Abstract
The Polycomb group (PcG) gene products form multimeric protein complexes and contribute to anterior-posterior (A-P) specification via the transcriptional regulation of Hox cluster genes. The Drosophila polyhomeotic genes and their mammalian orthologues, Phc1, Phc2, and Phc3, encode nuclear proteins that are constituents of evolutionarily conserved protein complexes designated class II PcG complexes. In this study, we describe the generation and phenotypes of Phc2-deficient mice. We show posterior transformations of the axial skeleton and premature senescence of mouse embryonic fibroblasts associated with derepression of Hox cluster genes and Cdkn2a genes, respectively. Synergistic actions of a Phc2 mutation with Phc1 and Rnf110 mutations during A-P specification, coimmunoprecipitation of their products from embryonic extracts, and chromatin immunoprecipitation by anti-Phc2 monoclonal antibodies suggest that Hox repression by Phc2 is mediated through the class II PcG complexes, probably via direct binding to the Hox locus. The genetic interactions further reveal the functional overlap between Phc2 and Phc1 and a strict dose-dependent requirement during A-P specification and embryonic survival. Functional redundancy between Phc2 and Phc1 leads us to hypothesize that the overall level of polyhomeotic orthologues in nuclei is a parameter that is critical in enabling the class II PcG complexes to exert their molecular functions.
The Polycomb group (PcG) genes were originally identified through their requirement in the maintenance of the stable repression of Hox genes during the development of Drosophila melanogaster (34, 38). The products of these Drosophila PcG genes form large multimeric protein complexes in the chromatin. It is thought that they stably repress the target genes by altering the configuration of the chromatin, as suggested by the synergistic genetic interactions between mutant alleles of different Drosophila PcG genes (15, 39).
PcG genes that are structurally and functionally related to those found in Drosophila have also been identified in mammals, where their products form at least two distinct functional complexes. One complex, designated the class I PcG complex, comprises the products of the embryonic endoderm development (Eed) (the orthologue of the Drosophila extra sex combs gene) and the Enx1 and Enx2 (the orthologues of the Drosophila enhancer of zeste gene) PcG genes. Since the SET domains of Enx1 and Enx2 function as histone methyltransferases, and Eed interacts with histone deacetylases, this complex is believed to alter chromatin structures by modifying core histone tails (35, 46). The second complex, designated class II, is closely related to the Polycomb repressive complex 1 (PRC1) in Drosophila and includes the products of the paralogues of another subset of PcG genes in HeLa cell extracts, designated hPRC-H (27, 39). This subset contains the following gene groups: Rnf110 (Mel18) and Bmi1; Cbx2 (M33), Cbx4 (Pc2), and Cbx8 (Pc3); Phc1 (rae28, mph1, or edr1), Phc2 (mph2 or edr2), and Phc3; and Ring1A and Rnf2 (Ring1B) (27). Apart from Rnf2 mutants, mice that are deficient in the individual components of class II PcG complexes display anterior shifts in the expression boundaries of Hox cluster genes in the paraxial mesoderm and neural tube and, in general, characteristically show posterior transformation of the axial skeleton. These mutant mice also invariably display severe combined immunodeficiency due to increased apoptosis and a lack of proliferative responses of hemopoietic cells via regulation of the Cdkn2a/p53 pathway (1, 2, 11, 13, 19, 21, 40, 41, 45, 47). There is accumulating biochemical and genetic evidence to indicate that the class II PcG complexes are compositionally and functionally conserved between flies and mammals. Nevertheless, many PcG genes have also diverged substantially, and most are either duplicated or triplicated in mammals (27, 39).
In Drosophila, the polyhomeotic (ph) locus consists of polyhomeotic-proximal (ph-p) and polyhomeotic-distal (ph-d) sequences, which have extensive homology. Three mammalian genes homologous to ph, Phc1, Phc2, and Phc3, have been identified (14, 16, 27, 32, 44). Database screening revealed that all three ph orthologues were evolutionarily conserved and expressed in various vertebrates, including humans, chickens, zebra fish, and fugu (Y. Murahashi and H. Koseki, unpublished data). Comparisons of the ph proteins and their vertebrate orthologues have shown that each has a single FCS finger [also called the (Cys)4-type Zn coordination domain] flanked by two additional conserved domains. The function of the upstream motif, described as homology domain I (HDI), is unknown. However, the downstream homology domain II (HDII), which is located at the C-terminal end, has been described as a sterile alpha motif (SAM) domain (also known as SEP or SPM) (4, 16). Recently, the FCS finger domain of Phc1 has been shown to encode an RNA binding motif and to regulate subnuclear localization when tested in Caenorhabditis elegans (50). The SAM domains are found at the C-terminal ends, not only of ph proteins and their mammalian orthologues, but also on the Drosophila Sex comb on the midleg (SCM) gene and its orthologues as a component of PcG proteins (8, 42). It has been shown that the SAM domains are able to self-associate, bind to other SAM domains, and form heterotypic interactions with non-SAM domain domain-containing proteins (23, 32, 38). Importantly, ph-SAM has been shown to form a helical polymer structure, which provides a possible mechanism for the extension of PcG complexes (23). This finding is in close agreement with the recent biochemical observation that Phc1 plays a pivotal role in mediating the PcG-dependent bridging of distant chromatin templates (26). Coimmunoprecipitation of mammalian ph orthologues from HeLa and U-2 OS cell extracts suggests that the heterophilic polymerization of the SAM domains of Phc1, Phc2, and Phc3 may be involved in mediating PcG-dependent regulatory mechanisms via higher-order chromatin structures (16, 27).
However, the molecular complicity among mammalian ph orthologues has not yet been addressed due, at least in part, to a lack of Phc2 or Phc3 mutant alleles. In this study, we describe the generation and phenotypes of Phc2-deficient mice. Using these mutants, we studied the biochemical and genetic evidence that supports the involvement of Phc2 as a functional component of class II PcG complexes during embryogenesis.
MATERIALS AND METHODS
Production of anti-Phc2 monoclonal antibody.
A partial cDNA fragment that encodes from N22 to the stop codon of Phc2 was subcloned into the pGEX vector to express a glutathione S-transferase-Phc2 fusion protein. This fusion protein was purified and injected several times into female BALB/c mice to generate a hybridoma, which was screened as described previously (5).
IP and immunoblotting.
Monoclonal antibodies to Phc2, Phc1, and Rnf2 (in 30 μl, 100 μl, and 30 μl of culture supernatant, respectively) and 5 μg of a polyclonal antibody to Rnf110 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA) were conjugated with 50% (vol/vol) protein G-Sepharose (25 μl) in 300 μl of buffer (20 mM HEPES [pH 7.8], 20 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, and 0.01% Triton X-100) at 4°C for at least 2 h (5, 28). A mouse embryo at 11.5 days postcoitus (dpc) was sonicated three times for 10 s in immunoprecipitation (IP) buffer (400 μl), consisting of 20 mM HEPES (pH 7.8), 10% (vol/vol) glycerol, 150 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, and 2 mM Pefabloc SC (Roche Molecular, Indianapolis, IN). After centrifugation, the supernatant was precleared with 50 μl of 50% (vol/vol) protein G-Sepharose for 60 min and then incubated with each of the three Sepharose-conjugated antibodies for 90 min at 4°C. The Sepharose-bound proteins were washed five times with 800 μl of IP buffer without Pefabloc SC, boiled in sodium dodecyl sulfate sample buffer, separated on 9% denaturing polyacrylamide gels, and subjected to immunoblot analysis.
Generation of Phc2-deficient mice.
To generate Phc2-deficient mice, a targeting vector was constructed (see Fig. 2A). This vector was introduced into R1 embryonic stem cells as described previously, and five homologous recombinants were obtained (31). Phc2+/− mice were backcrossed six times onto a C57BL/6 background, and homozygotes were generated by mating between heterozygotes. Phc2+/− mice were crossed with Phc1+/− and Rnf110+/− mice, after backcrossing to C57BL/6 a few times, to generate double heterozygotes. These Phc2+/−-Phc1+/− and Phc2+/−-Rnf110+/− mice were viable and fertile. In order to generate Phc2-Phc1 double homozygotes, double heterozygotes were crossed. For the generation of Phc2-Rnf110 double homozygotes, we first generated Phc2−/−-Rnf110+/− mice, which were again viable and fertile, and crossed them to either Phc2+/−-Rnf110+/− or Phc2−/−-Rnf110+/− mice. All animal experiments were carried out according to the in-house guidelines for the care and use of laboratory animals of the Riken Research Center for Allergy and Immunology, Yokohama, Japan.
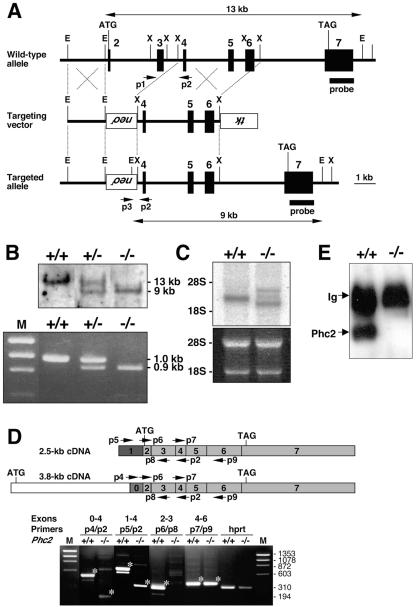
FIG. 2.
Disruption of the Phc2 gene in mice. (A) Diagram of the Phc2 locus, the targeting vector, and the targeted allele. The PGKneo and pMC1-tk expression cassettes were used for positive and negative selection, respectively. The relevant positions of the restriction sites (EcoRI, E; XhoI, X), locations of the external probe and PCR primers, and sizes of diagnostic fragments are indicated. (B) Southern (top) and PCR (bottom) analyses for genotyping. For Southern blotting, genomic DNA was digested by EcoRI and probed with the 3′ probe, as indicated in panel A. For PCR, a mixture of three primers (p1, p2, and p3 in panel A) was used. Lane M, molecular size markers. (C) Northern analysis of Phc2 expression in 11.5-dpc wild-type and homozygous embryos (top). Ethidium bromide staining of the same gel is shown below. (D) RT-PCR analysis of Phc2 expression. The locations of the PCR primers are indicated on both the 2.5- and 3.8-kb transcripts (top). The asterisks indicate specific PCR products. Note the presence of truncated Phc2 transcripts lacking exons 2 and 3. (E) Immunoprecipitation and immunoblot analyses of Phc2 expression in 11.5-dpc embryos.
Morphological analyses.
Skeletal preparations were prepared from either perinatal or late-gestational mice as described previously (22). In situ hybridization on tissue sections and whole-mount in situ hybridization were performed as described previously (22, 48).
Cell culture conditions.
Mouse embryonic fibroblasts (MEFs) were prepared from each genotype using 13.5- to 14.5-dpc fetuses and maintained according to a 3T9 protocol (20). The proliferation of MEFs was investigated at passage 5 by plating 104 cells per 60-mm-diameter dish in replicate cultures and then counting the cells from duplicate cultures every other day. This experiment was repeated four times, using MEFs derived from eight homozygous mutants and six wild types; almost identical results were obtained for each of the four repeats.
Chromatin immunoprecipitation (ChIP).
Using scissors, the embryonic tissues were minced on ice and then subjected to chemical cross-linking by incubating them in 1% formaldehyde in phosphate-buffered saline for 10 min at room temperature. After extensive sonication, the chromatin fractions were purified by CsCl isopycnic centrifugation (33); 10 mM NaCl and 0.1% NP-40 were added to the purified fractions in order to perform optimal immunoprecipitation of the anti-Phc2 antibody. Precleared protein extracts were then incubated with rocking, with 40 μl of anti-Phc2 culture supernatant per 200 μl lysate, at 4°C from 2 h to overnight. Immune complexes were captured by incubating them for 3 h with protein A-Sepharose beads.
To isolate genomic DNA from these immune complexes, the beads were treated with 50 μg/ml of RNase A at 37°C for 30 min, followed by incubation overnight with 500 μg/ml proteinase K-0.5% sodium dodecyl sulfate at 37°C. After being heated for several hours at 65°C to achieve reverse cross-linking, the supernatants were collected, extracted by phenol-chloroform, and concentrated by ethanol precipitation. Genomic DNA was also isolated from the original unfractionated chromatin by the same procedure described above and designated “input” DNA. To measure the DNA yield, aliquots of immunoprecipitated DNA were electrophoresed on an agarose gel next to serially diluted input DNA.
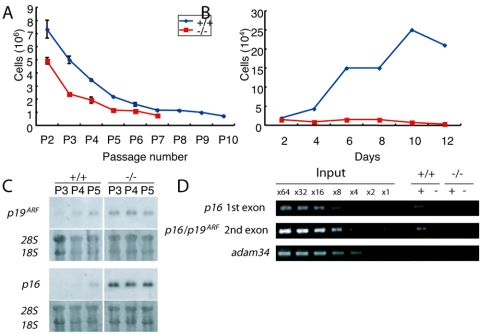
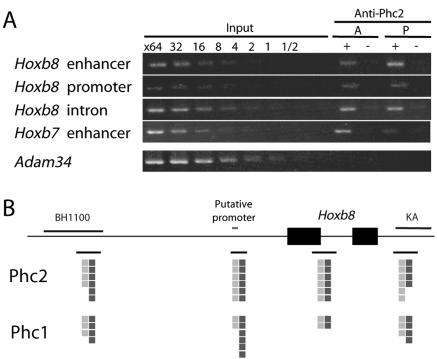
We carried out semiquantitative PCR using serially diluted input DNA and immunoprecipitated DNA as templates. The enrichment of DNA by immunoprecipitation was estimated from the band intensities of the gel images. This series of experiments were all performed at least three times and gave similar results. The following primer pairs were used in this study: p16 first exonic region, 5′-CGAACTCGAGGAGAGCCATC-3′ and 5′-ACACTCCTTGCCTACCTGAA-3′; common second exonic regions of p16/p19ARF, 5′-TCACGTAGCAGCTCTTCTGC-3′ and 5′-CAGCGGAACGCAAATATCGC-3′; Hoxb8 enhancer region (BH1100), 5′-GGGTATAAATTTCTGAAGGTTAAG-3′ and 5′-AGGGATGAGAAGGGCCGAGGG-3′; Hoxb8 promoter region, 5′-TATGACTACCTCGTTGTTTG-3′ and 5′-CAAAGACTGATGTGGGGGAGT-3′; Hoxb8 intronic region, 5′-CCCTGGATGCGCCCTCAAG-3′ and 5′-TCTCCACAGCCCCCATAAAAC-3′; Hoxb7 enhancer region (KA), 5′-CTCCTTCCCTTCTCTTGGGGGTCC-3′ and 5′-CAATGCTCACAGCGCGCATGC-3′; and Adam34 coding region, 5′-ATGAGTGGGACTAAGGCCCTG-3′ and 5′-GCGGTTATGATCTATTACTAC-3′.
Isolation of lymphocyte subpopulations at distinct stages of development.
Bone marrow cells derived from C57BL/6 mice were stained with allophycocyanin-labeled B220, phycoerythrin (PE)-labeled anti-CD43, and fluorescein isothiocyanate (FITC)-labeled anti-immunoglobulin M (IgM). Populations of B220+ CD43+ IgM− (pro-B), B220+ CD43− IgM− (pre-B), and B220dull CD43− IgM+ (immature B) cells were isolated with a Vantage fluorescence-activated cell sorter (BD Biosciences, Mountain View, CA). Similarly, thymocytes were stained with FITC-labeled CD4 and PE-labeled CD8 antibodies, and the CD4− CD8−, CD4+ CD8+, CD4+ CD8−, and CD4− CD8+ subpopulations were sorted. To isolate immature and mature B cells, splenocytes were stained with FITC-labeled B220 and PE-labeled AA4 antibodies, and B220+ AA4+ (immature B) and B220+ AA4− (mature B) cells were sorted. To purify the different mature-B-cell populations, splenocytes were stained with allophycocyanin-labeled B220, FITC-labeled CD21, and PE-labeled CD23 antibodies, and B220+ CD21− CD23+ (follicular B), B220+ CD21+ CD23dull (marginal-zone B), and B220+ CD21− CD23− (newly formed B) cells were sorted. To isolate germ center B cells, mice were immunized with 100 μg of 2,4-dinitrofluorobenzene-conjugated ovalbumin in alum. Twelve days after immunization, spleen cells were stained with PE-labeled B220, and FITC-labeled peanut agglutinin (PNA) and B220+ PNA+ cells were sorted using a high-speed cell sorter (Aria; BD Biosciences). The sorted cells were dissolved in Trizol reagent (Invitrogen, Carlsbad, CA), and total RNA was extracted according to Invitrogen's protocol.
RT-PCR analysis.
Mouse total-RNA panels were purchased from BD Biosciences. First-strand cDNA was synthesized from 2 μg of the total RNA using SuperScript III reverse transcriptase (RT) (Invitrogen) and random primers, according to the manufacturer's protocol. The forward and reverse primers for PCR were 5′-CCTACAAGTTCAAGCGTTCC-3′ and 5′-GTCCCTCATGTGCATGTCAG-3′ for mouse Phc2, 5′-GACAGGCTAGCTCCCCAAAC-3′ and 5′-GCTAGGGCCTGGCTAGAAGT-3′ for Phc1, and 5′-ATGGATGACGATATCGCT-3′ and 5′-ATGAGGTAGTCTGTCAGGT-3′ for β-actin genes. The amplification conditions were 95°C for 10 s, 60°C for 20 s, and 72°C for 1 min for 30 cycles for both Phc2 and Phc1 genes and 95°C for 5 s, 54°C for 10 s, and 72°C for 1 min for 25 cycles for β-actin genes.
RESULTS
Association of Phc2 with the hPRC-H-like complex during embryogenesis.
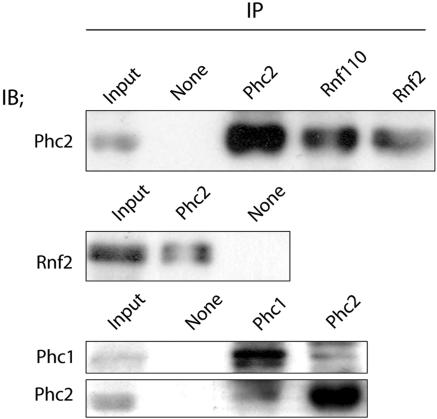
To address whether Phc2 is involved in a multimeric protein complex similar to hPRC-H during mouse embryogenesis (27), we raised a monoclonal antibody against a glutathione S-transferase-Phc2 (N22 to the stop codon) fusion protein (K. Isono and H. Koseki, unpublished data). We used whole-cell extracts from 11.5-dpc embryos for the coimmunoprecipitation experiments, as mammalian PcG complexes are considered to be functional at this gestational stage, at least as demonstrated by the derepression of Hox gene expression (1, 41, 45). The anti-Phc2 monoclonal antibody recognized an ∼40-kDa protein on the immunoblots of these 11.5-dpc embryonic extracts. The anti-Phc2 and goat anti-Rnf110 and anti-Rnf2 monoclonal antibodies were able to immunoprecipitate the Phc2 proteins from 11.5-dpc embryonic extracts (Fig. 1). Phc2 was not detected in the absence of these primary antibodies. Inclusion of Rnf2 in the Phc2 immunoprecipitates confirmed the result described above. The anti-Phc2 and -Phc1 monoclonal antibodies were both able to immunoprecipitate Phc1 and Phc2 in a reciprocal fashion. These results revealed the association of Phc2 with Rnf110, Rnf2, and Phc1 in 11.5-dpc embryos; this interaction has been shown previously in human osteosarcoma U-2 OS cells (16, 29, 49).
FIG. 1.
Association of Phc2 with other components of class II PcG complexes during embryogenesis. Antibodies used for IP and immunoblotting (IB) are indicated over each lane and beside each blot, respectively. Phc2 was coimmunoprecipited with Rnf110, Rnf2, and Phc1 from 11.5-dpc embryonic extracts. Original whole-cell extract was loaded on the lanes labeled “Input”; mock immunoprecipitation without primary antibody was performed as a negative control on the lanes labeled “None.”
Generation of a Phc2 mutant allele.
To examine the physiological roles of Phc2, we inactivated the Phc2 locus by replacing the second and third exons with a neomycin resistance (Neor) gene cassette (Fig. 2A). This replacement was expected to delete the polypeptide stretch from the start codon to E95, which encodes the HDI region from the 36-kDa isoform. Phc2 heterozygotes were backcrossed six times to C57BL/6 mice to eliminate the effects of genetic background, and then homozygotes were generated. Over 20 litters were genotyped either at birth or at weaning. Three genotypes were identified, which segregated into ratios according to the laws of Mendelian inheritance (Fig. 2B) (J. Shinga and H. Koseki, unpublished data). Phc2−/− mice were viable and fertile. Next, we examined the expression of Phc2 in the homozygotes (Fig. 2C). Northern blot analysis, using total cellular RNA from 11.5-dpc embryos, revealed three aberrant transcripts in the homozygotes, with only a 2.5-kb band present in the wild type. At least two differently spliced transcripts appeared to be generated from the Phc2 locus in the wild type, represented by a 2.5-kb and a very faint 3.8-kb transcript. Exons 2 to 7 were common to both, while exons 1 and 0 were unique to the 2.5- and 3.8-kb transcripts, respectively (49) (Fig. 2D, top). RT-PCR analysis identified the prospective 2.5- and 3.8-kb transcripts, which were missing the second and third exonic regions, as being expressed in the homozygous mutants (Fig. 2D, bottom). Since in these homozygotes, the putative start codon had been removed from the 2.5-kb transcript while the deletion of the second exonic region from the 3.8-kb transcripts resulted in the frameshift leading to premature termination, it is likely that Phc2 proteins were not produced or that a truncated protein starting from M128 of the 36-kDa isoform and lacking the HD1 and FCS finger might be expressed. Indeed, no signals were detected at around 40 kDa, or in the range of smaller molecular mass below, by immunoblot analysis, which used materials immunoprecipitated by the anti-Phc2 monoclonal antibody from 11.5-dpc embryos (Fig. 2E). Therefore, this Phc2 mutant allele may be null or encode a truncated C-terminal polypeptide that could not be detected by the anti-Phc2 monoclonal antibody.
Homeotic transformations of the axial skeleton concomitant with derepression of Hox genes in Phc2 mutants.
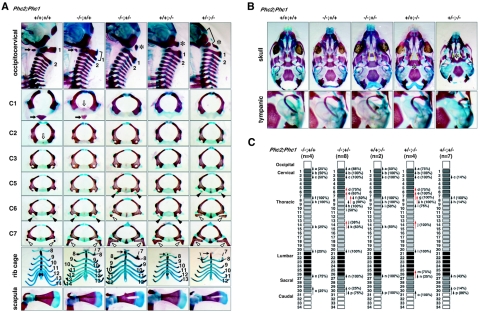
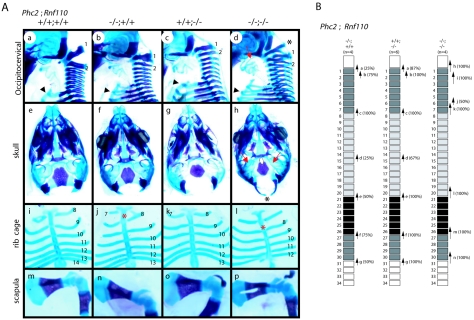
Common features shared by mammalian class II PcG mutants are homeotic transformations of the axial structures associated with derepression of Hox cluster genes and premature senescence of embryonic fibroblasts associated with the overexpression of Cdkn2a transcripts (19). Eighteen pups, obtained by mating homozygous males and heterozygous females, were analyzed. All homozygotes exhibited at least a few alterations to the axial skeleton that were characteristic of posterior transformations (Fig. 3A and C), whereas heterozygotes showed much lower penetrance (Shinga and Koseki, unpublished). These alterations were as follows: an ectopic bone floating between the occipital bones and the first cervical vertebra (C1); in the cervical region, an odontoid process (a characteristic feature of C2) was fused to C1, suggesting intermediate morphological features between C1 and C2 (Fig. 3A); and the dorsal part of C1 or C2 was often bifurcated in Phc2 homozygotes (Fig. 3A). The lateral view of mutant C2 was similar to that of wild-type C3. In all homozygotes, ectopic ribs were associated with C7 (Fig. 3A). In 67% of cases, the ectopic ribs were imperfect and fused to the middle part of the first ribs, while in the remainder, perfect ribs were formed that were directly associated with the cranially shifted sternum. The prominent spinous process characteristic of the second thoracic vertebra (T2) in the wild type was associated with T1 in all homozygotes. In the rib cage, the 7th ribs were detached from the sternum and the 13th ribs were missing or floating (Fig. 3A). Similarly, the thoracolumbar and lumbosacral boundaries were shifted anteriorly in the homozygotes. No significant changes were seen in the skull.
FIG. 3.
Skeletal alterations in Phc2−/− mice and gene dose-dependent skeletal alterations in Phc2-Phc1 compound mutants. (A) Lateral views of the occipitocervical region; overviews of individual C1, C2, C3, C5, C6, and C7 vertebrae; ventral views of rib cages; and overviews of scapulas. In the lateral views of the occipitocervical region, C1 and C2 are indicated numerically, and asterisks indicate the ectopic arch or piece. In Phc2+/−-Phc1−/− mice, a bracket indicates the segmentation of the exoccipital bone and an arrow indicates the anterior tuberculus of C1. In C1 and C2 vertebrae, closed and open arrows, respectively, indicate the anterior tuberculus and odontoid processes. In C5, C6, and C7, closed and open arrowheads, respectively, indicate the anterior tuberculus for vertebral arteries and ectopic ribs. In ventral views of the rib cages, the numbers of vertebrae to which ribs are attached are indicated. (B) Ventral views of skulls and tympanic bones. In the skull, a cleft in the secondary palate is emphasized by the use of yellow lines indicating the medial edges of the palatine. Note the absence of cartilaginous condensation at the center of the sphenoid in Phc2+/−-Phc1−/− mice. (C) Summary of posterior transformations. Each arrow represents the homeotic transformation of vertebrae. (a) Supraoccipital bone→C1, appearance of the ectopic arch or bone; (b) C1→C2, fusion of the odontoid process to the C1 vertebra; (c) C2→C3; (d) C5→C6, association of the anterior tuberculus with the C5 vertebra; (e) C6→T1, association of the cervical rib with the C6; (f) C7→T1, association of the cervical rib with C7; (g) C7→T2, prominent spinous process on C7; (h) T1→T2, prominent spinous process on T1; (i) T2→T3, lack of prominent spinous process in T2; (j) T6→T8, dissociation of the sixth rib from the sternum; (k) T7→T8, dissociation of the seventh rib from the sternum; (l) T13→L1, loss of the rib from the 20th vertebra; (m) L4→S1, formation of a sacroiliac joint in the 25th vertebra; (n) L6→S1, formation of a sacroiliac joint in the 26th vertebra; (o) S3→Ca1, appearance of the first caudal vertebra in the 29th vertebra; (p) S4→Ca1, appearance of the first caudal vertebra in the 30th vertebra.
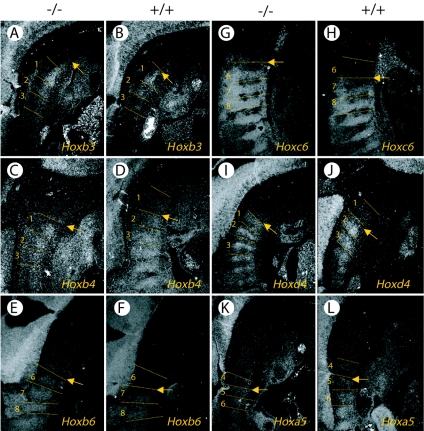
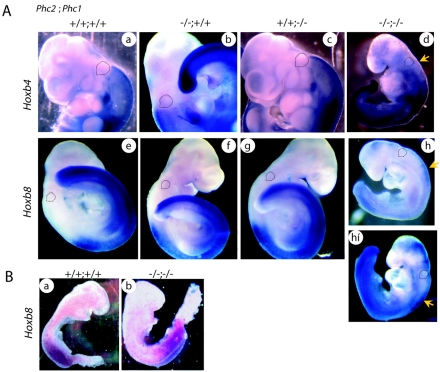
Next, we examined the phenotypic expression of Hoxb3, -b4, -b6, -c6, -d4, and -a5 (Fig. 4). The anterior boundary of Hoxb3 expression at the first prevertebra in the wild type was shifted cranially to reach the caudal part of the basioccipital bone anlage in Phc2−/− embryos. Similarly, the anterior boundaries of Hoxb4, -b6, -c6, and -d4 were shifted cranially in Phc2−/− embryos, while Hoxa5 was not. Therefore, this showed that Phc2 was involved in the anterior-posterior (A-P) specification of the vertebral column through the regulation of Hox gene expression, as well as other PcG proteins.
FIG. 4.
Changes in Hox gene expression in Phc2−/− mice. Expression of Hox genes in 11.5-dpc Phc2−/− (A, C, E, G, I, and K) and wild-type (B, D, F, H, J, and L) embryos. The expressions of Hoxb3 (A and B), Hoxb4 (C and D), Hoxb6 (E and F), Hoxc6 (G and H), Hoxd4 (I and J), and Hoxa5 (K and L) are shown. Several prevertebrae are numbered in each panel, and dotted lines indicate the segment boundaries. The arrows indicate anterior boundaries of Hox gene expression.
Premature senescence of MEFs in Phc2 mutants.
Phc2−/− MEFs exhibit defects in proliferation and premature senescence, as well as MEFs derived from M33, Bmi1, and Rnf110 mutants (11, 19) (H. Koseki, unpublished data). Growth curves of MEFs from wild-type and Phc2−/− genotypes were compared, using a strict 3T9 protocol (20) (Fig. 5A). Phc2−/− MEFs grew more slowly than wild-type MEFs, even in early passages; they also stopped dividing earlier (passage 7 versus passage 15, respectively). The proliferation of Phc2−/− MEFs was clearly affected at passage 5 (Fig. 5B). These observations indicated that Phc2−/− MEFs senesced more quickly than the wild-type MEFs. This prompted us to examine the expression of the two Cdkn2a genes, p16 and p19ARF, and we found that their expression was clearly derepressed in Phc2−/− MEFs, even as early as passage 3 (Fig. 5C). Therefore, premature senescence induced by the Phc2 mutation was shown to involve the activation of Cdkn2a gene products, as has been found for other PcG mutants (19). It is also known that some immunological defects seen in Bmi1 mutants are mediated by the overexpression of Cdkn2a products (19). We did not see significant changes in lymphocyte development in Phc2−/− mice.
FIG. 5.
Defects in the proliferation of Phc2−/− MEFs. (A) Cell proliferation using a 3T9 protocol. At 3-day intervals, the total numbers of wild-type and Phc2−/− MEFs per culture were determined. The error bars indicate standard errors of the mean. (B) Proliferation of MEFs from respective genotypes at passage 5. (C) Northern analysis of p16 and p19ARF transcripts in wild-type and Phc2−/− MEFs at passages 3, 4, and 5 (P3, P4, and P5). The same gel was stained with ethidium bromide to verify the loaded amounts. (D) Distribution of Phc2 proteins in the Cdkn2a genomic region in MEFs. Representative results show the association of the Phc2 protein with the second exonic region of Cdkn2a, using the adam34 coding region as a negative control. Immunoprecipitation of the chromatin was performed using the anti-Phc2 antibody (+); mock immunoprecipitation (−) was used as a negative control. Equivalent amounts of genomic DNA from different sources were adjusted and designated ×1.
We then addressed whether the Phc2 mutation had an impact on Cdkn2a expression by using ChIP to look at the physical association of Phc2 proteins with the gene (Fig. 5D). We compared the Phc2 association with the first and the second exonic regions of the p16 gene between the wild-type and Phc2-null MEFs (20). Anti-Phc2 immunoprecipitated a significant quantity of these genomic regions from the wild-type MEFs, but not from Phc2−/− MEFs. Therefore, it is likely that Phc2 regulates the transcription of Cdkn2a through a direct association with the chromatin.
Synergistic action of Phc2 with Phc1 and Rnf110 mutations.
Genetic interactions of mammalian PcG mutations have been reported previously using mice that were doubly deficient for either homologous (Rnf110-Bmi1) or nonhomologous (Bmi1-M33 and Rnf110-Rnf2) PcG genes (3, 6, 40). The expression of Hox genes was synergistically affected in Rnf110-Bmi1 and Bmi1-M33 double homozygotes, with the Rnf110-Bmi1 interaction much stronger in terms of Hox derepression. Together with the biochemical evidence, these observations were interpreted as follows. First, the Rnf110, Bmi1, M33, and Rnf2 proteins act in synergy to repress Hox genes by forming multimeric protein complexes. Second, homologous PcG gene products have overlapping functions and, to a certain extent, act in a mutually compensatory fashion, as expected from the Rnf110-Bmi1 double-mutant phenotypes. However, the mutant interactions of a mammalian ph homologue with either homologous or nonhomologous PcG genes have not been addressed. Therefore, in this study we generated Phc2-Phc1 and Phc2-Rnf110 double homozygotes in order to investigate their axial phenotypes. In particular, it was of interest to compare the impact of Phc2-Phc1 double deficiency on axial development with that of Rnf110-Bmi1, because Phc3, a product of the evolutionarily conserved third ph homologue involved in hPRC-H, is still present in the Phc2-Phc1 double mutants, while the inclusion of an additional Psc homologue has not been reported (27, 44). A comparison of the Phc2-Phc1 and Phc2-Rnf110 doubly deficient phenotypes might also allow us to evaluate the functional differences between homologous and nonhomologous interactions among the complexes.
Phc2-Phc1 compound heterozygotes, which were viable, fertile, and externally normal but which exhibited posterior transformations with full penetrance, were intercrossed to produce various compound mutants (Fig. 3C). As expected from the gestational and perinatal lethality of Phc1 homozygotes, no living Phc1−/− pups were found, irrespective of Phc2 genotypes at birth (41). Importantly, Phc2−/−-Phc1+/− fetuses were alive at 17.5 dpc but did not survive birth, while Phc2−/− single mutants were viable (Table 1). Therefore, the heterozygous loss of the Phc1 gene was shown to affect the survival of Phc2 homozygotes during the perinatal period. Phc2−/−-Phc1−/− embryos were lost in a progressive fashion, at an earlier gestational stage than Phc2+/+-Phc1−/−, Phc2+/−-Phc1−/−, or Phc2−/−-Phc1+/− fetuses, which survived at least until 17.5 dpc (Table 1). These results suggested that Phc1 and Phc2 mutations were synergistically affecting the survival of embryos in a gene dosage-dependent manner.
TABLE 1.
Offspring generated by crossing between Phc2-Phc1 compound heterozygotes
| Phc2-Phc1 genotype | No. (%) of offspring |
|||
|---|---|---|---|---|
| 9.5 dpc (n = 283) | 10.5 dpc (n = 89) | 11.5 dpc (n = 63) | 17.5 dpc (n = 63) | |
| +/+ +/+ | 19 (6.7) | 5 (5.6) | 6 (9.5) | 2 (3.2) |
| +/+ +/− | 38 (13.4) | 11 (12.4) | 7 (11.1) | 10 (15.9) |
| +/+ −/− | 27 (9.5) | 3 (3.4) | 2 (3.2) | 2 (3.2) |
| +/− +/+ | 38 (13.4) | 11 (12.4) | 11 (17.5) | 14 (22.2) |
| +/− +/− | 31 (11.0) | 8 (9.0) | 7 (11.1) | 4 (6.4) |
| +/− −/− | 36 (12.7) | 10 (11.2) | 8 (12.7) | 8 (12.7) |
| −/− +/+ | 12 (4.2) | 6 (6.7) | 2 (3.2) | 4 (6.4) |
| −/− +/− | 65 (23.0) | 24 (27.0) | 18 (28.6) | 19 (30.2) |
| −/− −/− | 13 (4.6) | 7 (7.9)a | 2 (3.2)b | 0 (0.0) |
| NDc | 4 (1.4) | 4 (4.5) | 0 (0.0) | 0 (0.0) |
Two out of seven had no heartbeat.
One out of two was absorbed.
ND, not determined.
Next, we compared the axial skeletal development of single and compound mutants. Axial skeletal alterations in Phc2 mutants were similar to but slightly milder than those seen in Phc1 mutants (Fig. 3A, B, and C); the most prominent differences were seen in the skull. The formation of an ectopic arch associated with the occipital bones, a cleft in the secondary palate, lack of ossification at the center of the presphenoid, and a partial split of the sphenoid bone were seen exclusively in Phc1-deficient mice, while changes to the vertebral column were similar in both mutant types. Importantly, skeletal defects due to the respective mutations were exaggerated by the heterozygous mutual loss of another gene. In Phc2−/−-Phc1+/− mice, we observed floating of the dorsal part of the occipital bones, a cleft in the secondary palate, association of anterior processes with C5, detachment of the sixth ribs from the sternum, and a hole in the scapula. These changes were never seen in either Phc1 single heterozygotes or Phc2 single homozygotes (Fig. 3A, B, and C). The same was true for Phc2+/−-Phc1−/− mice. Skeletal defects in the vertebral column and scapula were very similar to those seen in Phc2−/−-Phc1+/− mice, including an association of anterior processes with C5, association of imperfect ribs with C6, detachment of the sixth ribs from the sternum, and a hole in the scapula. Defects in the skull were also exaggerated compared to the Phc1 single mutant, as demonstrated by the obvious segmentation of the exoccipital bone and complete lack of the presphenoid and of the caudal half of the tympanic bone (Fig. 3A, B, and C). Therefore, it appears that homeotic transformations in the axial skeleton and defects in the skull and scapula were synergistically enhanced by Phc1 and Phc2 mutations.
Most of the Phc2−/−-Phc1−/− embryos were lost before the mid-gestational stage (Table 1). Phc2−/−-Phc1−/− embryos developed normally up to 8.5 dpc, but severe growth retardation and external abnormalities became progressively evident, and the embryos died from 9.5 dpc onwards. At 9.5 dpc, double-homozygous embryos could be distinguished from their littermates with other genotypes based solely on their size and external morphological features. Internally, further phenotypic distinctions could be made. In the cranial region, the first and second branchial arches were poorly developed. In the trunk and caudal regions, the somitic mesoderm was irregularly segmented; on average, approximately 20 somites were formed in double homozygotes while there were 25 in littermates with other genotypes, and the tail bud was shrunken. Therefore, axial elongation was progressively affected in Phc2−/−-Phc1−/− embryos. These phenotypes were very similar to those seen in Rnf110-Bmi1 doubly homozygous mice (3).
Because of the early lethality of Phc2−/−-Phc1−/− embryos, we examined the expression of Hox cluster genes at 9.5 dpc instead of skeletal development. The anterior boundaries of Hoxb4 and -b8 in single mutants were not shifted to any great extent in either of the single mutants compared with the wild type (Fig. 6A). In double homozygotes, Hoxb4 expression was slightly but significantly derepressed in the cranial region, while the transcription level in the expression domain was not changed (Fig. 6A, a to d). Hoxb8 expression was not only derepressed in the cranial region but also decreased in its expression domain (Fig. 6A, a to h and hí). Therefore, Phc2 gene products were shown to act in synergy to repress the expression of Hoxb4 and -b8 rostral to the expression domain and also to maintain Hoxb8 expression within its expression domain. Very similar changes have been reported in the Rnf110-Bmi1 double mutant (12). In addition, since the derepression of Hoxb6 in Rnf110-Bmi1 double homozygotes has been shown to occur progressively between 8.5 and 9.5 dpc (3), we also looked at Hoxb8 expression at the earlier gestational time point of 8.5 dpc. In our study, Hoxb8 expression was not significantly altered in the 8.5-dpc embryos (Fig. 6B). Therefore, based on these results, it appears that Phc1 and Phc2 are involved in maintaining the transcriptional status of Hoxb8 rather than in its early induction, as observed in Rnf110-Bmi1 double homozygotes (3, 12).
FIG. 6.
Expression of Hoxb4 and Hoxb8. (A) Hoxb4 (frames a to d) and Hoxb8 (frames e to h) expression in 9.5-dpc wild-type and Phc2 and Phc1 single homozygotes and double homozygotes; the genotypes are indicated across the top. Each specimen was subjected to chromogenic reaction for the same length of time. The specimen shown in frame h was subjected to a longer chromogenic reaction (frame hí). Dotted lines indicate the otic vesicles in this series of figures. The prospective anterior boundary of the expression is indicated by a yellow arrow in double homozygotes. All pictures for both Hoxb4 and Hoxb8 expressions were taken under the same magnification. (B) Hoxb8 expression in 8.5-dpc wild-type and double-homozygous embryos; the pictures were taken under the same magnification.
Following this, we looked at the genetic interactions between the Phc2 and Rnf110 mutations. Since Phc2−/−-Rnf110+/− mice were viable and fertile, they were intercrossed to generate double homozygotes. Phc2−/−-Rnf110−/− mice died during late gestation, around 18.5 dpc, while most Phc2 and Rnf110 single mutants were shown to grow to adulthood (1). Furthermore, skeletal defects in double homozygotes were stronger than in either of the respective single mutants and resembled those in Phc2+/−-Phc1−/− and Phc2−/−-Phc1+/− mice (Fig. 7). Several defects were observed: the occipital bones were clearly segmented to form an ectopic arch, while the basioccipital bone was not completely segmented; the middle parts of the skull base and scapula failed to undergo cartilaginous condensation; the upper horn of the hyoid bone was fused to the styloid process; and the entire rib cage was shifted anteriorly more perfectly than in either respective single mutant. Unlike the Phc2-Phc1 interactions, the skeletal phenotypes of the respective single homozygotes were not exaggerated by the heterozygous mutual loss of another gene (Shinga and Koseki, unpublished). Taken together, these results showed that the Phc2 mutation affected the survival and A-P patterning of the axial skeleton in synergy with the Rnf110 mutation, but to a lesser degree than was observed with the Phc1 mutation.
FIG. 7.
Gene dose-dependent skeletal alterations in wild-type and Phc2 and Rnf110 single homozygotes and double homozygotes. (A) The genotypes of the specimens are shown along the top of the frames. (Frames a to d) Lateral views of the occipitocervical region. The arrowheads indicate the ribs associated with the eighth vertebrae. C1 and C2 are indicated numerically, and asterisks indicate the ectopic arch or piece. (Frames d and h) An asterisk indicates an ectopic arch that represents perfect segmentation of the exoccipital bone. Red arrows indicate ectopic cartilaginous condensation bridging the hyoid bone and styloid process, while the dotted line indicates the lack of cartilaginous condensation between the occipital and sphenoid bones. (Frames e to h) Ventral views of the skull. (Frames i to l) Ventral views of rib cages. The numbers of vertebrae to which ribs are attached are indicated numerically. Asterisks indicate the sternums that are shifted anteriorly in Phc2 single (frame j) and double (frame l) homozygotes. (Frames m to p) Overviews of the scapula. (B) Summary of posterior transformations: (a) supraoccipital bone→C1, appearance of the ectopic arch or bone; (b) C1→C2, fusion of the odontoid process to the C1 vertebra; (c) C7→T1, association of the cervical rib with C7; (d) T7→T8, dissociation of the seventh rib from the sternum; (e) T13→L1, loss of the rib from the 20th vertebra; (f) L6→S1, formation of a sacroiliac joint in the 26th vertebra; (g) S4→Ca1, appearance of the first caudal vertebra in the 30th vertebra; (h) more perfect supraoccipital bone→C1 transformation, perfect segmentation of supra- and exoccipital bones; (i) C1→C3, overall structure similar to C3 rather than C2; (j) C5→C6, association of the anterior tuberculus with the C5 vertebra; (k) C6→T1, association of the cervical rib with C6; (l) T12→L1, loss of the rib from the 19th vertebra; (m) L4→S1, formation of a sacroiliac joint in the 25th vertebra; (n) S3→Ca1, appearance of the first caudal vertebra in the 29th vertebra.
Direct association of Phc2 with the Hoxb8 locus.
The significant impacts of the Phc2 mutation on the transcriptional regulation of Hox genes prompted us to investigate the physical association of Phc2 proteins with the Hoxb8 locus using ChIP (Fig. 8A). We dissected 11.5-dpc embryos into cranial and caudal parts, in which the transcription of Hoxb8 transcription is silent and active, respectively. Phc2 association was examined at the upstream enhancer (BH1100 region), putative promoter region, and first exon of Hoxb8 and the upstream enhancer region of Hoxb7 (KA region) (9). Phc2 was associated with these genomic regions irrespective of transcriptional status, as was the case with Phc1 (Fig. 8B). Therefore, it is likely that Phc2 and Phc1 regulate the transcription of Hoxb8 through a direct association with the chromatin.
FIG. 8.
Distribution of Phc2 proteins in the Hoxb8 genomic region at 11.5 dpc. (A) Representative results showing the association of the Phc2 protein with the Hoxb8 enhancer and promoter and intronic and Hoxb7 enhancer regions, with the adam34 coding region as a negative control. The positions of PCR fragments are shown in panel B. Tissues of 12.5-dpc embryos were dissected, and chromatin fractions were purified from transcriptionally silent anterior (A) and active posterior (P) tissues (mesoderm plus neurectoderm). Immunoprecipitation of the chromatin was performed by the anti-Phc2 antibody (+); mock immunoprecipitation (−) was used as a negative control. Equivalent amounts of genomic DNAs from different sources were adjusted and designated 1′. (B) The distribution of Phc2 in the Hoxb8 genomic region is schematically represented. The black and gray squares represent the degree of enrichment of immunoprecipitated genomic DNA in posterior and anterior tissues, respectively: each square represents a twofold enrichment in comparison with the unfractionated “input” DNA. The distribution of Phc1 is shown as a reference.
Analysis of the comparative expression of Phc2 and Phc1 in various tissues.
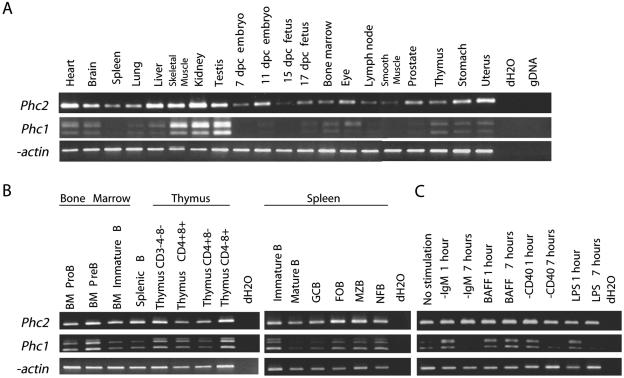
Although genetic evidence suggests that Phc2 and Phc1 gene products act in a redundant manner, it is also true that the single deficient phenotypes for Phc2 and Phc1 are not identical. In particular, overt defects in the hematopoietic lineage or cranial neural crest derivatives are seen exclusively in Phc1 mutants (41, 43). Importantly, Phc2 and Rnf110 mutations synergistically affect the proliferation of interleukin 7 receptor α-positive lymphoid precursor cells residing in the fetal gut, while single mutants are unaffected (T. Sato and H. Koseki, submitted for publication). This implies that the involvement of Phc2 during the proliferation of lymphoid precursors is dispensable and that latent defects in hematopoietic lineage development might presumably be fully compensated for by Phc1 and/or Phc3 in Phc2-deficient mice. It has therefore been suggested that the phenotypical differences between Phc2 and Phc1 single mutants might be attributable to tissue- and/or stage-specific variations in their expression or requirement (17). To address this issue, we compared the expression of Phc2 and Phc1 in various tissues by RT-PCR (Fig. 9A). Phc2 transcripts were detected as 383-bp PCR products and Phc1 as at least 346 and 502 bp due to the differential usage of exons. The expression of Phc2 and Phc1 was seen in all the tissues examined, although the levels varied. Tissue-specific variation of expression was noticeably more prominent in Phc1 than Phc2.
FIG. 9.
The expression of Phc2 and Phc1 in various tissues, lymphocyte subpopulations, and B lymphocytes induced by various extracellular stimuli. (A) The expression of Phc2 and Phc1 was examined by reverse transcriptase-PCR in various adult tissues and whole embryos at 7, 11, 15, and 17 days postcoitus. Distilled water (dH2O) and mouse genomic DNA (gDNA) were used as negative controls; β-actin was used to normalize the amounts of cDNA. (B) The expression of Phc2 and Phc1 was examined in developing B and T lymphocytes in the bone marrow and thymus, respectively, and B-cell subpopulations in naïve and immunized spleens. Mature splenic B cells were fractionated into germ center B (GCB), follicular B (FOB), marginal zone B (MZB), and newly formed B (NFB) subpopulations. Phc2 did not exhibit any variation in expression levels, in contrast to Phc1, where the level of expression was variable. (C) Expression of Phc2 and Phc1 in resting B lymphocytes upon activation by B-cell receptor engagement by α-IgM, BAFF stimulation, CD40 ligation by α-CD40, and LPS stimulation.
We extended our analysis to lymphocyte subpopulations in bone marrow, thymus, naïve and immunized spleen, and B lymphocytes activated by various signals, since the role of Phc2 was shown to be dispensable (Fig. 9B). Developing B and T lymphocytes in the bone marrow and thymus, respectively, and B-cell subpopulations in naïve and immunized spleens, were fractionated according to their developmental stages. Phc2 and Phc1 expression was seen in all fractions, and while Phc1 expression was variable, that of Phc2 was not. This developmental-stage-specific variation in Phc1 expression prompted us to examine whether the expression of Phc2 and Phc1 in resting B lymphocytes was regulated by various inductive signals mediated by B-cell receptor cross-linking, B-cell activation factor from the TNF family (BAFF) stimulation, CD40 ligation, or lipopolysaccharide (LPS) stimulation (Fig. 9C). The engagement of B-cell receptor by anti-IgM antibodies induces the survival and cell cycle entry of resting B cells (24); BAFF functions to maintain B-cell viability but does not induce S-phase entry of the cell cycle (28); CD40 ligation by anti-CD40 induces the survival, cell cycle progression, and up-regulation of a variety of adhesion molecules for cell-cell interaction (7); and LPS represents a T-independent antigen that has potent mitogenic activity on B cells (30). Interestingly, these signals significantly increased Phc1 expression within 1 hour, whereas they had no effect on Phc2. Up-regulation of Phc1 expression induced by BAFF persisted for at least 7 h, while induction by other signals was transient. Therefore, Phc1 expression appears to be regulated by various inductive stimuli, whereas Phc2 is expressed in a constitutive manner.
DISCUSSION
In this study, we have provided biochemical and genetic evidence to indicate that Phc2 is a functional component of class II PcG complexes during embryogenesis. Phc2 is constitutively involved in an hPRC-H-like complex with other constituents of class II PcG complexes and associates with the Hoxb8 genomic regions at 11.5 dpc. We have shown that the Phc2 mutant allele is involved in the mediation of the transcriptional repression of the Hox cluster genes in mid-gestational embryos and the Cdkn2a genes in MEFs; similar findings have been reported for other mammalian PcG mutants. The synergistic actions of the Phc2 mutation with the Phc1 and Rnf110 mutations during A-P specification demonstrate that Hox repression by Phc2 is mediated through the class II PcG complexes. Phc2-Phc1 double deficiency reveals that Phc2 and Phc1 act synergistically to maintain the repression of Hox genes but are not involved in early induction.
The genetic evidence provided by this study allows us to evaluate further the functional complicity of Phc2 with Phc1 in vivo. Although both Phc1 and Rnf110 associate and form complexes with Phc2, the Phc1 mutation exacerbated the Phc2 mutant phenotypes to a much greater extent than the Rnf110 mutation in terms of Hox derepression and embryonic survival. These different modes of genetic interaction are at least partly due to functional overlap and the mutually compensatory properties of Phc2 with Phc1, but not with Rnf110. Indeed, in vitro pull-down and yeast two-hybrid assays have shown that Phc2 and Phc1 interact with almost the same set of PcG proteins and that the primary structures of their functional domains are highly conserved (16, 18, 25, 36). This is also consistent with in vitro evidence that the inhibition of remodeling and transcription, and the recruitment of a free polynucleosomal array mediated by hPRC-H and PRC1 in the solution, is mostly substituted by mouse PRC core complexes (mPCCs) comprised exclusively of Phc1, M33, Rnf2, and either Rnf110 or Bmi1, although the original hPRC-H harbors Phc1, Phc2, and Phc3 (26, 27). Nevertheless, it is also true that the single deficient phenotypes for Phc2 and Phc1 are not identical. These phenotypical differences between Phc2 and Phc1 single mutants might be at least partly attributed to tissue- and/or stage-specific variations in their expression or requirement (17), a concept that is supported by the results of the comparative expression analyses of Phc2 and Phc1 in this study. Although Phc2 and Phc1 act in a highly compensatory manner, both mutants still exhibit very similar derepression of Hox cluster genes. This leads us to hypothesize that the overall quantity of ph orthologues in the nuclei may also be an important parameter for class II PcG complexes, allowing them to exert their molecular functions, at least at the Hox loci, as well as Psc orthologues (3). We were able to show by using ChIP that Phc2 and Phc1 consistently associate together with several Hoxb8 genomic regions. Furthermore, a strict dose-dependent requirement for Phc2 and Phc1 is seen during A-P specification of the axis.
Biochemical analyses suggest that the ability of mPCC to inhibit remodeling and transcription, as well as to bridge chromatin templates, might represent substantially separate functions requiring different subunits (26). While Phc1 plays a key role in bridging chromatin templates in vitro, it is dispensable for binding to a chromatin template and inhibiting its remodeling. Phc2-Phc1 double homozygotes reveal that the molecular processes mediated by Phc2 and Phc1 are essential for the maintenance of Hox repression by class II PcG complexes. The recruitment activity is expected to represent a long-range interaction or spreading of class II PcG complexes on the genome, which is likely to be mediated through interactions among PcG proteins. Therefore, class II PcG complexes that lack both Phc2 and Phc1 may fail to mediate PcG repression because of defects in long-range effects. The oligomerization of PcG complexes, mediated by heterotypic and/or homotypic interactions of the SAM domains of Phc2 and Phc1, plays a key role because the self-association activity of the SAM domain of SCM has been shown to be closely correlated with PcG repression in Drosophila (37). Given that mPCC-like complexes act as a fundamental unit of class II PcG complexes in vivo, oligomerization of mPCC-like complexes via the SAM domains of ph orthologues may be an essential process in the formation of functional class II PcG complexes on the chromatin (26, 36, 43). Consistent with this is the close association of Phc2 with Phc1, as demonstrated by both co-IP and ChIP analyses in our study and elsewhere (16). Interestingly, Phc3 remains present in Phc2-Phc1 double mutants, while the inclusion of an additional Psc homologue has not been reported in class II PcG complexes, although the double-mutant phenotypes for Phc2-Phc1 and Rnf110-Bmi1 are almost identical (3, 27, 44). This implies that Phc3 alone may not be quantitatively sufficient to allow the oligomerization of PcG complexes, or possibly excess amount of mPCCΔPhc2- or mPCCΔPhc1-like complexes may affect the oligomerization of mPCC-like complexes containing Phc3.
In this study, we also showed that both Phc2 and Phc1 gene products associate with the Hoxb8 genomic region within the transcriptionally active domain, as well as within the repressed loci. This finding is consistent with previous reports that the predominant subnuclear localization of several PcG proteins is in the perichromatin compartment, where most pre-mRNA synthesis takes place (10). It is possible that these results tie in with the recent discovery that class II PcG complexes are associated with a positive function. Null mutations in the Rnf110 and Phc1 loci have been shown to decrease the transcription level of endogenous Hoxb1 and to severely impair the transcription from Hox-LacZ reporters and knockin loci (12). In addition, a positive action of PcG complexes could explain the fall in gene expression levels within Hoxb8 expression domains, which has been reported to be around 9.5 dpc in Phc2-Phc1 and Bmi1-Rnf110 and double mutants (3, 12). Importantly, the transcriptional silencing of Hox-reporter loci in Rnf110 and Phc1 mutants is accompanied by DNA methylation of the promoter region (12). Therefore, the positive role of class II PcG gene products, mediated by their physical association with Hoxb8 genomic regions, may involve local suppression of the pathway leading to local DNA methylation. However, it is still conceivable that the PcG gene products act by repressing the expression of Hox transcription repressors. This possibility needs to be addressed in the future.
Acknowledgments
This project was supported by Special Coordination Funds for the Promotion of Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (H.K.).
This paper is dedicated to the memory of Shozo Sugimori, who was responsible for the care of the mutant animals used in this study. We are also grateful to T. Hasegawa and to M. Iida, M. Uchida, and R Moriizumi for their help.
REFERENCES
- 1.Akasaka, T., M. Kanno, R. Balling, M. A. Mieza, M. Taniguchi, and H. Koseki. 1996. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122:1513-1522. [DOI] [PubMed] [Google Scholar]
- 2.Akasaka, T., K. Tsuji, H. Kawahira, M. Kanno, K. Harigaya, L. Hu, Y. Ebihara, T. Nakahata, O. Tetsu, M. Taniguchi, and H. Koseki. 1997. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity 7:135-146. [DOI] [PubMed] [Google Scholar]
- 3.Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. [DOI] [PubMed] [Google Scholar]
- 4.Alkema, M. J., M. Bronk, E. Verhoeven, A. Otte, L. J. van 't Veer, A. Berns, and M. van Lohuizen. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 11:226-240. [DOI] [PubMed] [Google Scholar]
- 5.Atsuta, T., Y. Fujimura, H. Moriya, M. Vidal, T. Akasaka, and H. Koseki. 2001. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma 20:43-46. [DOI] [PubMed] [Google Scholar]
- 6.Bel, S., N. Core, M. Djabali, K. Kieboom, N. van der Lugt, M. J. Alkema, and M. van Lohuizen. 1998. Genetic interactions and dosage effects of Polycomb group genes in mice. Development 125:3543-3551. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, G. A., and B. S. Hostager. 2003. The CD40-CD154 interaction in B cell-T cell liaisons. Cytokine Growth Factor Rev. 14:297-309. [DOI] [PubMed] [Google Scholar]
- 8.Bornemann, D., E. Miller, and J. Simon. 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122:1621-1630. [DOI] [PubMed] [Google Scholar]
- 9.Charite, J., W. de Graaff, R. Vogels, F. Meijlink, and J. Deschamps. 1995. Regulation of the Hoxb-8 gene: synergism between multimerized cis-acting elements increases responsiveness to positional information. Dev. Biol. 171:294-305. [DOI] [PubMed] [Google Scholar]
- 10.Cmarko, D., P. J. Verschure, A. P. Otte, R. van Driel, and S. Fakan. 2003. Polycomb group gene silencing proteins are concentrated in the perichromatin compartment of the mammalian nucleus. J. Cell Sci. 116:335-343. [DOI] [PubMed] [Google Scholar]
- 11.Core, N., S. Bel, S. J. Gaunt, M. Aurrand-Lions, J. Pearce, A. Fisher, and M. Djabali. 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124:721-729. [DOI] [PubMed] [Google Scholar]
- 12.de Graaff, W., D. Tomotsune, T. Oosterveen, Y. Takihara, H. Koseki, and J. Deschamps. 2003. Randomly inserted and targeted Hox/reporter fusions transcriptionally silenced in Polycomb mutants. Proc. Natl. Acad. Sci. USA 100:13362-13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Mar Lorente, M., C. Marcos-Gutierrez, C. Perez, J. Schoorlemmer, A. Ramirez, T. Magin, and M. Vidal. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127:5093-5100. [DOI] [PubMed] [Google Scholar]
- 14.Dura, J. M., N. B. Randsholt, J. Deatrick, I. Erk, P. Santamaria, J. D. Freeman, S. J. Freeman, D. Weddell, and H. W. Brock. 1987. A complex genetic locus, polyhomeotic, is required for segmental specification and epidermal development in D. melanogaster. Cell 51:829-839. [DOI] [PubMed] [Google Scholar]
- 15.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunster, M. J., D. P. Satijn, K. M. Hamer, J. L. den Blaauwen, D. de Bruijn, M. J. Alkema, M. van Lohuizen, R. van Driel, and A. P. Otte. 1997. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol. Cell. Biol. 17:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunster, M. J., F. M. Raaphorst, K. M. Hamer, J. L. den Blaauwen, E. Fieret, C. J. Meijer, and A. P. Otte. 2001. Differential expression of human Polycomb group proteins in various tissues and cell types. J. Cell Biochem. 81:129-143. [DOI] [PubMed] [Google Scholar]
- 18.Hemenway, C. S., B. W. Halligan, and L. S. Levy. 1998. The Bmi-1 oncoprotein interacts with dinG and MPh2: the role of RING finger domains. Oncogene 16:2541-2547. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 20.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 21.Katoh-Fukui, Y., R. Tsuchiya, T. Shiroishi, Y. Nakahara, N. Hashimoto, K. Noguchi, and T. Higashinakagawa. 1998. Male-to-female sex reversal in M33 mutant mice. Nature 393:688-692. [DOI] [PubMed] [Google Scholar]
- 22.Kessel, M., and P. Gruss. 1991. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67:89-104. [DOI] [PubMed] [Google Scholar]
- 23.Kim, C. A., M. Gingery, R. M. Pilpa, and J. U. Bowie. 2002. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9:453-457. [DOI] [PubMed] [Google Scholar]
- 24.Kraus, M., M. B. Alimzhanov, N. Rajewsky, and K. Rajewsky. 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igα β heterodimer. Cell 117:787-800. [DOI] [PubMed] [Google Scholar]
- 25.Kyba, M., and H. W. Brock. 1998. The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev. Genet. 22:74-84. [DOI] [PubMed] [Google Scholar]
- 26.Lavigne, M., N. J. Francis, I. F. King, and R. E. Kingston. 2004. Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol. Cell. 13:415-425. [DOI] [PubMed] [Google Scholar]
- 27.Levine, S. S., A. Weiss, H. Erdjument-Bromage, Z. Shao, P. Tempst, and R. E. Kingston. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay, F., and J. L. Browning. 2002. BAFF: a fundamental survival factor for B cells. Nat. Rev. Immunol. 2:465-475. [DOI] [PubMed] [Google Scholar]
- 29.Miyagishima, H., K. Isono, Y. Fujimura, M. Iyo, Y. Takihara, H. Masumoto, M. Vidal, and H. Koseki. 2003. Dissociation of mammalian Polycomb-group proteins, Ring1B and Rae28/Ph1, from the chromatin correlates with configuration changes of the chromatin in mitotic and meiotic prophase. Histochem. Cell Biol. 120:111-119. [DOI] [PubMed] [Google Scholar]
- 30.Miyake, K. 2004. Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 31.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura, M., Y. Takihara, and K. Shimada. 1994. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: one of the early inducible clones encodes a novel protein sharing several highly homologous regions with a Drosophila polyhomeotic protein. Differentiation 57:39-50. [DOI] [PubMed] [Google Scholar]
- 33.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 34.Paro, R. 1995. Propagating memory of transcriptional states. Trends Genet. 11:295-297. [DOI] [PubMed] [Google Scholar]
- 35.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock, and J. Simon. 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17:6683-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson, A. J., D. R. Mallin, N. J. Francis, C. S. Ketel, J. Stamm, R. K. Voeller, R. E. Kingston, and J. A. Simon. 2004. Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167:1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pirrotta, V. 1997. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 7:249-258. [DOI] [PubMed] [Google Scholar]
- 39.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, M., Y. Mizutani-Koseki, Y. Fujimura, H. Miyagishima, T. Kaneko, Y. Takada, T. Akasaka, H. Tanzawa, Y. Takihara, M. Nakano, H. Masumoto, M. Vidal, K. Isono, and H. Koseki. 2002. Involvement of the Polycomb-group gene Ring1B in the specification of the anterior-posterior axis in mice. Development 129:4171-4183. [DOI] [PubMed] [Google Scholar]
- 41.Takihara, Y., D. Tomotsune, M. Shirai, Y. Katoh-Fukui, K. Nishii, M. A. Motaleb, M. Nomura, R. Tsuchiya, Y. Fujita, Y. Shibata, T. Higashinakagawa, and K. Shimada. 1997. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development 124:3673-3682. [DOI] [PubMed] [Google Scholar]
- 42.Tomotsune, D., Y. Takihara, J. Berger, D. Duhl, S. Joo, M. Kyba, M. Shirai, H. Ohta, Y. Matsuda, B. M. Honda, J. Simon, K. Shimada, H. W. Brock, and F. Randazzo. 1999. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation 65:229-239. [DOI] [PubMed] [Google Scholar]
- 43.Tomotsune, D., M. Shirai, Y. Takihara, and K. Shimada. 2000. Regulation of Hoxb3 expression in the hindbrain and pharyngeal arches by rae28, a member of the mammalian Polycomb group of genes. Mech. Dev. 98:165-169. [DOI] [PubMed] [Google Scholar]
- 44.Tonkin, E., D. M. Hagan, W. Li, and T. Strachan. 2002. Identification and characterisation of novel mammalian homologues of Drosophila polyhomeotic permits new insights into relationships between members of the polyhomeotic family. Hum. Genet. 111:435-442. [DOI] [PubMed] [Google Scholar]
- 45.van der Lugt, N. M., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag, H. te Riele, M. van der Valk, J. Deschamps, M. Sofroniew, M. van Lohuizen, and A. Berns. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757-769. [DOI] [PubMed] [Google Scholar]
- 46.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 47.Voncken, J. W., B. A. Roelen, M. Roefs, S. de Vries, E. Verhoeven, S. Marino, J. Deschamps, and M. van Lohuizen. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 100:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson, D. G., and M. A. Nieto. 1993. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225:361-373. [DOI] [PubMed] [Google Scholar]
- 49.Yamaki, M., K. Isono, Y. Takada, K. Abe, T. Akasaka, H. Tanzawa, and H. Koseki. 2002. The mouse Edr2 (Mph2) gene has two forms of mRNA encoding 90- and 36-kDa polypeptides. Gene 288:103-110. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., A. Christoforou, L. Aravind, S. W. Emmons, S. Van Den Heuvel, and D. A. Haber. 2004. The C. elegans Polycomb gene sop-2 encodes an RNA binding protein. Mol. Cell 14:841-847. [DOI] [PubMed] [Google Scholar]