Abstract
MDMX is a homolog of MDM2 that is critical for regulating p53 function during mouse development. MDMX degradation is regulated by MDM2-mediated ubiquitination. Whether there are other mechanisms of MDMX regulation is largely unknown. We found that MDMX binds to the casein kinase 1 alpha isoform (CK1α) and is phosphorylated by CK1α. Expression of CK1α stimulates the ability of MDMX to bind to p53 and inhibit p53 transcriptional function. Regulation of MDMX-p53 interaction requires CK1α binding to the central region of MDMX and phosphorylation of MDMX on serine 289. Inhibition of CK1α expression by isoform-specific small interfering RNA (siRNA) activates p53 and further enhances p53 activity after ionizing irradiation. CK1α siRNA also cooperates with DNA damage to induce apoptosis. These results suggest that CK1α is a functionally relevant MDMX-binding protein and plays an important role in regulating p53 activity in the absence or presence of stress.
The p53 tumor suppressor is critical for maintenance of genome stability and protection against malignant transformation. p53 is regulated by multiple signal pathways and mechanisms, allowing it to respond to a wide range of stress conditions and act as a tumor suppressor in many cell types (44). p53 turnover is regulated by MDM2, which binds p53 and functions as an ubiquitin E3 ligase to promote p53 ubiquitination and degradation by the proteasomes (46). Stress signals, such as DNA damage, induce p53 accumulation by phosphorylation (33). Mitogenic signals activate p53 by induction of the ARF tumor suppressor, which inhibits the ability of MDM2 to ubiquitinate p53 (46). Ubiquitination of p53 and MDM2 requires the C-terminal RING domain of MDM2, which is involved in recruitment of the ubiquitin-conjugating enzyme E2 (22). Efficient ubiquitination of p53 also requires the central acidic domain of MDM2, which is an important regulatory domain targeted by ARF and possibly other signals (3, 28).
MDMX is a recently identified homolog of MDM2 (39). MDMX shares strong homology to MDM2 at the amino acid sequence level and can bind to p53 and inhibit its transcription function in transient-transfection assays. MDMX alone does not promote p53 ubiquitination or degradation in vivo (41). Furthermore, expression of MDMX mRNA is not induced by DNA damage (39). MDM2 is well established as an important regulator of p53 activity during embryonic development. Knockout of MDM2 in mice results in embryonic lethality due to hyperactivation of p53 (30). Recent studies showed that MDMX-null mice also die in utero in a p53-dependent fashion, which can be rescued by crossing into the p53-null background (11, 29, 32). Therefore, MDMX is also an important regulator of p53 during embryonic development, having functions that cannot be substituted by endogenous MDM2.
The embryonic lethality of the MDMX-null mouse suggests that MDMX mainly functions as a p53 inhibitor. Recent evidence suggests that MDMX may cooperate with MDM2 to promote p53 degradation, consistent with the genetic data from mice (18). Human tumor cell lines with wild-type p53 often express high levels of MDMX (34), suggesting that MDMX may contribute to p53 inactivation during tumorigenesis. Therefore, regulation of MDMX expression and function may be an important mechanism for controlling p53. Several laboratories showed that MDMX can be ubiquitinated and degraded by MDM2 (7, 24, 31). Interestingly, ARF inhibits the ability of MDM2 to ubiquitinate p53 but stimulates MDM2 ubiquitination of MDMX (31). This suggests a novel mechanism by which ARF activates p53 function by selective regulation of MDM2 E3 ligase function toward different substrates. The ability of ARF to promote MDMX ubiquitination connects MDMX to mitogenic signaling pathways through MDM2 (31). DNA damage also promotes MDMX nuclear translocation and degradation by the proteasomes (26, 31), suggesting that MDMX is an important target during p53 DNA damage response.
It is likely that MDMX inhibition of p53 is regulated by additional factors. To search for novel MDMX regulators, we performed affinity purification of ectopically expressed MDMX from HeLa cells and identified casein kinase 1 alpha (CK1α) as an MDMX-binding protein. The casein kinase 1 family of serine/threonine protein kinases is highly conserved from Saccharomyces cerevisiae to human and has been genetically linked to the regulation of DNA repair, cell cycle progression, and cytokines in yeast (16). Mammalian CK1 isoforms have recently been found to regulate the circadian clock (10), regulate Wnt1 signaling by promoting beta-catenine degradation (1, 27), and regulate apoptosis by phosphorylation of Bid (9). A fraction of CK1α localizes to nuclear structures associated with mRNA splicing (17). CK1δ and CK1ɛ have been implicated in phosphorylation of murine p53 (but not human) N-terminal serines 6 and 9, although the functional significance is unclear (25). Threonine 18 of human p53 can also be phosphorylated by CK1α in vitro, provided that serine 15 is modified by other kinases, such as ATM (25, 36). Functional studies described in this report demonstrate that CK1α stimulates the ability of MDMX to bind and inactivate p53. Depletion of CK1α by small interfering RNA (siRNA) activates p53 in cells and cooperates with ionizing radiation to activate p53. These observations suggest that CK1α is an important regulator of MDMX function and plays a role in regulating the p53 pathway.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents.
MDMX/p53 double-null mouse embryo fibroblast 41.4 is a kind gift from Guillermina Lozano (32). H1299 (non-small cell lung carcinoma, p53-null), U2OS (osteosarcoma, wild-type p53), MCF7 (breast carcinoma, wild-type p53), and PA1 (ovarian carcinoma, wild-type p53) were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum. HCT116-p53+/+ and HCT116-p53−/− cells were provided by Bert Vogelstein and maintained in McCoy 5A medium with 10% fetal bovine serum. Human MDMX cDNA with an N-terminal myc epitope tag (myc-MDMX) was kindly provided by Donna George (38). A FLAG epitope tag (DYKDDDDK) was added to the C terminus of myc-MDMX by PCR to create double-tagged myc-MDMX-FLAG, which was inserted into the pCMV-neo-Bam vector to facilitate high-level expression (2). MDMX deletion mutants were generated by PCR amplification and subcloning. CK1α expression plasmid pCS2-CK1α2 was kindly provided by David Virshup (12). Kinase-deficient CK1α mutants pCS2-CK1α2-K46A, pCS2-CK1α2-D136N, and pCS2-CK1α2-D136N-K138E were created by site-directed mutagenesis using the QuikChange kit (Stratagene). Escherichia coli expression of glutathione S-transferase (GST)-CK1α fusion protein was achieved by PCR amplification of the entire CK1α open reading frame and cloning into pGEX-3X vector. CK1-7 was purchased from Seikagaku Co.
Western blotting.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and centrifuged for 5 min at 10,000 × g, and the insoluble debris was discarded. Cell lysate (10 to 50 μg protein) was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline (PBS) containing 5% nonfat dry milk and 0.1% Tween 20. The following antibodies were used: 3G9 for MDM2 (5), DO-1 for p53 (Pharmingen), 8C6 monoclonal antibody or a rabbit polyclonal serum for MDMX (26), and goat anti-CK1α antibody for CK1α (Santa Cruz Biotechnology). The filter was developed using horseradish peroxidase-conjugated secondary antibodies and ECL-plus reagent.
Affinity purification of MDMX-associated protein.
HeLa-S cells stably transfected with FLAG-tagged MDMX were grown as a suspension culture. Cells from a 500-ml culture (∼2 × 108 cells) were lysed in 10 ml lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF) and centrifuged for 5 min at 10,000 × g, and the insoluble debris was discarded. The lysate was precleared with 100-μl bed volume of protein A Sepharose beads for 30 min and then incubated with 50-μl bed volume of M2-agarose bead (Sigma) for 4 h at 4°C. The beads were washed extensively with lysis buffer, and MDMX and its associated proteins were eluted with 70 μl of 20 mM Tris, pH 8.0, 2% SDS, and 200 μg/ml FLAG epitope peptide (Sigma) for 15 min. The eluted proteins were fractionated on SDS-polyacrylamide gels and stained with Coomassie blue. Proteins copurified with MDMX were excised from the gel and subjected to protease digestion and peptide sequencing by mass spectrometry at the Harvard Microchemistry Laboratory.
Immunoprecipitation and in vitro binding assay.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF) and centrifuged for 5 min at 10,000 × g, and the insoluble debris was discarded. Cell lysate (200 to 500 μg protein) was immunoprecipitated using 100 μl 8C6 hybridoma supernatant against MDMX and protein A agarose beads for 4 h at 4°C. The beads were washed extensively with lysis buffer, boiled in SDS sample buffer, fractionated by SDS-PAGE, and analyzed by anti-CK1α or anti-p53 Western blotting using rabbit or goat polyclonal antibodies. To determine MDMX-p53 or MDMX-CK1α binding efficiency, H1299 cells in 10-cm plates were transfected with 5 μg of MDMX and p53 or CK1α plasmids by calcium phosphate precipitation and cells were lysed for immunoprecipitation (IP) after 48 h.
To map the CK1α-binding domain on MDMX in vitro, [35S]methionine-labeled MDMX deletion mutants were expressed using the TNT in vitro transcription/translation kit (Promega). Five microliters of the translation product was incubated with glutathione agarose beads loaded with ∼0.1 μg E. coli-produced GST-CK1α in PBS with 0.1% Triton X-100 for 2 h at 4°C. The beads were washed with PBS with 0.1% Triton X-100 and fractionated by SDS-PAGE, and bound MDMX mutants were detected by autoradiography.
Luciferase reporter assay.
Cells (50,000/well) were plated in 24-well plates and transfected with a mixture containing 10 ng p53-responsive BP100-luciferase reporter plasmid (15), 5 ng CMV-lacZ plasmid, 0.1 ng p53 expression plasmid, 20 ng MDMX plasmid, and 20 ng CK1α plasmid. To detect endogenous p53 activity, cells were transfected as described above except without p53 expression plasmid. Transfection was achieved using Lipofectamine PLUS reagents (Invitrogen), and cells were analyzed for luciferase and beta-galactosidase expression after 24 h. The ratio of luciferase/beta-galactosidase activity was used as an indicator of p53 transcription activity.
In vitro phosphorylation and amino acid analysis.
To detect MDMX phosphorylation by coprecipitated kinases, cell lysate (0.5 mg protein) was immunoprecipitated with 8C6 and protein A Sepharose beads (10-μl bed volume). The beads were washed with lysis buffer and suspended in 20 μl kinase buffer (25 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.1 mM dithiothreitol), 10 μCi [γ-32P]ATP (5,000 Ci/mmol) was added, and the suspension was incubated at 30°C for 30 min. To detect MDMX phosphorylation by recombinant CK1α, His6-MDMX (1 μg) was immunoprecipitated with 8C6 and the beads were incubated with 0.1 μg purified GST-CK1α and [γ-32P]ATP as described above. Phosphorylated MDMX was detected by SDS-PAGE and autoradiography after transfer to a nylon membrane. For phosphoamino acid analysis, a nylon membrane containing radiolabeled MDMX band was excised and incubated with 5.7 N HCl at 110°C for 1 h. The hydrolyzed MDMX residues were lyophilized, mixed with phosphoamino acid standards (Sigma), and analyzed by two-dimensional electrophoresis on thin-layer cellulose plate using a pH 1.9 buffer for the first dimension and pH 3.5 buffer for the second dimension on a HTLE-7002 apparatus (C.B.S Scientific).
In vivo phospholabeling and phosphopeptide analysis.
To detect MDMX phosphorylation in vivo, H1299 or 293T cells in 10-cm plates were transiently transfected with 10 μg MDMX and 5 μg CK1α expression plasmids. Forty hours after transfection, cells were washed with DMEM without phosphate and incubated with 32Pi (0.2 mCi/ml) in DMEM without phosphate for 4 h. Cell lysate was immunoprecipitated with 8C6 and analyzed by SDS-PAGE and autoradiography. An identical set of 8C6 IP samples was analyzed by MDMX Western blotting to confirm the protein expression level.
To analyze phosphorylation of MDMX by two-dimensional peptide mapping, nylon membranes containing radiolabeled MDMX bands from in vivo and in vitro labeling were excised and incubated with 50 ng endoproteinase Asp-N (Sigma) for 16 h in 50 mM ammonium bicarbonate at 37°C. An aliquot of the Asp-N-digested sample was further digested with 500 ng trypsin (Sigma) for 16 h at 37°C. MDMX peptides were oxidized with performic acid and resolved by electrophoresis on a thin-layer cellulose plate for 30 min at 1.0 kV in formic acid-glacial acetic acid-water (1:3.1:35.9; pH 1.9) using the HTLE-7002 apparatus. This was followed by chromatography in the second dimension in n-butyl alcohol-pyridine-glacial acetic acid-water (5:3.3:1:4) for 16 h. The phosphopeptides were visualized by autoradiography and eluted from the plate with electrophoresis buffer. The purified phosphopeptides were subjected to manual Edman degradation as described previously to identify the phosphorylation sites (35). Phosphopeptides were covalently coupled to Sequelon-AA disks (Applied Biosystems) and subjected to consecutive cycles of Edman degradation. The amount of 32P label released in each cycle was determined by liquid scintillation counting.
CK1α RNA interference (RNAi).
Cells were transfected with 200 nM control siRNA (AATTCTCCGAACGTGTCACGT) and CK1α siRNA (AATCTCAGAAGGCCAGGCATC) using Oligofectamine (Invitrogen) according to the instructions from the supplier. After 48 h of transfection, cells were irradiated with 10 Gy gamma radiation and analyzed for protein expression or luciferase activity after 6 h. For cell death assay, cells were transfected for 24 h and camptothecin was added to medium containing siRNA and incubated for an additional 24 h. The numbers of viable cells were compared using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Cell Titer kit; Promega).
RESULTS
Copurification of CK1α with MDMX.
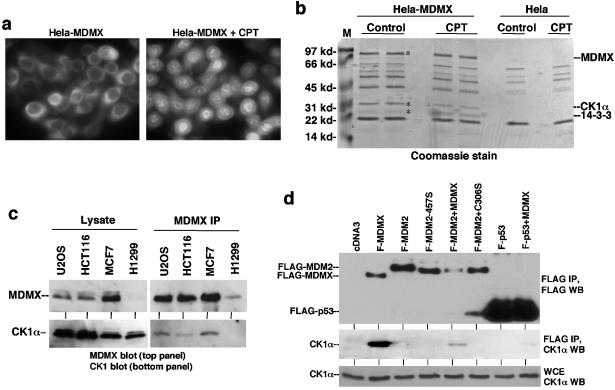
In order to determine whether MDMX interacts with novel cellular proteins, HeLa cells were stably transfected with FLAG-MDMX to obtain a sufficient amount of material for affinity purification. Immunofluorescence staining of transfected MDMX showed that the majority of MDMX was located in the cytoplasm. Treatment with the DNA-damaging agent camptothecin led to significant nuclear translocation of MDMX (Fig. 1a). When MDMX was purified by anti-FLAG immunoprecipitation from control and camptothecin-treated cells, two proteins reproducibly copurified with MDMX (Fig. 1b). By peptide sequencing by mass spectrometry, the 34-kDa protein was determined to be casein kinase 1 alpha, and the 24-kDa protein was determined to be 14-3-3τ. The efficiency of CK1α copurification judged from the intensity after Coomassie blue staining suggested that it is an abundant and stable binding partner of MDMX.
FIG. 1.
CK1α forms a complex with MDMX. (a) HeLa cells stably transfected with FLAG-tagged human MDMX were stained using anti-FLAG M2 antibody before or after treatment with 0.5 μM camptothecin (CPT) for 16 h. (b) Coomassie blue staining of affinity-purified MDMX and associated proteins. The two marked bands were identified as CK1α and 14-3-3τ by mass spectrometric peptide sequencing. Control purification was performed using HeLa cells to exclude background. The positions of molecular mass markers (M) (in kilodaltons) are shown to the left of the gel. (c) Cell lines expressing different levels of endogenous MDMX were analyzed by MDMX IP/CK1α Western blotting. The relative expression levels of MDMX and CK1α were determined by direct Western blotting of identical amounts of whole-cell extract from each cell line. (d) H1299 cells transfected with FLAG-tagged MDMX (F-MDMX), F-MDM2, and F-p53-281G mutant were analyzed by anti-FLAG IP and anti-CK1α Western blotting (WT) to detect coprecipitation of endogenous CK1α. Cotransfection of nontagged MDMX was tested for mediating formation of trimeric complexes. F-MDM2-457S is a RING domain mutant for control. C306S is an MDMX zinc finger mutant defective for CK1α binding. WCE, whole-cell extract.
The major function of 14-3-3 proteins is to bind to proteins with phosphorylated serine and threonine residues and regulate their functions through a wide range of mechanisms (42, 43). The binding efficiency of MDMX to 14-3-3 was increased after DNA-damaging treatment (Fig. 1b), suggesting that MDMX phosphorylation may be stimulated by DNA damage. The MDMX-14-3-3 interaction is currently under investigation and will not be addressed here, since current results suggest that it is not directly related to CK1α phosphorylation of MDMX.
To confirm the interaction between endogenous MDMX and CK1α, several tumor cell lines were immunoprecipitated with MDMX antibody 8C6, followed by anti-CK1α Western blotting. The results showed that endogenous MDMX in these cell lines was also coprecipitated with CK1α (Fig. 1c). The amount of CK1α coprecipitated by the 8C6 antibody was dependent on the expression levels of MDMX in different cell lines. Therefore, MDMX forms a specific complex with endogenous CK1α.
MDMX shares extensive homology to MDM2, which has been shown to interact with the CK1δ isoform (45). p53 has also been shown to bind CK1δ and CK1ɛ isoforms (25). To determine whether MDMX-CK1α interaction is specific, the binding efficiencies of CK1α to MDMX, MDM2, and p53 were directly compared. H1299 cells were transfected with FLAG-tagged MDM2, MDMX, and p53 expression plasmids, and coprecipitation of endogenous CK1α was determined by FLAG IP and CK1α Western blotting. Control Western blotting using FLAG antibody served as a control for the relative expression levels of MDM2, MDMX, and p53. Under these conditions, only MDMX showed significant binding to endogenous CK1α (Fig. 1d). MDM2 and p53 showed no binding to CK1α despite being expressed at higher levels. However, FLAG-MDM2 or FLAG-p53 coprecipitated CK1α when coexpressed with nontagged MDMX (Fig. 1d). As expected, the MDMX-C306S point mutant defective for CK1α binding (see below) did not mediate CK1α binding to MDM2 or p53. Therefore, MDMX interacts specifically with CK1α and can also target CK1α to MDM2 or p53.
p53 binding and inhibition by MDMX are stimulated by CK1a.
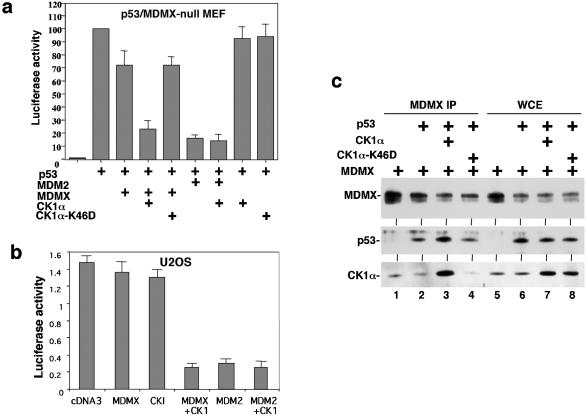
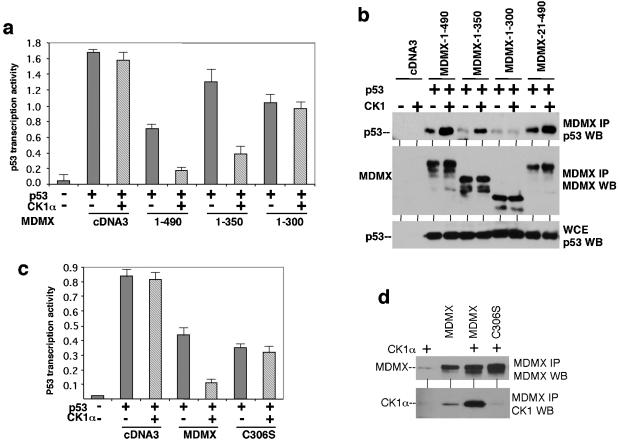
To investigate the function of MDMX-CK1α interaction, the effect of CK1α expression on p53 transcriptional activity was examined using mouse embryo fibroblasts derived from the MDMX/p53 double-null mouse. p53 cotransfection with the BP100-luciferase reporter containing a p53-binding site derived from the MDM2 P2 promoter resulted in strong activation of luciferase expression (15). Cotransfection with MDMX expression plasmid resulted in only weak inhibition of p53 function, whereas cotransfection with MDM2 resulted in more efficient inhibition of p53 (Fig. 2a). The weak effect of MDMX in p53 inhibition was typical in our experiments and consistent with other studies (18, 39). Cotransfection with CK1α expression plasmid significantly improved the ability of MDMX to inhibit p53 (Fig. 2a), whereas CK1α alone had negligible effect on p53 activity. MDM2 expression inhibited p53 more efficiently, but its effect was not significantly stimulated by CK1α overexpression (Fig. 2a). Therefore, CK1α specifically increased the ability of MDMX to inhibit p53 transcription function. A kinase-deficient CK1α mutant (K46A) was not able to stimulate MDMX function (Fig. 2a and c) (12), suggesting that CK1α may modulate MDMX function by phosphorylation of MDMX.
FIG. 2.
CK1α stimulates MDMX binding and inhibition of p53. (a) p53 activation of BP100-luciferase reporter was detected after transient transfection into MDMX/p53 double-null 41.4 mouse embryo fibroblasts (MEF). The inhibitory effects of MDMX and CK1α were determined by cotransfection. CK1α-K46D is a kinase-deficient mutant for control. (b) Endogenous p53 activity in U2OS cells were measured by transfection of BP100-luciferase. Inhibition by MDMX and CK1α was determined by cotransfection and luciferase assay. (c) Binding efficiency between MDMX and p53 was determined by transient cotransfection of H1299 cells with MDMX and p53 expression plasmids, followed by MDMX IP/p53 Western blotting. Whole-cell extracts (WCE) were analyzed to confirm similar levels of protein expression.
To test the effect of CK1α on endogenous p53 activity, U2OS cells were transfected with the BP100-luciferase reporter. Cotransfection with MDM2 plasmid resulted in strong inhibition of luciferase expression (Fig. 2b), indicating that the readout reflected endogenous p53 activity. MDMX cotransfection alone only weakly inhibited p53, while coexpression of CK1α stimulated the effect of MDMX by fourfold (Fig. 2b). CK1α alone also weakly inhibited the reporter, possibly due to the presence of endogenous MDMX in this cell line. In separate experiments, CK1α also stimulated the ability of MDMX to inhibit activation of GAL4-tk-luciferase reporter by GAL5-p53-1-52 fusion protein containing the p53 N-terminal 52 residues (data not shown). Therefore, CK1α regulates p53 transcription activation function by interacting with MDMX.
To determine how CK1α stimulated MDMX inhibition of p53, we examined the ability of CK1α to modulate MDMX ubiquitination and degradation by MDM2. The results indicated that CK1α overexpression did not have a significant effect in these assays (data not shown). However, when CK1α was coexpressed with MDMX, the ability of MDMX to bind p53 was significantly increased (Fig. 2c, compare lanes 2 and 3). The kinase-deficient CK1α-K46A mutant did not stimulate MDMX-p53 binding; it also did not coprecipitate with MDMX (Fig. 2c) (further addressed in Fig. 8). Ectopic expression of CK1α did not affect the expression level of p53 and MDMX in these transfection experiments, suggesting that the kinase worked by enhancing the binding between p53 and MDMX.
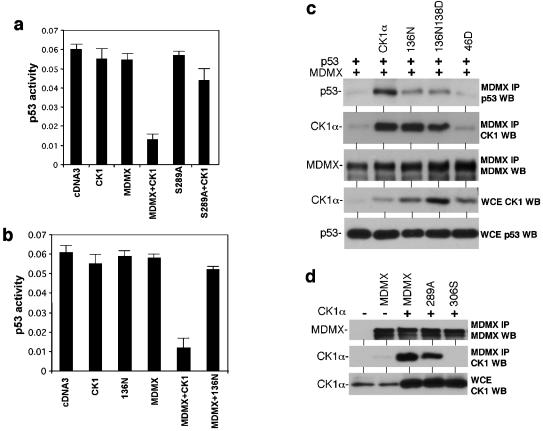
FIG. 8.
Phosphorylation of MDMX serine 289 by CK1α is required for functional regulation. (a) U2OS cells were transfected with BP100-luciferase reporter and expression plasmids for MDMX, MDMX-S289A mutants, and CK1α. Luciferase activity was determined after 24 h as an indicator of endogenous p53 activity. (b) U2OS cells were transfected with BP100-luciferase reporter and expression plasmids for MDMX, CK1α, and CK1α-D136N. The effects of the combinations on endogenous p53 activity were determined after 24 h. (c) H1299 cells were transfected with p53 and MDMX expression plasmids in combination with wild-type CK1α and CK1α with point mutations. The effects of the kinase-deficient mutants on p53-MDMX binding efficiency were determined after 48 h by MDMX IP and p53 Western blotting (WB). The ability of CK1α-D136N and CK1α-D136N-K138D mutants to bind MDMX was confirmed by MDMX IP and CK1α Western blotting. WCE, whole-cell extracts. (d) H1299 cells were transfected with MDMX mutant and CK1α expression plasmids. The ability of MDMX-289A to bind CK1α was determined by MDMX IP and CK1α Western blotting (WB). MDMX-306S served as a negative control.
Mapping of CK1α-binding site on MDMX.
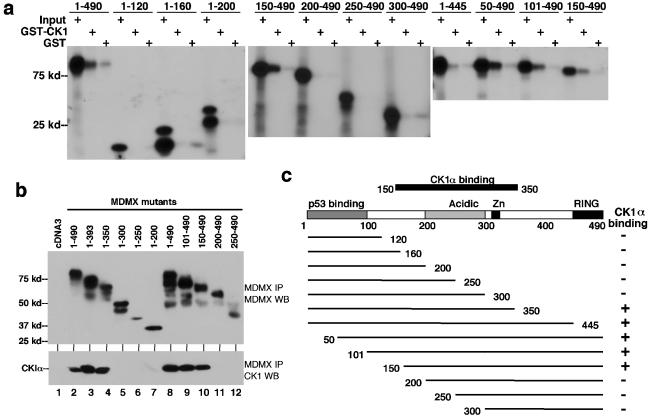
In order to test the specificity and elucidate the mechanism by which CK1α stimulates the ability of MDMX to inhibit p53, we mapped the region of MDMX important for binding to CK1α. A panel of MDMX N- and C-terminal truncation mutants were translated in vitro and incubated with glutathione beads loaded with E. coli-expressed GST-CK1α (Fig. 3a). The result of the in vitro binding showed that the integrity of region 150-350 of MDMX was important for binding to CK1α (Fig. 3c). The structural requirement for CK1α binding in vivo was also examined using the same panel of MDMX deletion mutants by cotransfection with CK1α expression plasmid into H1299 cells. Coprecipitation between MDMX and CK1α was determined by MDMX IP, followed by CK1α Western blotting. The results of the in vivo binding assay were identical to the results of the in vitro binding assay (Fig. 3b). The MDMX domain required for CK1α binding involved a rather large region, including the acidic domain and zinc finger. Deletions from both boundaries lead to complete loss of CK1α binding, suggesting that the three-dimensional structure formed by this region was important for interaction with CK1α. The importance of the MDMX zinc finger for CK1α binding was also confirmed using point mutations (see below).
FIG. 3.
Mapping of CK1α-binding region on MDMX. (a) MDMX deletion mutants were generated by in vitro translation and incubated with glutathione agarose beads loaded with GST-CK1α. Bound MDMX proteins were detected by SDS-PAGE and autoradiography. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (b) MDMX deletion mutants were transiently cotransfected with CK1α expression plasmid into H1299 cells. MDMX was immunoprecipitated using 8C6 (epitope located between positions 101 to 150) or a rabbit polyclonal serum (for mutants without the 8C6 epitope). Coprecipitated CK1α was detected by Western blotting (WB). Expression and immunoprecipitation of MDMX mutants were confirmed by Western blotting (top panel). (c) Diagram of MDMX deletion mutants and summary of binding results.
MDMX-CK1α interaction is required for regulation by CK1a.
To determine whether MDMX regulation by CK1α requires complex formation with CK1α, a panel of MDMX C-terminal deletion mutants that retained the p53-binding domain was used to test the response to CK1α regulation. The ability of these mutants to inhibit p53 transcriptional function in reporter assay was determined in the presence or absence of CK1α cotransfection. The results showed that the C-terminal RING domain of MDMX was not required for stimulation by CK1α; however, deletion of part of the CK1α-binding region, including the zinc finger, completely abolished the response to CK1α (Fig. 4a). As expected, the binding between zinc finger deletion mutant (1-300) and p53 was not stimulated by CK1α coexpression (Fig. 4b). Therefore, interaction between MDMXand CK1α is necessary for stimulating MDMX-p53 binding and inhibition.
FIG. 4.
CK1α binding to MDMX is required for p53 regulation. (a) p53 activation of BP100-luciferase reporter was detected after transient transfection into MDMX/p53 double-null 41.4 cells. The inhibitory effects of MDMX mutants and stimulation by CK1α were determined by cotransfection. (b) Binding efficiency between MDMX mutants and p53 was determined by transient cotransfection of H1299 cells with MDMX, p53, and CK1α expression plasmids, followed by MDMX IP/p53 Western blotting (WB). Whole-cell extracts (WCE) were analyzed to confirm similar levels of p53 expression. (c) MDMX-C306S zinc finger point mutant was not regulated by CK1α in a luciferase reporter assay similar to the results shown in panel a. (d) MDMX-C306S mutant did not bind CK1α after cotransfection into H1299 cells.
In order to specifically determine the role of the MDMX zinc finger motif in CK1α binding and regulation, a point mutant of the conserved cysteine residue (C306S) was created. MDMX-C306S was found to be nonresponsive to CK1α stimulation in p53 reporter gene assay (Fig. 4c). It was also deficient for CK1α binding in coimmunoprecipitation assay (Fig. 4d). Therefore, the MDMX zinc finger motif was critical for both physical and functional interactions with CK1α.
MDMX is a phosphorylation substrate for CK1a.
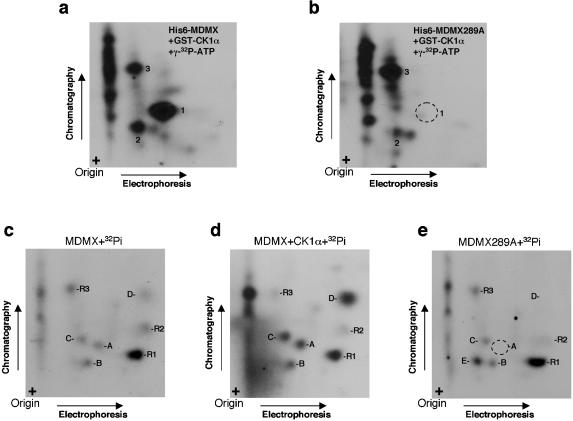
To test whether CK1α overexpression promotes MDMX phosphorylation, H1299 cells were transfected with MDMX and CK1α expression plasmids and labeled with [32P]phosphate. MDMX was immunoprecipitated from labeled cells, and incorporation of 32P was detected by autoradiography. The results showed that MDMX was phosphorylated in the absence of CK1α cotransfection and that CK1α overexpression led to a moderate increase in the level of 32P incorporation (Fig. 5a). This suggested that MDMX was phosphorylated on multiple sites by multiple kinases, and the effect of additional phosphorylation on CK1α sites was partially obscured by the background (see peptide mapping in Fig. 7). In an IP-kinase assay, MDMX was immunoprecipitated using 8C6 and incubated with [γ-32P]ATP. MDMX was phosphorylated in this reaction by a coprecipitating kinase. Addition of the CK1-specific inhibitor CKI-7 to the in vitro kinase reaction strongly inhibited the labeling (Fig. 5b) (6), suggesting that CK1α was a major MDMX-binding kinase and responsible for much of the in vitro labeling reaction. Treatment of cells by ionizing radiation or camptothecin before MDMX IP led to degradation of MDMX as described previously (31) but did not abrogate MDMX phosphorylation in vitro.
FIG. 5.
Phosphorylation of MDMX by CK1α. (a) H1299 cells were transiently transfected with MDMX and CK1α and incubated with 32Pi for 3 h. MDMX was immunoprecipitated by 8C6 and detected by autoradiography. Duplicate samples were analyzed by MDMX Western blotting (WB) to confirm expression. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (b) MCF7 cells were immunoprecipitated with 8C6, and the immunocomplex was incubated with [γ-32P]ATP. CKI-7 (70 μM) was used to inhibit CK1 in the reaction. Cells were treated with DNA-damaging agents (10 Gy gamma radiation or 1 μM camptothecin [CPT]) for 6 h. H1299 cells expressing very low-level MDMX were used as a control. Phosphorylation of MDMX was detected by autoradiography, and protein levels were determined by Western blotting. (c) His6-MDMX was expressed in E. coli, purified on a Ni2+-nitrilotriacetic acid column, and immunoprecipitated by 8C6. The immunocomplex was incubated with GST-CK1α and [γ-32P]ATP. Phosphorylation of MDMX was detected by autoradiography, and the multiple lower bands were confirmed to be truncated forms of GST-MDMX by Western blotting (WB).
FIG. 7.
Confirmation of serine 289 phosphorylation in vitro and in vivo. (a, b) Wild-type His6-MDMX or MDMX-289A mutant was phosphorylated by GST-CK1α and [γ-32P]ATP in vitro, followed by Asp-N digestion and two-dimensional peptide mapping. Mutation of serine 289 eliminated the major phosphopeptide spot 1. (c, d, e) 293T cells transfected with MDMX, MDMX and CK1α, and MDMX-289A mutant were labeled for 4 h with 32Pi. MDMX was immunoprecipitated with 8C6, digested with Asp-N, and analyzed by two-dimensional peptide mapping. Spot A migrated at the same position as the S289-containing peptide 1 in vitro, was enhanced by coexpression of CK1α, and was eliminated by the 289A mutation.
To further determine whether MDMX was a direct phosphorylation substrate for CK1α, His6-MDMX was expressed in E. coli, immunoprecipitated with 8C6, and incubated with purified GST-CK1α and [γ-32P]ATP. MDMX was phosphorylated by GST-CK1α, but not kinase-deficient GST-CK1α-K46A (Fig. 5c). Furthermore, no phosphorylation of immunoglobulin heavy or light chains by GST-CK1α was detected, suggesting phosphorylation of MDMX by CK1α was specific. These results demonstrated that MDMX is a substrate for CK1α.
Phosphorylation of MDMX serine 289 by CK1a.
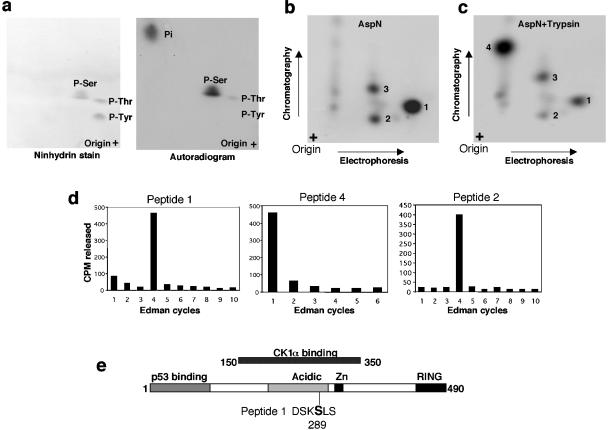
CK1α does not have a well-defined target consensus sequence but preferentially targets serine or threonine in the context of acidic amino acids or containing phosphorylated serine at the −3 position (13, 14, 40). The CK1α-binding region on MDMX includes the acidic domain rich in serine and threonine residues and presents numerous potential modification sites for CK1α. Initial efforts to identify phosphorylation sites by protease digestion and mass spectrometry or to target mutagenesis of conserved serine and threonine residues were uninformative. Therefore, a two-dimensional peptide mapping in combination with Edman degradation approach was employed.
Phosphoamino acid analysis of MDMX phosphorylated by CK1α in vitro showed predominant modification of serine residues (Fig. 6a). Digestion of in vitro-phosphorylated MDMX by endoproteinase Asp-N (cleaves before aspartic acid), followed by two-dimensional separation on thin-layer cellulose plates, showed one major spot (peptide 1) and two minor spots (peptides 2 and 3 [Fig. 6b]). Double digestions of MDMX with Asp-N and trypsin (cleaves after lysine) converted peptide 1 into peptide 4 (Fig. 6c). Edman degradation of recovered peptides showed that peptide 1 contains a phosphorylated residue at the fourth position from the N terminus. When peptide 1 was cleaved into peptide 4 by trypsin, the first position contained the phosphorylated residue. Inspection of the MDMX sequence indicated that the only serine residue that can fulfill these requirements is serine 289 (Fig. 6e). Peptide 2 is most likely an incomplete digestion product containing peptide 1, since it also contains radiolabeled residue at the fourth position (Fig. 6d), can be cleaved further by trypsin (Fig. 6c), and was lost after mutation of serine 289 (Fig. 7b). The minor phosphorylation site represented by peptide 3 was released after 13 cycles of Edman degradation, representing C-terminal serine 408. This site was not investigated further; because the in vivo metabolic labeling of MDMX did not generate such a peptide, it was likely an artifact of in vitro phosphorylation reaction.
FIG. 6.
Identification of in vitro CK1α phosphorylation site on MDMX. (a) His6-MDMX phosphorylated by GST-CK1α and [γ-32P]ATP in vitro was hydrolyzed by HCl, and phosphoamino acids were analyzed by two-dimensional electrophoresis. Ninhydrin stain indicates the positions of phosphoamino acid standards. P-Ser, phosphorylated serine. (b and c) His6-MDMX phosphorylated by GST-CK1α and [γ-32P]ATP was digested with Asp-N or Asp-N/trypsin. Phosphopeptides were analyzed by electrophoresis, followed by chromatography on a cellulose plate. (d) Phosphopeptides recovered from panels b and c were subjected to manual Edman degradation, and the release of radioactivity in each cycle was measured to determine the position of the phosphoamino acid from the N terminus. (e) Serine 289 was identified as the major CK1α phosphorylation site on the basis of Edman degradation results and sequence inspection.
To confirm that serine 289 was the CK1α phosphorylation site, a serine-to-alanine substitution was introduced. As expected, the MDMX-289A mutant failed to generate the phosphorylated peptide 1 by CK1α in vitro (compare Fig. 7a and b). To confirm that serine 289 was also phosphorylated in vivo by CK1α, phospholabeling using 32Pi was performed. As anticipated from the result of Fig. 5a, multiple phospholabeled peptides were detected after Asp-N digestion of immunoprecipitated MDMX from transfected cells (Fig. 7c). Spot A migrated at the same position as peptide 1 from in vitro-phosphorylated MDMX. Cotransfection of MDMX with CK1α stimulated the phosphorylation of at least four spots (spots A, B, C, and D compared to relatively unchanged reference spots R1, R2, and R3) (Fig. 7d). The MDMX-289A mutant did not generate spot A after in vivo labeling (Fig. 7e), consistent with this residue being responsible for the labeling of spot A.
These results indicated that serine 289 is the major CK1α phosphorylation site in vitro and is one of possibly several CK1α phosphorylation sites in vivo. Whether the other in vivo CK1α-regulated spots (B, C, and D) are due to direct modification by CK1α remains to be determined. The presence of several CK1α-independent phosphopeptides from in vivo labeling (R1, R2, and R3) is consistent with our recent identification of several DNA damage-regulated phosphorylation sites at the C-terminal region of MDMX (unpublished results), which are expected to be phosphorylated under the radiolabeling condition. The 289A mutation also caused intense labeling of a previously very weak spot (Fig. 7e, spot E), possibly forcing the kinase to modify a weak site.
Phosphorylation of MDMX by CK1α is required for functional regulation.
To determine whether phosphorylation of MDMX on serine 289 plays a role in CK1α regulation of MDMX-p53 functional interaction, the S289A mutant was cotransfected with CK1α into U2OS cells. Readout of endogenous p53 activity from the cotransfected BP100-luciferase reporter showed that the MDMX-S289A mutant was significantly less responsive to stimulation by CK1α (Fig. 8a). These results suggested that modification of serine 289 is critical for regulation by CK1α. It is possible that phosphorylation of additional sites directly or indirectly by CK1α in vivo may also play a contributing role, since the S289A mutant still retained a weak response.
Since CK1α copurified with MDMX at close to 1:1 molar ratio as judged by Coomassie blue staining (Fig. 1b), it raised the question of whether binding by the kinase alone is sufficient to affect MDMX-p53 interaction. The CK1α-K46D mutant did not bind MDMX (Fig. 2c and 8c), precluding such an analysis. Therefore, two additional CK1α mutants were generated with substitutions in the ATP-binding loop (CK1α-D136N and CK1α-D136N-K138E). The CK1α-D136N mutant was previously shown to have lost >90% activity against a peptide substrate (47). Both mutants were found to have completely lost the ability to phosphorylate MDMX in vitro, similar to CK1α-K46D (data not shown). In luciferase reporter assays, CK1α-D136N failed to promote p53 inhibition by MDMX (Fig. 8b). Furthermore, the ability of the CK1α-D136N and CK1α-D136N-K138E mutants to stimulate MDMX-p53 binding was also lost, despite retaining the ability to bind MDMX (Fig. 8c). A control experiment showed that MDMX-S289A retained the ability to bind CK1α, although at slightly reduced efficiency compared to the wild type (Fig. 8d). These results favor the interpretation that phosphorylation of MDMX serine 289 by CK1α is important for regulating its p53-binding function.
RNAi of CK1α activates p53.
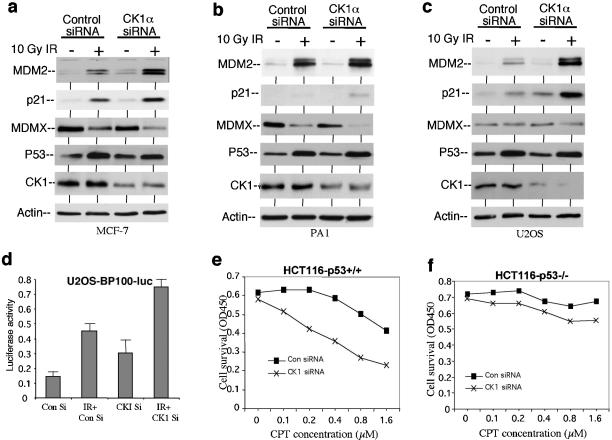
To determine whether endogenous CK1α plays a role in regulating p53 function, we transfected MCF7, PA1, and U2OS cells with a previously described isoform-specific siRNA against CK1α (27), which reduced the level of CK1α by two- to threefold. This partial down regulation also correlated with higher MDM2 and p21 expression after gamma irradiation (Fig. 9a, b, and c). Furthermore, the stably transfected p53-responsive BP100-luciferase reporter in these cells were also activated by the combination of CK1α siRNA and irradiation (Fig. 9d), suggesting that the siRNA induced p53 transcriptional activity, rather than altering the stability of MDM2 or p21 (20, 21, 38, 41). To rule out off-target effects of the siRNA, an additional CK1α isoform-specific siRNA targeting a different region of the CK1α sequence was also tested and generated similar results (data not shown). Therefore, these results suggest that endogenous CK1α plays a role in inhibiting p53 activity, possibly through interacting with MDMX. The level of MDMX decreased after DNA damage as previously reported (Fig. 9a and b) (24, 31). Recent experiments suggest that this is triggered by phosphorylation of the MDMX C-terminal region by an ATM kinase-dependent pathway and is not directly related to nor requires CK1α (unpublished results).
FIG. 9.
Inhibition of CK1α expression by RNAi activates p53. (a, b, c) Cells were transfected with control or CK1α-specific siRNA (200 nM) for 48 h and treated with 10 Gy gamma irradiation (IR) for 6 h. MDM2 and p21 levels were determined by Western blotting. (d) U2OS cells stably transfected with p53-responsive BP100-luciferase reporter were treated as in panel c, and luciferase activity was determined using identical amount of cell lysate. Con Si, control siRNA; IR + CKI Si, gamma irradiation and CK1α-specific siRNA. (e, f) HCT116 cells were transfected with control siRNA (Con siRNA) or CK1α-specific siRNA (CK1 siRNA) (200 nM) for 24 h and treated with camptothecin (CPT) at the indicated concentrations for 24 h. Cell survival was determined by the MTT assay. OD450, optical density at 450 nm.
Next, we tested whether increased activation of p53 by targeting CK1α cooperates with DNA-damaging agents in cell death induction. HCT116 cells were transfected with control siRNA and CK1α siRNA for 24 h, followed by treatment with the topoisomerase I inhibitor camptothecin for 24 h (gamma radiation was not used for this assay, since a single treatment did not induce significant cell death). The results showed that inhibiting CK1α expression by RNAi increased cell death induction by camptothecin (Fig. 9e). Furthermore, the effect of CK1α siRNA was largely dependent on the presence of p53 and was not observed using p53-deficient HCT116 cells (Fig. 9f). Similar results were also obtained using PA1 cells (data not shown). Interestingly, despite the expectation that targeting CK1α may lead to general toxicity due to its role in regulating other cellular targets, no significant cell death or growth arrest was observed in several cell lines treated with CK1α siRNA alone (Fig. 9e and data not shown). In related experiments, we found that the CKI-7 inhibitor also activated p53 and cooperated with gamma irradiation to activate p53 (data not shown). Although it will be difficult to establish specificity when using pharmacological inhibitors, the results were consistent with the CK1α siRNA experiments. Therefore, CK1α may be a potentially useful drug target for activation of p53 in cooperation with DNA-damaging drugs. The CK1α siRNA experiments also indicated that CK1α has a unique role in regulating MDMX and p53 that was not compensated by other CK1 isoforms.
DISCUSSION
The MDM2 homolog MDMX recently emerged as an important regulator of p53 and is likely to be targeted by signals that regulate p53 function. Results described in this report showed that MDMX forms a stable complex with CK1α. The high efficiency of CK1α copurification with MDMX in HeLa cells suggested that CK1α is a significant binding partner of MDMX. Importantly, our results identified a clear functional effect of CK1α in stimulating MDMX inhibition of p53 by increasing formation of MDMX-p53 complexes. Furthermore, pharmacological or siRNA-mediated inhibition of CK1α cooperated with DNA damage in the activation of p53 target genes and inducing cell death. Regulation of MDMX requires formation of stable MDMX-CK1α complexes and phosphorylation of MDMX by CK1α. Our results showed that CK1α binds specifically to MDMX, but not to MDM2 or p53. However, MDMX can target CK1α into complexes containing MDM2 or p53. This suggests that stable MDMX-CK1α complexes may have additional functions in the phosphorylation of other MDMX-interacting proteins.
We have identified serine 289 as the major CK1α phosphorylation site on MDMX in vitro. Mutation of this site significantly reduced the response to CK1α regulation. Phosphopeptide analysis confirmed that serine 289 is modified in vivo but also revealed that CK1α overexpression stimulates the phosphorylation of other sites on MDMX. It is not clear whether these sites are direct or indirect targets of CK1α in vivo. The complete absence of these phosphopeptides in the in vitro labeling reaction by recombinant CK1α suggests that other kinases may be responsible for their modification, which is influenced by CK1α. It is still not clear how phosphorylation of serine 289 stimulates MDMX-p53 binding. This residue is located in the central acidic domain and presumably not directly involved in interaction with p53. It is possible that the acidic domain has a regulatory function in p53 binding and that modification of serine 289 causes conformational change in the protein and affects the N-terminal p53-binding domain. Alternatively, phosphorylation of serine 289 may lead to secondary modifications that affect p53 binding. Identification of the additional in vivo phosphorylation sites may lead to better understanding of the regulation of MDMX.
Our results provide evidence that the CK1 family kinases are directly involved in regulating the p53 pathway by phosphorylation of MDMX. Recent studies showed that MDM2 is a target of the CK1δ isoform in vitro and in vivo (45). CK1δ modifies MDM2 on several serine residues in the acidic domain implicated in the regulation of p53 degradation (3). These residues become hypophosphorylated after DNA damage, leading to attenuation of MDM2-mediated p53 degradation (3). It will be important to determine whether MDMX targets CK1α to promote MDM2 phosphorylation in a manner similar to CK1δ and contributes to the regulation of MDM2. CK1α is an abundant kinase, and although there are reports of its regulation by DNA damage in Drosophila (37), it is unclear whether its enzyme activity is regulated by DNA damage in mammalian cells. Since DNA damage promotes degradation of MDMX (24, 31), it should also lead to reduced targeting of CK1α to MDM2 and thus reduce the level of MDM2 phosphorylation. The cooperative effect of CK1 inhibitor and gamma irradiation to activate p53 in cells suggests that CK1 family kinases are potential drug targets for sensitizing p53 to DNA damage.
Human CK1α is highly homologous to the HRR25 gene in Saccharomyces cerevisiae (57% identity within the catalytic core, 41% overall identity), which was originally identified as a gene involved in DNA repair (8, 19). Deletion of HRR25 gene in yeast prevents entry into meiosis and affects chromosomal segregation during mitosis (19). HRR25 also plays a role in regulating calcineurin-mediated stress response by phosphorylation of the transcription factor Crz1p and inhibiting its nuclear import (23). A previous study found that CK1α relocalized to the centrosome and kinetochore microtubules during mitosis in mammalian cells (4). p53 also has important roles in the maintenance of chromosome stability in mammalian cells. The interaction between MDMX and CK1α again raises the question of whether mammalian CK1α functions in DNA repair and maintenance of genomic stability, in part through its role in regulating MDMX and p53.
Acknowledgments
We thank David Virshup for providing the CK1α construct, William Lane for advice and help on protein identification by mass spectrometry, and Jin Cheng for advice on peptide mapping. We thank the Moffitt Molecular Biology Core for DNA sequence analyses. We are also grateful for Alicia Chen for help in manuscript preparation.
This work was supported by grants from the American Cancer Society and National Institutes of Health to J. Chen.
REFERENCES
- 1.Amit, S., A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah, and I. Alkalay. 2003. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman, J. L., S. D. Gross, M. R. Sussman, and R. A. Anderson. 1992. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc. Natl. Acad. Sci. USA 89:9454-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chijiwa, T., M. Hagiwara, and H. Hidaka. 1989. A newly synthesized selective casein kinase I inhibitor, N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase I from bovine testis. J. Biol. Chem. 264:4924-4927. [PubMed] [Google Scholar]
- 7.De Graaf, P., N. A. Little, Y. F. Ramos, E. Meulmeester, S. J. Letteboer, and A. G. Jochemsen. 2003. Hdmx protein stability is regulated by the ubiquitin ligase activity of mdm2. J. Biol. Chem. 278:38315-38324. [DOI] [PubMed] [Google Scholar]
- 8.DeMaggio, A. J., R. A. Lindberg, T. Hunter, and M. F. Hoekstra. 1992. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc. Natl. Acad. Sci. USA 89:7008-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desagher, S., A. Osen-Sand, S. Montessuit, E. Magnenat, F. Vilbois, A. Hochmann, L. Journot, B. Antonsson, and J. C. Martinou. 2003. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8:601-611. [DOI] [PubMed] [Google Scholar]
- 10.Eide, E. J., and D. M. Virshup. 2003. Casein kinase I: another cog in the circadian clockworks. Chronobiol. Int. 18:389-398. [DOI] [PubMed] [Google Scholar]
- 11.Finch, R. A., D. B. Donoviel, D. Potter, M. Shi, A. Fan, D. D. Freed, C. Y. Wang, B. P. Zambrowicz, R. Ramirez-Solis, A. T. Sands, and N. Zhang. 2002. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 62:3221-3225. [PubMed] [Google Scholar]
- 12.Fish, K. J., A. Cegielska, M. E. Getman, G. M. Landes, and D. M. Virshup. 1995. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J. Biol. Chem. 270:14875-14883. [DOI] [PubMed] [Google Scholar]
- 13.Flotow, H., P. R. Graves, A. Q. Wang, C. J. Fiol, R. W. Roeske, and P. J. Roach. 1990. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 265:14264-14269. [PubMed] [Google Scholar]
- 14.Flotow, H., and P. J. Roach. 1991. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 266:3724-3727. [PubMed] [Google Scholar]
- 15.Freedman, D. A., C. B. Epstein, J. C. Roth, and A. J. Levine. 1997. A genetic approach to mapping the p53-binding site in the MDM2 protein. Mol. Med. 3:248-259. [PMC free article] [PubMed] [Google Scholar]
- 16.Gross, S. D., and R. A. Anderson. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 17.Gross, S. D., J. C. Loijens, and R. A. Anderson. 1999. The casein kinase Ialpha isoform is both physically positioned and functionally competent to regulate multiple events of mRNA metabolism. J. Cell Sci. 112:2647-2656. [DOI] [PubMed] [Google Scholar]
- 18.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277:19251-19254. [DOI] [PubMed] [Google Scholar]
- 19.Hoekstra, M. F., R. M. Liskay, A. C. Ou, A. J. DeMaggio, D. G. Burbee, and F. Heffron. 1991. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science 253:1031-1034. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, M. W., M. S. Lindstrom, and S. J. Berberich. 2002. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J. Biol. Chem. 276:25336-25341. [DOI] [PubMed] [Google Scholar]
- 21.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22:6365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joazeiro, C. A., and A. M. Weissman. 2002. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 23.Kafadar, K. A., H. Zhu, M. Snyder, and M. S. Cyert. 2003. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes Dev. 17:2698-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai, H., D. Wiederschain, H. Kitao, J. Stuart, K. K. Tsai, and Z. M. Yuan. 2003. DNA damage-induced MDMX degradation is mediated by MDM2. J. Biol. Chem. 278:45946-45953. [DOI] [PubMed] [Google Scholar]
- 25.Knippschild, U., D. M. Milne, L. E. Campbell, A. J. DeMaggio, E. Christenson, M. F. Hoekstra, and D. W. Meek. 1997. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene 15:1727-1736. [DOI] [PubMed] [Google Scholar]
- 26.Li, C., L. Chen, and J. Chen. 2002. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol. Cell. Biol. 22:7562-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, C., Y. Li, M. Semenov, C. Han, G. H. Baeg, Y. Tan, Z. Zhang, X. Lin, and X. He. 2003. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837-847. [DOI] [PubMed] [Google Scholar]
- 28.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2001. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 29.Migliorini, D., E. L. Denchi, D. Danovi, A. Jochemsen, M. Capillo, A. Gobbi, K. Helin, P. G. Pelicci, and J. C. Marine. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 22:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oca Luna, R. M., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 31.Pan, Y., and J. Chen. 2003. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parant, J., A. Chavez-Reyes, N. A. Little, W. Yan, V. Reinke, A. G. Jochemsen, and G. Lozano. 2002. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29:92-95. [DOI] [PubMed] [Google Scholar]
- 33.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 34.Ramos, Y. F., R. Stad, J. Attema, L. T. Peltenburg, A. J. van der Eb, and A. G. Jochemsen. 2002. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 61:1839-1842. [PubMed] [Google Scholar]
- 35.Rowan, B. G., N. Garrison, N. L. Weigel, and B. W. O'Malley. 2000. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol. Cell. Biol. 20:8720-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakaguchi, K., S. Saito, Y. Higashimoto, S. Roy, C. W. Anderson, and E. Appella. 2000. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275:9278-9283. [DOI] [PubMed] [Google Scholar]
- 37.Santos, J. A., E. Logarinho, C. Tapia, C. C. Allende, J. E. Allende, and C. E. Sunkel. 1996. The casein kinase 1 alpha gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J. Cell Sci. 109:1847-1856. [DOI] [PubMed] [Google Scholar]
- 38.Sharp, D. A., S. A. Kratowicz, M. J. Sank, and D. L. George. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274:38189-38196. [DOI] [PubMed] [Google Scholar]
- 39.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Larr, P. Dekker, M. Bazuine, R. C. A. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 40.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, A. J. DeMaggio, M. F. Hoekstra, J. Blenis, T. Hunter, and L. C. Cantley. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stad, R., N. A. Little, D. P. Xirodimas, R. Frenk, A. J. van der Eb, D. P. Lane, M. K. Saville, and A. G. Jochemsen. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzivion, G., and J. Avruch. 2002. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061-3064. [DOI] [PubMed] [Google Scholar]
- 43.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins: bringing new definitions to scaffolding. Oncogene 20:6331-6338. [DOI] [PubMed] [Google Scholar]
- 44.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 45.Winter, M., D. Milne, S. Dias, R. Kulikov, U. Knippschild, C. Blattner, and D. Meek. 2004. Protein kinase CK1delta phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry 43:16356-16364. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175-186. [PubMed] [Google Scholar]
- 47.Zhu, J., F. Shibasaki, R. Price, J. C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]