. Treatment with any of the three prolyl hydroxylase inhibitors reliably reduced the hydroxylation of both proline 402 and proline 564. Despite treatment for 6 h, there is a degree of hydroxylated HIF-1α that is not degraded during this time course, suggesting that there are no “dehydroxylating” enzymes, that perhaps the hydroxylation reaction is irreversible and only nonspecific protein turnover can eliminate hydroxylated HIF-1α, or that protein turnover is dramatically reduced, allowing the hydroxylated protein to persist despite exposure to the prolyl hydroxylase inhibitors. In contrast to cells that lack VHL, hydroxylation of both sites is completely impaired by these inhibitors in cells with wild-type VHL and is only detected in normoxic cells treated with proteasome inhibitor (Fig. 6B). These data are consistent with peptide and recombinant protein studies, which demonstrated that hypoxia and agents that inhibit prolyl hydroxylation of both proline 402 and proline 564 lead to HIF-1α stabilization (9, 20, 21, 33). Furthermore, these data demonstrate that both proline 402 and 564 are hydroxylated in the endogenous full-length HIF-1α protein and that hydroxylation of both prolines is inhibited in response to these stabilizing stresses.
Finally, given that both hydroxylation sites are targeted by stabilizing stresses and are sufficient to direct at least some degradation but that they are not equivalent in terms of kinetics or their influences on one another, we investigated whether the hydroxylation state of either site is preferentially changed as oxygen levels decreased. RCC4 cells without functional VHL were treated for 6 to 24 h at various levels of hypoxia (2%, 0.5%, and ≥0.02% O2). Samples were also subjected to proteasome inhibition for the last hour of hypoxic treatment. Hydroxylated proline 564 was evident under conditions of mild hypoxia (0.5% and 2% O2) without the use of a proteasome inhibitor and completely abolished by anoxia (Fig. 6C). Hydroxylated proline 402 was detectable at 2% O2 but not at 0.5% or under conditions of anoxia. Consistent with our findings that disrupting either hydroxylation site reduces degradation (Fig. 5A), this failure of hydroxylation of proline 402 at 0.5% O2 results in significant HIF-1α stabilization. These results indicate that despite the predominant role of proline 564 compared to proline 402 under normoxic conditions, the regulation of hydroxylation of proline 402 is integral for determining the levels of stabilized HIF-1α as oxygen levels decrease.
DISCUSSION
This study examines the interaction of the two destruction domains of HIF-1α within the context of the full-length protein. Other reports have examined hydroxylation of these two degradation domains individually but never in relation to one another (12, 15, 20, 21, 33, 40). These previous studies employed synthesized peptides and recombinant fusion proteins in an in vitro setting, conditions which are not easily amenable to identification of an interaction between two degradation domains. In particular, our studies revealed a unique interaction of the two degradation domains of HIF-1α in the context of the full-length protein in vivo.
As the two hydroxylation sites are highly conserved evolutionarily as well as across HIF-α subunits, we sought to understand why there are two hydroxylation sites and how each site contributes to HIF-1α degradation. In other proteins, such as Sic1p, multiple degradation domains represent a threshold that must be crossed before the protein is destroyed (39, 54). These destruction motifs can also be functionally redundant signals as dual signals are necessary for destruction in other proteins, such as IκBκ and β-catenin (7, 10, 37, 45). For HIF-1α, these multiple destruction domains are not completely independent but instead are coordinately regulated, with the hydroxylation of one site affecting the second site unidirectionally. This interdependence of modification sites has been reported for p53 in response to genotoxic stress, but in the case of p53, these modifications are not directly related to degradation (47). We discovered that the two proline hydroxylation residues (P402 and P564) of HIF-1α are not equivalently hydroxylated (Fig. 1E, 2B, and 2C). Furthermore, the hydroxylation of proline 564 occurs first and is essential for the efficient hydroxylation of proline 402, demonstrating that the 402 ODD is dependent on the 564 ODD in a nonreciprocal fashion (Fig. 1E).
A similar preference for one ODD was described in a recent report from Bemis et al., which identified CSN5, a subunit of the COP9 signalosome, as a modulator of HIF-1α activity under both aerobic and hypoxic conditions (3). More specifically, CSN5 was found to be necessary for the aerobic stabilization of HIF-1α protein and was required for optimum hypoxia-induced HIF-1α stabilization. CSN5 specifically bound proline 564 but not proline 402. Furthermore, the interaction of CSN5 with proline 564 was independent of hydroxylation. CSN5 overexpression prevents the hydroxylation of proline 564, indicating that CSN5 activity inhibits the prolyl hydroxylase activity on proline 564, which in turn negatively affects the hydroxylation of proline 402.
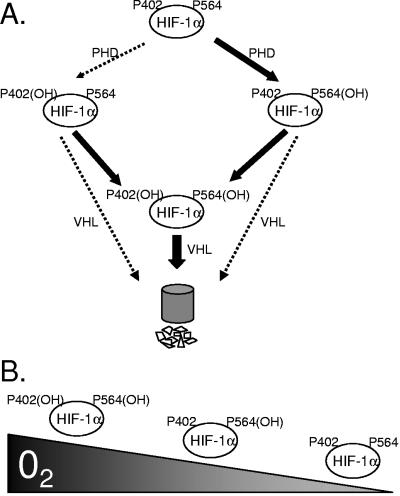
Our data suggest a model of carefully coordinated steps to ensure HIF-1α degradation under conditions of normoxia (Fig. 7A). Proline 564 is hydroxylated prior to proline 402 and increases the chances of hydroxylation of proline 402 (Fig. 1E). HIF-1α hydroxylated at proline 564 can then interact with either a PHD or VHL, resulting in hydroxylation of proline 402 or degradation, respectively. This singly hydroxylated molecule remains degradation competent in that VHL can, at a low frequency, interact to target HIF-1α for degradation. More likely, the HIF-1α protein hydroxylated at proline 564 enhances the interaction with a PHD and thus induces the hydroxylation of proline 402, resulting in two VHL recognition sites to ensure HIF-1α protein is targeted for degradation (Fig. 5).
FIG. 7.
Model of HIF-1α hydroxylation. (A) Under normoxic conditions, proline 564 of HIF-1α is recognized by a prolyl hydroxylase enzyme, leading to hydroxylation of proline 564. HIF-1α can now be recognized by VHL or by another hydroxylase. If VHL interacts, HIF-1α is degraded. Alternatively, hydroxylation of proline 564 enhances the hydroxylation of proline 402. Thus, a PHD can interact with HIF-1α protein hydroxylated at proline 564 to hydroxylate proline 402, providing VHL with two recognition sites to target HIF-1α protein for degradation. (B) As oxygen levels decrease, proline 402 hydroxylation is inhibited prior to proline 564 hydroxylation.
However, our data suggest that proline 402 is the sentinel hydroxylation site under hypoxic conditions. Under conditions of limiting oxygen concentrations, proline 402 is able to detect a decrease in oxygen tension more readily, priming HIF-1α to exquisitely respond to physiological changes in oxygen levels. As oxygen becomes limiting, proline 402 hydroxylation is inhibited prior to proline 564, and HIF-1α is stabilized and transcriptionally competent (Fig. 7B). Thus, it is possible that specific modulators of proline 402 or proline 564 could result in selective activation of HIF-responsive genes. As these sites are universal to HIF-1 and HIF-2 homologs, the potential for selective gene activation or modulation of the amplitude of gene activation should be examined.
Our results indicate specific recognition differences between the three prolyl hydroxylases for prolines 402 and 564 (Fig. 3 and 4). PHD1 and PHD2 appear to prefer proline 564 based primarily on its conformational location within the folded protein, whereas PHD3 is able to distinguish proline 564 based on sequence recognition, regardless of positioning. Differential regulation of these sites by the hydroxylases demonstrates yet another layer of the intricate modulation of HIF-1α protein that has evolved to regulate the expression of HIF targets. Berra et al. (5) and Appelhoff et al. (1) both demonstrated that PHD2 is the predominant prolyl hydroxylase responsible for maintaining low levels of HIF-1α protein under normoxic conditions. Taken with our data, this suggests that conformational recognition of HIF-1α by PHD2 may be the primary means of controlling HIF-1α protein under normoxic conditions. A recent report highlighted the concept that the contribution of a given PHD to HIF-1α regulation is directly related to the abundance of the PHD (1). Furthermore, Nakayama and colleagues identified Siah2 as a regulator of PHD1 and PHD3 abundance (38). Under conditions of hypoxia, it appears that PHD1 and PHD3 may be the critical prolyl hydroxylases responsible for maintaining HIF-1α protein at appropriate levels or ensuring the cell is able to respond rapidly to reoxygenation. Thus, under conditions of hypoxia, it appears that both conformation and sequence play roles in the hydroxylation of HIF-1α.
These studies highlight the need for future experiments to determine which prolyl hydroxylases are active under physiological conditions in vivo, including various oxygen tensions. Furthermore, a recent study by Baek et al. identified a novel protein, OS-9, which interacts with both HIF-1α and all three PHDs to promote hydroxylation of proline 564 (2). It will be important to understand how the PHDs themselves are regulated and how PHD2 differs from PHD1 and PHD3 in hydroxylating HIF under physiological conditions. The regulation of the degradation domains of HIF-1α by the prolyl hydroxylases demonstrates the sophisticated and complex control for oxygen homeostasis in eukaryotic cells.
Acknowledgments
We thank Eric Alemany and Sharon Clarke for all their technical support, Glenn Ruiz at ZYMED for excellent technical assistance, and Trent Watkins for critical review of the manuscript.
This investigation was supported by CA67166 (A.J.G.) and CA09302 (D.A.C. and P.D.S.), both awarded by the National Cancer Institute.
REFERENCES
- 1.Appelhoff, R. J., Y. M. Tian, R. R. Raval, H. Turley, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and J. M. Gleadle. 2004. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279:38458-38465. [DOI] [PubMed] [Google Scholar]
- 2.Baek, J. H., P. C. Mahon, J. Oh, B. Kelly, B. Krishnamachary, M. Pearson, D. A. Chan, A. J. Giaccia, and G. L. Semenza. 2005. OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha. Mol. Cell 17:503-512. [DOI] [PubMed] [Google Scholar]
- 3.Bemis, L., D. A. Chan, C. V. Finkielstein, L. Qi, P. D. Sutphin, X. Chen, K. Stenmark, A. J. Giaccia, and W. Zundel. 2004. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 18:739-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernad, R., H. van der Velde, M. Fornerod, and H. Pickersgill. 2004. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24:2373-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 22:4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahimi-Horn, C., N. Mazure, and J. Pouyssegur. 2005. Signalling via the hypoxia-inducible factor-1alpha requires multiple posttranslational modifications. Cell Signal. 17:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 267:1485-1488. [DOI] [PubMed] [Google Scholar]
- 8.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. A., P. D. Sutphin, N. C. Denko, and A. J. Giaccia. 2002. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J. Biol. Chem. 277:40112-40117. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., J. Hagler, V. J. Palombella, F. Melandri, D. Scherer, D. Ballard, and T. Maniatis. 1995. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 9:1586-1597. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 12.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson, K. S., L. A. McNeill, M. V. Riordan, Y. M. Tian, A. N. Bullock, R. W. Welford, J. M. Elkins, N. J. Oldham, S. Bhattacharya, J. M. Gleadle, P. J. Ratcliffe, C. W. Pugh, and C. J. Schofield. 2002. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277:26351-26355. [DOI] [PubMed] [Google Scholar]
- 15.Hirsila, M., P. Koivunen, V. Gunzler, K. I. Kivirikko, and J. Myllyharju. 2003. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278:30772-30780. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J., Q. Zhao, S. M. Mooney, and F. S. Lee. 2002. Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem. 277:39792-39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, L. E., E. A. Pete, M. Schau, J. Milligan, and J. Gu. 2002. Leu-574 of HIF-1alpha is essential for the von Hippel-Lindau (VHL)-mediated degradation pathway. J. Biol. Chem. 277:41750-41755. [DOI] [PubMed] [Google Scholar]
- 19.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 20.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 21.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 22.Jeong, J. W., M. K. Bae, M. Y. Ahn, S. H. Kim, T. K. Sohn, M. H. Bae, M. A. Yoo, E. J. Song, K. J. Lee, and K. W. Kim. 2002. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 111:709-720. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, B. H., E. Rue, G. L. Wang, R. Roe, and G. L. Semenza. 1996. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271:17771-17778. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 25.Kageyama, Y., M. Koshiji, K. K. To, Y. M. Tian, P. J. Ratcliffe, and L. E. Huang. 2004. Leu-574 of human HIF-1alpha is a molecular determinant of prolyl hydroxylation. FASEB J. 18:1028-1030. [DOI] [PubMed] [Google Scholar]
- 26.Katschinski, D. M., L. Le, D. Heinrich, K. F. Wagner, T. Hofer, S. G. Schindler, and R. H. Wenger. 2002. Heat induction of the unphosphorylated form of hypoxia-inducible factor-1alpha is dependent on heat shock protein-90 activity. J. Biol. Chem. 277:9262-9267. [DOI] [PubMed] [Google Scholar]
- 27.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 28.Lando, D., D. J. Peet, J. J. Gorman, D. A. Whelan, M. L. Whitelaw, and R. K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 30.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 31.Li, H., H. P. Ko, and J. P. Whitlock. 1996. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J. Biol. Chem. 271:21262-21267. [DOI] [PubMed] [Google Scholar]
- 32.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell, P. H., C. W. Pugh, and P. J. Ratcliffe. 2001. The pVHL-hIF-1 system. A key mediator of oxygen homeostasis. Adv. Exp. Med. Biol. 502:365-376. [PubMed] [Google Scholar]
- 35.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 36.McNeill, L. A., K. S. Hewitson, T. D. Claridge, J. F. Seibel, L. E. Horsfall, and C. J. Schofield. 2002. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem. J. 367:571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama, K., I. J. Frew, M. Hagensen, M. Skals, H. Habelhah, A. Bhoumik, T. Kadoya, H. Erdjument-Bromage, P. Tempst, P. B. Frappell, D. D. Bowtell, and Z. Ronai. 2004. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 117:941-952. [DOI] [PubMed] [Google Scholar]
- 39.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 40.Oehme, F., P. Ellinghaus, P. Kolkhof, T. J. Smith, S. Ramakrishnan, J. Hutter, M. Schramm, and I. Flamme. 2002. Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun. 296:343-349. [DOI] [PubMed] [Google Scholar]
- 41.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 42.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 43.Pugh, C. W., and P. J. Ratcliffe. 2003. The von Hippel-Lindau tumor suppressor, hypoxia-inducible factor-1 (HIF-1) degradation, and cancer pathogenesis. Semin. Cancer Biol. 13:83-89. [DOI] [PubMed] [Google Scholar]
- 44.Richard, D. E., E. Berra, E. Gothie, D. Roux, and J. Pouyssegur. 1999. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274:32631-32637. [DOI] [PubMed] [Google Scholar]
- 45.Rubinfeld, B., P. Robbins, M. El-Gamil, I. Albert, E. Porfiri, and P. Polakis. 1997. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275:1790-1792. [DOI] [PubMed] [Google Scholar]
- 46.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 47.Saito, S., H. Yamaguchi, Y. Higashimoto, C. Chao, Y. Xu, A. J. Fornace, Jr., E. Appella, and C. W. Anderson. 2003. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 278:37536-37544. [DOI] [PubMed] [Google Scholar]
- 48.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 49.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 50.Semenza, G. L. 2002. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8:S62-S67. [DOI] [PubMed] [Google Scholar]
- 51.Semenza, G. L. 1998. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 8:588-594. [DOI] [PubMed] [Google Scholar]
- 52.Semenza, G. L., M. K. Nejfelt, S. M. Chi, and S. E. Antonarakis. 1991. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc. Natl. Acad. Sci. USA 88:5680-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 54.Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 55.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]
- 57.Yasinska, I. M., and V. V. Sumbayev. 2003. S-nitrosation of Cys-800 of HIF-1alpha protein activates its interaction with p300 and stimulates its transcriptional activity. FEBS Lett. 549:105-109. [DOI] [PubMed] [Google Scholar]
- 58.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]