Abstract
Translation initiation factor eukaryotic translation initiation factor 4E (eIF4E) plays a key role in regulation of cellular proliferation. Its effects on the m7GpppN mRNA cap are critical because overexpression of eIF4E transforms cells, and eIF4E function is rate-limiting for G1 passage. Although we identified eIF4E as a c-Myc target, little else is known about its transcriptional regulation. Previously, we described an element at position −25 (TTACCCCCCCTT) that was critical for eIF4E promoter function. Here we report that this sequence (named 4EBE, for eIF4E basal element) functions as a basal promoter element that binds hnRNP K. The 4EBE is sufficient to replace TATA sequences in a heterologous reporter construct. Interactions between 4EBE and upstream activator sites are position, distance, and sequence dependent. Using DNA affinity chromatography, we identified hnRNP K as a 4EBE-binding protein. Chromatin immunoprecipitation, siRNA interference, and hnRNP K overexpression demonstrate that hnRNP K can regulate eIF4E mRNA. Moreover, hnRNP K increased translation initiation, increased cell division, and promoted neoplastic transformation in an eIF4E-dependent manner. hnRNP K binds the TATA-binding protein, explaining how the 4EBE might replace TATA in the eIF4E promoter. hnRNP K is an unusually diverse regulator of multiple steps in growth regulation because it also directly regulates c-myc transcription, mRNA export, splicing, and translation initiation.
eIF4E catalyzes the rate-limiting step in eukaryotic translation and was shown to be the least abundant of all translation initiation factors in cells (27). Although it binds to the 5′ cap structures on all mRNAs, eIF4E must exert specific effects on various mRNAs because it functions as an oncogene in transformation assays (18, 52). Translational control by eIF4E specifically regulates steps in cell proliferation that are frequently abnormal in malignant cells. For example, the yeast mutant expressing a temperature-sensitive eIF4E (cdc33) arrests in G1 at its restrictive temperature (7).
eIF4E's transforming function is likely to be clinically significant because eIF4E is overexpressed in a variety of malignant cells and tissues (15, 16, 53, 77). One critical role for eIF4E in oncogenesis may be as the downstream effector of the akt survival pathway that collaborates with various other oncogenes (83, 103). As such, it appears to work in part by preventing apoptotic cell death (54). Whether its overexpression is a cause or an effect of malignant transformation remains an intriguing and important question. In both head and neck tumors, as well as in rare breast cancers, amplifications of the eIF4E gene have been described (37, 92). However, noncytogenetic regulatory events appear to contribute to its overexpression in colon, lung, lymphoma, and bladder cancers (77). The nature of these regulatory events remains unknown.
Despite the growing realization of its potential importance in cancer, astonishingly little is known about mechanisms that regulate levels of eIF4E (57). Numerous studies have shown its functional regulation by various signal transduction pathways and by binding to its antagonist, the 4E binding protein (reviewed in references 11 and 15). However, measurements of its quantitative levels in cells have attracted much less attention (101). Early studies that demonstrated serum and heat shock regulation of initiation factors were hampered because eIF4E levels were too low to detect in two-dimensional gels (24-26). Tissue-specific changes in eIF4E mRNA have been demonstrated in Drosophila and zebra fish (31, 39). In mammalian systems, its levels vary during epithelial differentiation and cardiocyte growth (56, 102). Our initial report that serum induces eIF4E mRNA remains a critical demonstration that transcriptional regulation of eIF4E can play an important role in its overall regulation (80). Importantly, the promoter of the eIF4E gene contains two essential E boxes that bind to, and can be regulated by, the MYC oncogene (32, 46). However, an additional site at −25 in the promoter was equally important to the function of the eIF4E promoter (45). Its position at −25, where TATA sequences would be anticipated, suggested that eIF4E might be regulated by a unique basal promoter element. To better understand the regulation of eIF4E in normal and malignant tissues, we sought to further characterize this regulatory motif that we now designate the 4E basal element (4EBE).
As in the case of eIF4E, little is known about transcriptional regulation of other translation initiation factors. The abundance of translation initiation factors eIF1 and eIF4A exceeds the abundance of ribosomal proteins; both are therefore among the most abundant proteins in the cell (26, 101). The promoters for both follow the classic paradigm of strong promoters that contain TATA sites (14, 73). The promoters for eIF2α and eIF2β have also been analyzed in some detail. eIF2α and eIF2β are both less abundant than ribosomal proteins. Their promoters lack TATA sequences (9, 42), and they both share a palindromic sequence (TGCGCATGCGCA) that binds nuclear respiratory factor 1 (NRF-1). NRF-1 is thought to coordinate transcriptional regulation of nucleus-encoded respiratory proteins located in the mitochondria (9, 29, 47). Also called α-palindromic binding protein (α-PAL), NRF-1 avidly binds the palindromic sequence in the eIF2α promoter (8, 29). Overexpression of NRF-1 increases eIF2α and net protein synthesis, although it actually inhibits cell division through effects on E2F1 (30). Intriguingly, NRF-1 can heterodimerize with max, and the α-PAL sequence is an alternative-binding site for Myc (4, 30, 80). Finally, an inhibitory sequence in the first intron of eIF2α functions as an initiator (INR) element, and INR elements are repressed by Myc (67, 81, 88).
Nuclear run-on experiments first demonstrated a connection between Myc and transcription of translation initiation factors eIF4E and eIF2α (80). Since then, accumulating data have linked Myc regulation to ribosomal proteins and additional translation initiation factors and thereby to growth regulation in the cell (86). The c-Myc target gene database lists 16 translation initiation factors whose potential connections to Myc regulation have been suggested (105). Evidence for this linkage comes from chromatin immunoprecipitation, loss of expression in c-myc-null cells, gain of expression in primary liver cells, and increased expression resulting from conditional increases in Myc function (32, 48, 68, 72). The best evidence for a Myc connection is still found for eIF4E and eIF2α, although the rules associating Myc with regulation of these two factors remain ambiguous (105). Although Myc does not regulate these factors in all circumstances, its regulation of translation initiation is potentially important in cancer biology because a dominant inhibitor of translation initiation blocks transformation by Myc (55).
The location of the 4EBE at the site where TATA should be found raises the possibility that it is a functional basal promoter element. Two types of cis-acting DNA elements regulate eukaryotic promoters transcribed by RNA polymerase II genes: (i) basal or core promoter elements located near or at the transcription initiation site and (ii) upstream or regulatory promoter elements that are binding sites for various transcription activators and/or repressors (40). Utilization of the TATA-binding protein (TBP) is common to promoters transcribed by all RNA polymerases (106). TBP is present in a complex called TFIID containing TBP along with various TBP-associated factors (TAFs) (28, 107, 108). Although TBP is generally required for the transcription of most genes, the identification of TBP-related factors TRF1 and TRF2 and a TBP-free, TAF-containing complex, which lacks TBP but contains TAFs, suggests that there are alternative mechanisms by which transcription may be initiated in the case of certain genes (34, 74). Additional basal promoter elements have been identified that can be distinguished from TATA. Seminal work contributed by Smale demonstrated an additional basal element spanning the transcription initiation site that was called the INR element that functions as a TATA equivalent in many TATA-less genes (90). Initiator elements are especially important in cell growth control since they were identified as the δ element in the promoters of ribosomal protein genes (1, 22, 35, 84). Specific DNA-binding proteins, including YY1, TFII-I, USF, and E2F, bind initiator elements (2, 43, 44, 82).
In characterizing the eIF4E promoter, we identified protein species of 68 and 97 kDa that bound the 4EBE (45). This element, initially designated LS3 (5′-TTACCCCCCCTT), is highly conserved between mouse, rat, and human eIF4E promoters (45, 56). We first considered that this element might be a classic initiator because of its strategic location, the lack of a TATA, and its pyrimidine richness. However, the element did not cross-compete with known initiator sequences, was not transactivated by YY1, and was not supershifted by antibodies to any known initiator-binding proteins in electrophoretic mobility shift assay (EMSA) experiments (45). Although C-rich it also did not bind SP1 because SP1 oligonucleotides did not cross-compete for binding and an antibody to SP1 did not supershift complexes in EMSA experiments (45). Here we examine the role of this element, which we now designate the 4EBE. We report that this sequence functions as a basal promoter element in reporter gene experiments and binds the transcription factor hnRNP K that is known to interact with TBP (62). We go on to demonstrate that overexpression of hnRNP K causes an eIF4E-dependent cellular transformation that synergizes with Myc, identifying a new regulatory interaction that could play a role in human malignancies overexpressing eIF4E.
MATERIALS AND METHODS
Plasmids and plasmid construction.
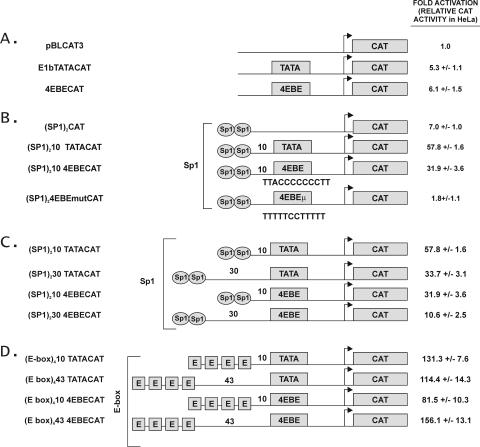
The E1bTATA CAT plasmid was used as the starting plasmid for construction of reporter plasmids because it lacks any known elements apart from its simple TATA. The sequence upstream of CAT in this reporter is shown in Fig. 1 and specifically does not include the initiator element from the E1b promoter. Initial constructs contained two Sp1 sites or four E-box elements upstream of TATA or 4EBE where the E1bTATA was replaced by 4EBE. The oligonucleotides used for all constructs are summarized in Fig. 1. Briefly, double-stranded oligonucleotides with the appropriate overhangs containing two Sp1 binding sites or four E-box elements were synthesized (Gibco-BRL) and annealed. In the cases where the E1bTATA was replaced by 4EBE, double-stranded oligonucleotides were synthesized containing the 4EBE sequence with two Sp1 sites or four E-box elements upstream of 4EBE. Diagrams of the various plasmids are included in Fig. 2. The E1bTATACAT plasmid was cut with the appropriate restriction enzymes and ligated with the double-stranded oligonucleotides. (E-box)410 TATACAT was used as the template for generating the plasmids (E-box)443 TATACAT, (E-box)4 10 4EBE CAT, and (E-box)443 4EBE CAT.
FIG. 1.
Oligonucleotides used in EMSA experiments.
FIG. 2.
4EBE can replace the TATA box in the E1b promoter. (A) The indicated promoter constructs were transfected into HeLa cells and analyzed for CAT activity as described in Materials and Methods. The schematic diagram identifies pBLCAT3 (no element), E1bTATACAT (TATA), and 4EBE CAT (4EBE). The CAT reporter activity is represented as the fold activation compared to CAT alone. All values are reported as the mean increase ± the standard error for three replicas in this and the subsequent panels. (B) The diagrammed activator/basal element promoter constructs were transfected into HeLa cells and analyzed for CAT activity. Constructs are illustrated in the center and included plasmids containing two Sp1 sites upstream of CAT or 10 bases upstream of TATA, 4EBE, or 4EBE μ in the E1bCAT plasmid [(Sp1)2CAT, (Sp1)210 TATACAT, (Sp1)210 4EBE CAT, and (Sp1)210 4EBEmutCAT]. The CAT reporter activity is represented as the fold activation compared to the CAT alone value taken from panel A. (C) Representative experiment showing the effect of distance between Sp1 and TATA or Sp1 and 4EBE on transcriptional activity. Briefly, exponentially growing HeLa cells in 100-mm dishes were transiently transfected with 10 μg of CAT reporter plasmids containing two Sp1 sites 10 or 30 bases upstream of TATA or 4EBE [(Sp1)210 TATACAT, (Sp1)230TATACAT, (Sp1)210 4EBE CAT, and (Sp1)230 4EBE CAT]. (D) Representative experiment showing the effect of distance between the E-box element and TATA compared to the E-box element and 4EBE on transcriptional activity. Briefly, exponentially growing HeLa cells in 100 mm dishes were transiently transfected with 10 μg of CAT reporter plasmids containing four E-box elements 10 or 43 bases upstream of TATA or 4EBE [(E-box)410 TATACAT, (E-box)443 TATACAT, (E-box)410 4EBECAT, and (E-box)443 4EBE CAT], along with 2 μg of human growth hormone plasmid as an internal control. Transfected cells were harvested 48 h posttransfection and assayed for CAT activity in all experiments.
For the constructs containing the linker scanning mutations in the eIF4E promoter, the p4ECAT plasmid that contains 403 nucleotides of proximal promoter sequence from the eIF4E genomic clone was used as the parental plasmid (45). In order to generate mutations in the 4EBE site in the native 4E promoter, the QuikChange site-directed mutagenesis kit was used (Stratagene). 5′ and 3′ primers harboring the mutations in the middle of the primers were synthesized and used for the generation of the mutations according to the manufacturer's instructions. The p4ECAT plasmid was used as the template. The various primers used for PCR-induced mutations are summarized in Fig. 1.
The expression plasmids pCEP4 (Invitrogen empty vector control [hygromycin selectable]), pCEPmyc (human Myc driven by cytomegalovirus), and pCEP4EBPμ(a constitutive dominant-inhibitory form of the 4E binding protein that blocks eIF4E function) were described previously (55). pcDNA-hnRNP K was created by using the cDNA for hnRNP K from an Image Clone purchased from Invitrogen that was then cloned into pcDNA3.1 (Invitrogen; G418 selectable) by using NotI and XhoI restriction sites. An hnRNP K retroviral expression vector was constructed by cloning the hnRNP K cDNA into the pBABE-puro vector by using an EcoRI-XhoI fragment from pcDNA-hnRNP K.
Terminal oligopyrimidine (TOP) reporter plasmids, including rpL32-green fluorescent protein (GFP) and a mutant rpL32-GH containing a C→A mutation in its cap to eliminate TOP function were a generous gift from O. Meyuhas (96).
Cells, transfections, and chloramphenicol acetyltransferase (CAT) assays.
HeLa cells were obtained from the American Type Culture Collection, Manassas, Va. Adherent cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FCS), antibiotics and l-glutamine. HO15 (myc−/−) and TGR (myc+/+) cells were obtained from John Sedivy (59) and were also grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, antibiotics, and l-glutamine. Rat 1A cells were isolated for their susceptibility to transformation in one step by Myc (93) and were obtained from Chi Dang.
For CAT assays, exponentially growing HeLa cells were transfected in 100-mm dishes by using standard calcium phosphate coprecipitations (85). Briefly, HeLa cells were transfected with 10 μg of CAT reporter plasmid and 2 μg of pSVtkhGH plasmid as internal control. Cells were fed with fresh medium 24 h after transfection and harvested for CAT activity 48 h posttransfection by a standard assay (46). The CAT activity in each case was normalized to human growth hormone levels in the media, as determined by using a commercial radioimmunoassay kit (Nichols Institute) according to the manufacturer's instructions. Transfections were typically performed in duplicate and repeated three times.
Rat1A cells were transfected by using Lipofectin reagents (Gibco-BRL) with pCEP4, pCEPmyc, and pCEP-4EBPμ, respectively, together with either pcDNA3.1 or pcDNA-hnRNP K as indicated in individual figure legends. Pooled transfectants were selected in the presence of 500 μg of Geneticin and 200 μg of hygromycin/ml. Since all colonies expressed the transfected proteins due to the double selection applied to all transfections, pooled transfectants were used in all studies.
Packaged retroviruses expressing hnRNP K or a GFP control were made with the EcoPac retroviral packaging vector; 293T cells were cotransfected with either pBABE-GFP or pBABE-hnRNP K by using standard calcium chloride techniques. Subconfluent HO15 cells were infected with 50% viral supernatants. Two days after infection, puromycin was added at a concentration of 2 μg/ml to select for infected cells, and puromycin selection was continued throughout the remainder of the experiment. After expansion, HO15 cells carrying either pBABE-GFP or pBABE-hnRNP K were seeded in 35-mm plates and allowed to become confluent. They were then serum starved for 48 h; half of the plates were stimulated with 10% fetal bovine serum for 20 h. DNA synthesis was measured by addition of [3H]thymidine (1 μCi of 50 mCi/mmol; MP Biomedicals) for the last 2 h of serum stimulation and is plotted as the mean and standard deviation for three determinations per condition. Cells were harvested by using a previously described method (75). Additional cells were expanded into 100-mm plates for polysomal analysis by the method described below.
For hnRNP K siRNA experiments, proliferating TGR or HO15 cells were transfected with 10 nM scrambled control duplex oligonucleotide (Dharmacon, Inc.) containing sense 5′-GCGCGCUUUGUAGGAUUCGtt-3′ and antisense 5′-CGAAUCCUACAAAGCGCGCtt-3′. These transfections were compared to 10 nM small interfering RNA (siRNA) for hnRNP K (Hrpk_1 siRNA; Ambion, Inc.) containing sense 5′-GGAACAAGCCUUUAAAAGAtt-3′ and antisense 5′-UCUUUUAAAGGCUUGUUCCtc-3′. Transfections were accomplished by using Oligofectamine (Invitrogen, Inc.) according to the manufacturer's instructions. mRNA and protein were harvested after 48 h and analyzed for the expression of hnRNP K and eIF4E as described below.
Additional siRNAs were used to block eIF4E expression in Rat1A cells. Proliferating Rat1A cells were transfected with 50 nM concentrations of the following siRNA duplexes from Ambion, Inc.: siRNA ID 56229 E1 sense (GGAUGGUAUUGAGCCUAUGtt), E1 antisense (CAUAGGCUCAAUACCAUCCtt); and siRNA ID 56139 E2 sense (GGUGGGCACUCUGGUUUUUtt) and E2 antisense (AAAAACCAGAGUGCCCACCtg). The cells were then compared to a nontargeting siRNA duplex for luciferase containing the following sequences (Dharmacon): nontargeting siRNA #2 sense, 5′-UAAGGCUAUGAAGAGAUACUU-3′; nontargeting siRNA #2 antisense, 5′-PGUAUCUCUUCAUAGCCUUAUU-3′.
These experiments were performed by using the same conditions as for the hnRNP K experiments. Protein was harvested after 48 h and analyzed for eIF4E and actin protein as described below. These siRNAs were then transfected into Rat1A cells expressing hnRNP K and transferred into soft agar after 48 h. Soft agar colony formation was evaluated 8 days later.
Protein purification and monitoring by Southwestern analysis.
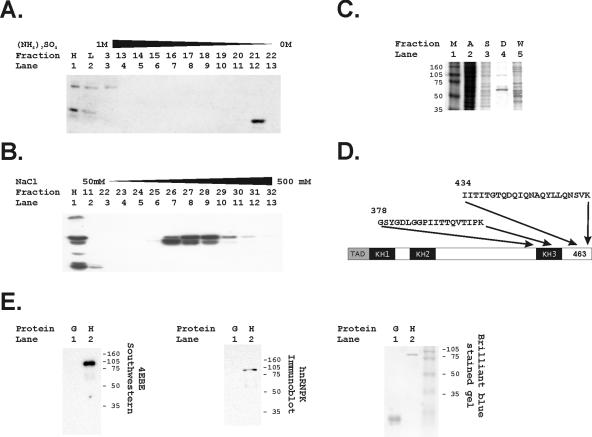
Nuclear extracts were prepared from at least 108 of HeLa cells growing in monolayer culture by a modification of the Dignam method (19, 20, 65). The extraction buffer was composed of 0.5% deoxycholate, 1% octyl-β-glucoside, 20 mM HEPES (pH 7.9), 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM dithiothreitol (DTT), and 25% glycerol. The initial crude extract was dialyzed overnight against hydrophobic exchange column buffer [1 M (NH4)2SO4, 20 mM HEPES (pH 7.2), 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT]. The precipitate resulting from dialysis against the 1 M (NH4)2SO4 was removed by centrifugation at 14,000 rpm for 5 min in a microcentrifuge. The supernatant was loaded on a phenyl Sepharose column and eluted by using a 1 to 0 M (NH4)2SO4 gradient over 10 column volumes. Then, 1-ml fractions were collected and the positive fractions were identified by using aliquots from the fractions in Southwestern analyses as described below. The positive fractions were dialyzed overnight in monoS column loading buffer (50 mM NaCl, 10 mM HEPES [pH 7.4], 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF). The loading preparation was cleared by centrifugation, and positive fractions were identified by Southwestern analysis after elution from the monoS column with a 50 to 500 mM NaCl gradient. The monoS-positive fractions were pooled and dialyzed overnight against affinity binding buffer (50 mM NaCl, 5 mM MgCl2, 20 mM HEPES [pH 7.4], 1 mM DTT, 1 mM EDTA, 0.2 mM PMSF, 5% glycerol). Affinity binding to a 1.2 mM concentration of a biotinylated 4EBE trimeric double-stranded oligonucleotide (CTAGATTCCTCTTACCCCCCCTTCTTCCTCTTACCCCCCCTTCTTCCTCTTACCCCCCCTTGG) was performed at 4°C for 20 min in the presence of 0.4 μg of salmon sperm DNA/μl. Next, we added 100 μl of streptavidin beads, and binding continued for an additional 6 h at 4°C with gentle mixing with continuous rotation. The streptavidin beads were then washed extensively with the affinity binding buffer and spun gently, and the bound material was eluted by boiling in standard Laemmli loading buffer. This preparation was run on a 10% denaturing polyacrylamide gel, and a single protein band was identified at 68 kDa by using colloidal blue staining.
Southwestern analyses were performed essentially as described previously (38, 89, 94, 100). Nuclear extracts (50 μg) from the indicated cell types were run on 10% denaturing polyacrylamide gels, electroblotted to nitrocellulose for 12 h, and allowed to dry. The filters were then immersed for 10 min in denaturation-renaturation buffer containing 6 M guanidine hydrochloride. Partial renaturation of immobilized proteins was effected by five successive incubations of the filters in buffer containing progressive (twofold) dilutions of guanidine hydrochloride and finally in buffer lacking the denaturant. Filters were blocked with 5% nonfat dry milk in binding buffer (50 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1 mM DTT) and were then washed twice with 0.25% nonfat dry milk in binding buffer. Filters were hybridized in binding buffer containing Klenow-labeled LS3 trimer oligonucleotide probe, 0.25% nonfat dry milk, and 1 μg of sonicated salmon sperm DNA per ml for 60 min at room temperature. Filters were then washed three times with binding buffer containing 0.25% nonfat dry milk alone and dried.
Recombinant gst-hnRNP K was purified by using standard methods from extracts of Escherichia coli transformed with pGEX-hnRNP K generously provided by David Levens (85, 98).
DNA-binding assays.
HeLa nuclear extracts were obtained and analyzed by using published methods (19, 97). Gel shift activity for 4EBE binding proteins was determined in EMSA binding buffer (25 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA, 200 mM glycine, 0.1% Tween 20, 0.55 mg of bovine serum albumin/ml, 10% glycerol) with 2 ng of poly(dI-dC)/μl as a nonspecific competitor. Complexes formed in binding buffer were resolved on a 4% nondenaturing polyacrylamide gel containing 0.5× TBE (0.045 M Tris-borate [pH 8.0], 0.5 mM EDTA) at 4°C. After labeling with [γ-32P]ATP with polynucleotide kinase, oligonucleotides were purified by polyacrylamide gel electrophoresis. Each binding reaction contained between 0.1 to 0.5 ng of labeled oligonucleotide. Competition experiments were performed by using the indicated molar excess of unlabeled oligonucleotides. Oligonucleotide sequences are provided in the relevant figures.
Chromatin immunoprecipitation assays.
We evaluated TGR cells during logarithmic growth, at confluence after 72 h in the absence of serum to arrest the cells, and 8 h after serum was added to the growth-arrested cells. Cells grown on 150-mm diameter plates were harvested and resuspended in growth medium. Cross-linking was performed by adding fresh 37% formaldehyde to a final concentration of 1.5%, followed by incubation at room temperature for 15 min with gentle agitation. The reaction was stopped by adding glycine to a final concentration of 0.125 M, followed by incubation for 5 min. Cells were collected by centrifugation at 1,500 × g for 5 min; the pellet was washed twice with ice-cold phosphate-buffered saline (120 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer [pH 7.4]), washed twice with immunoprecipitation buffer (IP buffer; 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris-HCl [pH 7.5], 0.5 mM DTT), and then resuspended with IP buffer supplemented with 0.5% sodium dodecyl sulfate (SDS) and protease inhibitor cocktail (Roche). Sonication was performed by Branson Sonifier 250 with a microtip at output 6 with 10-s bursts until the desired DNA fragment sizes were reached. The lysate was then subjected to centrifugation at 12,000 × g for 10 min. The supernatant was collected and diluted at least fivefold with IP buffer. For immunoprecipitation, antibody was added to the lysate, followed by incubation at 4°C for 1 h with agitation. Protein A-Sepharose beads (Amersham) were added to the mixture and further incubated for 1 h. The beads were collected by centrifugation at 1,500 × g for 30 s, washed twice with IP buffer, twice with high-salt IP buffer (500 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris-HCl [pH 7.5], 0.5 mM DTT), washed twice with stringent wash buffer (10 mM Tris-HCl [pH 7.5], 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and washed twice with TE buffer (10 mM Tris-HCl, 0.1 mM EDTA). The bound fraction was eluted by incubating the beads in elution buffer (TE with 1% SDS) at 65°C for 10 min. The cross-links were reversed by incubating the eluate at 65°C for at least 6 h. Samples were treated with proteinase K at 37°C for 1 h, phenol-chloroform extracted, and ethanol precipitated. The following primer sets were used in PCRs: pair 1, c-myc promoter (TACTCTACTCCAGCTCTGGAACG, forward) and c-myc promoter (ACCTACGACAGATGAGGTCTGAG, reverse); pair 2, nontranscribed locus AC120715 (ACCAGCCAGTCAGAGTTCAGG, forward) and nontranscribed locus AC120715 (GGTTTCATCATCCCGAGAC, reverse); pair 3, eIF4E promoter (CTCCACTTCCCAGAAGCCTCTTG, forward) and eIF4E promoter (CGGTTCCACAGTCGCCATCTTAG, reverse); and pair 4, 5S rRNA genes (GTCTACGGCCATACCACCCT, forward) and 5S rRNA genes (AAAGCCTACAGCACCCGGTA, reverse). Aliquots of the samples were assayed by PCR at 95°C for 30 s, 59°C for 45 s, and 72°C for 30 s for 35 cycles and then fractionated on an agarose gel.
Expression analyses for mRNA and protein.
Levels of expression of myc, hnRNP K, eIF4E, and actin mRNAs were analyzed by using total cellular RNA from the indicated transfectants that was size fractionated (10 μg/lane) on formaldehyde agarose gels, transferred to Hybond-N nylon matrices, and cross-linked by using UV light (10). Filters were hybridized in a rapid hybridization solution (Rapidhyb; Amersham) at 65°C with c-myc, hnRNP K, eIF4E, or actin cDNA fragments α32-P labeled by the Klenow reaction by using random priming. For Western analyses, cells were lysed in Laemmli loading buffer. A total of 10 μg of protein sample was subjected to SDS-10% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane. Membranes were hybridized with anti-hnRNP K (sc-25373; Santa Cruz), anti-eIF4E (BD610270; Becton Dickinson), anti-Myc (SC41; Santa Cruz), or anti-actin (MAB1501R; Calbiochem) as indicated and detected by enhanced chemiluminescence (Amersham). For RNA blots of the polysomal and subpolysomal fractions, RNA was extracted from the polysomal and subpolysomal fractions by using TRIzol reagent, and 50% of the harvested RNA was analyzed by using standard RNA blotting techniques (85).
Polysomal profile analysis.
One 100-mm diameter plate containing the indicated stable Rat1A transfectants was harvested for each polysomal analysis. Confluent transfectants were harvested and lysed in 300 μl of RSB (10 mM NaCl, 10 mM Tris-HCl [pH 7.4], 15 mM MgCl2) containing 100 μg of heparin/ml, 1.2% Triton X-100, and 0.12% deoxycholate (60, 96). Nuclei were pelleted for 3 min in a microcentrifuge at 4°C. The 300-μl extract was layered over 11.5 ml of a 15 to 45% (wt/wt) sucrose gradient with a 0.5-ml cushion of 45% sucrose. The gradients were centrifuged at 37,000 rpm for 2.5 h in an SW 41 (Beckman) rotor at 4°C. After centrifugation, the A260 was continuously monitored and recorded across the gradient.
For the polysomal rpL32 transient reporter experiments, Rat 1A cells were transfected in 100-mm plates with plasmids carrying either the 5′-untranscribed region of rpL32-GFP or the same untranscribed region lacking the consensus 5′-terminal oligopyrimidine element designated rpL32(−1C>A)-GH. Transfected cells were grown to confluence for 72 h, and during the last 24 h cells were grown in medium lacking serum. For RNA blots of the polysomal and subpolysomal fractions, RNA was extracted from the polysomal and subpolysomal fractions by using TRIzol reagent, and 50% of the harvested RNA was probed for the GFP or GH cDNAs in the reporter genes by standard RNA blotting techniques (49).
Cell characterization.
Protein content per cell was determined by lysis of a known number of cells in ELB lysis buffer and measurement of the protein content of an aliquot of these lysates by using a kit from Bio-Rad.
Subconfluent transfectants were labeled with 10 μM bromodeoxyuridine cell-labeling reagent (Amersham Pharmacia) for 30 min and harvested for cell cycle analysis. Cells were fixed in 80% ethanol for at least 1 h, incubated in anti-bromodeoxyuridine antibody (Becton Dickinson) for 30 min and exposed to anti-mouse-fluorescein secondary antibody (Vector Labs) for 30 min. Cells were resuspended in propidium iodide (70 μg/ml) supplemented with RNase A (25 μg/ml) and analyzed. DNA content was measured by using a FACScan cytometer (Becton Dickinson). The mean and standard error were determined for each cell type in two repetitions of experiments with three independent assays per cell type.
Clonogenicity of the Rat1A transfectants in soft agar was performed as described previously (93). For the eIF4E siRNA experiments, Rat1A cells expressing hnRNP K were transfected with the nontargeting siRNA #2 from Dharmacon, and the two siRNAs for eIF4E from Ambion; untransfected control Rat1A cells expressing hnRNP K were included in the analysis of soft agar clone formation. Transfection was allowed to proceed for 48 h when the cells were treated with trypsin and replated in the soft agar. Colony numbers were scored 8 days later. For the pCEP4EBPμ experiments, cells were transfected for 48 h and then placed in selection medium for an additional 48 h. They were then transferred to soft agar and colonies were scored 14 days later.
RESULTS
4EBE can replace TATA in a heterologous promoter.
We previously reported that the eIF4E gene has a unique transcription initiation site despite its lack of known basal promoter elements (46). Using linker scanning mutants of the eIF4E promoter to identify regions that were important for its expression, we found a novel DNA element spanning positions −28 to −16, which we originally designated LS3 (5′-TTACCCCCCCTT), that was crucial for eIF4E expression (45). Due to the strategic location of LS3 approximating that of TATA, we first tested its function as a potential counterpart of a core promoter element. We replaced the TATA-box in the E1bCAT promoter with the element present in LS3 (here renamed 4EBE) and tested it for transcriptional activity in HeLa cells in transient-transfection assays. Placing 4EBE sequences upstream of the CAT reporter resulted in an increase in transcriptional activity equivalent to that of the TATA alone (Fig. 2A).
4EBE supports activated transcription.
We next evaluated the potential for 4EBE to support activated transcription by placing two binding sites for the activator Sp1 about 10 bases upstream of either TATA or 4EBE in the E1bCAT plasmid. As shown in Fig. 2B, both TATA and 4EBE can mediate Sp1-activated transcription, although TATA-mediated Sp1 transcription was stronger than 4EBE-mediated Sp1 transcription. Mutating the 4EBE element (5′-TTACCCCCCCTT to 5′-TTTTTCCTTTTT) resulted in a dramatic decrease in transcription activity similar to the absence of 4EBE or TATA. These results demonstrated that 4EBE could mediate transcriptional activity of the upstream activator Sp1 in transient-transfection reporter gene assays. TATA appeared more efficient in its ability to mediate activation by Sp1 when the binding sites for Sp1 were placed in relatively close proximity to TATA and was in general a stronger basal promoter element than 4EBE at this distance.
Effect of spacing between TATA/4EBE and upstream activators on transcription.
Gene expression is regulated by interactions between the trans-acting factors that bind various cis-acting DNA elements in the promoter of a gene. The spacing of cis-acting elements in a promoter plays an important role in determining the ability of the trans-acting factors to interact with each other. In order to test the effect of spacing on the interactions between 4EBE and various upstream activating sequences, we placed two binding sites for Sp1 10 or 30 bases upstream of the TATA or 4EBE in the E1bCAT plasmid and four E-box elements 10 or 43 bases upstream of the E1bTATA or 4EBE (Fig. 2D). As predicted, both E1bTATA and 4EBE interacted best with Sp1 when Sp1 binding sites were close to the TATA or 4EBE. Although the optimal spacing between basal elements and E-boxes is not known, we chose 43 bases because 4EBE is 43 nucleotides from the proximal E-box element in the native eIF4E promoter. As shown in Fig. 2D, both TATA and 4EBE mediated transactivation by E-box binding proteins, but 4EBE was most active when separated from the E-box by its native spacing interval.
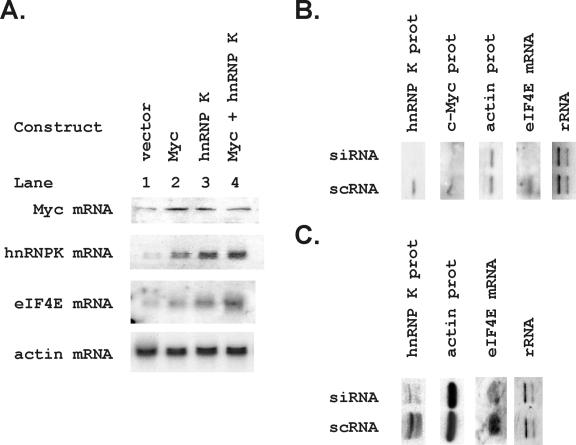
The poly(C)-binding transcription factor hnRNP K binds to the 4E-binding element.
We purified proteins binding to the 4E-binding element by using a three-step purification procedure that is described in the methods section and included a DNA affinity step (Fig. 3). Purification was monitored by using Southwestern analysis. As the first step, hydrophobic exchange column chromatography through a 1 to 0 M (NH4)2SO4 gradient provided a significant enrichment for the 68-kDa band revealed by Southwestern analysis (Fig. 3A). As a second step, either anion-exchange chromatography or cation-exchange chromatography yielded very similar results (Fig. 3B) and could be used interchangeably. A final DNA affinity step yielded a single protein band at 68 kDa (Fig. 3C), which was identified as the poly(C)-binding transcription factor by microsequencing of the isolated protein band (Fig. 3D). hnRNP K binds a C-rich CT element in the promoter of the c-myc gene and has been shown to transactivate through binding to polypyrimidine tract DNA elements (36, 62, 95, 98). We generated a gst-hnRNP K fusion protein and found that this purified hnRNP K binds 4EBE (Fig. 3E).
FIG. 3.
Purification of the 68-kDa protein binding to the 4EBE by DNA affinity chromatography identifies hnRNP K. (A) Hydrophobic exchange chromatography was performed as the initial step in protein purification. Fractions identifying the 68-kDa species binding to the 4EBE were identified by Southwestern analyses as described in Materials and Methods. Lane 1 contains a positive control HeLa nuclear extract (H), and lane 2 contains an aliquot of the HeLa extract (L) before loading on the column. (B) The second step in purification used anion exchange chromatography. Fractions identifying the 68-kDa species binding to the 4EBE were identified by Southwestern analyses as described in Materials and Methods. The positive control HeLa extract was again included for comparison (H). (C) Two sequential bindings to a DNA affinity chromatography matrix using a trimeric 4EBE sequence purified a 68-kDa band as demonstrated in this brilliant blue-stained acrylamide gel. Size markers (M, lane 1), an aliquot of the positive fraction from the hydrophobic exchange column (A, lane 2), an aliquot of the positive fraction from the monoS column (S, lane 3), and an aliquot of the DNA affinity wash (W, lane 5) are compared to the protein bound to the 4EBE-DNA affinity matrix (D) shown in lane 4. (D) The 68-kDa protein band was submitted for sequencing and the indicated amino acid sequences were identified. These correspond uniquely to hnRNP K. (E) Bacterially synthesized gst-hnRNP K was run in a standard Southwestern analysis, and the trimeric 4EBE probe bound to the gst-hnRNP K (H) but not to glutathione S-transferase alone (G). This band was confirmed to be hnRNP K by an immunoblot analysis with anti-hnRNP K. Finally, the gel was also stained with brilliant blue to demonstrate loading of the species tested for binding to the 4EBE.
Nucleotides in 4EBE important for binding and transcription.
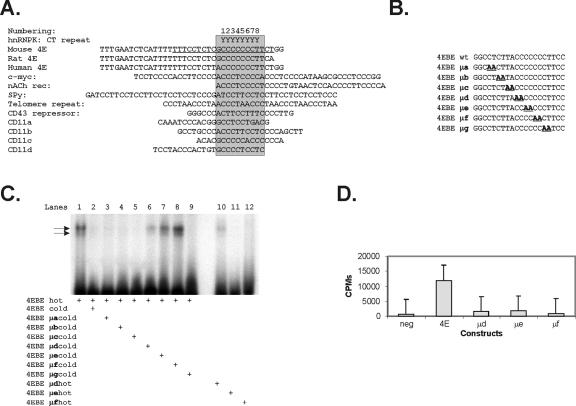
Known hnRNP K binding sites are remarkable for their polypyrimidine tracks that follow a single purine (Fig. 4A). To assess the similarity of the hnRNP K site to our 4EBE, two nucleotides at a time were mutated in the 4EBE sequence (5′-CTCTTACCCCCCCTT), where the sequence indicated in boldface was previously shown to be important for binding activity and the 5′ sequence CTC was shown to enhance binding in EMSAs (46) (sequences are presented in Fig. 4B). This defined the nucleotides in the 4EBE sequence that are essential and sufficient for binding in EMSAs. As shown in Fig. 4C, cold wild-type 4EBE oligonucleotide (lane 2) and cold oligonucleotides for 4EBE mutants μa, μb, μc, and μg (lanes 3, 4, 5, and 9, respectively) were able to compete 4EBE in an EMSA, implying that these nucleotides were dispensable for binding activity. The 4EBE mutant μd partially competed 4EBE (lane 6). The 4EBE mutants μe and μf (lanes 7 and 8) did not compete with the 4EBE oligonucleotide. Furthermore, the 4EBE mutants that did not compete, 4EBE μe and μf, did not show any binding activity in an EMSA when they were themselves radiolabeled. 4EBE μd did show partial binding (lane 10), again correlating with partial competition with cold μd in lane 6. These data suggest that the nucleotides 4 to 9 in the 4EBE sequence 5′-TTACCCCCCCTT (shown in boldface) are essential for binding in EMSAs. We further tested the effect of the same mutations on transcriptional activity of 4EBE in transient-transfection assays. As shown in Fig. 4D the 4EBE mutants μd, μe, and μf abrogated the transcriptional activity of the native eIF4E promoter in reporter assays.
FIG. 4.
Nucleotides in 4EBE sufficient and necessary for binding and transcription. (A) Diagram of 4EBE and comparison to promoter sites known to interact with hnRNP K (76) (12, 21, 51, 87, 95). hnRNP K binding sites are highlighted in promoter sequences previously shown to be regulated by hnRNP K. These are further compared to the 4EBE from human, rat, and mouse eIF4E promoters. (B) Schematic of mutations made in 4EBE to evaluate the function of the polypyrimidine tract shared between the 4EBE and known hnRNP K binding sites. (C) Representative EMSA depicting the nucleotides in 4EBE (5′-CTCTTACCCCCCCTT) sufficient for binding. Radioactively labeled 4EBE oligonucleotide shows two binding activities in an EMSA (lane 1 [arrows identify binding activity]). The binding to 4EBE is competed by adding 500-fold excess of cold wild-type 4EBE oligonucleotide (lane 2) or 500-fold excess of cold 4EBE mutant oligonucleotides μa, μb, μc, and μg (lanes 3, 4, 5, and 9, respectively). (D) Representative experiment correlating binding to 4EBE with transcriptional activity in transient-transfection assays in HeLa cells. Briefly, mutations in the 4EBE sequence (μd, μe, and μf) that did not show binding in EMSA were generated in the native 4E promoter, as described in Materials and Methods. The CAT reporter plasmids for these mutants were subsequently transfected into exponentially growing HeLa cells, along with the human growth hormone as an internal control for transfection efficiency. The cells were harvested 48 h posttransfection and assayed for CAT activity as described in the text.
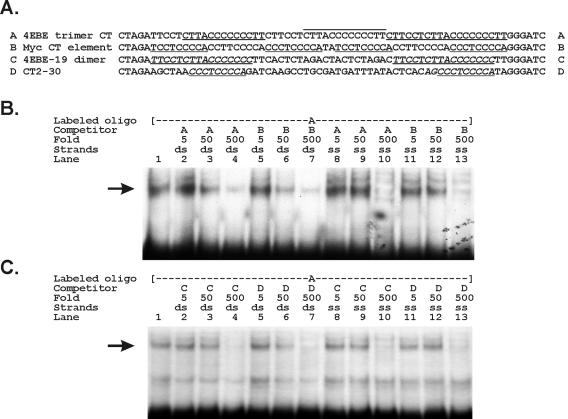
The myc CT element (CTE) is a six-mer repeat of a core CT element (line B in Fig. 5A) (98). To further compare 4EBE with the myc CTE, we performed competition experiments between 4EBE probes and unlabeled myc-CT element competitors in EMSAs (Fig. 5). A labeled 4EBE probe (oligonucleotide A) was competed at equivalent concentrations of both the cold myc CTE competitor (oligonucleotide B) and cold 4EBE itself (Fig. 5B, compare lanes 7 and 4). We then tested competition with single-stranded versions of these oligonucleotides because hnRNP K binds single-stranded myc CTE sequences (6, 50, 97, 98). We again demonstrated equivalent competition between the myc CTE and the 4EBE when both were evaluated as single-stranded cold competitors (Fig. 5B, compare lanes 13 and 10). Tomonaga and Levens further showed that hnRNP K tightly binds dimers of the core myc CT elements when separated by more than 12 nucleotides (oligonucleotide D). We designed a matching dimeric oligonucleotide substituting the 4EBE sequence for the myc CTE (oligonucleotide C). Using unlabeled versions of the dimeric 4EBE and myc CTE as competitors, we again found equivalent cross competitions to the labeled 4EBE probe (Fig. 5C, compare lanes 7 and 4). We also tested these unlabeled competitors in single-stranded forms and again found similar cross competition between the dimeric 4EBE and myc CTE (Fig. 5C, compares lanes 13 and 10).
FIG. 5.
The endogenous binding activities to the 4EBE and the myc CTE are indistinguishable in EMSAs. (A) We compared DNA binding to the eIF4E polypyrimidine element with binding to the myc CTE element first described as the DNA-binding site for hnRNP K (95). The sequences of the DNA oligonucleotides tested in this experiment are labeled as A to D. We compared two configurations of the 4EBE (sequences A and C) with identical configurations of the myc hnRNP K CTE binding site (sequences B and D). We compared a trimer of the eIF4E CT element to a complete myc CT element of the identical length (sequences A and B, respectively). We further compared dimers of each individual element separated by the myc CTE optimal distance (sequences C and D). The CT repeats in oligonucleotides A and B are highlighted by underlining and overlining. The CT elements in oligonucleotides C and D are highlighted by underlining and italics. (B) EMSA findings with HeLa whole-cell extracts bound to labeled oligonucleotide A. Binding was competed for by the indicated trimeric oligonucleotides A or B at the indicated fold excess concentrations. Finally, we compared competition with double-stranded oligonucleotides (ds) or single-stranded oligonucleotides (ss). (C) EMSA findings with HeLa whole-cell extracts bound to labeled oligonucleotide A. Binding was competed for by the indicated dimeric oligonucleotides C or D at the indicated fold excess concentrations. We further compared competition with double-stranded oligonucleotides (ds) or single-stranded oligonucleotides (ss).
A rabbit polyclonal anti-C-terminal hnRNP K has been shown to immunoprecipitate chromatin complexes containing hnRNP K in inducibly transcribed loci (71). These experiments confirmed the in vivo binding of hnRNP K to the actively transcribed c-myc promoter. Using this same antibody we compared the presence of hnRNP K at the eIF4E promoter to a nontranscribed locus (GenBank accession number AC120715), the 5S rRNA region and to the c-myc promoter (Fig. 6). Overall, this antibody specifically immunoprecipitated chromatin containing the eIF4E promoter, indicating binding of hnRNP K to the eIF4E gene in vivo. First, no signal was seen for AC120715, eIF4E or c-myc in the arrested cells, and a nonspecific band was detected in all three immunoprecipitation conditions for the 5S rRNA signal in these quiescent cells. In the serum stimulated and growing cells the 5S rRNA and nontranscribed locus bands were seen in equivalent and therefore nonspecific, amounts in the lanes containing nonimmune rabbit serum, anti-hnRNP K antibody, or anti-hnRNP K with a blocking peptide. In contrast, the c-myc and eIF4E PCR signals were markedly higher in the anti-hnRNP K lanes than in either the rabbit serum or blocking peptide control lanes when cells were serum stimulated or actively growing. Comparing the absence of anti-hnRNP K eIF4E or c-myc chromatin immunoprecipitation signals in the arrested cells to their presence in the serum-stimulated and growing cells suggests that hnRNP K is recruited to these loci in response to signals regulating cell proliferation.
FIG. 6.
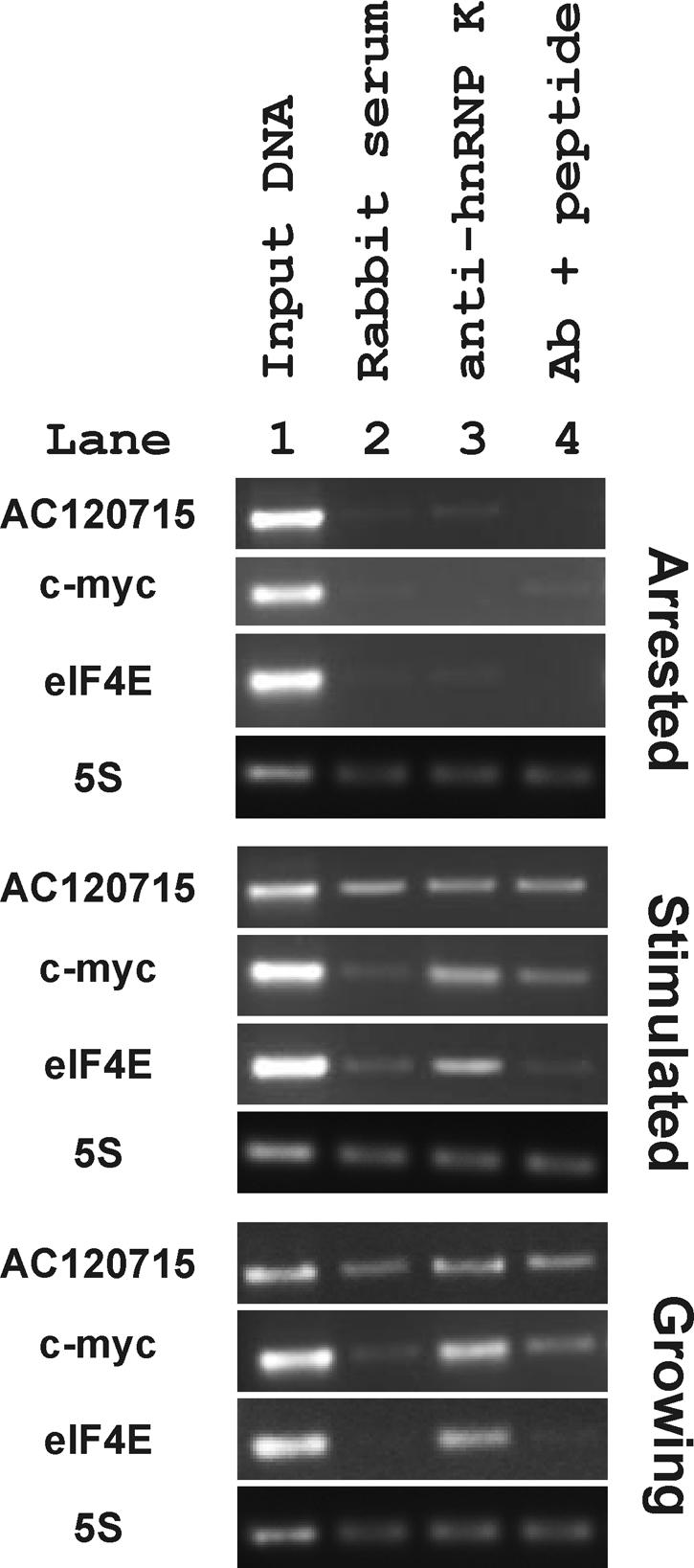
Chromatin immunoprecipitation demonstrates binding of hnRNP K to the eIF4E promoter. Cell lysates from formaldehyde-treated TGR cells in the indicated growth conditions were immunoprecipitated with nonimmune rabbit serum (lane 2) or with anti-hnRNP K antibody that was coincubated with (lane 4) or without (lane 3) the peptide used to generate the antibody in rabbits (99). DNA was purified from the eluted complex and used as a template in PCRs with primers to a nontranscribed rat locus (AC120715), the rat c-myc promoter (c-myc), the eIF4E promoter (eIF4E), and the 5S rRNA region (5S). PCR products were separated by agarose gel electrophoresis and visualized with ethidium bromide. Input DNA was used as a positive control in lane 1.
We transfected constructs expressing myc, hnRNP K, and their combination into Rat 1A cells to evaluate the effect of hnRNP K on eIF4E expression. Pooled transfectants expressing exogenous myc, hnRNP K, and myc plus hnRNP K were analyzed in Northern blots to demonstrate expression of the introduced constructs (Fig. 7A) and evaluate their effect on eIF4E mRNA. Both Myc and hnRNP K increased eIF4E mRNA as predicted by their proposed interactions with its promoter, and the two together produced a larger increase in eIF4E than either alone. We then evaluated the ability of a siRNA for hnRNP K to decrease eIF4E mRNA expression (Fig. 7B and C). In myc wild-type (TGR = myc+/+) cells, our siRNA successfully decreased hnRNP K protein levels compared to scrambled oligonucleotide (Fig. 7B) and decreased eIF4E mRNA levels. Since knockdown of hnRNP K could have regulated eIF4E mRNA by indirect effects on Myc, which is known to be responsive to hnRNP K (Fig. 7B), we also evaluated the effect of the hnRNP K knockdown in myc-null cells (Fig. 7C, HO15 = myc−/−). Importantly, eIF4E mRNA levels were decreased by the hnRNP K siRNA in both the myc wild type and the myc-null cells.
FIG. 7.
Overexpression of hnRNP K and hnRNP K knockdowns regulate endogenous eIF4E mRNA. (A) Rat 1A cells were transfected with vector alone, myc, hnRNP K, or the combination of hnRNP K and myc. Pooled transfectants were selected in neomycin and hygromycin and grown to confluence. mRNA was harvested, and 5 μg was run in a Northern analysis. The same blot was probed for the indicated genes, including c-myc, hnRNP K, eIF4E, and actin. The probes used for the c-myc and hnRNP K blots were the human full-length cDNAs used to construct the expression vectors to increase the specificity of detection of the transfected sequences. The actin and eIF4E probes were mouse full-length cDNA probes. (B and C) TGR (myc+/+) (B) and HO15 (myc−/−) (C) cells were transiently transfected with an siRNA oligonucleotide for hnRNP K. Expression of hnRNP K protein and eIF4E mRNA were analyzed 48 h after transfection in confluent transfected cells. Loading controls include actin protein and the rRNA bands on an ethidium bromide-stained gel. The levels of hnRNP K protein, myc protein, and eIF4E mRNA were compared in cells transfected with an siRNA for hnRNP K (si) to those transfected with a scrambled siRNA (sc).
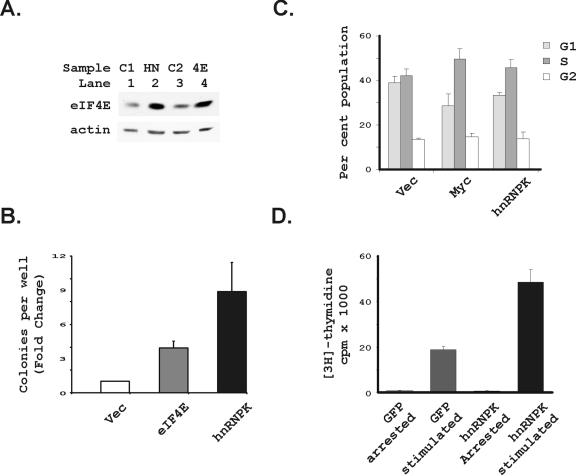
Both hnRNP K and eIF4E transform cells (52, 58). To address the potential significance of hnRNP K's regulation of eIF4E, we first compared the transforming potential of each gene in Rat1A transfectants (Fig. 8). eIF4E protein levels in Rat1A cells transfected with eIF4E (55) were very similar to their levels in Rat1A cells transfected with hnRNP K (Fig. 8A). We then found that hnRNP K transformed the target Rat1A cells somewhat more potently than eIF4E, suggesting that it has multiple effects on transformation in addition to any potential effect mediated by eIF4E (Fig. 8B). eIF4E is known to promote entry into S phase (18). To address hnRNP K's effects on cell cycle regulation, we therefore evaluated the ability of hnRNP K to accelerate passage through G1 into S phase (Fig. 8C), as has been observed for eIF4E overexpression previously (17, 18). As would be predicted if it regulates translation initiation through eIF4E, hnRNP K decreased the proportion of cells in the G1 phase of the cell cycle and increased those in the S phase. We also introduced hnRNP K into myc−/− cells by using a retroviral expression vector (Fig. 8D). In this case, DNA synthesis was measured shortly after infection to examine the acute effects of hnRNP K expression in the absence of c-myc. We found significantly increased DNA synthesis in hnRNP K-infected cells compared to a GFP-expressing vector after serum stimulation (Fig. 8D).
FIG. 8.
Overexpression of hnRNP K transforms cells and enhances passage through G1. (A) eIF4E protein levels were compared between Rat1A cells transfected with hnRNP K and cells transfected with an expression construct for eIF4E (55). Vector control transfected cells (C1) for the hnRNP K transfectants, the hnRNP K transfectants (HN), vector control transfected cells for the eIF4E transfectants (C2), and the eIF4E transfectants (4E) are shown. Immunoblots for eIF4E and actin are shown. (B) The cells presented in panel A were then evaluated for transformation in a standard soft agar assay. Plotted is the mean and standard error of the fold change in cells per well for six wells each in four separate repetitions of the experiment comparing transfected constructs to vector control cells for each repetition (n = 24 for each plot). The hnRNP K and eIF4E transfectants are indicated. (C) Rat1A cells transfected with the indicated constructs were analyzed by fluorescence-activated cell sorting for DNA content. Plotted is the percentage of cells in the G1, S, and G2 phases for three separate determinations for each of the two times the experiment was repeated, together with the standard error of the mean for these determinations. Cells studied were transfected with the indicated expression vectors, including the vector control (vec), myc, and hnRNP K. These determinations were made for actively proliferating subconfluent cells. (D) Myc−/− cells were infected with retroviruses expressing either GFP or hnRNP K. The infected cells were grown to confluence and were then growth arrested over 48 h by removing serum from the culture medium. DNA synthesis was measured as described in Materials and Methods in the subsequent absence (arrested) or presence (stimulated) of serum for 20 h. hnRNP K infection alone did not enhance DNA synthesis in the growth arrested cells but significantly increased DNA synthesis after cells were stimulated to initiate division.
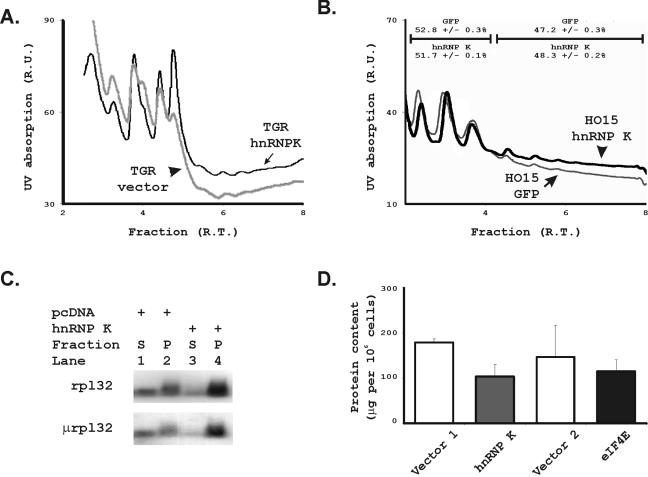
Altering eIF4E levels should result in altered rates of translation initiation if hnRNP K's regulation of eIF4E is biologically significant. To evaluate the effect of hnRNP K overexpression on global translation initiation rates, we compared the net quantity of mRNAs contained in the actively translating polysomal fractions of Rat 1A cells overexpressing hnRNP K to those transfected with the vector alone (Fig. 9A). hnRNP K produced a significant shift of the profile toward an increased fraction of mRNAs in polysomes. Since hnRNP K could exert this effect through regulation of c-myc, we further analyzed the effects of hnRNP K expression in myc−/− cells where we again found that hnRNP K increased the proportion of mRNAs present in actively translating, polysomal fractions of cellular mRNAs (Fig. 9B). To evaluate hnRNP K's effects on a single mRNA that is known to be translationally regulated, we then assessed the effect of hnRNP K on the translation rate of ribosomal protein L32 (rpL32) in a standard translational reporter gene experiment (Fig. 9C). After transient transfection of hnRNP K, a reporter gene under the translational control of the 5′ untranslated leader sequence of rpL32 shifted into the polysomal fraction as expected. The 5′ untranslated leader of rpL32 contains a 5′ terminal oligopyrimidine (TOP) sequence as expected for a ribosomal protein. We were concerned that the TOP sequence resembles an hnRNP K binding site and that this shift might represent direct binding of hnRNP K to the TOP reporter. We therefore also tested hnRNP K's effects on a mutated version of the rpL32 leader sequence that has lost its TOP function and found that hnRNP K again had an equal effect in enhancing its translation rate. Finally, we previously showed that loss of eIF4E function in cells transfected with a dominant-inhibitory 4E binding protein (4EBPμ) results in a paradoxical increase in cell size (55). This appears to be related to disproportionate effects of eIF4E to enhance translation of mRNAs involved in cell division over those involved in cell growth. We found no change in cell size in any of our Rat 1A transfectants in response to hnRNP K. We therefore evaluated the protein content per cell and found that hnRNP K actually decreased protein content as predicted if it had disproportionate effects on mRNAs that enhance cell division over cell growth (Fig. 9D). A similar decrease in protein content per cell was observed for the cells overexpressing eIF4E alone. Thus, although net translation initiation is enhanced by hnRNP K, this effect may select for specific, cell cycle-promoting mRNAs as we have observed with eIF4E.
FIG. 9.
Overexpression of hnRNP K alters translation rates. (A) Polysomal profiles of Rat 1A cells transfected with hnRNP K (dark black line) were compared those containing an empty vector control (gray line). Cells were grown to confluence for 48 h in medium lacking serum, and cytoplasmic extracts were run on standard sucrose gradients to separate mRNAs in monosomal versus polysomal fractions. The y axis identifies UV absorption in relative units, and the x axis identifies fractions of the gradient in relative time from least dense on the left to most dense on the right. (B) Polysomal profiles of myc−/− cells infected with pBABE-hnRNP K (dark black line) were compared to those infected with pBABE-GFP (lighter gray line). Cells were again evaluated at confluence in the absence of serum stimulation. Axes are as described in panel A. We further indicate the mean and standard error of the percentage of RNAs found in the subpolysomal versus polysomal fractions for two replicas for each analysis. (C) To confirm the global patterns of polysomal fractionation, we then analyzed the fractionation of reporter genes coupled to the translationally regulated leader sequence of ribosomal protein L32. We compared a reporter construct containing the 5′TOP from ribosomal protein L32 (rpL32) coupled to a GFP cDNA with a reporter construct containing the inactivated TOP sequence from rpL32 coupled to a growth hormone cDNA. These two constructs were transfected into Rat1A cells in the absence or presence of pcDNA-hnRNP K. The cells were grown to confluence over 72 h and serum was withdrawn for the last 48 h before harvest. Polysomal (P) and subpolysomal (S) pooled fractions were isolated by using sucrose density gradients. The mRNAs contained in each pool were isolated from the cells transfected either with (hnRNP K+) or without (pcDNA+) the hnRNP K expression vector. The pooled polysomal and subpolysomal RNAs were then blotted in standard RNA blots and probed for the GFP (rpl32) and growth hormone (μrpl32) reporters. The ratio of the intensity of the signal in the polysomal fraction (P) compared to the subpolysomal fraction (S) demonstrates increased translation initiation rates. (D) Subconfluent, proliferating Rat 1A cells from Fig. 7 transfected with eIF4E and hnRNP K were harvested, and the protein content per cell was measured for each construct.
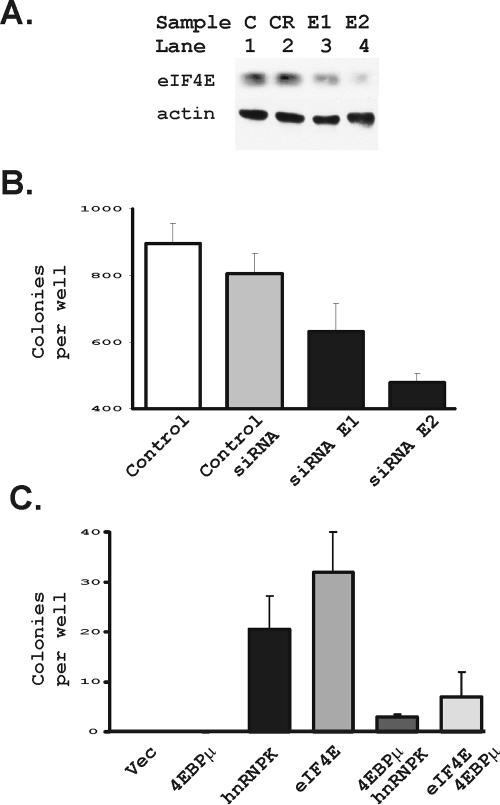
Last, we sought to determine whether hnRNP K's effects on eIF4E were required for it to transform cells (Fig. 10). We first tested two siRNA oligonucleotides that could block expression of eIF4E protein in Rat1A cells (Fig. 10A). We transfected these siRNAs into Rat1A cells overexpressing hnRNP K and compared transformation efficiencies to the same cells that were untransfected or were transfected with a control siRNA for luciferase (Fig. 10B). As shown, the eIF4E siRNAs decreased formation of anchorage independent colonies by Rat1A cells overexpressing hnRNP K.
FIG. 10.
hnRNP K transforms cells through effects on eIF4E expression. (A) Rat1A cells were transfected with a control RNA (CR) and two siRNAs for eIF4E (E1 and E2) as described in Materials and Methods. Protein extracts were made 48 h after transfection and evaluated for eIF4E and actin levels by immunoblotting. An untransfected control lane is also included (lane C). (B) Rat1A cells expressing hnRNP K from Fig. 7 above were transfected with the control siRNA and the two siRNAs from panel A. Additional hnRNP K-expressing cells were examined without transfecting them (control). At 48 h after transfection the cells were seeded for a soft agar assay. The mean and standard error of colonies per well, evaluated 8 days after the cells were put in soft agar for each condition, were plotted. (C) Rat1A transfectants were evaluated for transformation in a standard soft agar assay. Shown are cells transfected with a vector control, a dominant inhibitor of eIF4E function (4EBPμ), hnRNP K, eIF4E, the combination of the dominant inhibitor of eIF4E, together with hnRNP K, and the combination of eIF4E, together with the dominant inhibitor of eIF4E, as indicated in the figure. Plotted is the mean and standard deviation of the cells per well for four wells each in two separate repeats of the experiment.
We had previously developed a dominant inhibitory form of the 4E binding protein (4EBP) that blocks eIF4E function, which we designated 4EBPμ (55). In that study, 4EBPμ's function was shown to depend on its interactions with eIF4E. We had used that construct to show that eIF4E function was required for Myc transformation. We therefore applied the same approach to confirming the importance of eIF4E function in transformation by hnRNP K (Fig. 10C). We found that transformation by hnRNP K depended on its effects on eIF4E because cotransfection of a 4EBPμ that constitutively blocks eIF4E function blocked soft agar transformation by hnRNP K (Fig. 10C). The transforming efficiencies of eIF4E and hnRNP K were more equivalent in this individual experiment than when data from four different experiments were evaluated in Fig. 8B. Nevertheless, the 4EBPμ had a similar capacity to block transformation by both eIF4E and hnRNP K overexpression.
DISCUSSION
Very little is known about the mechanisms underlying the transcriptional regulation of eIF4E and other translation factors despite the fact that the levels of these translation factors play an important role in cellular growth and differentiation and in cancer. Ribosomal protein genes in yeast are regulated as a function of cell growth by DNA factor(s) that are required for activation and coordinated regulation of the whole family of genes coding for the translational apparatus (41, 49). Much less is known about the coordination of synthesis of the growth apparatus of mammalian cells. We were therefore especially interested in characterizing the second element in the eIF4E promoter that was essential for its regulation (45).
We had previously demonstrated the required role of the 4EBE by using loss-of-function studies. We therefore evaluated its potential function as a dominant expression element by comparing it to TATA sequences that it appears to replace. This is especially important because basal promoter elements determine the range of transcriptional activators that regulate individual promoters (13). The regulation of eIF4E levels in the cell represents a particular physiologic challenge because the range of normal expression levels of eIF4E is very narrow. With the exception of mRNAs that use internal ribosomal entry sites, eIF4E is required for translation initiation of nearly all mRNAs. It must therefore be expressed ubiquitously in all cells at some undefined minimum level. In contrast, as little as a threefold increase in eIF4E levels is sufficient to derepress translation of at least a subset of mRNAs that can transform cells to a malignant phenotype (7, 91).
Using standard reporter gene analyses we show here that the 4EBE can function like TATA to drive reporter gene expression, and its activity can be enhanced by either SP1 or E-box elements (Fig. 2). Our reporter experiments do not eliminate the possibility that the 4EBE might also be functioning to bind a classic transcriptional activator. Further in vitro studies will be required to prove this point, but our reporter gene studies make 4EBE an interesting candidate to be an additional basal control element (40). The simple structure of the eIF4E promoter now includes a required E-box element and a promoter element that appears to replace TATA. To better understand this simple form of regulation, we then sought to identify the factor(s) that might bind to the 4EBE.
We identified hnRNP K as a candidate 4EBE binding protein by using standard protein purification methodology. Known interactions of hnRNP K with other promoters, especially c-myc, make it an especially interesting candidate eIF4E regulatory factor. eIF4E is a candidate myc target gene (86), and hnRNP K is one of the most important transcriptional controls of the c-myc promoter (36, 62, 95, 98). The coincidence of hnRNP K regulation of both myc and eIF4E promoters, together with the candidate role of myc in regulating eIF4E, would suggest that complex combinatorial interactions between myc and hnRNP K could be especially important in controlling eIF4E levels. Indeed, we initially showed that the eIF4E regulatory factor increases in cells in direct relationship to myc levels, as might be expected if this factor can regulate myc or is regulated by myc (45). Michelotti et al.'s demonstration that hnRNP K directly interacts with the TBP (62) fits our demonstration that the 4EBE is a core promoter element in the eIF4E promoter. Using EMSAs, additional reporter gene experiments and chromatin immunoprecipitation we demonstrated that the polypyrimidine track in the 4EBE functioned like that in the myc promoter and that both elements functioned identically as DNA-binding elements (Fig. 4 to 6).
We then transfected hnRNP K singly and together with Myc to assess the functional significance of hnRNP K as a 4E regulatory factor (Fig. 7). eIF4E mRNA levels increased in response to hnRNP K in pooled, selected transfectants overexpressing hnRNP K. Moreover, this increase was further augmented if both myc and hnRNP K were transfected into the cells. We then identified a siRNA that could knock down hnRNP K protein levels. In siRNA knockdown experiments we used TGR cells that are an immortalized diploid rat cell line so that we might also evaluate hnRNP K loss in the corresponding myc-null isolate (HO15) (59). Transient transfection of an hnRNP K siRNA markedly inhibited hnRNP K protein levels and decreased eIF4E mRNA levels 48 h after transfection. The loss of eIF4E expression in response to loss of hnRNP K might be an indirect effect of the parallel myc knockdown in myc wild-type cells since hnRNP K is known to regulate c-myc. We therefore repeated these experiments in myc-null cells. The hnRNP K knockdown had less effect on eIF4E mRNA levels in the myc-null cells, either due to a change from myc dependence in the TGR cells where the siRNA also affected eIF4E regulation by c-myc or because they are slower growing and therefore harder to optimally transfect. Nevertheless, we again saw decreases in eIF4E mRNA 48 h after introduction of the hnRNP K siRNA.
We have previously shown that eIF4E leads to preferential effects on mRNAs that regulate cell division over those that regulate cell growth as one potential explanation for its ability to transform cells (78, 79). This appears to be a mechanism through which cell growth can be coordinated with cell division. We would predict that an eIF4E regulatory factor should phenocopy some effects of eIF4E itself. We therefore first compared transformation by eIF4E to hnRNP K and found that hnRNP K was a somewhat more potent transforming agent at similar levels of eIF4E (Fig. 8A and B). hnRNP K overexpression also caused the same kinds of changes in cell proliferation patterns as those seen for eIF4E because its overexpression led to a decreased proportion of cells in the G1 phase of the cell cycle and accelerated entry into S phase (Fig. 8C and D).
hnRNP K is a multifunctional protein known to translationally repress several mRNAs (5), especially those involved in erythrocyte differentiation (33, 69). For example, the DICE element in the 15-lipoxygenase mRNA is silenced by hnRNP K in actively proliferating erythroid precursors. Our findings suggest that, in contrast, hnRNP K should actually enhance translation initiation through increased eIF4E. To resolve these apparent differences, we analyzed global translation initiation rates in hnRNP K-overexpressing cells by using polysomal profiles. We compared the amounts of mRNA in polysomal versus nontranslating fractions in Rat 1A and myc−/− cells expressing increased amounts of hnRNP K to control cells (Fig. 9A and B). hnRNP K expression caused a clear shift toward increased mRNAs in the polysomal fraction that is actively translating mRNA. We also assessed the effect of hnRNP K on a 5′TOP reporter construct (Fig. 9C), which also showed enhanced translation initiation through a generalized enhancement of translation initiation. Thus, hnRNP K globally stimulates translation initiation, as one would expect of a factor that regulates eIF4E. Its repression of the DICE element is likely therefore a narrower function unique to that mRNA. We have shown that loss of eIF4E function has more effect on mRNAs regulating cell division than cell growth by effects on cell size and protein content (55). We therefore compared the cellular protein content of cells overexpressing either hnRNP K or eIF4E and found similar effects on cellular protein content; our findings were again consistent with the idea that both of them enhance expression of mRNAs involved in cell division in preference to those controlling cell growth (Fig. 9D).
Finally, we assessed the significance of eIF4E regulation by hnRNP K as a potential contributor to eIF4E's role in malignancy. Averaged over four experiments, hnRNP K more efficiently transformed cells than equivalent amounts of eIF4E expressed on its own (Fig. 8B). However, blocking eIF4E activity blocked transformation by hnRNP K by using two different methods to inhibit eIF4E (Fig. 10). Thus, although eIF4E is certainly not the only target of hnRNP K that functions in its ability to transform cells, it is an important one.
Taken together, our data strongly suggest that hnRNP K is a likely candidate eIF4E regulatory factor (4ERF). Purification of hnRNP K by 4EBE binding, demonstration of in vitro-synthesized hnRNP K binding to the 4EBE, demonstration that 4EBE binding is identical to myc CTE binding, regulation of endogenous eIF4E levels by altering hnRNP K levels, and similar cellular behavior of cells overexpressing hnRNP K and eIF4E are all most consistent with this view.
hnRNP K is more abundant than most typical transcription factors, and it affects nearly every step in transcription and translation (5). Moreover, it is an unusual transcription factor since it also binds to single-stranded DNA (ssDNA) and single-stranded RNA. Its functions as a candidate transcription factor need to be interpreted in this light. Its binding to TBP is consistent with our proposal that it could act to recruit TBP to the −25 region of the eIF4E promoter. In this view, the 4EBE likely functions as the equivalent of a basal promoter element. In contrast, the presence of a transactivation domain in the hnRNP K N terminus is more consistent with the view that hnRNP K might be acting as another activating protein, although the transactivation domain in hnRNP K is relatively weak (62, 98). Perhaps the most intriguing possibility is that hnRNP K acts indirectly to promote melting of promoter regions by virtue of its preferential binding to ssDNA (97). In this view, hnRNP K acts as a structural activating protein that regulates DNA conformation to promote transcription (6). Such ssDNA binding properties would allow hnRNP K to act on its own without requiring interactions with other parts of the transcription apparatus. ssDNA binding proteins are known to relieve the torsional stress of transcription and so enhance the rate of transcription by this alternative mechanism (50). Although we observed ssDNA binding by hnRNP K to the 4EBE in the eIF4E promoter, it was not as strong as was described for the myc CT-element by the Levens group. Clarification of the relative importance of these three mechanisms to hnRNP K's regulation of eIF4E will require additional exploration of all three mechanisms. Other proteins are also known to bind similar single-stranded polypyrimidine tracks, including several that bind to polypyrimidine elements in the c-myc promoter, and most function both as transactivators and to regulate DNA conformation (64). Any of these additional proteins are potential additional or alternative eIF4E regulatory factor candidates that will require additional future assessments. These proteins include the far upstream sequence binding protein (23), the Pur proteins (3, 87), the cellular nucleic acid binding protein (63), and myc single-stranded proteins (66). Finally, the nature of the 97-kDa protein that we originally identified remains unknown from these initial purification studies (45).
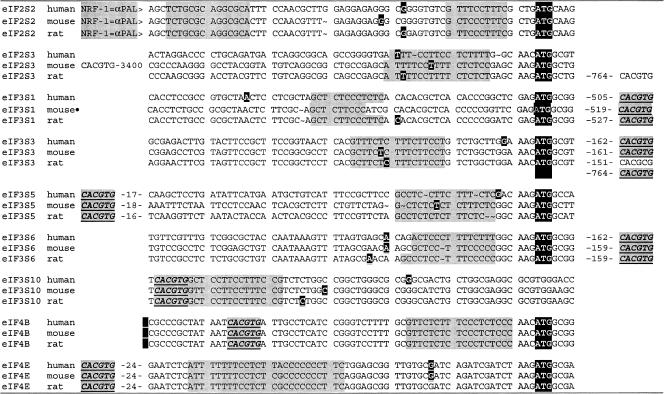
Overexpression of hnRNP K produced changes in global translation initiation rates, suggesting the possibility that the polypyrimidine sequence in the eIF4E promoter might be a unifying regulatory motif for additional translation initiation factors. This would be a particularly interesting result, given how little we understand mechanisms by which translation initiation factor synthesis might be coordinated. To evaluate this possibility, we inspected the proximal promoter regions of all 38 human translation initiation factors and found that eight additional promoters contained polypyrimidine stretches analogous to the sequence in eIF4E (Fig. 11). All nine polypyrimidine stretches were conserved in human, mouse, and rat genomes. All fell within 20 nucleotides of either the transcription initiation site or the translation initiation codon. Moreover, the location of c-Myc binding sequences within 10 to 500 bp of these polypyrimidine stretches in nearly all of these same promoters suggests that the E-box-polypyrimidine combination in the eIF4E promoter could be a regulatory motif that might regulate additional translation initiation factors.
FIG. 11.
Polypyrimidine elements in mammalian translation initiation factor promoters. The genomic sequences of thirty-eight human translation initiation factor genes (eIFs) were identified by using LocusLink (104). Five thousand nucleotides 5′ to and 3′ to the transcription initiation site of each gene were downloaded into Clone Manager Suite 7 for further manipulation (Scientific and Educational Software, Durham, N.C.). The immediate promoter regions of all of these sequences were inspected for sequences containing stretches of eight or more pyrimidines that were less than 20 nucleotides from the transcription initiation site or the translation initiation codon. Thirteen human eIFs revealed such sequences. Two thousand nucleotides surrounding the transcription initiation site of the 13 candidates were then used in a BLAST search of the mouse and rat genomes. Promoter regions were considered to be confirmed if this region showed >70% homology, and it fell within a calculated CpG island in all three species. The polypyrimidine stretches were conserved between mouse, rat, and human genomes in nine of the confirmed promoter candidates as shown. Listed are the 70 nucleotides containing the proximal promoter sequences from all three species for the nine candidates containing conserved polypyrimidine stretches, and the polypyrimidine stretches are identified by gray highlighting. The transcription initiation sites identified in the genome databases were confirmed or modified by a BLAST comparison of each genomic sequence against available expressed sequence tag sequences for each species. Indicated as white letters against a black background are either the site listed in LocusLink as the transcription initiation site, the site mapping of the 5′ end of the majority of all of the available ests for the indicated species, or the 5′-most expressed sequence tag sequence if no consensus 5′ end was obvious. The translation initiation codon (ATG) is identified for the eight candidates where it is positioned close to the transcription initiation site. Myc binding sites (CACGTG) were identified by locating Pm1I sites in each promoter and are shown in boldface, underlined italics with gray highlighting. Remarkably, Myc binding sites were located at extremely short distances from the initiation site of seven of the nine candidate promoters; four were within 50 nucleotides, two were at about 150 nucleotides, and one was about 500 nucleotides distant. A nuclear respiratory factor 1α palindromic sequence (NRF-1=α PAL) binding site that also binds max and may be an alternative Myc binding site is identified in the eIF2S2 promoters. LocusLink identifications include eIF2S2 (human 8894, mouse 67204, and rat 296302), eIF2S3 (1968, 26905, and 299027), eIF3S1 (8669, 78655, and 311371), eIF3S3 (8667, 68135, and 299899), eIF3S5 (8665, 66085, and 293427), eIF3S6 (3646, 16341, and 299872), eIF3S10 (8661, 75705, and 300253), eIF4B (1975, 75705, and 300253), and eIF4E (1977, 13684, and 117045). The eIF3S1 mouse promoter sequences are assumed based on their homology to the rat and human sequences. We found no mouse cDNA or EST sequence that matched this sequence, and it was not annotated as the promoter sequence in the genome database.
Identification of hnRNP K as the leading 4ERF candidate suggests a variety of additional directions for further study. Its role in human cancers merits further clinical studies to see whether abnormalities in hnRNP K expression, distribution, or signaling function coincide with overexpression of eIF4E (70). The nature of its interactions with Myc will need further elucidation to determine whether it interacts with the regulation of other genes transactivated by Myc. The coincidence of c-Myc binding sites and polypyrimidine stretches in additional translation initiation factors makes this a particularly important issue. Why hnRNP K might play such a prominent role in eIF4E regulation or global regulation of translation initiation is equally interesting. hnRNP K is downstream of multiple signal transduction pathways where integration between signaling cascades and net translation capacity of the cell might provide a significant evolutionary advantage (5). Moreover, hnRNP K has a unique KNS-mediated nuclear importation signal that is dependent on RNA polymerase II transcription (61). This dependence of hnRNP K nuclear localization on polymerase II activity might provide a mechanism to adjust eIF4E and other translation initiation factors to net mRNA copy numbers in the cell. Regardless, the identification of hnRNP K as a 4ERF will certainly guide further explorations of mechanisms that govern eIF4E levels in the cell.
Acknowledgments
This work was supported by PHS grant RO1-CA63117 (M.L., S.M., L.C., and E.S.). K.K. was supported by NIDDK training grant T32 DK007191. M.R. was supported by NIH NRSA training grant 5T32 CA009216-24. M.J.P. is supported by the Cutaneous Biology Research Center through the Massachusetts General Hospital/Shiseido Co., Ltd., agreement.
We thank Marc Wathelet and Tom Maniatis, Department of Molecular and Cellular Biology, Harvard University, for kindly providing the E1bCAT plasmid; Karol Bomsztyk for the generous provision of antibody 54 to the hnRNP K protein; and O. Meyuhas for the 5′TOP translation reporter plasmids.
REFERENCES
- 1.Atchison, M. L., O. Meyuhas, and R. P. Perry. 1989. Localization of transcriptional regulatory elements and nuclear factor binding sites in mouse ribosomal protein gene rpL32. Mol. Cell. Biol. 9:2067-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azizkhan, J. C., D. E. Jensen, A. J. Pierce, and M. Wade. 1993. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit. Rev. Eukaryot. Gene Expr. 3:229-254. [PubMed] [Google Scholar]
- 3.Bergemann, A. D., Z. W. Ma, and E. M. Johnson. 1992. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol. 12:5673-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell, T. K., J. Huang, A. Ma, L. Kretzner, F. W. Alt, R. N. Eisenman, and H. Weintraub. 1993. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 13:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein multiple processes. Bioessays 26:629-638. [DOI] [PubMed] [Google Scholar]
- 6.Braddock, D. T., J. L. Baber, D. Levens, and G. M. Clore. 2002. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 21:3476-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, C., N. Nakayama, M. Goebl, K. Tanaka, A. Toh-e, and K. Matsumoto. 1988. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:3556-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chau, C. M., M. J. Evans, and R. C. Scarpulla. 1992. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2α, and tyrosine aminotransferase: specific interaction of purified NRF-1 with multiple target genes. J. Biol. Chem. 267:6999-7006. [PubMed] [Google Scholar]
- 9.Chiorini, J. A., S. Miyamoto, S. J. Harkin, and B. Safer. 1999. Genomic cloning and characterization of the human eukaryotic initiation factor-2β promoter. J. Biol. Chem. 274:4195-4201. [DOI] [PubMed] [Google Scholar]
- 10.Chirgwin, J. M., A. E. Przybyla, R. J. MacDonald, and W. J. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease activity. Biochemistry 18:5294-5299. [DOI] [PubMed] [Google Scholar]
- 11.Clemens, M. J. 2004. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene 23:3180-3188. [DOI] [PubMed] [Google Scholar]
- 12.Da Silva, N., A. Bharti, and C. S. Shelley. 2002. hnRNP-K and Pur(alpha) act together to repress the transcriptional activity of the CD43 gene promoter. Blood 100:3536-3544. [DOI] [PubMed] [Google Scholar]
- 13.Das, G., C. S. Hinkley, and W. Herr. 1995. Basal promoter elements as a selective determinant of transcriptional activator function. Nature 374:657-660. [DOI] [PubMed] [Google Scholar]
- 14.Davis, W., Jr., and R. M. Schultz. 1998. Molecular cloning and expression of the mouse translation initiation factor eIF-1A. Nucleic Acids Res. 26:4739-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Benedetti, A., and J. R. Graff. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23:3189-3199. [DOI] [PubMed] [Google Scholar]
- 16.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 17.De Benedetti, A., S. Joshi-Barve, C. Rinker-Schaeffer, and R. E. Rhoads. 1991. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol. Cell. Biol. 11:5435-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Benedetti, A., and R. E. Rhoads. 1990. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc. Natl. Acad. Sci. USA 87:8212-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 21.Du, Q., I. N. Melnikova, and P. D. Gardner. 1998. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J. Biol. Chem. 273:19877-19883. [DOI] [PubMed] [Google Scholar]
- 22.Dudov, K. P., and R. P. Perry. 1986. Properties of a mouse ribosomal protein promoter. Proc. Natl. Acad. Sci. USA 83:8545-8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan, R., L. Bazar, G. Michelotti, T. Tomonaga, H. Krutzsch, M. Avigan, and D. Levens. 1994. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8:465-480. [DOI] [PubMed] [Google Scholar]
- 24.Duncan, R., and J. W. Hershey. 1984. Heat shock-induced translational alterations in HeLa cells. Initiation factor modifications and the inhibition of translation. J. Biol. Chem. 259:11882-11889. [PubMed] [Google Scholar]
- 25.Duncan, R., and J. W. Hershey. 1983. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 258:7228-7235. [PubMed] [Google Scholar]
- 26.Duncan, R., and J. W. Hershey. 1985. Regulation of initiation factors during translational repression caused by serum depletion: abundance, synthesis, and turnover rates. J. Biol. Chem. 260:5486-5492. [PubMed] [Google Scholar]
- 27.Duncan, R., S. C. Milburn, and J. W. Hershey. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control: heat shock effects on eIF-4F. J. Biol. Chem. 262:380-388. [PubMed] [Google Scholar]
- 28.Dynlacht, B. D., T. Hoey, and R. Tjian. 1991. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66:563-576. [DOI] [PubMed] [Google Scholar]
- 29.Efiok, B. J., J. A. Chiorini, and B. Safer. 1994. A key transcription factor for eukaryotic initiation factor-2 alpha is strongly homologous to developmental transcription factors and may link metabolic genes to cellular growth and development. J. Biol. Chem. 269:18921-18930. [PubMed] [Google Scholar]
- 30.Efiok, B. J., and B. Safer. 2000. Transcriptional regulation of E2F-1 and eIF-2 genes by alpha-pal: a potential mechanism for coordinated regulation of protein synthesis, growth, and the cell cycle. Biochim. Biophys. Acta 1495:51-68. [DOI] [PubMed] [Google Scholar]
- 31.Fahrenkrug, S. C., M. O. Dahlquist, K. J. Clark, and P. B. Hackett. 1999. Dynamic and tissue-specific expression of eIF4E during zebra fish embryogenesis. Differentiation 65:191-201. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habelhah, H., K. Shah, L. Huang, A. Ostareck-Lederer, A. L. Burlingame, K. M. Shokat, M. W. Hentze, and Z. Ronai. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3:325-330. [DOI] [PubMed] [Google Scholar]
- 34.Hansen, S. K., S. Takada, R. H. Jacobson, J. T. Lis, and R. Tjian. 1997. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91:71-83. [DOI] [PubMed] [Google Scholar]
- 35.Hariharan, N., D. E. Kelley, and R. P. Perry. 1989. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 3:1789-1800. [DOI] [PubMed] [Google Scholar]
- 36.Hay, N., J. M. Bishop, and D. Levens. 1987. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1:659-671. [DOI] [PubMed] [Google Scholar]
- 37.Haydon, M. S., J. D. Googe, D. S. Sorrells, G. E. Ghali, and B. D. Li. 2000. Progression of eIF4e gene amplification and overexpression in benign and malignant tumors of the head and neck. Cancer 88:2803-2810. [DOI] [PubMed] [Google Scholar]
- 38.Hendrickson, W., and R. Schleif. 1985. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl. Acad. Sci. USA 82:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez, G., R. Diez del Corral, J. Santoyo, S. Campuzano, and J. M. Sierra. 1997. Localization, structure and expression of the gene for translation initiation factor eIF-4E from Drosophila melanogaster. Mol. Gen. Genet. 253:624-633. [DOI] [PubMed] [Google Scholar]
- 40.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 41.Huet, J., P. Cottrelle, M. Cool, M. L. Vignais, D. Thiele, C. Marck, J. M. Buhler, A. Sentenac, and P. Fromageot. 1985. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 4:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humbelin, M., B. Safer, J. A. Chiorini, J. W. Hershey, and R. B. Cohen. 1989. Isolation and characterization of the promoter and flanking regions of the gene encoding the human protein-synthesis-initiation factor 2 alpha. Gene 81:315-324. [DOI] [PubMed] [Google Scholar]
- 43.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javahery, R., A. Khachi, K. Lo, B. Zenzie-Gregory, and S. T. Smale. 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston, K. A., M. Polymenis, S. Wang, J. Branda, and E. V. Schmidt. 1998. Novel regulatory factors interacting with the promoter of the gene encoding the mRNA cap binding protein (eIF4E) and their function in growth regulation. Mol. Cell. Biol. 18:5621-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, R. M., J. Branda, K. A. Johnston, M. Polymenis, M. Gadd, A. Rustgi, L. Callanan, and E. V. Schmidt. 1996. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol. Cell. Biol. 16:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 48.Kim, S., Q. Li, C. V. Dang, and L. A. Lee. 2000. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl. Acad. Sci. USA 97:11198-11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouzine, F., J. Liu, S. Sanford, H. J. Chung, and D. Levens. 2004. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 11:1092-1100. [DOI] [PubMed] [Google Scholar]
- 51.Lacroix, L., H. Lienard, E. Labourier, M. Djavaheri-Mergny, J. Lacoste, H. Leffers, J. Tazi, C. Helene, and J. L. Mergny. 2000. Identification of two human nuclear proteins that recognize the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 28:1564-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 53.Li, B. D., L. Liu, M. Dawson, and A. De Benedetti. 1997. Overexpression of eukaryotic initiation factor 4E (eIF4E) in breast carcinoma. Cancer 79:2385-2390. [PubMed] [Google Scholar]
- 54.Li, S., D. M. Perlman, M. S. Peterson, D. Burrichter, S. Avdulov, V. A. Polunovsky, and P. B. Bitterman. 2004. Translation initiation factor 4E blocks endoplasmic reticulum-mediated apoptosis. J. Biol. Chem. 279:21312-21317. [DOI] [PubMed] [Google Scholar]
- 55.Lynch, M., C. Fitzgerald, K. A. Johnston, S. Wang, and E. V. Schmidt. 2004. Activated eIF4E-binding protein slows G1 progression and blocks transformation by c-myc without inhibiting cell growth. J. Biol. Chem. 279:3327-3339. [DOI] [PubMed] [Google Scholar]
- 56.Makhlouf, A. A., A. M. Namboodiri, and P. J. McDermott. 2001. Transcriptional regulation of the rat eIF4E gene in cardiac muscle cells: the role of specific elements in the promoter region. Gene 267:1-12. [DOI] [PubMed] [Google Scholar]
- 57.Mamane, Y., E. Petroulakis, L. Rong, K. Yoshida, L. W. Ler, and N. Sonenberg. 2004. eIF4E-from translation to transformation. Oncogene 23:3172-3179. [DOI] [PubMed] [Google Scholar]
- 58.Mandal, M., R. Vadlamudi, D. Nguyen, R. A. Wang, L. Costa, R. Bagheri-Yarmand, J. Mendelsohn, and R. Kumar. 2001. Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J. Biol. Chem. 276:9699-9704. [DOI] [PubMed] [Google Scholar]
- 59.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 60.Meyuhas, O., Y. Blierman, P. Pierandrei-Amaldi, and F. Amaldi. 1996. Analysis of polysomal RNA, p. 65-81. In P. Krieg (ed.), A laboratory guide to RNA: isolation, analysis and synthesis. Wiley-Liss, New York, N.Y.
- 61.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michelotti, E. F., G. A. Michelotti, A. I. Aronsohn, and D. Levens. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16:2350-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michelotti, E. F., T. Tomonaga, H. Krutzsch, and D. Levens. 1995. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 270:9494-9499. [DOI] [PubMed] [Google Scholar]
- 64.Michelotti, G. A., E. F. Michelotti, A. Pullner, R. C. Duncan, D. Eick, and D. Levens. 1996. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16:2656-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miltenberger, R. J., K. A. Sukow, and P. J. Farnham. 1995. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol. Cell. Biol. 15:2527-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Negishi, Y., Y. Nishita, Y. Saegusa, I. Kakizaki, I. Galli, F. Kihara, K. Tamai, N. Miyajima, S. M. Iguchi-Ariga, and H. Ariga. 1994. Identification and cDNA cloning of single-stranded DNA binding proteins that interact with the region upstream of the human c-myc gene. Oncogene 9:1133-1143. [PubMed] [Google Scholar]
- 67.Noguchi, M., S. Miyamoto, T. A. Silverman, and B. Safer. 1994. Characterization of an antisense Inr element in the eIF-2 alpha gene. J. Biol. Chem. 269:29161-29167. [PubMed] [Google Scholar]
- 68.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 69.Ostareck-Lederer, A., D. H. Ostareck, C. Cans, G. Neubauer, K. Bomsztyk, G. Superti-Furga, and M. W. Hentze. 2002. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22:4535-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostrowski, J., and K. Bomsztyk. 2003. Nuclear shift of hnRNP K protein in neoplasms and other states of enhanced cell proliferation. Br. J. Cancer 89:1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ostrowski, J., Y. Kawata, D. S. Schullery, O. N. Denisenko, and K. Bomsztyk. 2003. Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res. 31:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi, Y., M. A. Gregory, Z. Li, J. P. Brousal, K. West, and S. R. Hann. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431:712-717. [DOI] [PubMed] [Google Scholar]
- 73.Quinn, C. M., A. P. Wiles, T. El-Shanawany, I. Catchpole, T. Alnadaf, M. J. Ford, S. Gordon, and D. R. Greaves. 1999. The human eukaryotic initiation factor 4AI gene (EIF4A1) contains multiple regulatory elements that direct high-level reporter gene expression in mammalian cell lines. Genomics 62:468-476. [DOI] [PubMed] [Google Scholar]
- 74.Rabenstein, M. D., S. Zhou, J. T. Lis, and R. Tjian. 1999. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. USA 96:4791-4796.10220372 [Google Scholar]
- 75.Ravitz, M. J., S. Yan, K. D. Herr, and C. E. Wenner. 1995. Transforming growth factor beta-induced activation of cyclin E-cdk2 kinase and down-regulation of p27Kip1 in C3H 10T1/2 mouse fibroblasts. Cancer Res. 55:1413-1416. [PubMed] [Google Scholar]
- 76.Ritchie, S. A., M. K. Pasha, D. J. Batten, R. K. Sharma, D. J. Olson, A. R. Ross, and K. Bonham. 2003. Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 31:1502-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosenwald, I. B. 2004. The role of translation in neoplastic transformation from a pathologist's point of view. Oncogene 23:3230-3247. [DOI] [PubMed] [Google Scholar]
- 78.Rosenwald, I. B., R. Kaspar, D. Rousseau, L. Gehrke, P. Leboulch, J. J. Chen, E. V. Schmidt, N. Sonenberg, and I. M. London. 1995. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and posttranscriptional levels. J. Biol. Chem. 270:21176-21180. [DOI] [PubMed] [Google Scholar]
- 79.Rosenwald, I. B., A. Lazaris-Karatzas, N. Sonenberg, and E. V. Schmidt. 1993. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol. 13:7358-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenwald, I. B., D. B. Rhoads, L. D. Callanan, K. J. Isselbacher, and E. V. Schmidt. 1993. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc. Natl. Acad. Sci. USA 90:6175-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roy, A. L., C. Carruthers, T. Gutjahr, and R. G. Roeder. 1993. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature 365:359-361. [DOI] [PubMed] [Google Scholar]
- 82.Roy, A. L., H. Du, P. D. Gregor, C. D. Novina, E. Martinez, and R. G. Roeder. 1997. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 16:7091-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruggero, D., L. Montanaro, L. Ma, W. Xu, P. Londei, C. Cordon-Cardo, and P. P. Pandolfi. 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10:484-486. [DOI] [PubMed] [Google Scholar]
- 84.Safrany, G., and R. P. Perry. 1995. The relative contributions of various transcription factors to the overall promoter strength of the mouse ribosomal protein L30 gene. Eur. J. Biochem. 230:1066-1072. [DOI] [PubMed] [Google Scholar]
- 85.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 86.Schmidt, E. V. 2004. The role of c-myc in regulation of translation initiation. Oncogene 23:3217-3221. [DOI] [PubMed] [Google Scholar]
- 87.Shelley, C. S., J. M. Teodoridis, H. Park, O. C. Farokhzad, E. P. Bottinger, and M. A. Arnaout. 2002. During differentiation of the monocytic cell line U937, Pur alpha mediates induction of the CD11c beta 2 integrin gene promoter. J. Immunol. 168:3887-3893. [DOI] [PubMed] [Google Scholar]
- 88.Silverman, T. A., M. Noguchi, and B. Safer. 1992. Role of sequences within the first intron in the regulation of expression of eukaryotic initiation factor 2 alpha. J. Biol. Chem. 267:9738-9742. [PubMed] [Google Scholar]
- 89.Singh, H., J. H. LeBowitz, A. S. Baldwin, Jr., and P. A. Sharp. 1988. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell 52:415-423. [DOI] [PubMed] [Google Scholar]
- 90.Smale, S. T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta 1351:73-88. [DOI] [PubMed] [Google Scholar]
- 91.Smith, M. R., M. Jaramillo, Y. L. Liu, T. E. Dever, W. C. Merrick, H. F. Kung, and N. Sonenberg. 1990. Translation initiation factors induce DNA synthesis and transform NIH 3T3 cells. New Biol. 2:648-654. [PubMed] [Google Scholar]
- 92.Sorrells, D. L., D. R. Black, C. Meschonat, R. Rhoads, A. De Benedetti, M. Gao, B. J. Williams, and B. D. Li. 1998. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann. Surg. Oncol. 5:232-237. [DOI] [PubMed] [Google Scholar]
- 93.Stone, J., T. de Lange, G. Ramsay, E. Jakobovits, J. M. Bishop, H. Varmus, and W. Lee. 1987. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol. 7:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swick, A. G., and M. D. Lane. 1992. Identification of a transcriptional repressor down-regulated during preadipocyte differentiation. Proc. Natl. Acad. Sci. USA 89:7895-7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takimoto, M., T. Tomonaga, M. Matunis, M. Avigan, H. Krutzsch, G. Dreyfuss, and D. Levens. 1993. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 268:18249-18258. [PubMed] [Google Scholar]
- 96.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21:8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tomonaga, T., and D. Levens. 1996. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. USA 93:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tomonaga, T., and D. Levens. 1995. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem. 270:4875-4881. [DOI] [PubMed] [Google Scholar]
- 99.Van Seuningen, I., J. Ostrowski, and K. Bomsztyk. 1995. Description of an IL-1-responsive kinase that phosphorylates the K protein. Enhancement of phosphorylation by selective DNA and RNA motifs. Biochemistry 34:5644-5650. [DOI] [PubMed] [Google Scholar]
- 100.Vinson, C. R., K. L. LaMarco, P. F. Johnson, W. H. Landschulz, and S. L. McKnight. 1988. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 2:801-806. [DOI] [PubMed] [Google Scholar]
- 101.von der Haar, T., and J. E. McCarthy. 2002. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 46:531-544. [DOI] [PubMed] [Google Scholar]
- 102.Walsh, D., P. Meleady, B. Power, S. J. Morley, and M. Clynes. 2003. Increased levels of the translation initiation factor eIF4E in differentiating epithelial lung tumor cell lines. Differentiation 71:126-134. [DOI] [PubMed] [Google Scholar]
- 103.Wendel, H. G., E. De Stanchina, J. S. Fridman, A. Malina, S. Ray, S. Kogan, C. Cordon-Cardo, J. Pelletier, and S. W. Lowe. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428:332-337. [DOI] [PubMed] [Google Scholar]
- 104.Wheeler, D. L., D. M. Church, R. Edgar, S. Federhen, W. Helmberg, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, E. Sequeira, T. O. Suzek, T. A. Tatusova, and L. Wagner. 2004. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 32(database issue):D35-D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zenzie-Gregory, B., A. Khachi, I. P. Garraway, and S. T. Smale. 1993. Mechanism of initiator-mediated transcription: evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol. Cell. Biol. 13:3841-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou, Q., T. G. Boyer, and A. J. Berk. 1993. Factors (TAFs) required for activated transcription interact with TATA box-binding protein conserved core domain. Genes Dev. 7:180-187. [DOI] [PubMed] [Google Scholar]
- 108.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]