Abstract
The divergently transcribed DIT1 and DIT2 genes of Saccharomyces cerevisiae, which belong to the mid-late class of sporulation-specific genes, are subject to Ssn6-Tup1-mediated repression in mitotic cells. The Ssn6-Tup1 complex, which is required for repression of diverse sets of coordinately regulated genes, is known to be recruited to target genes by promoter-specific DNA-binding proteins. In this study, we show that a 42-bp negative regulatory element (NRE) present in the DIT1-DIT2 intergenic region consists of two distinct subsites and that a multimer of each subsite supports efficient Ssn6-Tup1-dependent repression of a CYC1-lacZ reporter gene. By genetic screening procedures, we identified DFG16, YGR122w, VPS36, and the DNA-binding proteins Rim101 and Nrg1 as potential mediators of NRE-directed repression. We show that Nrg1 and Rim101 bind simultaneously to adjacent target sites within the NRE in vitro and act as corepressors in vivo. We have found that the ability of Rim101 to be proteolytically processed to its active form and mediate NRE-directed repression not only depends on the previously characterized RIM signaling pathway but also requires Dfg16, Ygr122w, and components of the ESCRT trafficking pathway. Interestingly, Rim101 was processed in bro1 and doa4 strains but was unable to mediate efficient repression.
The yeast Saccharomyces cerevisiae can respond to changes in nutrient availability and environmental conditions by switching from one growth form to another. Extracellular cues are interpreted by various signal transduction pathways that act by both independent and convergent mechanisms to coordinate morphogenetic events. The different regulatory cascades and the interplay between them ultimately modulate the activity of target transcription factors and progression through the cell cycle. Haploid cells that are starved for glucose and diploid cells that are starved for nitrogen switch from growth as single budding cells to growth as long filaments of connected cells, with haploid cells becoming particularly adept at invasive growth and penetration into solid agar medium (reviewed in reference 63). Starvation of a diploid MATa/MATα cell for an essential nutrient in the absence of glucose and in the presence of a nonfermentable carbon source directs the cell into the pathway for spore formation (reviewed in reference 40). The nutritional signaling leads to arrest of the starved cell at G1 and contributes in a diploid to the expression of two key regulatory genes, IME1 and IME2, which encode a transcriptional activator and a protein kinase, respectively (reviewed in reference 43). These regulators set in motion the sporulation program that consists of meiotic DNA replication, recombination, the two meiotic divisions, and encapsulation of each haploid nucleus within a multilayered spore wall (reviewed in reference 48). This coordinated series of genetic and morphological events depends on the sequential expression of temporally distinct classes of sporulation-specific genes (18, 68).
The regulated expression of some classes of sporulation-specific genes is mediated, at least in part, by their repression in mitotic cells. For example, Ume6 binds to the promoter region of many early meiotic genes and prevents their expression in mitotic cells by recruitment of the Sin3-Rpd3 histone deacetylase complex and the Isw2 chromatin remodeling complex (reviewed in reference 43). Similarly, the DNA-binding protein Sum1 prevents expression of a middle class of sporulation-specific genes in mitotic cells by recruitment of Hst1, a NAD-dependent histone deacetylase (57, 95). The DIT1 and DIT2 genes, which belong to the mid-late class of sporulation-specific genes (15), are subject to Ssn6-Tup1-mediated repression in mitotic cells (27). The Ssn6-Tup1 complex, which is required for repression of diverse sets of coordinately regulated genes, is recruited to target genes by promoter-specific DNA-binding proteins (reviewed in reference 78). Multiple mechanisms, including recruitment of histone deacetylases, repositioning of nucleosomes, and interference with the transcriptional machinery, contribute to Ssn6-Tup1-mediated repression (for an example, see references 35 and 99).
We previously identified a 76-bp transcriptional regulatory element, termed NREDIT, that mediates Ssn6-Tup1-dependent repression of the DIT1 and DIT2 genes in mitotic cells (27). In this study, we show that NREDIT is a bipartite operator that depends on Rim101 and Nrg1 as mediators of repression. Both of these C2H2 Zn-finger-containing, DNA-binding proteins have been shown to depend on Tup1-Ssn6 for repression (50, 64). Rim101 is the S. cerevisiae homolog of PacC, which has been extensively characterized in Aspergillus nidulans as a DNA-binding transcriptional activator and repressor of alkaline- and acid-induced genes, respectively (reviewed in references 65 and 66). RIM101 was first identified in S. cerevisiae on the basis of its mutant phenotype as a positive regulator of early meiotic gene expression (80) and subsequently shown to behave as a positive regulator of haploid invasive growth (52) and of alkaline-pH-induced gene expression (51). More recently, Lamb and Mitchell (50) have shown that Rim101 mediates repression of two genes that encode the transcription factors Nrg1 and Smp1. Nrg1, which had been previously characterized as a negative regulator of glucose-repressed genes and genes involved in filamentous growth (47, 64, 92, 100), was shown to be a negative regulator of genes involved in ion homeostasis (50). Smp1, a MEF2-like transcription factor, which has been recently identified as a target of the stress-activated Hog1 kinase (20), was implicated as a negative regulator of genes involved in invasive growth and sporulation (50). It is clear that there is a complex interplay among signaling pathways that regulate the activity of transcription factors in response to environmental signals. In this study, we also show that the ability of Rim101 to be proteolytically processed to its active form and act as a repressor at the negative regulatory element (NRE) not only depends the previously characterized pH-responsive RIM signaling pathway (reviewed in references 2, 65, and 66) but also requires Dfg16, Ygr122w, and components of the ESCRT trafficking pathway.
MATERIALS AND METHODS
Media, growth conditions, and genetic methods.
Rich medium contained 1% yeast extract, 2% Bacto-peptone, and 2% glucose. Minimal medium contained 0.7% yeast nitrogen base without amino acids and auxotrophic supplements as required. Yeast strains were grown at 30°C. Mating, sporulation, and tetrad analysis were carried out using standard methods (76). The lithium acetate method (33) was used for yeast transformations.
Plasmids.
For these studies, we used pLG312Bgl and pLG312n, which were derived from pLGΔ312 (37), as the parental plasmids for assaying DNA fragments for operator function. pLGΔ312 is a high-copy URA3-containing plasmid that contains a CYC1-lacZ reporter gene. pLG312Bgl (27, 37) and pLG312n contain an XhoI-SalI-BglII-SalI-XhoI polylinker sequence and an XhoI-BglII-KpnI-SalI polylinker sequence, respectively, between the CYC1 upstream activation sequences (UASs) and the TATA box of the reporter gene. pLG312n was constructed by annealing the synthetic oligonucleotides MCS TOP (5′-TCGAGAGATCTGGTACCGTCGAC-3′) and MCS BOT (5′-TCGAGTCGACGGTACCAGATCTC-3′) (XhoI overhangs are indicated in boldface type) after the 5′ ends had been phosphorylated and then cloning the double-stranded oligonucleotide between the XhoI sites of pLG312(Bgl). Sequencing of the region spanning the polylinker revealed that the downstream insert/vector junction, expected to be TCGA, was 5′-CCCA-3′. The pLG312Bgl- and pLG312n-derived plasmids are referred to as pLG and pLGn, respectively.
pLG+NRE76, pLG+NRE44, pLG+NRE53, pLGΔSS, which contains a CYC1-lacZ reporter gene lacking its UASs, and p(−537)DIT1-lacZ, which contains a DIT1-lacZ translational fusion gene, have been described previously (27, 28). pLGΔSS+NREXX plasmids, which contain NREXX-lacZ fusion genes lacking the CYC1 UASs, were obtained from the corresponding pLGn+NREXX plasmids by removal of the SmaI-to-XhoI fragment that spans the UASs. pLG+NRE30(+), pLG+NRE30(−), and pLG + 3×NRE30 were constructed by annealing the synthetic oligonucleotides PAC-T (5′-GATCCGGGTTCTCTTGCCAAGAAAAAATAAAAAGG-3′) and PAC-B (5′-GATCCCTTTTTATTTTTTCTTGGCAAGAGAACCCG-3′) (BglII overhangs are indicated in boldface type) after their 5′ termini had been phosphorylated and then cloning the double-stranded oligonucleotide into the BglII site of pLG312Bgl. pLG+NRE30m and pLG+3×NRE30m were constructed in a similar manner with the synthetic oligonucleotides T19T (5′-GATCCGGGTTCTCTTGCCTAGAAAAAATAAAAAGG-3′) and B21A (5′-GATCCCTTTTTATTTTTTCTAGGCAAGAGAACCCG-3′). The following plasmids were constructed in a similar manner with pLGn312 as vector. The synthetic oligonucleotides NRE42T (5′-GATCCCATAAATAAAAGGGTTCTCTTGCCAAGAAAAAATAAAAAGG-3′) and NRE42B (5′-GATCCCTTTTTATTTTTTCTTGGCAAGAGAACCCTTTTATTTATGG-3′) were used to make pLGn+NRE42. The synthetic oligonucleotides NRE25-T (5′-GATCCATAAATAAAAGGGTTCTCTTGCC-3′) and NRE25-B (5′-GATCGGCAAGAGAACCCTTTTATTTATG-3′) were used to make pLGn+NRE25, pLGn+2×NRE25, and pLG+3×NRE25. The synthetic oligonucleotides NRE22D-T (5′-GATCCTTGCCAAGAAAAAATAAAAAG-3′) and NRE22D-B (5′-GATCCTTTTTATTTTTTCTTGGCAAG-3′) were used to make pLGn+NRE22D, pLGn+2×NRE22D, pLGn+ 3×NRE22D, and pLGn+4×NRE22D. pLG+NRE42m-1 was made with NRE42m-1-T (5′-GATCCCCATAAATAAACTGTATCTCTTGCCAAGAAAAAATAAAAAGG-3′) and NRE42m-1-B (5′-GATCCCCCTTTTTATTTTTTCTTGGCAAGAGATACAGTTTATTTATGG-3′). The region spanning the insert and polylinker sequence was sequenced in all plasmids to verify the sequence of the insert and to determine the orientation and number of inserts present.
pRIM101 was constructed by subcloning a 3.5-kb EcoRV fragment from pCF5, a genomic library plasmid that complemented the frd5-1 mutant, into pRS313. pKR41 was constructed to allow replacement of the RIM101 gene with the RIM101.HA3 allele (52), which encodes a version of Rim101 that contains three hemagglutinin (HA) epitopes after codon 473. This plasmid was constructed in two steps. First, a 4.8-kb PstI-KpnI fragment containing the RIM101.HA3 allele with 500 bp of upstream and 2.3 kbp of downstream sequence was purified from plasmid pWL41 (52) and cloned between the same sites in pBS SK(+) to yield plasmid pKR39. A PCR-generated 2.9-kbp fragment spanning the LEU2 gene with NheI sites at its ends was then cloned into the unique NheI site of pKR39 to generate pKR41. This places the LEU2 gene within the genomic sequence downstream of the RIM101.HA3 gene. A 7.3-kbp DNA fragment purified from pKR41 that had been digested with PstI and NcoI and that contained the RIM101.HA3::LEU2 sequence was used for integrative transformation of various yeast strains.
The pGAD424 (6) derivative pGAD424-RIM101(1289), which allows expression in yeast of a chimeric protein containing the activation domain of GAL4 and the DNA-binding region of Rim101, was constructed as follows. A DNA fragment containing the Rim101 coding region from residues 1 to 289 was amplified by PCR from a RIM101-containing plasmid template with a forward primer that added an EcoRI recognition site just upstream of the initiator ATG and a reverse primer that added a stop codon and a SalI recognition site just after codon 289. The gel-purified PCR product was digested with EcoRI and SalI and cloned between the corresponding sites of pGAD424 to generate an in-frame fusion gene. The pET21a derivative, pET21a-RIM101(1-289), which can be used to direct in vitro synthesis and bacterial expression of Rim101(1-289), was constructed by purifying the RIM101-containing EcoRI-to-SalI fragment from pGAD424-RIM101(1-289) and cloning this fragment between the corresponding sites of pET21a. pMAL-c2-Nrg1 for bacterial expression of an MBP-Nrg1 fusion protein was constructed as follows. An NdeI-BamHI fragment containing the Nrg1 coding region with an NdeI recognition site embedded in its initiator ATG codon and a BamHI recognition site just after its stop codon was recovered from pET16b-Nrg1. The NdeI-BamHI fragment was then cloned between the EcoRI and BamHI sites of pMAL-c2 after the NdeI and EcoRI sites of the insert and the vector, respectively, had been filled in.
Yeast strains.
Y102, Y104, Y108, Y170, KRY302, KRY308, and KRY318 were derived from the W303 strains described previously (39). Y102, a MATa rim101Δ::URA3 strain, was obtained by transformation of W303-1A with a 1.8-kb BglII-SalI, rim101Δ::URA3-containing fragment that had been purified from pSS179C (provided by A. Mitchell). Replacement of the wild-type RIM101 locus with the rim101Δ::URA3 allele was confirmed in Ura+ transformants by Southern blot and PCR analyses. Y104 is a rim101Δ::ura3 derivative of Y102 that was obtained by growing Y102 to saturation in liquid yeast extract-peptone-dextrose medium and then selecting for cells that could grow in the presence of 5-fluoroorotic acid (10). Y104 retained the same physical map as Y102 through the RIM101 locus as assessed by Southern blot and PCR analyses. Y108 is a MATa frd5-1 strain that was recovered by phenotypic analysis of the haploid progeny obtained on sporulation of the diploid created by mating Yfrd5-1 (28) with W303-1A.
Y170, a MATa tup1-Δ1::TRP1 strain, was obtained by transformation of yeast cells with a PCR-generated DNA fragment containing the tup1-Δ1::TRP1 allele of strain RTY148 (provided by R. Trumbly) (93). The primers were TUP1F2 (5′-TTCAGCTCCTTGACTTGTGC-3′) and TUP1R (5′-GAAACACAGGAAAAGGAGGG-3′). A Trp+ transformant of the diploid strain LP112, which had been confirmed by PCR analysis to contain the tup1-Δ1::TRP1 allele, was sporulated, and Y170 was derived from a MATa Trp+ spore whose progeny had a slow-growth and flocculent phenotype (93). Y169 is the corresponding MATα tup1-Δ1::TRP1 strain.
KRY302, a MATα nrg1::kanMX4 strain, was obtained by transformation of W303-1B with a PCR-generated fragment containing the nrg1Δ::kanMX4 allele from the S288c deletion array strain. The rim101Δ::natMX4 allele in strain KRY308 (MATa rim101Δ::natMX4) was generated by PCR with a derivative of pAG25 as template and F1 and R1 primers with RIM101-specific sequence upstream of the start codon and downstream of the stop codon, respectively (34, 53). KRY318, a MATa rim101Δ::natMX4 nrg1Δ::kanMX4 strain, was derived as a haploid progenitor obtained by mating KRY302 with KRY308.
Strains from the Saccharomyces cerevisiae deletion collection were obtained from Brenda Andrews (Department of Medical Genetics and Microbiology, University of Toronto) and are derivatives of the S288c strain BY4741 (MATa LYS2 ura3Δ his3Δ leu2Δ met15Δo). In some experiments, BY4742 (MATα lys2Δ ura3Δ his3Δ leu2Δ; obtained from Brenda Andrews) was used as the wild-type strain. These strains contain a Ty insertion in the HAP1 gene that results in reduced activity of the CYC1 UAS (30). We found, however, that a colony overlay assay (see below) provided sufficient sensitivity to monitor expression of a CYC1-lacZ reporter gene. The presence of the designated deletion allele and the absence of the corresponding wild-type allele were confirmed for strains from the deletion collection containing the following alleles: nrg1Δ::kanMX4, rim101Δ::kanMX4, dfg16Δ::kanMX4, ygr122wΔ::kanMX4, vps27Δ::kanMX4, vps23Δ::kanMX4, vps28Δ::kanMX4, vps37Δ::kanMX4, vps22Δ::kanMX4, vps25Δ::kanMX4, vps36Δ::kanMX4, snf7Δ::kanMX4, vps20Δ::kanMX4, vps2Δ::kanMX4, vps24Δ::kanMX4, vps4Δ::kanMX4, doa4Δ::kanMX4, bro1Δ::kanMX4, and rim13Δ::kanMX4.
KRY111, a BY4741-derived strain that contains the RIM101(1-531).3HA-HIS3MX6 allele, was constructed by transformation of BY4741 with a PCR product generated with plasmid pFA6-3HA-kanMX6 as template and an F2 primer with RIM101-specific sequence ending at codon 531 and an R1 primer with gene-specific sequence downstream of the RIM101 stop codon (53). KRY114, a MATα strain containing the RIM101(1-531).3HA-HIS3MX6 allele, was obtained from a haploid spore segregant of a diploid obtained by mating KRY111 and BY4742. xxx::kanMX4 strains containing the RIM101(1-531).3HA-HIS3MX6 allele were isolated as haploid spore segregants of diploid strains obtained by mating KRY114 and the corresponding MATa xxx::kanMX4 strain from the deletion array. Strains expressing a full-length version of RIM101 tagged with three HA epitopes after codon 473 were constructed by transforming the appropriate MATa xxx::kanMX4 strain from the deletion array with a 7.3-kb PstI-NcoI fragment of plasmid pKR41 (see above) and selecting Leu+ transformants. The genotype of all strains was confirmed by PCR at both the RIM101 locus and the appropriate kanMX4 loci.
β-Galactosidase assays.
Liquid β-galactosidase assays were carried out as described previously (39), with some minor changes. Modifications included washing cells of flocculent strains such as Y169 with a solution containing 20 mM Tris-HCl (pH 7.5) and 10 mM EDTA. For some experiments, cells were broken by vortexing in the presence of glass beads with an Eppendorf shaker (model 5432) for 15 min at 4°C, and for some experiments, the cell lysates were clarified by centrifugation for 5 min at 10,000 × g at 4°C. β-Galactosidase activity is expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside (ONPG) cleaved per minute per milligram of protein at 28°C.
A colony overlay assay was used as a qualitative measure of reporter gene activity. For this assay, colonies that had grown on agar-containing medium were overlaid with a solution containing 0.5 M potassium phosphate (pH 7.0), 0.1% sodium dodecyl sulfate (SDS), 6% dimethyl formamide, 0.5% agar, and 0.3 to 0.5 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. After the agar had solidified, the plates were incubated at 30°C. The images of the plates are scans acquired with an Espon Perfection 610 scanner and transferred into Adobe Photoshop.
Cloning and identification of FRDX genes.
Cloning of FRD1, FRD2, FRD4, and FRD5 was performed essentially as described previously (28). In brief, each of the mutant strains Yfrd1-1, Yfrd2-1, Yfrd4-1, and Yfrd5-1 (28) containing the pLG+NRE76 reporter plasmid was transformed with a p366-based (CEN4 ARS1) yeast genomic library (American Type Culture Collection; a gift of Neil Macpherson and B. Andrews; [described in reference 71]). Colonies derived from ∼2 × 104 to 3 × 104 transformants were overlaid with X-Gal-containing agar, and cells were recovered from those colonies that remained white after 18 h of incubation at 30°C. Plasmid DNA was then recovered from these strains in MC1066, a leuB strain of Escherichia coli (16), and reintroduced into the mutant strains carrying pLG+NRE76. The region spanned by the inserts of plasmids that maintained repression of the reporter gene was deduced by comparison of sequence obtained at the vector-insert junctions with the Saccharomyces Genome Database. The open reading frame (ORF) within each insert that was responsible for complementation was then identified by a combination of Tn1000 (γδ) transposon-mediated mutagenesis and subcloning procedures.
Screen of yeast deletion array for strains defective in NRE-mediated repression.
A variation of the synthetic genetic array method (84) was used to identify mutant strains that were defective in NRE-mediated repression. A MATα strain containing either the plasmid-borne CYC1-NRE42-lacZ reporter gene or the CYC1-3×NRE25-lacZ reporter gene was mated with each strain in the gene deletion array by robotic pinning. The resulting diploids were carried through appropriate sporulation and selection steps as described previously (84) to isolate haploid progeny containing both the reporter gene and a deletion allele. These strains were then tested by an X-Gal colony overlay assay for their ability to mediate NRE-dependent repression of the lacZ reporter gene. The single screen of the deletion array with the CYC1-NRE42-lacZ reporter gene identified 293 strains as potentially having a defect in NRE-mediated repression; the two screens with the CYC1-3×NRE25-lacZ reporter gene identified 879 mutant strains. Of these potential positives, 58 strains were selected as candidates for retesting. These included the 11 strains that were identified in all three screens and an additional 14 strains that were identified in both screens with the CYC1-3×NRE25-lacZ reporter gene. The remaining 33 strains were chosen based on the phenotype of the mutant strain or because the deletion was in a gene that encoded a known or predicted transcription factor or had an appropriate expression pattern as listed in the Saccharomyces Genome Database. Upon retesting, the strain with a deletion of NRG1 was the only strain to show a significant loss of repression of the CYC1-3×NRE25-lacZ reporter; strains with a deletion of DGF16 or YGR122w were the best candidates for having defects in repression of the CYC1-NRE42-lacZ reporter gene.
EMSA.
Proteins for use in electrophoretic mobility shift assays (EMSAs) were either synthesized in vitro or expressed in E. coli. Bacterial expression of Rim101(1-289), MBP, and MBP-Nrg1 was carried out essentially as described previously (73). In brief, pET21a-RIM101(1-289), pMal-c2, and pMal-c2-Nrg1 were transformed into the Escherichia coli strain BL21(DE3), and 1 ml of a culture grown overnight was used to inoculate 100 ml of LBA medium (1% NaCl, 1% tryptone, 0.5% yeast extract with 100 μg of ampicillin per ml) containing 0.05 mM ZnSO4 and also supplemented with 2% glucose for the pMAL-c2- and pMAL-c2-Nrg1-containing cells. Mid-log-phase cells were incubated for 4 h at 37° after the addition of isopropyl-β-d-thiogalactopyranoside to 1 mM. The cells were then harvested and resuspended in 4 ml of buffer A (20 mM HEPES, pH 7.4, 5 mM MgCl2, 0.05 mM ZnSO4, 10% glycerol, 250 mM NaCl, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride) plus protease inhibitors (Mini-complete minus EDTA; supplied by Roche and used as suggested by the manufacturer). Cells were broken by sonication, and the soluble fraction was used to provide Rim101(1-289). MBP and MBP-Nrg1 were affinity purified from the soluble fraction by batch absorption to 12 ml of an amylose-Sepharose slurry for 2 h at 4°. The resin was then packed into a column, and the bound protein was eluted with buffer containing 50 mM maltose. Aliquots of the eluate fractions were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) analysis to identify the MBP- and MPB-Nrg1-containing fractions. Rim101(1-289) was also synthesized in vitro with the use of the TnT coupled transcription-translation system (Promega) programmed with pET21a-RIM101(1-289) DNA as a template for transcription.
The standard 20-μl reaction mixture for EMSAs contained 10 mM HEPES (pH 7.9), 125 mM NaCl, 2.5 mM MgCl2, 25 μM ZnSO4, 5% glycerol, 2 μg of bovine serum albumin, 2 μg of poly(dI-dC) · poly(dI-dC), and 4 μg of salmon sperm DNA. The indicated radiolabeled DNA, nonlabeled competitor DNAs, and protein samples were added and the reaction mixtures were incubated for 10 min at room temperature before being applied to a 6% polyacrylamide gel. Probes were made by annealing complementary oligonucleotides that generated a 5′ overhang(s) and filling in the resulting 5′ overhang(s) with the Klenow form of DNA polymerase in the presence of [α-32P]dCTP. The 5′ overhangs of annealed competitor DNAs were filled in with cold XTPs.
MBP-Nrg1 affinity purification of Rim101(1-531).3HA from yeast lysates.
Cells of strain KRY114, which contains the RIM101(1-531).3HA allele, were grown in 250 ml yeast extract-peptone-dextrose medium to mid-log phase, harvested, and resuspended in 1 ml lysis buffer (30 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol, 0.1% NP-40, and 1 mM phenylmethylsulfonyl fluoride plus protease inhibitors). Cells were broken open by vortexing in the presence of 300 μl of glass beads for eight 1-min bursts separated by 1 min on ice. Samples were spun at 4°C for 20 min at 23,500 × g, and the supernatant was transferred to a new tube and respun for an additional 20 min. Protein concentration was determined using the Bradford assay (13) with bovine serum albumin as the standard. Extracts were stored at −70°C.
Preparation of bacterial lysates containing MBP and MBP-Nrg1 and batch binding of these proteins to amylose-Sepharose were carried out as described above for purification of proteins for use in EMSA reactions. The resin was then packed into columns, and the columns were washed with 15 ml of buffer A and then equilibrated with 15 ml of lysis buffer. KRY114-derived extract (see above) containing 5 mg of protein was loaded over each column in a total volume of 10 ml of lysis buffer. The columns were then washed with 15 ml of lysis buffer, and bound protein was eluted with 15 ml lysis buffer containing 50 mM maltose. SDS-PAGE was used to identify MBP- and MBP-Nrg1-containing fractions. Aliquots of these fractions were again subjected to SDS-PAGE and transferred to nitrocellulose for Western blotting with a horseradish peroxidase (HRP)-conjugated anti-HA monoclonal antibody (Santa Cruz) as described previously (72). An ECL chemiluminescence system (New England Biolabs) was used for detection. The filters were then stripped and reprobed with anti-MBP (New England Biolabs) as the primary antibody and an HRP-conjugated secondary antibody.
Lysate preparation and Western blot analysis to monitor in vivo processing of Rim101.HA3.
Strains containing the RIM101.HA3::LEU2 allele, which encodes a full-length, internally HA-tagged Rim101 protein, were grown in yeast extract-peptone-dextrose medium to mid-log phase. Cells were fixed by adding 1 ml ice-cold 100% trichloroacetic acid to 9 ml of culture. Following a 20-min incubation on ice, the cells were pelleted, washed with 1 ml ice-cold 1 M Tris-HCl, pH 8.0, and resuspended in 50 μl 2× SDS-PAGE sample buffer. The samples were vortexed in the presence of 50 μl glass beads for six 30-second bursts interrupted by chilling on ice. An additional 50 μl 2× sample buffer was added to the samples after they had been boiled for 5 min, and the samples were again vortexed for two 15-s bursts. After a brief centrifugation of these cell lysates, the protein in 4- to 10-μl aliquots of the supernatants was fractionated on a 9-cm-long, 8% SDS-polyacrylamide gel and transferred to nitrocellulose. Western blotting was performed with an HRP-conjugated anti-HA monoclonal antibody as described above.
RESULTS
FRD5 is identical to RIM101.
We previously carried out a classic genetic screen to identify genes that are required for NREDIT-mediated mitotic repression. This study defined five function in repression of DIT (FRD) complementation groups, referred to as FRD1 through FRD5, that appeared to contribute specifically to NREDIT-mediated repression of a heterologous reporter gene (28). We identified FRD3 as SPE3, the gene encoding spermidine synthase, and showed that spermidine contributes to NREDIT-mediated repression (28). With the goal of identifying a direct regulator of NREDIT-mediated repression, we have recovered candidate genes for FRD1, FRD2, FRD4, and FRD5 by following the same approach (28). Briefly, we first introduced the reporter plasmid pLG+NRE76 into cells of strains containing the frd1-1, frd2-1, frd4-1, or frd5-1 allele. This plasmid contains a CYC1-NREDIT-lacZ reporter gene which has the 76-bp NREDIT sequence inserted between the UASs and the TATA box of the parental CYC1-lacZ gene (27). Thus, lacZ is not expressed in wild-type FRDX strains due to NRE-mediated repression but is expressed in mutant frdX strains. We then transformed each mutant strain with a CEN-based yeast genomic library and screened for restoration of repression of the reporter gene by an X-Gal overlay assay of colonies (see Materials and Methods). Complementing plasmids were recovered, and the sequences at the junctions of the genomic inserts were compared with the Saccharomyces Genome Database to identify the cloned region that potentially contained the wild-type FRDX gene. To determine which of the several ORFs within each insert was responsible for complementation, we used a combination of transposon-mediated mutagenesis and subcloning procedures (see Materials and Methods). This led to the tentative identification of FRD1 as DFG16, FRD2 as the ORF YGR122w, FRD4 as VPS36, and FRD5 as RIM101. DFG16, which encodes a predicted 619-residue protein with multiple putative transmembrane domains, is implicated in cell wall assembly (56) and filamentous growth (59). YGR122w encodes a predicted 412-residue protein with no known homolog. Vps36 is a component of the ESCRT II endosomal membrane-associated complex that is involved in trafficking of proteins to the vacuole (4). Rim101 is the S. cerevisiae homolog of PacC, which has been extensively characterized in Aspergillus nidulans as a DNA-binding transcriptional activator and repressor of genes induced under alkaline and acidic conditions, respectively (reviewed in reference 66). Frd5/Rim101 appeared to be the best candidate among the FRDX-encoded products to be a direct regulator of DIT gene expression. Indeed, inspection of NREDIT for the sequence 5′-TGCCAAGA-3′, which has been shown to be a high-affinity recognition sequence for PacC (25, 83), revealed an identical sequence, which we refer to as PacCDIT, extending from nucleotide (nt) −484 to nt −477 within NREDIT (Fig. 1A; +1 is the start site of transcription of the DIT1 gene).
FIG. 1.
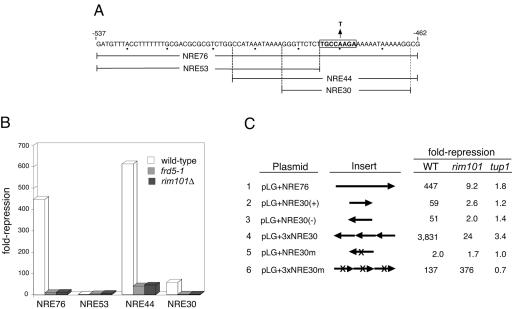
A RIM101-dependent operator element within NREDIT. (A) The sequence of the 76-bp NREDIT, from nt −537 to −462 upstream of the DIT1 gene, is given in the top line and denoted as NRE76. The PacCDIT sequence is boxed and annotated to show the A-to-T mutation at nt −480. The sequences spanned by NRE53, NRE44, and NRE30 are also shown. (B) Strains W303-1A (wild type, open bars), Y108 (frd5-1, gray bars), and Y104 (rim101Δ, black bars) were separately transformed with pLG312, pLG+NRE76, pLG+NRE53, and pLG+NRE30(+). Repression (n-fold) of gene expression is given as the ratio of β-galactosidase activity measured in crude extracts of cells in which the plasmid-borne CYC1-lacZ reporter gene had no insert to the activity measured in cells from the same strain in which the plasmid-borne CYC1-lacZ reporter gene contained the indicated NRE site inserted between the UASCYC1 and the TATA box (see Materials and Methods). (C) Strains W303-1A (wild type [WT]), Y104 (rim101Δ), and Y170 (tup1Δ) were separately transformed with each of the plasmids indicated in the first column and pLG312. The arrowheads in the schematic of the inserts given in the second column denote the orientation of the inserts in the plasmid-borne CYC1-lacZ reporter genes relative to the DIT1 gene. NRE30m and 3×NRE30m are identical to NRE30(−) and 3×NRE30, respectively, with the exception that the A residue at nt −480 has been mutated to a T residue and the orientation of the inserts is reversed in 3×NRE30m relative to 3×NRE30. Repression (n-fold) given in the last three columns refers to the ratio of β-galactosidase activity measured in cells containing pLG312 to the activity measured in cells of the same strain containing the plasmid-borne CYC1-lacZ reporter with the indicated insert. The activities reported in this figure and in Fig. 2 are the average activities obtained from three or more independent cultures analyzed at the same time. Each experiment was repeated from one to five times; although absolute values varied between experiments, the relative levels of β-galactosidase activities were similar.
To confirm that RIM101 was indeed FRD5 and not a low-copy suppressor, we constructed and sporulated a diploid MATa/MATα rim101Δ::URA3/frd5-1 ura3/ura3 strain. After testing the spore progeny of tetrads to confirm that the Ura phenotype segregated 2 Ura+:2 Ura−, we introduced pLG+NRE76 into the Ura− progeny and assayed for expression of β-galactosidase by a colony overlay assay. In all but 1 of 20 tetrads analyzed, the two Ura− spores gave colonies in which expression of the CYC1-NREDIT-lacZ reporter gene was derepressed. On the basis of this low frequency of Frd+ Ura− progeny, we concluded that frd5-1 was an allele of RIM101.
Potential target site for Rim101 within NRE30.
We next compared the repression activity of various segments of NREDIT in wild-type, frd5-1, and rim101Δ::ura3 strains. These sequences included NRE76, NRE53, NRE44, and NRE30 (Fig. 1A) (27, 28). The putative Rim101 target site, PacCDIT, is present in NRE76, NRE44, and NRE30 but not in NRE53. As observed previously, NRE76 and NRE44 directed potent repression when inserted into the CYC1-lacZ reporter gene, NRE30 served as a modest operator, and NRE53 was ineffective in repression (27, 28) (Fig. 1B). NRE76- and NRE30-mediated repression was reduced to almost background levels in strains containing either the frd5-1 or the rim101Δ::ura3 allele of RIM101, whereas NRE44 retained a low level of repression in the mutant strains (Fig. 1B).
We found that Rim101-dependent repression mediated by NRE30 was independent of the orientation of the insert, was TUP1-dependent, and was dramatically enhanced by multimerization (Fig. 1C, lines 2 to 4). A single copy of NRE30 led to ∼60-fold repression, whereas a trimer of NRE30 (3×NRE30) directed ∼4,000-fold repression. NRE30- and 3×NRE30-mediated repression was sensitive to mutation of an A residue to a T residue at nt −480 within the PACDIT site (Fig. 1C, lines 5 and 6). This mutation was previously shown to abolish in vitro binding of Aspergillus PacC to its target site (25, 83).
Curiously, we found that both 3×NRE30 and 3×NRE30m mediated significant TUP1-dependent repression, 24- and 376-fold, respectively, in a rim101Δ strain (Fig. 1C, lines 4 and 6). This observation led us to hypothesize that there was a distinct Tup1-dependent operator site adjacent to, or partially overlapping, the PacCDIT site that recruited an unidentified repressor, which we refer to as factor X (see below). As a possible explanation for the observation that the level of repression mediated by 3×NRE30m was ∼3-fold higher in the rim101Δ strain than in the wild-type strain, we speculated that mutation of the PacCDIT site interfered with repression mediated by the putative Rim101-factor-X heterodimer in the wild-type strain. This effect would be obviated in the rim101Δ strain. It is also possible that the expression of factor X is enhanced on deletion of RIM101. We cannot readily account for the surprisingly high level of repression mediated by 3×NRE30m relative to 3×NRE30 in the rim101Δ strain; it is possible, however, that the mutation has uncovered a cryptic binding site for an additional repressor.
NRE42 is bipartite.
To further explore the notion that NREDIT contained tandem operator elements, we tested a series of overlapping fragments for their ability to reduce expression of the CYC1-lacZ reporter gene (data not shown). None of the fragments that we tested provided significant operator function when present in single copy; however, multiple copies of the fragment termed NRE25, which extended from nt −505 to nt −481, or the fragment termed NRE22D, which extended from nt −486 to nt −464 and encompassed the PacCDIT site, gave significant repression (Fig. 2A and B). The observation that multimerization of operator sites enhanced repression suggested that binding of proteins to the sites was cooperative or that factors recruited to the sites acted cooperatively to mediate repression.
FIG. 2.
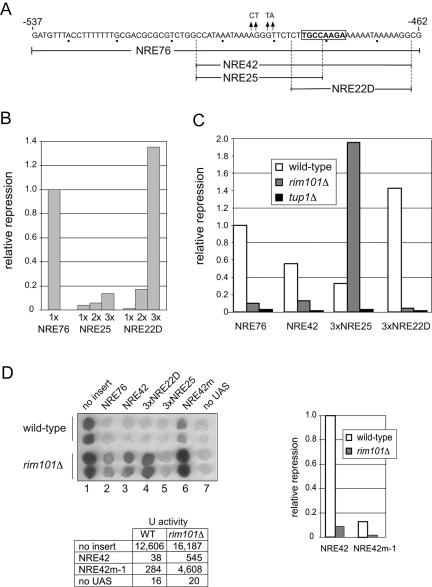
A RIM101-independent operator site within the bipartite NREDIT. (A) The sequence of NRE76 is given in the top line with the PacCDIT site boxed, and the four base-pair mutations present in NRE42m are indicated by arrows. The sequences spanned by NRE42 (nt −505 to nt −464), NRE25 (nt −505 to nt −481), and NRE22D (nt −486 to nt −464) are shown. (B) Comparison of expression levels of plasmid-borne CYC1-NRE-lacZ reporter genes in wild-type cells. Repression (n-fold) of gene expression was measured as the ratio of β-galactosidase activity in cells in which the plasmid-borne CYC1-lacZ reporter gene had no insert to the activity in cells from the same strain in which the plasmid-borne CYC1-lacZ reporter gene contained NRE76, 1×NRE25, 2×NRE25, 3×NRE25, 1×NRE22D, 2×NRE22D, or 3×NRE22D inserted between the UASCYC1 and the TATA box. Relative repression is the ratio of the repression (fold) mediated by the indicated insert normalized to the repression (fold) mediated by NRE76. (C) Comparison of expression levels of plasmid-borne CYC1-NRE-lacZ reporter genes in wild-type (W303-1A; open bars), rim101Δ (Y104; gray bars), and tup1Δ (Y170; black bars) cells. Relative repression values were obtained as described above for panel B. (D) Introduction of mutations upstream of the PacCDIT site reduces NRE42-mediated repression in wild-type and rim101Δ cells. Colonies derived from wild-type cells and rim101Δ cells containing pLG312 (no insert), pLG+NRE76, pLGn+NRE42, pLGn+3×NRE25, pLGn+3×NRE22D, pLGn+NRE42m, or pLGΔSS (no UAS) were overlaid with X-Gal-containing agar (upper left panel). The image is a scan of the colonies after incubation at 30°C. The table gives units of β-galactosidase activity in wild-type (WT) and rim101Δ cells containing pLG312 (no insert), pLGn+NRE42, pLGn+NRE42m-1, or pLGΔSS (no UAS). Relative repression values calculated from these data as described above are given in the bar graph.
We next compared the effect of NRE42, which spans the sequence represented by NRE25 and NRE22D (Fig. 2A), 3×NRE25, and 3×NRE22D on expression of the CYC1-lacZ reporter gene in wild-type, rim101Δ, and tup1Δ strains. Repression mediated by NRE42, 3×NRE25, and 3×NRE22D was Tup1 dependent, and repression mediated by NRE42 and 3×NRE22D, which contained the PacCDIT site, was in part or almost fully Rim101 dependent, respectively (Fig. 2C). In contrast, repression mediated by 3×NRE25 was significantly enhanced in the rim101Δ strain (Fig. 2C), an observation reminiscent of our previous finding that 3×NRE30m-mediated repression was higher in the rim101Δ strain than in the wild-type strain (Fig. 1C).
These data suggested that NREDIT contained at least two distinct operator sites, one within the sequence spanned by NRE25 and one within the sequence spanned by NRE22D. As a test for the presence of an operator site within NRE42 that was distinct from the PacCDIT site, we constructed a CYC1-NRE42m-1-lacZ reporter gene that contained four base-pair mutations in the region of NRE42 specific to the NRE25 sequence (Fig. 2A). As a qualitative test for the extent of repression, we used an X-Gal colony overlay assay. Colonies of control wild-type cells containing the CYC1-lacZ reporter gene became dark blue after being overlaid with X-Gal-containing agar (Fig. 2D). In contrast, colonies of wild-type cells that contained NRE76, NRE42, 3×NRE22D, or 3×NRE25 inserted into the reporter gene remained white or became only pale blue, and cells that contained the CYC1-NRE42m-1-lacZ reporter gene became light blue (Fig. 2D). As expected, mutation of RIM101 affected repression mediated by 3×NRE22D but not repression mediated by 3×NRE25. The modest level of repression that was maintained by NRE42 in the rim101Δ strain was lost on the introduction of mutations into the NRE25-specific region (Fig. 2D). To confirm that the visual impression obtained on inspection of colonies that had been overlaid with X-Gal-containing agar was representative of in vivo activities of the reporter gene, we also measured β-galactosidase activity in lysates of control cells and cells containing NRE42 and NRE42m-1 inserted into the reporter gene. β-Galactosidase activities in extracts from cells containing the plasmid-borne CYC1-lacZ reporter gene with no insert or with NRE42 or NRE42m-1 inserted into the reporter gene and a reporter gene lacking a UAS showed a good correlation with the qualitative overlay assays (Fig. 2D). Overall, these data were consistent with the idea that NRE42 contained a RIM101-dependent operator site within the NRE22D-specific region and a RIM101-independent, factor X-dependent operator site within the NRE25-specific region.
Rim101 binds to NRE30 in vitro and in vivo.
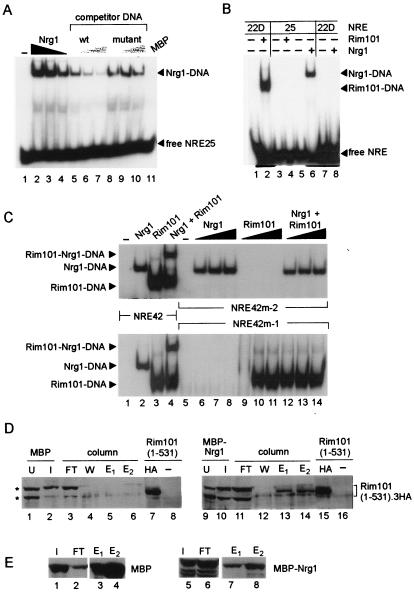
We next tested the notion that Rim101 binds directly to the PacCDIT site. First, we monitored the ability of an in vitro-synthesized polypeptide that spanned residues 1 to 289 of Rim101, which includes the zinc fingers, to bind to the PacCDIT site. As assessed by an EMSA, the Rim101(1-289) polypeptide formed a complex with a radioactively labeled double-stranded oligonucleotide containing the sequence represented by NRE22D (Fig. 3, lane 2). Formation of this protein-DNA complex was reduced on the addition of an increasing amount of the unlabeled double-stranded NRE22D oligonucleotide (Fig. 3, lanes 3 to 6) but not on addition of a mutated version of this double-stranded oligonucleotide (Fig. 3, lanes 7 to 10).
FIG. 3.
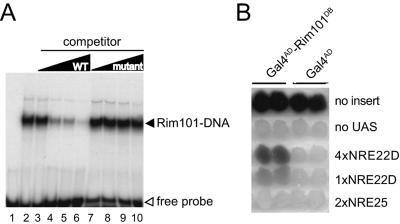
Rim101 binds to the PacCDIT site in vitro and in vivo. (A) A truncated version of Rim101 extending from residue 1 to residue 289 was synthesized in vitro and tested for its ability to bind to a radiolabeled NRE22D-containing oligonucleotide by EMSA as described in Materials and Methods. An equivalent amount of Rim101(1-289) protein was incubated with a radiolabeled NRE22D-containing double-stranded oligonucleotide (5′-GATCCTTGCCAAGAAAAAATAAAAAGGATC-3′ [the PacCDIT site is shown in boldface type]) in the absence of competitor DNA (lane 2) and in the presence of a 10-, 50-, 100-, or 250-fold excess of nonlabeled NRE22D-containing double-stranded oligonucleotide (lanes 3 to 6) or a nonlabeled mutant version of this double-stranded oligonucleotide (lanes 7 to 10) as specific and nonspecific competitors, respectively. The mutant oligonucleotide had a 1-bp change within the PacCDIT site, changing the sequence from 5′-TGCCAAGA-3′ to 5′-TGCCTAGA-3′. The reaction of lane 1 contained only the probe DNA. WT, wild type. (B) A Gal4AD-Rim101(1-289) fusion protein expressed in vivo activates an NRE22D-containing reporter gene. W303-1B was transformed with pACTII-Rim101(1-289) (Gal4AD-Rim101DB) or pACTII (Gal4AD), as indicated at the top of the panel. As indicated on the right of the panel, these transformants were then transformed with pLG312 (no insert); pLGΔSS (no UAS); pLGΔSS+4×NRE22D, which contains a 4×NRE22D-lacZ gene reporter gene; pLGΔSS+1×NRE22D, which contains a 1×NRE22D-lacZ reporter gene; or pLGΔSS+2×NRE25, which contains a 2×NRE25-lacZ reporter gene. Colonies representing two distinct transformants of each strain were grown up and overlaid with X-Gal-containing agar to monitor β-galactosidase expression.
We attempted to use a combined in vivo chromatin cross-linking in vitro immunoprecipitation approach to test for in vivo binding of HA-tagged Rim101 to the intergenic region of the DIT1-DIT2 genes. However, we discovered that we could readily detect DIT1-containing DNA in anti-HA immunoprecipitates from lysates of control cells that expressed Rim101-HA but that had not been treated with a cross-linker (data not shown). As we could not exclude the possibility that the Rim101-DNA association occurred in vitro after lysis of the cells, we turned to a different assay to detect in vivo DNA binding. We constructed a chimeric gene that expressed the activation domain of Gal4 fused to the DNA-binding region of Rim101 and tested the ability of this protein to activate expression of a lacZ reporter gene that lacked a UAS but contained the PacCDIT operator site. As assessed by an X-Gal overlay assay of colonies, the chimeric Gal4AD-Rim101(1-289) fusion protein directed significant expression of a 4×NRE22D-lacZ reporter gene and a lower level of expression of a 1×NRE22D-lacZ reporter gene (Fig. 3B). The fusion protein, however, did not promote expression of a 2×NRE25-lacZ reporter gene (Fig. 3B). We concluded that the zinc finger region of Rim101 directed binding of the Gal4AD-Rim101(1-289) fusion protein to the NRE22D sequence, but not to the NRE25 sequence, in vivo.
NRE22D-mediated repression requires the RIM signaling pathway.
The signal transduction pathway that regulates the activity of PacC/Rim101 in response to environmental pH has been extensively characterized in fungi and yeast (reviewed in references 65 and 66). In S. cerevisiae, this pathway consists of two transmembrane proteins, Rim9 and Rim21, which have been postulated to be pH sensors; Rim8 of unknown function; and Rim20, which may act as an adaptor between the protease, Rim13, and its substrate, Rim101 (96). Mutation of any of the RIM genes results in a shared set of phenotypes including reduced expression of IME1, reduced sporulation efficiency, cold sensitivity, defects in haploid invasive growth and growth at alkaline pH, and failure to proteolytically process Rim101 (29, 52, 79, 85, 96). We found that RIM8, RIM9, RIM13, RIM20, and RIM21 were all required for repression of the CYC1-3×NRE22D-lacZ reporter gene (Fig. 4A), consistent with the notion that it is the Rim13-processed form of Rim101 that serves as a negative regulator at NRE22D.
FIG. 4.
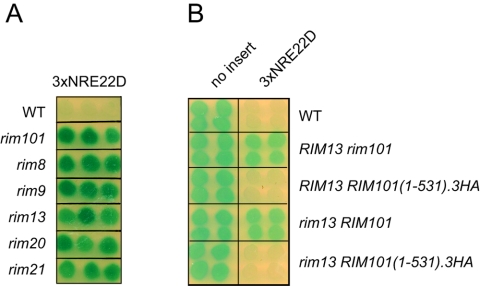
Rim101-dependent, 3×NRE22D-mediated repression requires the RIM signaling pathway. (A) The wild-type (WT) strain or the indicated rim deletion strains from the yeast deletion collection, as denoted on the left-hand side of the panel, were transformed with pLGn+3×NRE22D, and β-galactosidase expression was monitored by a colony overlay assay. (B) Expression of Rim101(1-531)0.3HA restores NRE22D-mediated repression to a rim13 strain. Wild-type, rim101Δ, RIM101(1-531).3HA-HIS3MX6, rim13Δ, and rim13Δ RIM101(1-531)0.3HA-HIS3MX6 strains, as denoted on the right-hand side of the panel, were transformed with pLG312n (no insert) or pLGn+3×NRE22D, and β-galactosidase expression was monitored by a colony overlay assay.
Although the exact site of Rim13-directed cleavage in Rim101 is not known, expression of a truncated version of Rim101 that extends to residue 531 bypasses the requirement for the RIM signaling pathway to generate active Rim101 (52). We found that a rim13 strain in which the wild-type RIM101 gene had been replaced with a RIM101(1-531).3HA allele recovered the ability to repress the CYC1-3×NRE22D-lacZ reporter gene (Fig. 4B). We therefore concluded that proteolytic cleavage of Rim101 under the control of the RIM signaling pathway is required for repression through the multimerized NRE22D element.
A screen of the array of yeast strains with deletions of nonessential genes identifies NRG1 as a contributor to NRE25-mediated repression.
To identify additional genes that might contribute to NREDIT-mediated repression, particularly, candidate genes for mediating repression through the RIM101-independent NRE25 subsite, we screened the array of viable yeast deletion strains (32). The array was interrogated once for genes that contribute to repression through NRE42 and twice for genes that contribute to repression through NRE25 by using a variation of the synthetic genetic array method (84) as follows. A MATα strain containing either the plasmid-borne CYC1-NRE42-lacZ reporter gene or the CYC1-3×NRE25-lacZ reporter gene was mated with each strain in the gene deletion array (see Materials and Methods). The resultant diploids were sporulated, and haploid progeny that contained the plasmid-borne reporter gene were selected. This new array of plasmid-containing haploid deletion strains was then tested by an X-Gal colony overlay assay for strains that were defective in repression of the lacZ reporter gene. This led to the identification of NRG1 as a potential contributor to NRE25-mediated repression and reidentified DFG16 and YGR122w as contributors to NRE42-mediated repression (see Materials and Methods).
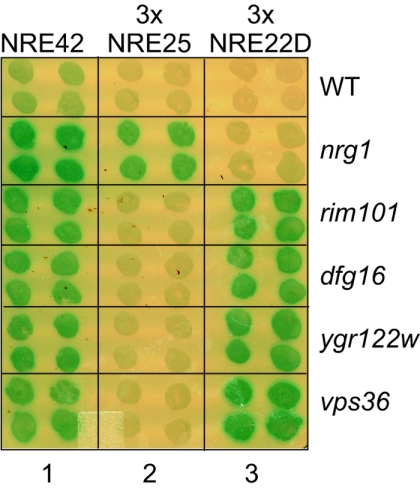
We next compared the effect of mutation of the genes that we had identified in this screen and our previous screen (28) for their roles in NRE42-, 3×NRE22D-, and 3×NRE25-mediated repression. As expected, efficient repression of the CYC1-NRE42-lacZ reporter gene required all five genes: NRG1, RIM101/FRD5, DFG16/FRD1, YGR122w/FRD2, and VPS36/FRD4 (Fig. 5, column 1). We found that NRG1 contributed specifically to repression of the CYC1-3×NRE25-lacZ reporter gene (Fig. 5, column 2) and that DFG16/FRD1, YGR122w/FRD2, and VPS36/FRD4 were additional contributors to RIM101/FRD5-directed repression of the CYC1-3×NRE22D-lacZ reporter gene (Fig. 5, column 3). None of the mutant strains led to increased expression of a control UAS-less lacZ reporter gene (data not shown). We also tested a strain deleted for NRG2, which encodes an Nrg1-related protein (92); this mutant strain appeared to maintain repression of the CYC1-3×NRE25-lacZ reporter gene (data not shown).
FIG. 5.
NRG1 is required for 3×NRE25- but not 3×NRE22D-mediated repression; RIM101, DFG16, YGR122w, and VPS36 are required for 3×NR22D- but not 3×NRE25-mediated repression. Colonies of the BY4741-derived deletion strains, as indicated on the right side of the panel, that contained pLGn+NRE42, pLGn+3×NRE25, or pLGn+3×NRE22D, as indicated at the top of the panel, were overlaid with X-Gal-containing agar. WT, wild type.
Nrg1 binds to a target site within NRE25.
Nrg1 is a DNA-binding, Tup1-dependent repressor of transcription in both S. cerevisiae and C. albicans (14, 60, 64), making it a good candidate for being a direct mediator of NRE25-dependent repression. Indeed, by comparison with the partially characterized C4T- or C3TC-containing target site for Nrg1 from S. cerevisiae (64) and the better-characterized target sequence (A/C)(A/C/G)C3T for Nrg1 from C. albicans (60), NRE25 contains a potential Nrg1-binding site, AACCCT, between nt −489 and −494.
We tested the ability of a recombinant MBP-Nrg1 fusion protein purified from E. coli to bind to NRE25 in vitro. As assessed by EMSA, MBP-Nrg1 formed a specific complex with a radioactively labeled double-stranded oligonucleotide containing the NRE25 sequence (Fig. 6A). Formation of this protein-DNA complex was competed by the addition of increasing amounts of the unlabeled double-stranded oligonucleotide containing the wild-type NRE25 sequence but not by the addition of increasing amounts of a mutated version of this double-stranded oligonucleotide (Fig. 6A). The observation that an Nrg1-NRE25 protein-DNA complex formed in vitro supported the idea that Nrg1 was a direct mediator of NRE25-directed repression in vivo.
FIG. 6.
Formation of Nrg1-NRE25 and Nrg1-Rim101-NRE42 protein-DNA complexes in vitro as assessed by EMSA and interaction of Nrg1 and Rim101 in vitro as assessed by affinity chromatography. (A) Nrg1 binds to NRE25 in vitro. Bacterially expressed MBP-Nrg1 was affinity purified and incubated with a radiolabeled double-stranded oligonucleotide containing NRE25 (5′-CCATAAATAAAAGGGTTCTCTTGCC-3′) prior to electrophoresis on a nondenaturing 6% polyacrylamide gel. The reactions of lanes 4 to 11 contained equivalent amounts of MBP-Nrg1; the reactions of lanes 5 to 7 contained increasing amounts of a nonlabeled NRE25-containing double-stranded oligonucleotide as specific competitor DNA; and the reactions of lanes 8 to 10 contained increasing amounts of a nonlabeled mutant NRE25-containing double-stranded oligonucleotide as a nonspecific competitor (the wild-type sequence 5′-AGGGT-3′ [indicated in boldface type in the sequence given above] was changed to 5′-CTGTA-3′). The reaction of lane 1 contained no protein, and the reaction of lane 11 contained bacterially expressed MBP. The reactions of lanes 2 and 3 contained 5- and 2.5-fold of the amount of MBP-Nrg1 present in the reaction of lane 4. (B) Rim101(1-289) does not interact with the NRE25 site, and MBP-Nrg1 does not interact with the NRE22D site. An EMSA was performed with bacterially expressed Rim101(1-289) present in the soluble fraction of a crude cell lysate (lanes 2 and 4) or affinity-purified bacterially expressed MBP-Nrg1 (lanes 6 and 8) that had been incubated with a radiolabeled oligonucleotide containing NRE22D (Fig. 3) (lanes 2 and 8) or NRE25 (lanes 4 and 6). The reactions of lanes 1, 3, 5, and 7 contained probe only. (C) An Nrg1-Rim101-DNA complex forms in vitro with NRE42 but not with NRE42m-2 or NRE42m-1. Bacterially expressed MBP-Nrg1 (lanes 2, 4, 6 to 8, and 12 to 14) or bacterially expressed Rim101(1-289) (lanes 3, 4, and 9 to 14) was incubated with a radiolabeled oligonucleotide containing NRE42 (see Materials and Methods) (lanes 1 to 4), NRE42m-2 (5′-CCATAAATAAAAGGGTTCTCTTGCCTAGAAAAAATAAAAAGGCC-3′ [the mutation is in boldface type]) (lanes 5 to 14, upper panel) or NRE42m-1 (see Materials and Methods) (lanes 5 to 14, lower panel). The reactions of lanes 1 and 5 contained no protein. The reactions of lanes 6 to 8 and lanes 12 to 14 (upper panel) contained increasing amounts of MBP-Nrg1; the reactions of lanes 9 to 11 and lanes 12 to 14 (lower panel) contained increasing amounts of Rim101(1-289). Only the portions of the autoradiograms representing the protein-DNA complexes are shown. (D) Interaction between Nrg1 and Rim101 in vitro. Western blot analysis with HRP-conjugated anti-HA antibodies of samples from an affinity chromatography experiment (see Materials and Methods) is shown. A lysate from induced bacterial cells expressing MBP (lanes 2 to 6) or MBP-Nrg1 (lanes 10 to 14) was incubated with amylose-Sepharose to prepare an affinity resin to capture Rim101 from a yeast cell lysate. The following samples were analyzed: an aliquot of uninduced bacterial cells (U, lanes 1 and 9) and an aliquot of the soluble fraction of a lysate prepared from induced bacterial cells (I, lanes 2 and 10) that contained a vector for expression of MBP (lanes 1 and 2) or MBP-Nrg1 (lanes 9 and 10); an aliquot of a flowthrough fraction after the bacterial lysate-resin (amylose-Sepharose) mixture had been loaded into a column (FT, lanes 3 and 11); an aliquot of a wash fraction from the columns after a lysate prepared from yeast cells expressing Rim101(1-531).3HA had been run through the columns (W, lanes 4 and 12); aliquots of resin-bound protein that had been eluted with maltose (E1 and E2, lanes 5, 6, 13, and 14); an aliquot of the lysate prepared from yeast cells expressing Rim101(1-531).3HA (lanes 7 and 15) or Rim101 (lanes 8 and 16). The asterisks on the left denote bacterial proteins that cross-reacted with the anti-HA antibody; the position of Rim101(1-531).3HA is denoted on the right. (E) The filter of panel C was stripped and reprobed with anti-MBP antibodies as the primary antibody and an HRP-conjugated secondary antibody. Lanes 1 through 8 correspond to lanes 2, 3, 5, 6, 10, 11, 13, and 14, respectively, of panel D.
The specificities of subsite binding by Nrg1 and Rim101 were confirmed in an EMSA experiment that demonstrated that the MBP-Nrg1 fusion protein, which formed a protein-DNA complex with the NRE25 probe (Fig. 6B, lane 6), did not form a complex with the NRE22D probe (Fig. 6B, lane 8). Similarly, the Rim101(1-289) polypeptide, which formed a protein-DNA complex with the NRE22D probe (Fig. 6B, lane 2), did not form a protein-DNA complex with the NRE25 probe (Fig. 6B, lane 3).
Nrg1 and Rim101 bind simultaneously to their adjacent target sites within the NRE42 operator.
To confirm that Rim101 and Nrg1 could act as coregulators at the NRE42 operator, we tested these proteins for their ability to bind simultaneously to NRE42 in vitro. Incubation of a radioactively labeled double-stranded oligonucleotide spanning the NRE42 sequence with bacterially expressed MBP-Nrg1 and Rim101(1-289) generated a protein-DNA complex of slower electrophoretic mobility (Fig. 6C, lane 4) than complexes formed with the same NRE42 probe and either MBP-Nrg1 alone (Fig. 6C, lane 2) or Rim101(1-289) alone (Fig. 6C, lane 3). We concluded that Rim101 and Nrg1 could bind simultaneously to their adjacent target sites within the NRE42 operator element.
We also carried out EMSAs with radioactively labeled NRE42 oligonucleotides that contained a mutation within the Rim101 target site (NRE42m-2) (Fig. 6C, lanes 5 to 14, upper panel) or mutations within the Nrg1 target site (NRE42m-1) (Fig. 6C, lanes 5 to 14, lower panel). As expected, MBP-Nrg1 formed a complex with the NRE42m-2 probe but not with the NRE42m-1 probe, and Rim101(1-289) formed a complex with the NRE42m-1 probe but not with NRE42m-2 probe (Fig. 6C, lanes 5 to 11). Incubation of the NRE42m-2 probe with both MBP-Nrg1 and Rim101(1-289) generated an MBP-Nrg1-NRE42m-2 complex only (Fig. 6C, lanes 12 to 14, upper panel). Conversely, incubation of the NRE42m-1 probe with both proteins generated a Rim101-NRE42m-1 complex only (Fig. 6C, lanes 12 to 14, lower panel). This experiment suggested that Rim101 and Nrg1 were being recruited individually to NRE42. We note, however, that because we had found that expression of Rim101(1-531) in bacteria was poor (data not shown), the EMSA cobinding experiments described above were carried with a bacterially expressed version of Rim101 that contained only its DNA-binding amino-terminal portion. It is possible that this truncated version of Rim101 lacked a region required for interaction with Nrg1.
As an alternative test for an interaction between Nrg1 and Rim101, we used a pull-down assay to determine whether bacterially expressed Nrg1 could interact with Rim101(1-531) present in a yeast lysate. MBP and an MBP-Nrg1 fusion protein that had been expressed in E. coli were captured on amylose-Sepharose beads, and these resins were used as affinity ligands. Aliquots of yeast lysates prepared from cells expressing Rim101(1-531).3HA were loaded onto columns that contained immobilized MBP or MBP-Nrg1. After the columns had been washed, bound protein was eluted with maltose-containing buffer and analyzed by Western blot. Rim101(1-531).3HA could be readily detected in the eluate of the column that contained immobilized MBP-Nrg1 (Fig. 6C, lanes 13 and 14) but not in the eluate of the control column that contained immobilized MBP (Fig. 6C, lanes 5 and 6). Thus, it is possible that binding of Rim101 and Nrg1 to their adjacent target sites in NREDIT could be assisted by prior formation of a Rim101-Nrg1 heterodimer.
The extracts of bacterial cells expressing MBP or MBP-Nrg1 contained proteins that cross-reacted with the anti-HA antibody and had mobilities similar to Rim101(1-531).3HA (Fig. 6C, lanes 1 to 3 and 9 to 11); however, these cross-reacting proteins did not bind to the amylose-containing resin (Fig. 6C, lanes 3 and 11). A control blot showed that MBP bound more efficiently to amylose-Sepharose than did MBP-Nrg1 (Fig. 6D, lanes 1, 2, 5, and 6); this accounted for the much larger amount of MBP relative to MBP-Nrg1 that eluted from the columns (Fig. 6D, lanes 3, 4, 7, and 8).
Nrg1 and Rim101 act as corepressors in vivo.
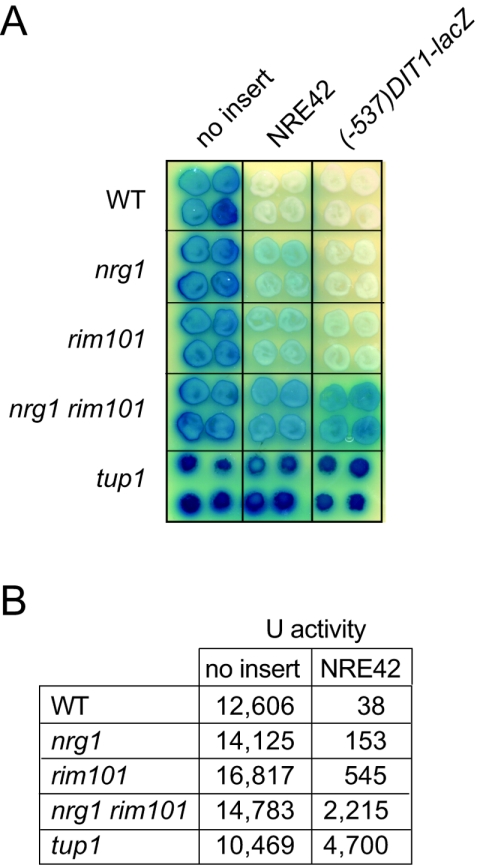
In our initial experiments, we used 3×NRE22D and 3×NRE25 as distinct operator elements to define potential roles for RIM101 and NRG1 in mediating mitotic repression of the DIT1 gene in vivo. To test for the roles of Nrg1 and Rim101 as corepressors at NREDIT, we compared expression of the CYC1-NRE42-lacZ reporter gene and a (−537)DIT1-lacZ translational fusion gene in nrg1Δ, rim101Δ, nrg1Δ rim101Δ, and tup1Δ strains. We have previously shown that mitotic repression of the translational fusion gene (−537)DIT1-lacZ, which contains DIT1 sequence from nt −537 to nt +53, is Tup1 dependent (27) but not necessarily Rim101 dependent (28). As assessed by a qualitative X-Gal colony overlay assay (Fig. 7A) and by measurement of β-galactosidase activity in cell extracts (Fig. 7B), the nrg1Δ and rim101Δ strains maintained partial repression of the CYC1-NRE42-lacZ reporter gene, whereas the nrg1Δ rim101Δ and tup1Δ strains were inefficient at maintaining repression. In contrast, efficient repression of the (−537)DIT1-lacZ reporter gene appeared to be maintained in the absence of either Rim101 or Nrg1 (Fig. 7A), with significant expression occurring only in the nrg1Δ rim101Δ and tup1Δ strains (Fig. 7A). This redundancy between Nrg1- and Rim101-mediated repression in the context of the (−537)DIT1-lacZ reporter gene may explain why Bogengruber and coworkers concluded in a previous study that RIM101 did not contribute to mitotic repression of the DIT genes (11).
FIG. 7.
Expression of the CYC1-NRE42-lacZ reporter gene and the (−537)DIT1-lacZ reporter gene is higher in an nrg1 rim101 strain than in either an nrg1 strain or a rim101 strain. (A) Colonies of wild-type (WT) (W303-B), nrg1Δ (KRY302), rim101Δ (KRY308), and nrg1Δ rim101Δ (KRY318) cells and tup1Δ (Y169) cells containing pLG312n, a plasmid with a CYC1-lacZ reporter gene (no insert); pLGn+NRE42, a plasmid with a CYC1-NRE42-lacZ reporter gene; and p(−537)DIT1-lacZ, a plasmid with a (−537)DIT1-lacZ reporter gene, were overlaid with X-Gal-containing agar. (B) The table gives units of β-galactosidase activity measured in extracts from cells containing pLG312 (no insert) or pLGn+NRE42 (NRE42).
In summary, the experiments of Fig. 6 and 7 have demonstrated that an Nrg1-Rim101-NRE42 ternary complex can assemble in vitro (Fig. 6C) and that both proteins are required to mediate efficient repression in the context of the CYC1-NRE42-lacZ reporter gene in vivo (Fig. 7). These data support a model in which Nrg1 and Rim101 bind to adjacent target sites within NREDIT and act together to efficiently recruit the Ssn6-Tup1 corepressor complex to the DIT1 promoter.
The ESCRT pathway is involved in NRE22D-mediated repression.
YGR122w/FRD2 and VPS36/FRD4 contribute to NRE22D- but not NRE25-mediated repression (Fig. 5). Both these genes have connections with the endosomal sorting complex required for transport (ESCRT), which consists of three multicomponent complexes referred to as ESCRT I, ESCRT II, and ESCRT III (reviewed in reference 70). The ESCRT pathway directs biosynthetic cargo, such as vacuolar hydrolases, from the Golgi to their destination in the vacuole and diverts endocytosed proteins, such as cell surface receptors and transporters, from a recycling pathway to the vacuole for degradation. Although YGR122w is an uncharacterized ORF, its product has been shown to have a two-hybrid interaction with Snf7 (42, 87). SNF7 was initially identified based on its role in glucose regulation of the SUC2 gene (86, 88) and was subsequently found to be identical to VPS32, one of 10 class E vacuolar protein sorting (VPS) genes that function in the ESCRT pathway (5, 46). Snf7/Vps32, which interacts with Vps20 to form the ESCRT IIIA subcomplex, also shows two-hybrid interactions with Rim13 and Rim20 (12, 42, 87, 96). Although Rim20 does not appear to have a role in the ESCRT pathway, Bro1, which is related to Rim20 and also interacts with Snf7/Vps32, functions in this pathway (62). Finally, VPS36/FRD4 is a component of ESCRT II, which recruits ESCRT III to the endosomal membrane (3, 4).
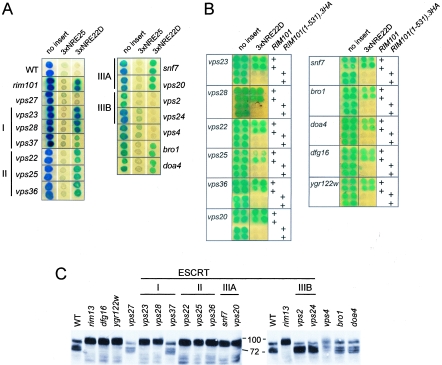
These intriguing connections between Rim101-dependent NRE22D-mediated repression and the ESCRT trafficking pathway led us to test additional components of the ESCRT pathway for a possible role in NREDIT-mediated repression. We introduced our series of reporter genes into various strains taken from the deletion array and monitored expression of the reporter gene by an X-Gal colony overlay assay (Fig. 8A). Mutation of VPS23 and VPS28, two of three genes encoding the components of the ESCRT I complex, led to a significant loss in repression of the plasmid-borne CYC1-3×NRE22D-lacZ reporter gene but not the CYC1-3×NRE25-lacZ reporter gene (Fig. 8A). All three components of the ESCRT II complex (Vps22, Vps25, and Vps36) and the two components of the ESCRT IIIA subcomplex (Snf7/Vps32 and Vps20) were also specifically required for repression of the CYC1-3×NRE22D-lacZ reporter gene (Fig. 8A). In contrast, VPS24 and VPS2, which encode the components of the ESCRT IIIB subcomplex; VPS27, which is required for initiating assembly of the endosomal ESCRT pathway; and VPS4, which is required for disassembly of the ESCRT complexes, were not required for NRE22D-mediated repression (Fig. 8A). Mutation of BRO1 or DOA4, which encodes a deubiquitinating enzyme that removes ubiquitin attached to cargo proteins, led to defective repression of the CYC1-3×NRE22D-lacZ reporter gene (Fig. 8A). Of several other genes that contribute to intracellular trafficking that we tested, none was involved in NRE22D-mediated repression (data not shown). These included, for example, SLA1 and END3, which encode components that contribute to cytoskeleton dynamics and act in a complex with Pan1 as an endocytic targeting adaptor (41, 82); PEP12, a class D VPS gene that encodes a multifunctional syntaxin that is required for all known trafficking pathways into the multivesicular body (MVB) (31); VPS34, which encodes a phosphatidylinositol-3-kinase that is involved in Golgi-to-MVB trafficking (77); FAB1, which encodes a phosphatidylinositol-3-phosphate-5-kinase that is involved in MVB-to-vacuole trafficking (77); and PEP4, which encodes a major vacuolar hydrolyase, proteinase A (89). However, many trafficking proteins have functional counterparts, which would necessitate the analysis of double mutants to reveal their roles. Overall, these results suggest that a portion of the ESCRT pathway has been specifically coopted to regulate Rim101-mediated repression.
FIG. 8.
Involvement of ESCRT components, Dfg16, and Ygr122w in 3×NRE22D-mediated repression and processing of Rim101. (A) Colonies of the BY4741-derived deletion strains, as indicated on the sides of the panels, containing pLG312n (no insert), pLGn+3×NRE25, or pLGn+ 3×NRE22D, as indicated above the panels, were overlaid with X-Gal-containing agar. I, II, IIIA, and IIIB refer to ESCRT complexes. (B) Repression of the CYC1-3×NRE22D-lacZ reporter gene can be restored in the mutant strains by expression of a truncated version of Rim101. For each mutant strain identified on the left of the panel, the top two rows of colonies were from cells that contained the wild-type (WT) RIM101 gene, and the bottom two rows of colonies were from cells that contained the RIM101(1-531).3HA-HIS3MX6 allele, as indicated on the right. The cells also contained pLG312n (no insert) or pLGn+3×NRE22D, as indicated above the panels. For panels A and B, composite images were prepared from scans of plates that had been overlaid with X-Gal-containing agar after color development. Each plate had control colonies consisting of wild-type cells and rim101Δ cells containing pLG312n or pLG+3×NRE22D to ensure equivalent color development. (C) Western blot of an SDS-polyacrylamide gel containing aliquots of crude lysates prepared from the indicated BY4741-derived deletion strains that expressed a full-length internally HA-epitope-tagged Rim101 protein. The blot was probed with HRP-conjugated anti-HA antibodies.
We next tested whether the RIM101(1-531).3HA allele would complement the repression defects observed in dfg16, ygr122w, and the vps mutant strains. Indeed, expression of Rim101(1-531).3HA, whose activity is independent of Rim13-mediated processing, restored repression of the CYC1-3×NRE22D-lacZ reporter gene in the dfg16, ygr122w, vps23, vps28, vps22, vps25, vps36, snf7, vps20, bro1, and doa4 strains (Fig. 8B).
Contribution of the ESCRT pathway to proteolytic processing of Rim101.
To determine whether the components of the ESCRT pathway acted before or after the processing step of Rim101, we monitored the status of epitope-tagged Rim101 in wild-type and mutant cells by Western blot analysis. We first compared the ability of two integrated alleles of RIM101, RIM101.HA2, and RIM101.HA3 to support repression of the CYC1-3×NRE22D-lacZ reporter gene. Rim101.HA2, which contains nine HA epitopes following codon 313, and Rim101.HA3, which contains three HA epitopes following codon 473, are processed and are able to complement some aspects of Rim101 function (50, 52, 96). We found, however, that only Rim101.HA3 could mediate repression of the CYC1-3×NRE22D-lacZ reporter gene (data not shown). This suggested that the HA epitopes present within Rim101.HA2 interfered with the ability of Rim101 to mediate NRE22D-directed repression without affecting some of its other functions.
Visualization of Rim101.HA3 processing by Western blot analysis showed that Rim101 existed as both full-length and processed forms in the wild-type strain and, as expected, in only the full-length form in a rim13 strain (Fig. 8C, lanes 1 and 2). The nonprocessed form of Rim101.HA3 that was present in the rim13 strain appeared to consist of two isoforms of similar size, and the processed Rim101.HA3 that was present in the wild-type strain appeared to be present in three forms (see Discussion). Rim101.HA3 was not processed in the dfg16, ygr122w, vps23, vps28, vps22, vps25, vps36, snf7, and vps20 strains and was processed in the vps27, vps37, vps2, vps24, vps4, bro1, and doa4 strains (Fig. 8C). Thus, the mutant strains that failed to process Rim101.HA3 were defective in 3×NRE22D-mediated repression, and the mutant strains that processed Rim101.HA3 supported repression of the reporter gene, with two exceptions. The exceptional strains, bro1 and doa4, contained processed Rim101.HA3 but were nonetheless defective in 3×NRE22D-mediated repression. Although the dispensability of BRO1 for Rim101 processing has been noted previously (96), this is the first report of its requirement for Rim101-mediated repression. Our data suggest that components of the ESCRT pathway have a novel role, be it directly or indirectly, in promoting Rim101 processing and activity and that they serve this role in the absence of Vps27, ESCRT IIIB, and Vps4.
DISCUSSION
We previously described NREDIT as a promoter element that mediates Ssn6-Tup1-dependent repression of the DIT1 and DIT2 genes during mitotic growth as well as contributing to their activation during sporulation (27). In this study, we have investigated further the mechanism by which NREDIT mediates repression. Our data show that NREDIT consists of two subsites (Fig. 2) and that the C2H2 zinc-finger-containing proteins Rim101 and Nrg1 bind to these adjacent sites and act together to mediate efficient repression (Fig. 3, 6, and 7). We have found that proteolytic processing of Rim101 to its active repressor form not only depends on the components of the well-characterized RIM pathway but also requires Dfg16, a putative membrane-spanning protein implicated in cell wall integrity, Ygr122w, and seven proteins that act in the ESCRT trafficking pathway (Fig. 4, 5, and 8).
Mutation of RIM101 in S. cerevisiae was initially described as leading to poor growth of cells at low temperature, a smooth colony morphology, inefficient sporulation of diploid strains due to reduced expression of IME1, and inability of haploid strains to switch to invasive growth (79). Characterization of the pH response pathway in A. nidulans and the recognition that the components of this pathway are conserved in S. cerevisiae and other fungi implicated Rim101, the homolog of A. nidulans PacC, as a regulator of pH-inducible genes (reviewed in references 65 and 66). Although rim101 strains grow poorly in alkaline media (29, 51), processing of Rim101 does not appear to be as tightly regulated by pH in S. cerevisiae as is the case for PacC in A. nidulans, nor do pH-regulated genes appear to be direct targets of Rim101 (51, 52). In fact, profiles of pH-regulated gene expression and the identification of genes that contribute to growth at alkaline pH have highlighted the role of genes involved in iron and copper homeostasis in tolerance to alkaline pH (17, 51, 74, 75). Copper and iron have reduced solubility at alkaline pH and become limiting for growth (74). Although there is no evidence at this time that genes involved in copper and iron homeostasis or their transcriptional regulators, Mac1 (49) and Atf1 (98), are directly regulated by Rim101 in S. cerevisiae, there is a suggestion that genes involved in the uptake of siderophores in A. nidulans are regulated by both PacC and the iron-responsive regulator SreA (24).
Lamb and Mitchell (50) found that SMP1 and NRG1 are direct targets of Rim101-mediated repression and that deletion of SMP1 restores haploid invasive growth to a rim101 mutant, thus establishing a link between Rim101 and cell differentiation. These investigators also established a link between Rim101 and growth at alkaline pH by showing that Nrg1 contributes to repression of ENA1 (50), which encodes a P-type ATPase that contributes to ion homeostasis by exporting Na+ and Li+ (38). ENA1 is also regulated by a second C2H2 Zn finger transcription factor, the calcineurin-regulated Crz1 (58, 75, 90). Although, as noted above, Rim101 has been found to repress NRG1 gene expression (50), this repression must be limited in its efficiency in wild-type cells growing in rich medium. We found that these cells contain sufficient Nrg1 to mediate repression of our CYC1-3×NRE25-lacZ reporter gene.
Our finding that Rim101 was an inefficient repressor when acting from a single site may account for the fact that only a few direct targets of Rim101-mediated repression have been identified (50). Because both Rim101 and Nrg1 can individually mediate Ssn6-Tup1-directed repression (50, 64; our data), we speculate that the two DNA-binding proteins act cooperatively as a heterodimer to establish a potent Ssn6-Tup1 repression complex at NREDIT. Since each protein could also mediate efficient repression from a multimer of its target site, it is possible that the proteins might also act as homodimers at these sites. Most C2H2-type zinc finger transcription factors have multiple zinc fingers and bind to DNA as monomers. Engineered dimers, however, have been studied as a means to direct cooperative DNA recognition (94). Further investigation is needed to elucidate the molecular details of the interaction between Nrg1 and Rim101.
Whereas processing of full-length PacC to its repressor form in A. nidulans requires two distinct proteolytic cleavages (21), processing of Rim101 in S. cerevisiae is thought to require only one Rim13-dependent cleavage (96). Our Western blot analyses, however, have revealed that our wild-type strain contained several processed isoforms of Rim101 that differed only slightly in mobility (Fig. 8). Rim101 that accumulated in the mutant strains that did not generate active Rim101 appeared as a tightly spaced doublet. Close inspection of autoradiograms suggested that the form with the slowest mobility differed from the form that had been considered to be residual full-length Rim101 in the wild-type strain (Fig. 8). This raises the possibility that there may be additional processing step(s) or posttranslational modification(s) of Rim101. We cannot exclude the possibility, however, that the multiple forms that we have noted represent an artifact of the HA-tagged Rim101 protein or in vitro proteolysis.
We have found that processing of Rim101 depends not only on the previously characterized components of the RIM pathway but also on components of the ESCRT pathway. This pathway has been dissected into three multicomponent complexes that act sequentially to deliver proteins from the late endosome to the vacuole (reviewed in references 36 and 44). The ESCRT I complex consists of Vps23, Vps28, and Vps37; the ESCRT II complex consists of Vps22, Vps25, and Vps36; and ESCRT III consists of two subcomplexes, IIIA and IIIB, which contain Snf7 and Vps20, and Vps24 and Vps2, respectively. Many cell surface proteins that are destined for internalization are tagged by monoubiquitination and packaged into clathrin-coated vesicles for delivery to the sorting endosome. From this compartment, the proteins can be recycled to the plasma membrane or routed on to the vacuole. In the latter case, the proteins are packaged into intralumenal vesicles by the action of the ESCRT pathway. Vps27, which serves as a gatekeeper for entry into this pathway, is enriched at endosomal membranes by virtue of its interaction with phosphatidyl-3-phosphate (45). Membrane-associated Vps27 recruits both monoubiquitinated cargo and the ESCRT I complex, facilitating transfer of captured cargo to ESCRT I (8, 9, 45). The cargo is then sequentially transferred from ESCRT I to ESCRT II to ESCRT III. At this point, debuiquitination of cargo proteins by Doa4 (1, 22, 54) is required for their deposition into intralumenal vesicles formed by the invagination and budding off of endosomal membranes to form the MVB (reviewed in references 36 and 44). The vesicles are then delivered to the vacuole by fusion of the MVB with the vacuole. Disassembly and recycling of the ESCRT complex requires Vps4, an AAA-ATPase (3, 4). Bro1, which was previously shown to act in concert with ESCRT III and to associate with Snf7/Vps32 (12, 61, 62), has recently been shown to recruit Doa4 to endosomes (55). This has led to a model in which the ESCRT IIIA component Snf7/Vps32 recruits Bro1, which in turn recruits Doa4 (55). Deubiquitination of cargo proteins is essential for maintenance of cellular ubiquitin levels (1, 81). In our study, we have found that with the exception of Vps37, all components of ESCRT I, ESCRT II, and ESCRT IIIA were required for proteolytic processing of Rim101 and Rim101-mediated repression. In contrast, Vps27, which is critical for assembly of the ESCRT pathway, ESCRT IIIB, and the ESCRT disassembly component Vps4 were not required. Interestingly, Rim101 was processed in bro1 and doa4 strains but was unable to mediate efficient repression. Because Rim20 and Bro1 are related proteins and both interact with Snf7 (see above), it is possible that a Bro1-Snf7 interaction is required to displace Rim20 from a putative Snf7-Rim20-Rim13-Rim101 processing complex (96). This exchange might be required to promote release of processed Rim101. The requirement for Doa4 to generate active Rim101 after it has been processed suggests that Rim101 might be ubiquitinated and that removal of the ubiquitin by Doa4 is required to enable Rim101-mediated repression. We note, however, that the activity of Rim101(1-531).3HA does not require DOA4. Future studies will lead to a better understanding of the role of the ESCRT pathway in Rim101 processing and the requirement for Bro1 and Doa4 to generate active Rim101.
Interestingly, ion resistance has been noted previously to depend on a portion of the ESCRT pathway. Eguez and colleagues (23) found that resistance of cells to high levels of Ca2+ and Li2+ required all components of ESCRT I, ESCRT II, and ESCRT IIIA but did not require Vps27, ESCRT IIIB, or Vps4. This observation was confirmed by Bowers and colleagues (12), with the exception that in their study, the ESCRT I component Vps37 was not required for ion resistance. The association between a subset of components of the endosomal sorting pathway and resistance to high ion levels can now be explained by the observation that the same subset of ESCRT components is required for processing of Rim101 and full expression of ENA1 (50, 51). While this paper was in preparation, Xu and colleagues (97) also demonstrated a role for the ESCRT pathway in processing of Rim101.
The ESCRT complex is notable in that it drives inward invagination of the limiting membrane of the late endosome in a poorly understood manner that involves enrichment for lipids that promote curvature within membranes. Ultimately, vesicles bud off into the endosomal lumen to form the MVB. It is intriguing that both the ESCRT machinery and Rim101-regulated processes, such as filamentation and spore wall formation, involve membrane remodeling. Recent studies have revealed that the human ESCRT machinery contributes to budding of RNA viruses at the plasma membrane (reviewed in references 67 and 91). Viral proteins that associate with the plasma membrane serve a Vps27-like function in recruiting the ESCRT complex to sites of viral assembly (reviewed in reference 19). This raises the intriguing possibility that a family of membrane-associated adaptors might serve to recruit the ESCRT machinery to various locations. This in turn leads us to speculate that the putative transmembrane proteins Dfg16, identified in this study as a contributor to Rim101 processing, Rim9, and Rim21 may assemble at the cell surface to form a complex that promotes recruitment of a partial ESCRT pathway. This ESCRT-derived pathway may then deliver Rim101 to Rim13 for processing. External environmental and nutritional cues could regulate this pathway by directly or indirectly controlling the activity of the membrane proteins. Ygr122w is an Snf7/Vps32-interacting, non-ESCRT protein that we have identified as also being a contributor to Rim101 processing. We speculate that it could serve this role as an additional member of the previously postulated Rim20-Snf7-containing scaffold that has been proposed to regulate Rim101 processing by presenting Rim101 to Rim13 (96).
We do not yet understand how the DIT1 and DIT2 genes are activated during sporulation. Our previous studies suggested that several cis-acting elements, including the NRE, contribute to activation (27). It is possible that Nrg1 and/or Rim101 is replaced by sporulation-specific activators; alternatively, they may be modified to serve as activators (11). Both Nrg1- and Ssn6-containing complexes have been shown to serve in activation of transcription under certain circumstances (7, 26, 69). Although PacC acts directly as both a transcriptional repressor and a transcriptional activator in A. nidulans (reviewed in reference 66), it remains unclear whether Rim101 can serve as an activator in S. cerevisiae. Indeed, the reason for the initial genetic identification of RIM101 as a putative activator of IME1, a regulator of entry into the sporulation pathway, has yet to be explained. Finally, it is not clear whether Rim101, be it in a full-length or a processed form, has a role at late times of sporulation.
Acknowledgments
We thank Joshua Silver for technical assistance, Brenda Andrews for the use of her robotic pinning instruments, and Aaron Mitchell for plasmids containing RIM101.HA2 and RIM101.HA3.
This work was supported by a Canadian Institute of Health Research grant (MOP-6826) to J.S. K.R. was supported in part by a University of Toronto fellowship.
REFERENCES
- 1.Amerik, A. Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11:3365-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arst, H. N., and M. A. Penalva. 2003. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 19:224-231. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 5.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 7.Berkey, C. D., V. K. Vyas, and M. Carlson. 2004. Nrg1 and Nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot. Cell 3:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilodeau, P. S., J. L. Urbanowski, S. C. Winistorfer, and R. C. Piper. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534-539. [DOI] [PubMed] [Google Scholar]
- 9.Bilodeau, P. S., S. C. Winistorfer, W. R. Kearney, A. D. Robertson, and R. C. Piper. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 11.Bogengruber, E., T. Eichberger, P. Briza, I. W. Dawes, M. Breitenbach, and R. Schricker. 1998. Sporulation-specific expression of the yeast DIT1/DIT2 promoter is controlled by a newly identified repressor element and the short form of Rim101p. Eur. J. Biochem. 258:430-436. [DOI] [PubMed] [Google Scholar]
- 12.Bowers, K., J. Lottridge, S. B. Helliwell, L. M. Goldthwaite, J. P. Luzio, and T. H. Stevens. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194-210. [DOI] [PubMed] [Google Scholar]
- 13.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 14.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 16.Casadaban, M. J., A. Martinez-Arias, S. K. Shapira, and J. Chou. 1983. Beta-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 100:293-308. [DOI] [PubMed] [Google Scholar]
- 17.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 19.Clague, M. J., and S. Urbe. 2003. Hrs function: viruses provide the clue. Trends Cell Biol. 13:603-606. [DOI] [PubMed] [Google Scholar]
- 20.de Nadal, E., L. Casadome, and F. Posas. 2003. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 23:229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diez, E., J. Alvaro, E. A. Espeso, L. Rainbow, T. Suarez, J. Tilburn, H. N. Arst, Jr., and M. A. Penalva. 2002. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 21:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupre, S., and R. Haguenauer-Tsapis. 2001. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell. Biol. 21:4482-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eguez, L., Y. S. Chung, A. Kuchibhatla, M. Paidhungat, and S. Garrett. 2004. Yeast Mn2+ transporter, Smf1p, is regulated by ubiquitin-dependent vacuolar protein sorting. Genetics 167:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisendle, M., H. Oberegger, R. Buttinger, P. Illmer, and H. Haas. 2004. Biosynthesis and uptake of siderophores is controlled by the PacC-mediated ambient-pH regulatory system in Aspergillus nidulans. Eukaryot. Cell 3:561-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espeso, E. A., J. Tilburn, L. Sanchez-Pulido, C. V. Brown, A. Valencia, H. N. Arst, Jr., and M. A. Penalva. 1997. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 274:466-480. [DOI] [PubMed] [Google Scholar]
- 26.Fragiadakis, G. S., D. Tzamarias, and D. Alexandraki. 2004. Nhp6 facilitates Aft1 binding and Ssn6 recruitment, both essential for FRE2 transcriptional activation. EMBO J. 23:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friesen, H., S. R. Hepworth, and J. Segall. 1997. An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friesen, H., J. C. Tanny, and J. Segall. 1998. SPE3, which encodes spermidine synthase, is required for full repression through NREDIT in Saccharomyces cerevisiae. Genetics 150:59-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futai, E., T. Maeda, H. Sorimachi, K. Kitamoto, S. Ishiura, and K. Suzuki. 1999. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260:559-568. [DOI] [PubMed] [Google Scholar]
- 30.Gaisne, M., A. M. Becam, J. Verdiere, and C. J. Herbert. 1999. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36:195-200. [DOI] [PubMed] [Google Scholar]
- 31.Gerrard, S. R., B. P. Levi, and T. H. Stevens. 2000. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic 1:259-269. [DOI] [PubMed] [Google Scholar]
- 32.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 33.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 35.Green, S. R., and A. D. Johnson. 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15:4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317-323. [DOI] [PubMed] [Google Scholar]
- 37.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 38.Haro, R., B. Garciadeblas, and A. Rodriguez-Navarro. 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291:189-191. [DOI] [PubMed] [Google Scholar]
- 39.Hepworth, S. R., L. K. Ebisuzaki, and J. Segall. 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honigberg, S. M., and K. Purnapatre. 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116:2137-2147. [DOI] [PubMed] [Google Scholar]
- 41.Howard, J. P., J. L. Hutton, J. M. Olson, and G. S. Payne. 2002. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 157:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 44.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 45.Katzmann, D. J., C. J. Stefan, M. Babst, and S. D. Emr. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kranz, A., A. Kinner, and R. Kolling. 2001. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 899-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Labbe, S., Z. Zhu, and D. J. Thiele. 1997. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272:15951-15958. [DOI] [PubMed] [Google Scholar]
- 50.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850-1856. [DOI] [PubMed] [Google Scholar]
- 52.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 54.Losko, S., F. Kopp, A. Kranz, and R. Kolling. 2001. Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12:1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luhtala, N., and G. Odorizzi. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell, S. B. Southard, C. I. Horenstein, J. Chen-Weiner, A. F. Ram, J. C. Kapteyn, T. W. Roemer, D. H. Vo, D. C. Bondoc, J. Hall, W. W. Zhong, A. M. Sdicu, J. Davies, F. M. Klis, P. W. Robbins, and H. Bussey. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendizabal, I., A. Pascual-Ahuir, R. Serrano, and I. F. de Larrinoa. 2001. Promoter sequences regulated by the calcineurin-activated transcription factor Crz1 in the yeast ENA1 gene. Mol. Genet. Genomics 265:801-811. [DOI] [PubMed] [Google Scholar]
- 59.Mosch, H. U., and G. R. Fink. 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145:671-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikko, E., A. M. Marini, and B. Andre. 2003. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J. Biol. Chem. 278:50732-50743. [DOI] [PubMed] [Google Scholar]
- 62.Odorizzi, G., D. J. Katzmann, M. Babst, A. Audhya, and S. D. Emr. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893-1903. [DOI] [PubMed] [Google Scholar]
- 63.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 64.Park, S. H., S. S. Koh, J. H. Chun, H. J. Hwang, and H. S. Kang. 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Penalva, M. A., and H. N. Arst, Jr. 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425-451. [DOI] [PubMed] [Google Scholar]
- 66.Penalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 68.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 69.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 70.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 71.Rose, M. D., and J. R. Broach. 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-230. [DOI] [PubMed] [Google Scholar]
- 72.Rowland, O., and J. Segall. 1998. A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol. 18:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowland, O., and J. Segall. 1996. Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5 S RNA gene. J. Biol. Chem. 271:12103-12110. [DOI] [PubMed] [Google Scholar]
- 74.Serrano, R., D. Bernal, E. Simon, and J. Arino. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279:19698-19704. [DOI] [PubMed] [Google Scholar]
- 75.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Arino. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 76.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 77.Simonsen, A., A. E. Wurmser, S. D. Emr, and H. Stenmark. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485-492. [DOI] [PubMed] [Google Scholar]
- 78.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 79.Su, S. S., and A. P. Mitchell. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su, S. S., and A. P. Mitchell. 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21:3789-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang, H. Y., A. Munn, and M. Cai. 1997. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4294-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 85.Treton, B., S. Blanchin-Roland, M. Lambert, A. Lepingle, and C. Gaillardin. 2000. Ambient pH signalling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH. Mol. Gen. Genet. 263:505-513. [DOI] [PubMed] [Google Scholar]
- 86.Tu, J., L. G. Vallier, and M. Carlson. 1993. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics 135:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 88.Vallier, L. G., and M. Carlson. 1991. New SNF genes, GAL11 and GRR1 affect SUC2 expression in Saccharomyces cerevisiae. Genetics 129:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Den Hazel, H. B., M. C. Kielland-Brandt, and J. R. Winther. 1996. Review: biosynthesis and function of yeast vacuolar proteases. Yeast 12:1-16. [DOI] [PubMed] [Google Scholar]
- 90.Viladevall, L., R. Serrano, A. Ruiz, G. Domenech, J. Giraldo, A. Barcelo, and J. Arino. 2004. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 279:43614-43624. [DOI] [PubMed] [Google Scholar]
- 91.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 92.Vyas, V. K., S. Kuchin, and M. Carlson. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams, F. E., and R. J. Trumbly. 1990. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:6500-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolfe, S. A., R. A. Grant, and C. O. Pabo. 2003. Structure of a designed dimeric zinc finger protein bound to DNA. Biochemistry 42:13401-13409. [DOI] [PubMed] [Google Scholar]
- 95.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamaguchi-Iwai, Y., R. Stearman, A. Dancis, and R. D. Klausner. 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15:3377-3384. [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]
- 100.Zhou, H., and F. Winston. 2001. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]