Abstract
Millions of people worldwide suffer from Parkinson’s disease (PD), a neurodegenerative disorder marked by motor symptoms such as tremors, bradykinesia, and stiffness. Accurate early diagnosis is crucial for effective management and treatment. This article presents a novel review of Machine Learning (ML) and Deep Learning (DL) techniques for PD detection and progression monitoring, offering new perspectives by integrating diverse data sources. We examine the public datasets recently used in studies, including audio recordings, gait analysis, and medical imaging. We discuss the preprocessing methods applied, the state-of-the-art models utilized, and their performance. Our evaluation included different algorithms such as support vector machines (SVM), random forests (RF), convolutional neural networks (CNN). These algorithms have shown promising results in PD diagnosis with accuracy rates exceeding 99% in some studies combining data sources. Our analysis particularly showcases the effectiveness of audio analysis in early symptom detection and gait analysis, including the Unified Parkinson’s Disease Rating Scale (UPDRS), in monitoring disease progression. Medical imaging, enhanced by DL techniques, has improved the identification of PD. The application of ML and DL in PD research offers significant potential for improving diagnostic accuracy. However, challenges like the need for large and diverse datasets, data privacy concerns, and data quality in healthcare remain. Additionally, developing explainable AI is crucial to ensure that clinicians can trust and understand ML and DL models. Our review highlights these key challenges that must be addressed to enhance the robustness and applicability of AI models in PD diagnosis, setting the groundwork for future research to overcome these obstacles.
Keywords: Parkinson’s disease, Deep learning, Machine learning, Artificial intelligence
Introduction
PD is a chronic, degenerative neurological disorder that has a major influence on mood, mobility, sleep patterns, and general wellbeing causing motor and non-motor symptomps. The latter worsen as it advances. It is caused by the loss of cells in the brain’s area that controls movement and balance that can produce the neurotransmitter dopamine. The illness affects around ten million people globally. By 2050, there will likely be 20 million people living with PD worldwide [1]. While there isn’t a cure, there are treatments and drugs that help lessen the affects. The main symptoms are trembling, rigid muscles, and trouble speaking. This illness is a major cause of dependency and impairment. Dementia is another typical side effect of Parkinson’s disease. PD can cause a range of cognitive problems that affect one’s memory, language, and thinking skills. In its later stages, PD may even develop to dementia or additional disorders of a similar nature. Emotional changes are also prevalent; people may suffer irritation, anxiety, and depression early in the disease, though therapies are available to assist manage these concerns. As the illness progresses, problems with chewing and swallowing, increasing the risk of malnutrition and raising the possibility of choking or drooling can occur due to weakening of the mouth muscles. Patients frequently have sleep difficulties; they frequently wake up during the night, have nightmares, or fall asleep during the day. Urinary urgency, constipation, and blood pressure swings that result in lightheadedness upon standing are other possible symptoms. Many also report significant fatigue, muscle and joint pain [2]. Although it usually affects older individuals, younger people might also experience it. Compared to women, men receive diagnoses more often. Genetics may be involved in Parkinson’s disease since those with a family history of the disorder are more likely to get it, even if the exact etiology of the illness is still unknown. Air pollution and certain chemical exposure are two examples of environmental factors that appear to raise the risk [3]. One to two individuals out of every 1000 are affected by PD. The frequency of the disease is rising with age, affecting 1% of those over the age of 60 [4].
Motivation
PD represents a significant and growing public health challenge worldwide, with an increasing prevalence and a substantial burden on affected individuals, caregivers, and healthcare systems. According to a study by The Global Burden of Disease (GBD) in 195 countries and territories, Parkinson’s disease is the fastest-growing among neurological disorders in terms of prevalence, death and disability [5]. Despite decades of research, there are still critical gaps in our understanding of PD pathology, diagnosis, and treatment. PD poses significant diagnostic challenges in its early stages due to the subtlety and variability of its symptoms [6]. A recent research study [7] supported by the Parkinson’s Foundation [8] in 2022 indicates that approximately 90,000 individuals receive a diagnosis of PD annually in the United States. This marks a significant 50% rise from the previously projected rate of 60,000 diagnoses per year.[9] Traditional diagnostic methods, largely reliant on clinical assessments and medical imaging, often lead to late or inaccurate diagnoses. In recent years, DL and ML have opened new horizons in medical diagnostics, offering innovative tools for early and accurate detection of PD. To address these challenges, this article provides an overview of the most recent studies and upcoming advancements, and prospective chances for PD identification using artificial intelligence techniques.
Contribution
This paper provides a comprehensive overview of recent advancements in the application of ML and DL techniques for PD detection. We review the state-of-the-art literature from different perspectives:
Availability of PD Databases: we evaluate the availability and characteristics of PD datasets used in recent research.
Data Preprocessing Techniques: we assess the data preprocessing techniques applied to diverse data modalities in the selected papers. By analyzing methods used to enhance data quality, handle missing values, and normalize features, we highlight the importance of preprocessing steps in optimizing algorithms for PD detection.
ML and DL approaches for PD: we discuss several approaches, including supervised and unsupervised learning, convolutional neural networks (CNNs), and recurrent neural networks (RNNs), examining how they have been employed in analyzing clinical data, genetic information, neuroimaging, and even data from wearable devices. By reviewing the architectures, training strategies, and model optimization techniques, we identify trends and innovations in DL and ML approaches for diagnosing PD from diverse data sources. The review also critically evaluates the current state-of-the-art models, highlighting their strengths, limitations, and the challenges faced in the deployment of these technologies in clinical settings.
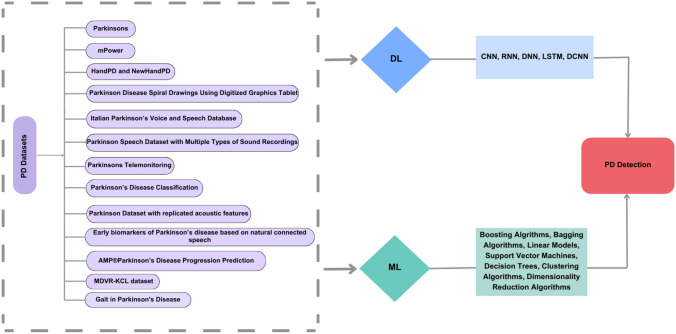
The structure of the paper follows a systematic review approach. We organize the discussion into distinct sections, starting with an overview of available PD-related datasets, followed by an assessment of data preprocessing techniques and a detailed analysis of ML and DL architectures employed for PD detection. We present key observations and findings derived from our review, identify research gaps, and offer recommendations for future studies. The paper concludes with a synthesis of the reviewed literature and its implications for advancing PD detection using ML and DL techniques. Figure 1 presents a taxonomy of these methodologies.
Fig. 1.
Taxonomy of ML and DL approaches for PD detection
Parkinson datasets
This section outlines the publicly available datasets on PD that were employed:
Parkinsons [10]: This dataset, available on the UCI Machine Learning Repository, consists of a CSV file where each row represents a biomedical voice recording. There are 197 instances in total from 31 individuals 23 of whom have PD. Max Little of the University of Oxford created this public dataset in collaboration with the National Centre for Voice and Speech, Denver, Colorado, which recorded the speech signals. It includes some metadata about the individuals in the dataset, such as age and gender.
Italian Parkinson’s Voice and Speech Database [11]: The Italian Parkinson’s Voice and Speech dataset comprises audio recordings collected by Dimauro et al. [11] from participants in Italy, including 15 healthy individuals aged 19–29 years, 22 healthy individuals aged 60–77 years, and 28 PD patients aged 40–80 years. The dataset includes recordings of participants performing different vocal tasks, such as reading a phonemically balanced text with a pause in between, executing specific syllables (’pa’ and ’ta’), and phonating different vowels (’a’, ’e’, ’i’, ’o’, ’u’). Additionally, participants are asked to read phonemically balanced words and phrases after a brief pause. The dataset is available in both WAV audio format and XLSX spreadsheet format. [12]
Parkinson Speech Dataset with Multiple Types of Sound Recordings [13]: At the Department of Neurology in Cerrahpasa Faculty of Medicine, Istanbul University, multiple types of sound recordings were collected from 40 individuals, comprising 20 healthy individuals (10 females and 10 males) and 20 individuals diagnosed with PD, with a distribution of 6 females and 14 males. The data contains a variety of sound samples, including words, numerals, sustained vowels, and brief sentences. Regression analysis can be performed using this dataset as it contains the UPDRS score assigned to each patient by a certified medical practitioner.
Parkinsons Telemonitoring [14]: This data, stored in ASCII CSV format, encompasses diverse biomedical voice measurements gathered from 42 individuals diagnosed with early-stage PD. They participated in a six-month trial assessing a telemonitoring device for remotely monitoring symptom progression, with recordings automatically obtained in their homes. Within the dataset, columns include participant ID, age, gender, time interval from the baseline recruitment date, motor UPDRS, total UPDRS, and 16 biomedical voice measures. Each row corresponds to one of the 5,875 voice recordings collected from these participants. The primary goal of the dataset is to predict the motor_UPDRS and total UPDRS scores based on the 16 voice measures.
Parkinson’s Disease Classification [15]: The dataset comprises information collected from 188 PD patients (107 males and 81 females), aged between 33 and 87 years, from the Department of Neurology at Cerrahpaşa Faculty of Medicine, Istanbul University. Additionally, the control group included 64 healthy participants (23 males and 41 females), aged between 41 and 82 years. It comprises 756 records with voice measurements including Pitch Period Entropy (PPE), Detrended Fluctuation Analysis (DFA), Recurrence Period Density Entropy (RPDE), and features related to shimmer, harmonics-to-noise ratio, energy, and MFCC coefficients. It also includes statistical parameters like skewness, kurtosis, entropy, and descriptors from time-frequency representations (TQWT) of energy and entropy. Gender is also included in the CSV file of the dataset, where each row is classified with a label of either 1 or 0, indicating the presence or absence, respectively, of PD.
Parkinson Disease Spiral Drawings Using Digitized Graphics Tablet [16]: The database comprises records from 62 PD patients and 15 healthy individuals, all sourced from the Department of Neurology at Cerrahpasa Faculty of Medicine, Istanbul University. It encompasses three types of handwriting assessments using a graphics tablet: the Static Spiral Test (SST), Dynamic Spiral Test (DST), and Stability Test on Certain Point (STCP). Data collected include motor performance metrics, tremor measurements, and patient demographics. It also contains image files of the spirals drawn by the PD patients.
Early biomarkers of Parkinson’s disease based on natural connected speech [17]: The dataset includes demographic and clinical information on 30 patients with untreated early-stage PD, 50 patients with REM sleep behavior disorder (RBD) and 50 healthy control subjects (HC). All patients were clinically assessed by a neurologist experienced in movement disorders and underwent speech analysis. Speech characteristics were automatically analyzed. Data are provided in xls and csv format, organized into sections with specific attributes, and each subject is identified by a unique code indicating its clinical diagnosis. The dataset is used for supervised learning tasks such as classification and regression, and comprises 130 instances in total.
Parkinson Dataset with replicated acoustic features [18]: This dataset comprises acoustic features extracted from three voice recording replications of sustained /a/ phonation for each of the 80 subjects, of which 40 have PD. The data’s nature is dependent within subjects but independent across subjects. With 240 instances for 80 subjects, the dataset includes many parameters such as subject ID, recording number, status (0 for Healthy, 1 for PD), gender (0 for Man, 1 for Woman), pitch and amplitude perturbation measures, harmonic-to-noise ratio measures, MFCC, recurrence period density entropy, detrended fluctuation analysis, pitch period entropy, and glottal-to-noise excitation ratio.
PPMI [19]: The Parkinson’s Progression Markers Initiative (PPMI), a long-term research project started in 2010, is a global partnership between The Michael J. Fox Foundation and academic scientists and industry organizations. The primary aim of PPMI is to identify essential biological markers associated with the onset and progression of PD. Through extensive collaboration, PPMI has established a comprehensive open-access dataset and biosample repository where clinical data receives new entries nightly, and a full database update occurs every Sunday. Monthly, imaging data is integrated into the database separately. the data can be accessed after completing the registration process and electronically signing the data use agreement.
AMP-Parkinson’s Disease Progression Prediction [20]: this database is composed of several files containing comprehensive information regarding PD patients and their clinical data, aggregated protein expression frequencies, and mass spectrometry data at the peptide level. The trainpeptides.csv file offers insights into peptide sequences, abundances, and their association with UniProt IDs and patient visit details. Meanwhile, the trainproteins file aggregates protein expression frequencies, and the trainclinicaldata provides UPDRS scores and medication details.
mPower [21]: In the initial six months of the mPower study, 960 individuals joined and completed a minimum of 5 self-administered active assessments aimed at assessing PD symptoms (such as speeded tapping, gait/balance, phonation, or memory) in non-clinical environments.
HandPD and NewHandPD [21]: The HandPD dataset consists of handwritten exams collected from two groups: the Healthy Group and the Patient Group, with the latter consisting of individuals affected by PD. These exams were gathered at Botucatu Medical School, São Paulo State University - Brazil. Each exam involves filling out a form containing four spirals and four meanders, which are then extracted and stored as “jpg” image files. The dataset includes 92 individuals, divided into 18 healthy individuals and 74 patients, with demographic characteristics such as gender, age, and handedness. There are a total of 736 images in the dataset, with 368 images for each drawing category (spirals and meanders). The NewHandPD dataset, an improved version of HandPD, comprises 66 individuals divided into Healthy and Patient groups. Each individual was asked to complete 12 exams, including spirals, meanders, circled movements, and left and right-handed diadochokinesis. Handwritten dynamics were recorded using a smart pen, resulting in a dataset containing images and signals for each individual. The NewHandPD dataset is more balanced and includes 264 images and 792 signals, labeled for easy access to each exam instance.
MDVR-KCL dataset [22] Jaeger et al. gathered the MDVR-KCL dataset back in September 2017 at King’s College London. They collected voice recordings using mobile devices from a total of 37 individuals, consisting of both PD patients (16 participants) and HC (21 participants). Each person recorded themselves reading the same text passage. These recordings, saved in wav format, were then labeled with information such as the Hoehn & Yahr (H&Y) score, UPDRS II part 5, and UPDRS III part 18 scale ratings, which help determine the stage of the disease. Expert assessments were used to assign these ratings.
Gait in Parkinson’s Disease [23]: 279 gait measures from 93 patients with idiopathic PD and 73 healthy controls are provided in this dataset available on PhysioNet[23]. The recordings capture how these individuals walk at their usual pace for about two minutes on level ground using eight sensors placed under each foot to measure force in Newtons over time, digitized at a rate of 100 Hz. The force values were normalized to each participant’s body weight percentage. The average age of the patients was 66 years, with 59% being male. The database comprises data from three different studies (abbreviated as Ga, Ju, Si). Additionally, it contains demographic information, assessments of disease severity in Hoehn & Yahr staging and/or the UPDRs. Table 1 summarizes these datasets used in the studies.
Table 1.
Public PD Datasets
| Dataset | Description | Data modality | References |
|---|---|---|---|
| Parkinsons[10] | 197 biomedical voice recordings in CSV format, collected from 31 individuals, including 23 with PD, by Max Little and the National Centre for Voice and Speech. | CSV | [24–27] |
| Italian Parkinson’s voice and speech database [12] | Audio recordings from 37 healthy participants and 28 PD patients, collected by Giovanni Dimauro et al.[11]. | WAV & XLSX | [28] |
| Parkinson speech dataset with multiple types of sound recordings[13] | 1040 sound recordings from 40 individuals, including 20 healthy and 20 with PD, collected at Istanbul University. | WAV | [29, 30] |
| Parkinsons telemonitoring[14] | Biomedical voice measurements from 42 individuals with early-stage PD, with 5,875 recordings in ASCII format. | ASCII | [31] |
| Parkinson’s disease classification[15] | 756 voice recordings of 188 PD patients and 64 healthy subjects from Istanbul University. | CSV | [32] |
| PPMI[19] | A longitudinal study providing clinical, imaging, and biospecimen data accessible after registration. | Various | [33] |
| mPower[21] | 3711 records of tapping activity and data from memory, walking, and voice activities. | Various | [31] |
| MDVR-KCL dataset[22] | Voice recordings from 37 participants, including 16 PD patients and 21 healthy controls. | WAV | [34, 35] |
| Gait in Parkinson’s disease [23] | 279 gait measures from 93 patients with idiopathic PD and 73 healthy controls, capturing walking patterns. | CSV | [36] |
| Parkinson disease spiral drawings using digitized graphics tablet [16] | Records from 62 PD patients and 15 healthy individuals, with handwriting assessments using a graphics tablet. | Digital images | [16] |
| Early biomarkers of Parkinson’s disease based on natural connected speech [17] | Dataset includes demographic and clinical information on 30 patients with early-stage PD and 50 healthy controls. | XLS & CSV | [17] |
| Parkinson dataset with replicated acoustic features [18] | Acoustic features from three voice recording replications for 80 subjects, 40 with PD. | CSV | [18] |
| AMP-Parkinson’s disease progression prediction [20] | Comprehensive clinical data on PD patients, including protein expression and mass spectrometry data. | Various | [20] |
| HandPD and NewHandPD [21] | Handwritten exams from healthy and PD patients, with image files and signals collected during various tests. | Digital images | [21] |
Data preprocessing techniques
Preprocessing data is a crucial step before building a pipeline. It reduces inconsistencies caused by missing values or errors. Multiple techniques are used for data preprocessing in ML and DL [37]: normalization [38], standardization, feature scaling, PCA [39] for dimensionality reduction, SMOTE [40], undersampling for handling imbalanced data and data augmentation methods using different techniques such as GANs [41–43].
The methods that were in this study included filtering to remove noise and normalization in order to standardize the data range of the GRF signals obtained by these wearable sensors. From here, they extracted sixteen time-domain and seven frequency-domain features. These features were transformed into vectors that characterized participants and differentiated those with PD from healthy controls. Feature vectors were combined with values from UPDRs and Hoehn and Yahr scale, which then served to train a regressor or classifier in order to develop predictive models. In an identical manner, test samples were converted into feature vectors for evaluation.
In the preprocessing phase of Sharanyaa et al. [27], two techniques were employed. Firstly, standardization was performed using mean and standard deviation values to prepare the data for subsequent feature scaling. Specifically, out of the 23 attributes, 13 were selected, and standardization feature scaling was executed using R studio. Secondly, normalization was applied to address the varying numeric range of attribute values in the dataset. This normalization step utilized the min-max normalization formula in R studio, ensuring that the feature values were scaled between 0 and 1. This process enhances data integrity, reduces redundancy, and prepares the normalized attribute values for input into the classifier models.
Uchitomi et al. [44] investigated the effectiveness of 11 data augmentation methods in classifying mild Parkinson’s disease using DL techniques with time-series data obtained from inertial measurement units. They utilized a dataset comprising time-series gait data collected through a shank-worn inertial measurement unit. By applying numerous magnitude-domain and time-domain transformation methods, along with combinations thereof, to augment the dataset, they enhanced the classification process. Notably, the rotation method within the magnitude-domain transformations significantly improved classification accuracy and F1-score by 5.5% and 5.9%, respectively, relative to baseline classification. This highlights the importance of selecting appropriate preprocessing techniques to achieve better performance in mild Parkinson’s disease diagnosis.
Nilashi et al. [45] employed Expectation-Maximization (EM) clustering to manage large datasets, while Principle Component Analysis (PCA) is utilized for data noise reduction. Addressing missing data, K-Nearest Neighbors (KNN)[46] is integrated into the method.
In the proposed method, the focus is on addressing missing data and enabling online learning through two distinct steps. Initially, KNN is utilized to handle missing data. Subsequently, the method is enhanced for incremental learning with Incremental PCA (IPCA) and Incremental DBN-ANFIS. Furthermore, the method is designed to accommodate new data by employing incremental techniques to update prediction models. This iterative process aims to streamline computation time for UPDRS prediction based on features present in the PD dataset.
For Rahman et al. [24], the dataset [10], sourced from UCI, undergoes several preprocessing steps before modeling. Initially, label encoding is applied to convert categorical values into numerical representations. To address label imbalance, a random oversampling technique is employed. Subsequently, MinMaxScaler normalizes the features between -1 and 1, leveraging the random oversampling method. Dimensionality reduction is achieved through principal component analysis (PCA), retaining 95% of the variance. Finally, the dataset is split into training and testing subsets, with an 80-20 ratio.
Zanini and Colombini [47] suggest two novel methods for data augmentation, employing Deep Convolutional Generative Adversarial Networks (DCGANs) and Style Transfer techniques. These approaches aim to enhance Parkinson’s disease electromyography (EMG) signals using two separate EMG databases.
In the preprocessing phase of [48], the Chaotic Chebyshev Zebra Optimization (CCZO) algorithm is devised by combining chaotic Chebyshev mapping with zebra optimization, enhancing the randomization criteria for obtaining global best solutions. Essential features are extracted from the EEG signals using a bandpass filter, and then feature mapping and classification are performed using CCZO_Residual_GhostNet. This method effectively combines the capabilities of ResNet-152 and GhostNet while optimizing the loss function for improved classification accuracy. Additionally, Ghost modules are introduced to capture additional features and patterns in the input data, enhancing the network’s discriminative abilities. Through this approach, the proposed method outperforms conventional PD classification techniques such as 2D-CNN, CSP+KNN, DWT+SVM, and Channelwise-CNN in terms of accuracy, sensitivity, specificity, and F-Score.
In the research conducted by Zahid et al. [49], the speech signals associated with PD were initially transformed into spectrograms before being input into the classifier. This conversion involved dividing the digitally sampled data into overlapping segments in the time domain. Fourier transforms were then applied to these segments to compute the spectral amplitude of each segment. Consequently, each segment corresponded to a vertical line in the resulting image, effectively capturing the frequency content of the speech signal.
Subramaniam et al [50] employed median filtering as a preprocessing technique to reduce image noise. This method proved effective results in noise reduction and edge preservation. It particularly excelled in eliminating “salt and pepper” noise. During median filtering, each pixel’s value was iteratively replaced with the median value of its neighboring pixels, covering the entire image pixel by pixel.
For Hasan et al. [51], the T2 weighted MRI scans were collected from the PPMI data repository and converted from Dicom to 3-channel JPG format. Data cleaning was performed to eliminate extremely dark MRI scans and achieve a bias-free classification. After cleaning, the dataset was balanced by considering 86 subjects each for PD and HC. Subsequently, the dataset was partitioned into training and validation subsets using 4-fold cross-validation. Data augmentation techniques, including zoom and brightness adjustments, were applied to increase the number of samples in each class of the training data. Finally, the MRI images were downsized to 224x224 pixels to reduce space requirements and training time.
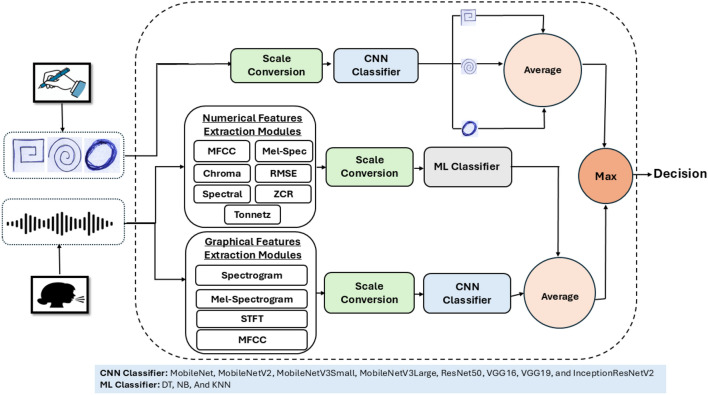
Yousif et al. [34] worked on enhancing the quality and diversity of both the handwritten images of NewHandPD dataset [21] and speech signals of MDVR-KCL dataset [22]. For the images, techniques like data equalization, augmentation, and scale conversion were employed. This included ensuring a balance in the number of records across different classes through data equalization, which involved identifying the class with the most records and randomly increasing the records in other classes until they match the highest number. Additionally, images were augmented with multiple transformations such as 15% shifting in the width and height, 15% shearing, 15% zooming, horizontal and vertical flipping, 25% rotation, and. For the MDVR-KCL dataset [22], 281 features were extracted by combining the “SpontaneousDialogue” and “ReadText” voice recordings. They used a variety of techniques to analyze audio recordings, extracting both numerical and graphical features. For numerical features, 16 techniques were used, including Mel-frequency Cepstral Coefficients (MFCC), chroma-based and spectral-based methods, Zero-crossing Rate (ZCR), Tonnetz techniques, and Root Mean Square Error (RMSE). These techniques capture different aspects of the audio signal, such as its frequency content, energy distribution, tonal characteristics, and temporal dynamics. In addition, graphical representations of the voice records were created using techniques like spectrogram, Mel-spectrogram, and Short-time Fourier Transform (STFT), providing insights into the acoustic characteristics of the recordings, including frequency changes over time and spectral envelopes. Table 2 provides an overview of the preprocessing techniques utilized in these studies.
Table 2.
Overview of preprocessing techniques for PD detection
| Refs. | Dataset | Technique |
|---|---|---|
| [27] | UCI [10] | Standardization (mean, standard deviation), normalization (min-max) |
| [44] | NewHandPD [52] | Magnitude-domain and time-domain transformation methods |
| [45] | Parkinsons Telemonitoring [14] | Expectation-Maximization clustering, PCA, KNN |
| [24] | UCI [10] | Label encoding, oversampling, normalization (MinMaxScaler), PCA |
| [47] | NinaPro [53] & private dataset | DCGAN, Style transfer |
| [48] | UCSD [54] | Bandpass filter for feature extraction from the EEG signals. |
| [49] | PC-GITA [55] | Transformation to Spectrograms, Segmentation, Fourier Transform |
| [50] | PPMI | Median filtering |
| [51] | PPMI | Data augmentation (zoom, brightness adjustments), resizing to 224x224 |
| [34] | NewHandPD dataset [21] | MDVR-KCL dataset [22] |
| [34] | NewHandPD dataset [21] & MDVR-KCL dataset [22] | For images [21]: Transformation (15% shifting in the width and height, 15% shearing, 15% zooming, horizontal and vertical flipping, 25% rotation, adjusting brightness). For audio signals [22]: MFCC, ZCR, Tonnetz, spectrogram, Mel-spectrogram, STFT, graphical representations of the voice records. |
ML approches for PD
Different ML algorithms were employed to diagnose PD. This section reviews the ML techniques used, summarized in Table 3.
Table 3.
Performance of ML models for PD detection
| References | Dataset | Model | Acc | Precision | F1-score | Recall | Specificity | AUC | MSE |
|---|---|---|---|---|---|---|---|---|---|
| [56] | NewHandPD [52] | SVM | 90.01% | – | – | 89.01% | 93.01% | – | – |
| [62] | Speech dataset from UCI [65] | XGBoost | 98.75% | 96.50% | 96.73% | 93.41% | – | 93.33% | - |
| [25] | Parkinsons [10] | EESDPD | 93.20% | 93.75% | 95.60% | 97.6% | – | – | – |
| [63] | PPMI [19] | PLR | 89.30% | – | – | – | – | 93.90% | – |
| [64] | UCI [65] | Extra Tree Classifier | 92.56% | 95.89% | 94.95% | 94.56% | – | 97.78% | – |
| [66] | PPMI [19] | ANN | – | – | – | 90.01% | 56.01% | – | – |
| [67] | mPower[21] | RWSL | – | – | – | – | – | – | 2.76% |
| [26] | Parkinsons [10] | MASS-PCNN | 91.10% | 97.80% | 99.50% | 94.70% | – | – | – |
| [27] | Parkinsons [10] | KNN | 90.20% | 90.01% | 89.50% | 89.30% | – | – | – |
| [69] | PPMI [19] | SVM-MFW | 98.30% | – | – | 98.51% | 97.90% | – | – |
In this study [36], Aşuroğlu et al. performed hybrid supervised learning analysis using the Locally Weighted Random Forest on the dataset [23] consisting of gait signals from PD patients and healthy controls. This model aimed to estimate UPDRs and forecast symptom severity using the H&Y scale. The findings showed a correlation with clinical evaluations. The model performed best with a correlation value of 0.895 for UPDRS, resulting in a mean absolute error of 4.462 and a root mean squared error of 7.382. On the H&Y scale, the performance was higher, with a correlation value of 0.960 and classification accuracy of 99.0% and specificity of 99.5%.
Nadella et al. [56] used The NewHandPD [52] dataset with a 60-40 split for training and testing. they employed ML algorithms such as Logistic Regression (LR) [57], Decision Trees (DT) [58], RF [59], SVM [60], and Naive Bayes (NB) [61]. The best-performing model was RF in the Spiral class, with 92% accuracy, 79% sensitivity, and 100% specificity.
In the study of Shrivastava et al. [62], discretizing the voice biomarker data have helped improve the performance of the classification algorithms: XGBoost, KNN, and NB, in detecting PD based on voice characteristics. When applied to the discretized PD data in percentage split test mode (70% training, 30% test), XGBoost emerged as the most accurate classifier, achieving an accuracy of 98.75%, along with 96.5% of precision, 96.73% of F1-score, 93.41% of recall, and AU-ROC of 93.33%.
Biswas et al. [25] proposed an ensemble-oriented model for diagnosing Parkinson’s disease, labeled as the Ensembled Expert System for Diagnosis of Parkinson’s Disease (EESDPD). Initially, it employed the SFFS feature selection technique to eliminate unnecessary and redundant features from the data [10]. Subsequently, the preprocessed data undergoes processing through a simple stacking ensemble technique, leveraging the diverse assumptions and robust predictive nature of RF and XGBoost models. A Meta-Model is then applied to offer a coherent interpretation of the predictions generated by the base models, with logistic regression being chosen for PD classification task. The method achieved promising results, with an accuracy of 93.20%, precision of 93.75%, recall of 97.6%, and an F1-score of 95.6%.
In Zhang et al. study [63], the researchers aimed to enhance PD detection by organizing PD-related factors by their cost and accessibility and integrating them into risk predictions using PPMI dataset [19]. They employed eight common ML models to comprehensively assess PD risk, leveraging the Shapley Additive Explanations (SHAP) method to explore factor contributions. Their findings revealed that models incorporating demographic variables, hospital admission exams, clinical assessments, and polygenic risk scores achieved superior predictive performance, with no added accuracy from invasive biomarkers. Penalized logistic regression and XGBoost emerged as the most accurate algorithms, with the former achieving an AUC of 94%. Notably, olfactory function and polygenic risk scores were consistently influential predictors.
Divya et al. [64] focused on utilizing ML algorithms to assess fluctuations in vocal patterns for PD detection. Leveraging parameter extraction and XGBoost classifiers, they utilized audio datasets available in the UCI [65] database. The research highlighted the potential of voice biomarkers for early PD diagnosis. XGBoost excelled in vocal-based PD detection, achieving an accuracy of 91.79%, an AUC of 96.67%, a recall of 95.67%, precision of 93.73%, and an F1 score of 94.54%. Other ML models, including LR, NB, SVM, Catboost Classifier, LightGBM, KNN, Extra Trees Classifier, and Gradient Boosting Classifier, were also assessed.
Alexander et al. [66] selected 630 patients with a minimum of 12 months of follow-up from PPMI [19]. They were categorized into two groups based on clinically significant worsening in UPDRS-II scores: one group with worsening (404 patients) and another without worsening (226 patients). A shallow artificial neural network (ANN) with a sigmoidal activation function and 10 hidden neurons was constructed to forecast clinically significant changes in Quality of Life (QoL) over one year. This model was developed through four stages. the ANN was developed solely utilizing UPDRS-II scores to predict whether a patient would experience clinical worsening at the 12-month follow-up. The proposed model utilized UPDRS-II scores from baseline, 2 months, and 4 months to forecast clinical deterioration at the 12-month mark. The data was divided into 70% for training (440 instances), 15% for validation (95 instances), and 15% for testing (95 instances). Both reducing and increasing the number of hidden neurons led to decreased accuracy, with no improvement observed for models with more than 10 hidden neurons. Thus, the final model was established with 10 hidden. Predictors included UPDRS-II scores at baseline, 2 months, and 4 months.
Alenezi et al. [67] used data from the mPower study [21] focusing their analysis on patients who completed a minimum of 30 tapping tasks before and after medication intake, resulting in 57 patients meeting this requirement. Among these patients, 16 individuals demonstrated significant medication response based on hypothesis testing suggested by previous literature by Neto et al. [68]. The dataset comprised 3711 tapping activity records collected before and after medication administration. Subsequently, a ranked dataset was created, encompassing data within a 3-day window before and after medication, resulting in 1284 valid ranked pairs. Labeled samples included those with available MDS-UPDRS measurements. The training and evaluation of the model were executed through 5-fold cross-validation, where RWSL, RankSVM, and ridge regression models were assessed based on mean squared error (MSE) and predictive correlation metrics. RWSL demonstrated the lowest MSE (with a mean value of 0.0276) and the highest predictive correlation (with an average value of 75.79), indicating its superior stability compared to the other models.
The data [10] used in Akila et al.’s study [26] involved voice features. To address class imbalance, random over-sampling was employed. A novel multi-agent salp swarm (MASS) algorithm extracted relevant features that were passed to a novel Parkinson classification neural network (PCNN) model. The MASS-PCNN model performance was: 99.1% accuracy, 97.8% precision, 94.7% recall, and 0.995 F1-score.
Sharanyaa et al. [27] aimed to discern PD using acoustic voice attributes [10] and ML algorithms: NB, LR, KNN, and RF. The best model was KNN (n=5), with an accuracy of 90.2%, a recall of 89.3%, and an F1-score of 89.5%, outperforming parametric models like NB and LR.
Bollipo et al. [69] introduced a novel ML technique, termed SVM-MFW, for classifying and predicting PD onset. Utilizing heterogeneous data from PPMI[19], the proposed model surpassed previous state-of-the-art techniques in both classification and prediction tasks. Specifically, the model achieved an accuracy of 98.3% in less computational time and low cross-entropy values.
DL approaches for PD
Table 4 gives an overview of the DL techniques utilized for detecting PD.
Table 4.
Performance of DL models for PD detection
| References | Dataset | Model | Acc | Precision | F1-score | Recall | Specificity | AUC | RMSE |
|---|---|---|---|---|---|---|---|---|---|
| [44] | NewHandPD[52] | CNN | 86.40% | 84.80% | 88.10% | 91.70% | – | – | – |
| [45] | Parkinsons Telemonitoring [14] | DBN+ANFIS | – | – | – | – | – | – | 54.40% |
| [24] | UCI [65] | XGBoost | 92.18% | 93.75% | 96.01% | 98.01% | – | 96.01% | – |
| [71] | PPMI[19] | CSSFOA ZF-Net | 94.50% | – | – | - | 95.70% | 93.90% | – |
| [48] | UCSD [54] | CCZO_Residual_GhostNet | 98.76% | – | 99.01% | - | 98.95% | – | – |
| [28] | Italian dataset [12] | CNN-LSTM | 97.01% | – | –- | – | – | – | – |
| [49] | PC-GITA [55] | AlexNet | 99.70% | – | – | – | – | – | – |
| [50] | PPMI [19] | CNNs | 96.01% | – | – | – | 97.01% | 99.01% | – |
| [51] | MRI(PPMI [19]) | EfficientNet-V2 | 99.23% | 98.90% | 99.22% | 99.55% | 98.91% | – | – |
In this study [70], Aşuroğlu et al. developed a computational framework that uses a deep learning model for gait analysis to accurately predict symptom levels. They used Ground Reaction Force sensors to extract features from sensor data, which were subsequently processed by a hybrid model that included Convolutional Neural Networks and Locally Weighted Random Forest. The results obtained were a correlation coefficient of 0.897, with a mean absolute error of 3.009 and a root mean square error of 4.556. Classification was also performed, and the framework achieved 99.5% accuracy, 98.7% sensitivity, and 99.1% specificity, surpassing many previous studies.
Ali et al. [30] proposed a two-stage approach that first employed a regularized linear SVM to refine the extracted feature set, followed by a DNN for classification. Two datasets [10, 13] were used to evaluate the performance of the proposed framework. An accuracy of 100% was obtained using Leave-one-subject-out (LOSO) cross-validation on the 1st dataset and 97.5% accuracy using k-fold cross-validation on the 2nd dataset.
Uchitomi et al. [44] constructed a CNN model architecture composed of four 2D convolutional layers including batch normalization (BN), Rectified Linear Unit (ReLU) activation layers, maxpooling layers, and two fully connected layers included in the architecture: one with 4096 units and another with 64 units. Finally, the output layer consists of a softmax layer with 2 units, representing the two categories, which correspond to patients with mild Parkinson’s disease and healthy elderly individuals. This particular CNN model utilized a configuration of 300 epochs, a batch size of 64, and the Adam optimizer during training. Additionally, the learning rate was set to 0.0001, with a weight decay of 0.000001. CNN achieved the best results with the rotation (R) method, yielding the highest F1-score (88.1%), accuracy (86.4%), recall (91.7%), and precision (84.8%)
The study of Nilashi et al. [45] aims to develop an integrated approach for PD diagnosis, employing ensemble learning techniques for online learning from extensive clinical datasets. Utilizing Deep Belief Network (DBN) and Neuro-Fuzzy approaches, prediction models are constructed for UPDRS diagnosis, a crucial aspect of PD management, with incremental machine learning strategies implemented for enhanced efficiency. Real-world PD data is evaluated, showcasing enhanced UPDRS prediction accuracy and reduced time complexity compared to existing methods. The dataset consists of 5875 records with 16 features and two outputs, including data from 28 males and 14 females, with Total-UPDRS and Motor-UPDRS serving as targets for prediction. The proposed method, leveraging Expectation-Maximization (EM), Deep Learning (DL), and Adaptive Neuro-Fuzzy Inference System (ANFIS) techniques, demonstrates improved accuracy and reduced computational complexity compared to previous approaches. The DBN+ANFIS method achieves the best results in test for Motor-UPDRS prediction, with an RMSE of 0.544, while the EM+DBN+ANFIS method excels for Total-UPDRS prediction, achieving an RMSE of 0.513.
Rahman et al. [24] employed DL and ML models, including XGBoost, AdaBoost, LightGBM, CatBoost, Gradient Boosting, RF, Ridge, DT, LR, K Neighbors, SVM - Linear Kernel, NB, and DNNs. The best performance was achieved by the three-layer DNN (DNN2) with an AUC of 96%, accuracy of 95%, and F1 score of 96%, along with 98% precision and 95% recall.
In this study [71], Dharani and Thamilselvan introduced a novel approach, termed the Chronological Smart Sunflower Optimization Algorithm (CSSFOA), for PD classification based on 2 voice datasets and signal samples [72] and [15]. Following feature selection, PD classification is conducted using the ZF-Net [33], where the CSSFOA algorithm, created by combining the chronological concept with the SFOA [73] and SFO [74] algorithms, fine-tunes the network weights. ZF-Net offers an advantage by yielding improved classification performance. Within this study, ZF-Net receives inputs from the selected features, derived from a concatenated vector, enabling the classification of these features as either normal or diseased. PD classification is performed using the ZF-Net, trained with the proposed CSSFOA to enhance classification performance. The ZF-Net incorporates various layers such as max-pooling, shareable convolutional, fully connected, and dropout layers. Its architecture involves passing selected features through convolutional layers (109x109x96), followed by max-pooling (54x54x96) and lambda layers (with identical specifications). Subsequently, additional convolutional layers (12x12x256, 10x10x384, 6x6x256) are employed alongside max-pooling (2x2x256) and flattening (1x1024). Three dense layers (1x4096) lead to the output layer, distinguishing between normal and abnormal conditions. ZFNet utilizes filters and reduced stride values. The softmax layer, positioned as the final layer, transforms scores into likelihoods for each voice data category. Modified linear units (ReLU) behind hidden and pooling layers aid in enhancing convergence rates for the network. The fitness function employs Mean Square Error (MSE) to ascertain the best solution. Experimental results demonstrate that the CSSFOA_ZF-Net algorithm achieved superior testing accuracy of 94,5%, with a sensitivity of 91,9% and specificity of 95,7%.
The proposed model [48] utilizes EEG signals and employs the CCZO_Residual_GhostNet for feature mapping and classification. This hybrid classifier, combining ResNet-152 and GhostNet architectures, uses the CCZO algorithm to optimize the loss function. It achieves impressive results with 98.76% accuracy, 98.59% sensitivity, 98.95% specificity, and a 99% F-score.
Pandey et al. [28] introduced a hybrid CNN-LSTM approach for detecting PD using voice signals [11]. Spectrogram images serve as input for the classifier, which effectively analyzes voice signals. Evaluated with data from 22 healthy individuals and 28 PD patients [75], the model achieved the highest accuracy of 97.01% with the vowel /i/, followed by 94.82% for /e/, 95.39% for /o/, and 92.49% for /u/.
The AlexNet model used by Zahid et al. [49] incorporated pre-trained convolutional layers on ImageNet data and three layers for PD data, utilizing transfer learning. RF, multilayer perceptron, and transfer learning achieved the highest accuracies of 99%, 99.7%, and 72%, respectively. For vowel /o/, accuracies were 99%, 99.6%, and 76%. For read text, accuracies were 97.8%, 99%, and 91%, while for monologues, accuracies were 97%, 99.3%, and 86.36%.
Leveraging the PPMI dataset, Subramaniam et al. [50] employed CNNs to discern PD from HC using MRI images. The CNN architecture included five convolutional layers with varying numbers of filters (16, 32, and 64), ReLU activation, and five max pooling layers. A flatten layer preceded the dense layers, which used ReLU activation in the primary layer and softmax activation in the final layer. The model achieved approximately 96% accuracy, 97% specificity, and 97% sensitivity, with an AUC of 99%.
The EfficientNet-V2 [76] used by Hasan et al. [51] begins with 224x224x3 images processed through layers with 32 filters and 3x3 kernel sizes. It incorporates six MBConv blocks with inverted residual connections and depthwise separable convolutions, progressively increasing filters and expansion ratios. Dropout is applied for regularization, and features are adjusted dynamically with SiLU activation and SE techniques. The model achieved an average accuracy of 99.13%, maintaining precision, recall, f1-score, sensitivity, and specificity levels exceeding 98.50% in each fold.
Yousif et al. [34] evaluated various pre-trained CNN models and ML algorithms for PD classification using handwritten images from the NewHandPD dataset [52] and speech signals from the MDVR-KCL dataset [22]. Models such as ResNet50, VGG16 [77], VGG19 [77], MobileNet [78], MobileNetV2 [78], MobileNetV3Small [78], MobileNetV3Large [78], and InceptionResNetV2 [79], originally trained on ImageNet [80], were fine-tuned and augmented via transfer learning. The Aquila Optimizer was used during training. For speech signals, ML algorithms including DT, SVM, NB, and KNN were utilized with preprocessing layers for scaling and variance thresholding. VGG19 achieved the best accuracy of 99.75% on the NewHandPD dataset, while combining numerical features with KNN and SVM achieved a peak accuracy of 99.94% on the MDVR-KCL dataset [22]. Integrating graphical features with the VGG19 structure led to a perfect accuracy of 100%. Figure 2 represents a summary of this approach.
Fig. 2.
The proposed approach for PD diagnosis in [34]
Discussion
We saw that ML and DL algorithms have demonstrated good results in PD diagnosis: using DL, Hasan et al. [51] trained EfficientNet-V2 on MRI scans from PPMI dataset after cleaning and processing data augmentation techniques including zoom and brightness. They achieved better performance (99.13% of accuracy, 98.98% of precision, a sensitivity of 99.28%, a specificity of 98.98% and 99.13% in F1-Score) in classifying PD compared to other CNN and DL models such as [81–83], across various evaluation metrics. Zahid et al. [49] used AlexNet as a feature extractor on audio recordings of the dataset [55] and then, pretrained a MLP to detect the presence of PD with an accuracy of 99.7%. Bollipo et al. used SBR values of DaTSCANs, biomarkers data and demographic data of the PPMI database. They proposed a model for the classification and prediction of early PD. Using incremental SVM[84] and modified Frank-Wolfe[85] method (SVM-MFW), the authors obtained an accuracy of 98.3%, 97.9% of specificity and a cross-entropy of 0.134. Their model is light in terms of CPU computational time. Datasets play an important role in building a reliable AI system. One of the major challenges in healthcare particularly in PD datasets is that there isn’t much data available. Yousif et al.[34] proposed a framework that uses both handwritten images and audio recordings for PD diagnosis, they reported that none of the available data have voice and handwriting data for the same patient. In the medical field, there is a lack of data sharing from both patients and hospitals for security, anonymity and privacy issues. To address this issue, Federated Learning [86] can be used.
Healthcare datasets in general have certain characteristics such as imbalance and noise that could affect the reliability and generalization of a model and its possibility to give good results fairly without being biased. Thanks to data augmentation techniques and Generative models like GANs and VAEs [87, 88], data issues can be addressed thus the robustness and performance of AI models can be improved by employing these techniques.
Another challenge is the ambiguity in the decision-making of DL models. This is very important in the field of healthcare to promote the application of these models in real data by medical practitioners and patients. The employment of Explainable AI (XAI) will ensure confidence in the use of ML and DL algorithms in healthcare [89, 90]. Table 5 compares the performance of ML and DL models in PD detection.
Table 5.
Comparative analysis of ML and DL models for PD detection
| Model Type | References | Dataset | Model | Acc | Precision | F1-score | Recall | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|---|
| ML Models | [44] | NewHandPD[52] | CNN | 86.40% | 84.80% | 88.10% | 91.70% | – | – |
| [45] | Parkinsons Telemonitoring [14] | DBN+ANFIS | – | – | – | – | - | 54.40% | |
| [24] | UCI [65] | XGBoost | 92.18% | 93.75% | 96.01% | 98.01% | - | 96.01% | |
| [71] | PPMI[19] | CSSFOA ZF-Net | 94.50% | – | – | – | 95.70% | 93.90% | |
| [48] | UCSD [54] | CCZO_Residual_GhostNet | 98.76% | – | 99.01% | – | 98.95% | – | |
| [28] | Italian dataset [12] | CNN-LSTM | 97.01% | – | – | – | – | – | |
| [49] | PC-GITA [55] | AlexNet | 99.70% | – | – | – | – | – | |
| [50] | PPMI [19] | CNNs | 96.01% | – | – | – | 97.01% | 99.01% | |
| [51] | MRI(PPMI [19]) | EfficientNet-V2 | 99.23% | 98.90% | 99.22% | 99.55% | 98.91% | – | |
| DL Models | [56] | NewHandPD[52] | SVM | 90.01% | – | - | 89.01% | 93.01% | – |
| [62] | Speech dataset from UCI [65] | XGBoost | 98.75% | 96.50% | 96.73% | 93.41% | – | 93.33% | |
| [25] | Parkinsons [10] | EESDPD | 93.20% | 93.75% | 95.60% | 97.6% | – | – | |
| [63] | PPMI [19] | PLR | 89.30% | – | –- | – | – | 93.90% | |
| [64] | UCI [65] | Extra Tree Classifier | 92.56% | 95.89% | 94.95% | 94.56% | – | 97.78% | |
| [66] | PPMI [19] | ANN | – | – | – | 90.01% | 56.01% | – | |
| [67] | mPower[21] | RWSL | – | – | – | – | – | 2.76% | |
| [26] | Parkinsons [10] | MASS-PCNN | 91.10% | 97.80% | 99.50% | 94.70% | – | – | |
| [27] | Parkinsons [10] | KNN | 90.20% | 90.01% | 89.50% | 89.30% | – | – | |
| [69] | PPMI [19] | SVM-MFW | 98.30% | – | – | 98.51% | 97.90% | – |
Conclusion
Recent advances in ML and DL have transformed PD diagnosis, providing more accurate and objective methods. Studies show that ML and DL can diagnose PD from various data sources, such as EEG signals [91], MRI images [33, 92, 93] and speech [32]. Leveraging neural networks such as ANNs and CNNs, these approaches have shown superior performance in identifying PD patients from HC. For instance, [48, 94] leveraged EEG signals to categorize PD patients with great accuracy, while [95] employed CNNs on MRI images to detect structural alterations associated with PD. Furthermore, [96] used speech data to find voice biomarkers associated with PD. In this work, we comprehensively reviewed the recent existing literature utilizing ML and DL methodologies for PD analysis. Our synthesis encompassed also an overview of preprocessing methodologies utilized alongside the public datasets employed in these studies.
Author contributions
H.R wrote the main manuscript text and conducted the literature search. M.A provided conceptual guidance and supervision throughout the manuscript preparation. All authors reviewed the manuscript.
Funding
This research was enabled in part by support provided by the Natural Sciences and Engineering Research Council of Canada (NSERC), funding reference number RGPIN-2024-05287.
Data availability
Not applicable.
Code availability
Not applicable.
Materials availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no Competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Europe P. Parkinson’s Statistics. 2024. https://parkinsonseurope.org/facts-and-figures/statistics/. Accessed 2 Oct 2024.
- 2.Mayo Clinic. Parkinson’s Disease. 2024. https://www.mayoclinic.org/diseases-conditions/parkinsons-disease/symptoms-causes/syc-20376055. Accessed 2 Oct 2024.
- 3.Organization WH. Parkinson disease. Online, 2024. https://www.who.int/news-room/fact-sheets/detail/parkinson-disease. Accessed 2 Oct 2024.
- 4.Tysnes O-B, Storstein A. Epidemiology of Parkinson’s disease. J Neural Transm. 2017;124:901–5. [DOI] [PubMed] [Google Scholar]
- 5.Dorsey ER, Elbaz A, Nichols E, Abbasi N, Abd-Allah F, Abdelalim A, Adsuar JC, Ansha MG, Brayne C, Choi J-YJ, et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2018;17(11):939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolosa E, Garrido A, Scholz SW, Poewe W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021;20(5):385–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willis A, Roberts E, Beck J, Fiske B, Ross W, Savica R, Van Den Eeden S, Tanner C, Marras CM. Incidence of Parkinson disease in North America. NPJ Parkinson’s Dis. 2022;8(1):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkinson’s Foundation https://www.parkinson.org/. Accessed 14 June 2024.
- 9.Parkinson’s Foundation: Understanding Parkinson’s: Statistics. https://www.parkinson.org/understanding-parkinsons/statistics. Accessed 14 June 2024.
- 10.Little M, McSharry P, Hunter E, Spielman J, Ramig L. Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. Nature Precedings, 2008;1–1 [DOI] [PMC free article] [PubMed]
- 11.Dimauro G, Di Nicola V, Bevilacqua V, Caivano D, Girardi F. Assessment of speech intelligibility in Parkinson’s disease using a speech-to-text system. IEEE Access. 2017;5:22199–208. 10.1109/ACCESS.2017.2762475. [Google Scholar]
- 12.Dimauro G, Girardi F. Italian Parkinson’s voice and speech. IEEE Dataport 2019. 10.21227/aw6b-tg17
- 13.Kursun O, Sakar B, Isenkul M, Sakar C, Sertbas A, Gurgen F. Parkinson Speech Dataset with Multiple Types of Sound Recordings. UCI Machine Learning Repository 2014. 10.24432/C5NC8M [DOI] [PubMed]
- 14.Tsanas A, Little M. Parkinsons Telemonitoring. UCI Machine Learning Repository. 2009. 10.24432/C5ZS3N
- 15.Sakar C, Serbes G, Gunduz A, Nizam H, Sakar B. Parkinson’s Disease Classification. UCI Machine Learning Repository 2018. 10.24432/C5MS4X
- 16.Isenkul M, Sakar B, Kursun O, et al. Improved spiral test using digitized graphics tablet for monitoring parkinson’s disease. In: The 2nd International Conference on E-health and Telemedicine (ICEHTM-2014), 2014;5:171–175
- 17.Hlavnika J, Tykalov T, Onka K, Rika E, Rusz J, J, J. Early biomarkers of Parkinson’s disease based on natural connected speech. UCI Machine Learning Repository. 2017. 10.24432/C5W02Q
- 18.Prez C. Parkinson Dataset with replicated acoustic features. UCI Machine Learning Repository. 2019. 10.24432/C5701F
- 19.Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S, et al. The Parkinson progression marker initiative (ppmi). Prog Neurobiol. 2011;95(4):629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsch L, Dane S, Adam S, Dardov V. AMP®-Parkinson’s Disease Progression Prediction. Kaggle 2023. https://kaggle.com/competitions/amp-parkinsons-disease-progression-prediction. Accessed 9 June 2024.
- 21.Bot BM, Suver C, Neto EC, Kellen M, Klein A, Bare C, Doerr M, Pratap A, Wilbanks J, Dorsey E, et al. The Mpower study, Parkinson disease mobile data collected using researchkit. Sci Data. 2016;3(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeger H, Trivedi D, Stadtschnitzer M. Mobile device voice recordings at king’s college London (mdvr-kcl) from both early and advanced Parkinson’s disease patients and healthy controls. Zenodo 2019
- 23.PhysioNet: Gait in Parkinson’s Disease. 2008. 10.13026/C24H3N. Accessed 25 Sep 2024.
- 24.Rahman S, Hasan M, Sarkar AK, Khan F. Classification of Parkinson’s disease using speech signal with machine learning and deep learning approaches. Eur J Electr Eng Comput Sci. 2023;7(2):20–7. [Google Scholar]
- 25.Biswas SK, Nath Boruah A, Saha R, Raj RS, Chakraborty M, Bordoloi M. Early detection of Parkinson disease using stacking ensemble method. Comput Methods Biomech Biomed Engin. 2023;26(5):527–39. [DOI] [PubMed] [Google Scholar]
- 26.Akila B, Nayahi J. Parkinson classification neural network with mass algorithm for processing speech signals. Neural Comput Appl. 2024;36:1–17. [Google Scholar]
- 27.Sharanyaa S, Renjith PN, Ramesh K. Classification of Parkinson’s disease using speech attributes with parametric and nonparametric machine learning techniques. In: 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS). IEEE, pp. 2020;437–442.
- 28.Pandey PVK, Sahu SS, Karan B, Mishra SK. Parkinson disease prediction using CNN-LSTM model from voice signal. SN Comput Sci. 2024;5(4):381. [Google Scholar]
- 29.Benba A, Jilbab A, Hammouch A. Analysis of multiple types of voice recordings in cepstral domain using MFCC for discriminating between patients with Parkinson’s disease and healthy people. Int J Speech Technol. 2016;19:449–56. [Google Scholar]
- 30.Ali L, Javeed A, Noor A, Rauf HT, Kadry S, Gandomi AH. Parkinson’s disease detection based on features refinement through l1 regularized SVM and deep neural network. Sci Rep. 2024;14(1):1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Z, Lu H, Zhang T, Guo X, Gao L. Remote assessment of Parkinson’s disease symptom severity based on group interaction feature assistance. Int J Mach Learn Cybern. 2023;15:1–24. [Google Scholar]
- 32.Hossain MA, Amenta F. Machine learning-based classification of Parkinson’s disease patients using speech biomarkers. J Parkinson’s Dis (Preprint). 2024;14:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beheshti I, Ko JH. Predicting the occurrence of mild cognitive impairment in Parkinson’s disease using structural MRI data. Front Neurosci. 2024;18:1375395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yousif NR, Balaha HM, Haikal AY, El-Gendy EM. A generic optimization and learning framework for Parkinson disease via speech and handwritten records. J Ambient Intell Humaniz Comput. 2023;14(8):10673–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamba R, Gulati T, Jain A, Rani P. A speech-based hybrid decision support system for early detection of Parkinson’s disease. Arab J Sci Eng. 2023;48(2):2247–60. [Google Scholar]
- 36.Aşuroğlu T, Açıcı K, Erdaş ÇB, Toprak MK, Erdem H, Oğul H. Parkinson’s disease monitoring from gait analysis via foot-worn sensors. Biocybern Biomed Eng. 2018;38(3):760–72. [Google Scholar]
- 37.Albahra S, Gorbett T, Robertson S, D’Aleo G, Kumar SVS, Ockunzzi S, Lallo D, Hu B, Rashidi HH. Artificial intelligence and machine learning overview in pathology & laboratory medicine: a general review of data preprocessing and basic supervised concepts. In: Seminars in Diagnostic Pathology. Amsterdam: Elsevier; 2023. p. 71–87. [DOI] [PubMed] [Google Scholar]
- 38.Singh D, Singh B. Investigating the impact of data normalization on classification performance. Appl Soft Comput. 2020;97: 105524. [Google Scholar]
- 39.Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Statist. 2010;2(4):433–59. [Google Scholar]
- 40.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. Smote: synthetic minority over-sampling technique. J Artif intell Res. 2002;16:321–57. [Google Scholar]
- 41.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. Advances in neural information processing systems 2014;27
- 42.Motamed S, Rogalla P, Khalvati F. Data augmentation using generative adversarial networks (GANs) for GAN-based detection of pneumonia and covid-19 in chest x-ray images. Inf Med Unlocked. 2021;27: 100779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan C, Chen M, Wang X, Wang J, Huang B. A review on data preprocessing techniques toward efficient and reliable knowledge discovery from building operational data. Front Energy Res. 2021;9: 652801. [Google Scholar]
- 44.Uchitomi H, Ming X, Zhao C, Ogata T, Miyake Y. Classification of mild Parkinson’s disease: data augmentation of time-series gait data obtained via inertial measurement units. Sci Rep. 2023;13(1):12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilashi M, Abumalloh RA, Yusuf SYM, Thi HH, Alsulami M, Abosaq H, Alyami S, Alghamdi A. Early diagnosis of Parkinson’s disease: a combined method using deep learning and neuro-fuzzy techniques. Comput Biol Chem. 2023;102: 107788. [DOI] [PubMed] [Google Scholar]
- 46.Cover TM, Hart PE. Nearest neighbor pattern classification. IEEE Trans Inf Theory. 1967;13(1):21–7. [Google Scholar]
- 47.Anicet Zanini R, Luna Colombini E. Parkinson’s disease EMG data augmentation and simulation with DCGANs and style transfer. Sensors. 2020;20(9):2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shankar S, et al. CCZO residual GhostNet: Parkinson disease classification using optimized deep learning technique. Int J Electr Comput Eng Syst. 2024;15(3):275–83. [Google Scholar]
- 49.Zahid L, Maqsood M, Durrani MY, Bakhtyar M, Baber J, Jamal H, Mehmood I, Song O-Y. A spectrogram-based deep feature assisted computer-aided diagnostic system for Parkinson’s disease. IEEE Access. 2020;8:35482–95. [Google Scholar]
- 50.Sangeetha S, Baskar K, Kalaivaani P, Kumaravel T. Deep learning-based early parkinson’s disease detection from brain mri image. In: 2023 7th International Conference on Intelligent Computing and Control Systems (ICICCS), IEEE. 2023;490–495.
- 51.Hasan MM, Alfaz N, Alam MAM, Rahman A, Shakhawat HM, Rahman S. Detection of parkinson’s disease from t2-weighted magnetic resonance imaging scans using efficientnet-v2. In: 2023 26th International Conference on Computer and Information Technology (ICCIT), IEEE. 2023;1–6
- 52.Pereira CR, Weber SAT, Hook C, Rosa GH, Papa JP. Deep learning-aided Parkinson’s disease diagnosis from handwritten dynamics. In: Proceedings of the SIBGRAPI 2016 - Conference on Graphics, Patterns and Images, 2016;340–346
- 53.Atzori M, Gijsberts A, Castellini C, Caputo B, Hager A-G, Elsig S, Giatsidis G, Bassetto F, Müller H. Electromyography data for non-invasive naturally-controlled robotic hand prostheses. In: Scientific data, vol. 1. New York: Nature Publishing Group; 2014. p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rockhill AP, Jackson N, George J, Aron A, Swann NC. UC San Diego Resting State EEG Data from Patients with Parkinson’s Disease. OpenNeuro 2021. 10.18112/openneuro.ds002778.v1.0.5
- 55.Orozco-Arroyave JR, Arias-Londoño JD, Vargas-Bonilla JF, González-Rátiva MC, Nöth E. New Spanish speech corpus database for the analysis of people suffering from Parkinson’s disease. In: Calzolari N, Choukri K, Declerck T, Loftsson H, Maegaard B, Mariani J, Moreno A, Odijk J, Piperidis S (eds). Proceedings of the Ninth International Conference on Language Resources and Evaluation (LREC’14). European Language Resources Association (ELRA), Reykjavik, Iceland 2014. pp. 342–347. http://www.lrec-conf.org/proceedings/lrec2014/pdf/7_Paper.pdf
- 56.Nadella Y, Ratnala VK, Vijayashree J, Ladi R. Parkinson’s disease detection using handwriting datasets. J Data Acquis Process. 2023;38(3):155. [Google Scholar]
- 57.Cox DR. Regression models and life-tables. J Roy Stat Soc: Ser B (Methodol). 1972;34(2):187–202. [Google Scholar]
- 58.Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106. [Google Scholar]
- 59.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 60.Cortes C, Vapnik VN. Support-vector networks. Mach Learn. 1995;20(3):273–97. [Google Scholar]
- 61.Lewis DD. Naive (bayes) at forty: the independence assumption in information retrieval. In: Durga R, editor. Machine learning: ECML-98 1398. Berlin: Springer; 1998. p. 4–15. [Google Scholar]
- 62.Shrivastava A, Chakkaravarthy M, Asif Shah M. A novel approach using learning algorithm for Parkinson’s disease detection with handwritten sketches’. Cybern Syst. 2022;55:1–17. [Google Scholar]
- 63.Zhang J, Zhou W, Yu H, Wang T, Wang X, Liu L, Wen Y. Prediction of Parkinson’s disease using machine learning methods. Biomolecules. 2023;13(12):1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Divya J, Radhakrishnan P, Pavithra G, Gopatoti A, Baburao D, Krishnamoorthy R. Detection of parkinson disease using machine learning. In: 2023 International Conference on Inventive Computation Technologies (ICICT), IEEE. 2023;53–57.
- 65.UCI Machine Learning Repository. http://archive.ics.uci.edu/ml. Accessed 3 June 2024.
- 66.Alexander TD, Nataraj C, Wu C. A machine learning approach to predict quality of life changes in patients with Parkinson’s disease. Ann Clin Transl Neurol. 2023;10(3):312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alenezi FD, Shi H, Li J. A ranking-based weakly supervised learning model for telemonitoring of Parkinson’s disease. IISE Trans Healthcare Syst Eng. 2022;12(4):322–36. [Google Scholar]
- 68.Chaibub Neto E, Bot BM, Perumal T, Omberg L, Guinney J, Kellen M, Klein A, Friend SH, Trister AD. Personalized hypothesis tests for detecting medication response in parkinson disease patients using iphone sensor data. In: Biocomputing 2016: Proceedings of the Pacific Symposium, World Scientific. 2016;273–284. [PubMed]
- 69.Bollipo LM, KV, K. Fast and robust supervised machine learning approach for classification and prediction of Parkinson’s disease onset. Comput Methods Biomech Biomed Eng Imaging & Visualization. 2021;9(6):690–706. [Google Scholar]
- 70.Aşuroğlu T, Oğul H. A deep learning approach for Parkinson’s disease severity assessment. Heal Technol. 2022;12(5):943–53. [Google Scholar]
- 71.Dharani M, Thamilselvan R. Hybrid optimization enabled deep learning model for Parkinson’s disease classification. Imagin Sci J. 2024;72(2):167–82. [Google Scholar]
- 72.mPower: Mobile Parkinson’s Disease Voice dataset. https://www.synapse.org/#!Synapse:syn4993293/wiki/247861. Accessed 3 June 2024.
- 73.Sattar D, Salim R. A smart metaheuristic algorithm for solving engineering problems. Eng Comput. 2021;37(3):2389–417. [Google Scholar]
- 74.Fu L, Feng Y, Majeed Y, Zhang X, Zhang J, Karkee M, Zhang Q. Kiwifruit detection in field images using faster r-CNN with ZFNet. IFAC-PapersOnLine. 2018;51(17):45–50. [Google Scholar]
- 75.Dimauro G, Di Nicola V, Bevilacqua V, Caivano D, Girardi F. Assessment of speech intelligibility in Parkinson’s disease using a speech-to-text system. IEEE Access. 2017;5:22199–208. [Google Scholar]
- 76.Tan M, Chen QV. Efficientnetv2: Smaller models and faster training. 2021. arXiv preprint arXiv:2104.00298. Accessed 10 Sept 2024.
- 77.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. CoRR arXiv:1409.1556. Accessed 10 Sept 2024.
- 78.Andrew G, Howard MZ, Bo Chen DK, Weijun Wang TW, Marco Andreetto HA. Efficient convolutional neural networks for mobile vision applications. 2017. CoRR arXiv:1704.04861. Accessed 10 Sepember 2024.
- 79.Christian Szegedy SI, Vanhoucke V. Inception-v4, inception-resnet and the impact of residual connections on learning. 2017. CoRR arXiv:1602.07261
- 80.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition, IEEE. 2009;248–255.
- 81.Dipro SH, Islam M, Al Nahian A, Azad MS, Chakrabarty A, Reza T. A federated learning based privacy preserving approach for detecting parkinson’s disease using deep learning. In: 2022 25th International Conference on Computer and Information Technology (ICCIT), IEEE. 2022; pp. 139–144.
- 82.Wang W, Lee J, Harrou F, Sun Y. Early detection of Parkinson’s disease using deep learning and machine learning. IEEE Access. 2020;8:147635–46. [Google Scholar]
- 83.Khanna K, Gambhir S, Gambhir M. A novel technique for classifying Parkinson’s disease using structural MRI scans. Multimed Tools Appl. 2023;82(29):46011–36. [Google Scholar]
- 84.Cauwenberghs G, Poggio T. Incremental and decremental support vector machine learning. Advances in neural information processing systems 2000;13
- 85.Nanculef R, Frandi E, Sartori C, Allende H. A novel frank-wolfe algorithm. Analysis and applications to large-scale SVM training. Inf Sci. 2014;285:66–99. [Google Scholar]
- 86.Xu J, Glicksberg BS, Su C, Walker P, Bian J, Wang F. Federated learning for healthcare informatics. J Healthcare Inf Res. 2021;5:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsu W-N, Zhang Y, Glass J. Unsupervised domain adaptation for robust speech recognition via variational autoencoder-based data augmentation. In: 2017 IEEE Automatic Speech Recognition and Understanding Workshop (ASRU), IEEE. pp. 2017;16–23.
- 88.Jeong J, Jeong H, Kim H-J. An autoencoder-based numerical training data augmentation technique. In: 2022 IEEE International Conference on Big Data (Big Data), IEEE. 2022;5944–5951.
- 89.Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, García S, Gil-López S, Molina D, Benjamins R, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible ai. Inf Fus. 2020;58:82–115. [Google Scholar]
- 90.Adadi A, Berrada M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access. 2018;6:52138–60. [Google Scholar]
- 91.Ramaiah A, Balasubramanian PD, Appathurai A, Muthukumaran NA. Detection of parkinson’s disease via clifford gradient-based recurrent neural network using multi-dimensional data. REVUE ROUMAINE DES SCIENCES TECHNIQUES’SÉRIE ÉLECTROTECHNIQUE ET ÉNERGÉTIQUE. 2024;69(1):103–8. [Google Scholar]
- 92.Ahalya R, Nkondo GF, Snekhalatha U. Automated detection of Parkinson’s disease based on hybrid CNN and quantum machine learning techniques in MRI images. Biomed Eng Appl Basis Commun. 2024;36:2450005. [Google Scholar]
- 93.Li C, Hui D, Wu F, Xia Y, Shi F, Yang M, Zhang J, Peng C, Feng J, Li C. Automatic diagnosis of Parkinson’s disease using artificial intelligence base on routine t1-weighted MRI. Front Med. 2024;10:1303501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao S, Dai G, Li J, Zhu X, Huang X, Li Y, Tan M, Wang L, Fang P, Chen X, et al. An interpretable model based on graph learning for diagnosis of Parkinson’s disease with voice-related EEG. NPJ Digit Med. 2024;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, He N, Zhang C, Zhang Y, Wang C, Huang P, Jin Z, Li Y, Cheng Z, Liu Y, et al. An automatic interpretable deep learning pipeline for accurate Parkinson’s disease diagnosis using quantitative susceptibility mapping and t1-weighted images. Hoboken: Wiley Online Library: Technical report; 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Cesare MG, Perpetuini D, Cardone D, Merla A. Machine learning-assisted speech analysis for early detection of Parkinson’s disease: a study on speaker diarization and classification techniques. Sensors. 2024;24(5):1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.
Not applicable.