Abstract
The 8-oxo-7,8-dihydrodeoxyguanosine (8oxoG), a major mutagenic DNA lesion, results either from direct oxidation of guanines or misincorporation of 8oxodGTP by DNA polymerases. At present, little is known about the mechanisms preventing the mutagenic action of 8oxodGTP in Saccharomyces cerevisiae. Herein, we report for the first time the identification of an alternative repair pathway for 8oxoG residues initiated by S. cerevisiae AP endonuclease Apn1, which is endowed with a robust progressive 3′→5′ exonuclease activity towards duplex DNA. We show that yeast cell extracts, as well as purified Apn1, excise misincorporated 8oxoG, providing a damage-cleansing function to DNA synthesis. Consistent with these results, deletion of both OGG1 encoding 8oxoG-DNA glycosylase and APN1 causes nearly 46-fold synergistic increase in the spontaneous mutation rate, and this enhanced mutagenesis is primarily due to G · C to T · A transversions. Expression of the bacterial 8oxodGTP triphosphotase MutT in the apn1Δ ogg1Δ mutant reduces the mutagenesis. Taken together, our results indicate that Apn1 is involved in an S. cerevisiae 8-oxoguanine-DNA glycosylase (Ogg1)-independent repair pathway for 8oxoG residues. Interestingly, the human major AP endonuclease, Ape1, also exhibits similar exonuclease activity towards 8oxoG residues, raising the possibility that this enzyme could participate in the prevention of mutations that would otherwise result from the incorporation of 8oxodGTP.
Endogenous oxidative DNA lesions are most abundant and inevitable as cells generate reactive oxygen species through aerobic respiration. The occurrence of oxidized bases in DNA can result from either direct oxidation or misincorporation of oxidized deoxyribonucleoside monophosphate by DNA polymerases from the nucleotide pool (23, 24). These oxidized bases are mutagenic and therefore must be removed by efficient DNA repair processes that include base excision repair (BER) and the nucleotide incision repair (NIR) pathways, as well as by a process involving the sanitization of a pool of oxidized deoxyribonucleotidetriphosphates (dNTPs) (14, 43). In Escherichia coli, three enzymes, formamidopyrimidine-DNA glycosylase (Fpg), adenine-DNA glycosylase (MutY), and a nucleotidase (MutT; 8oxodGTP triphosphatase), play important roles in counteracting the mutagenic effect of the oxidized residue 8-oxo-7,8-dihydrodeoxyguanosine (8oxoG), which forms stable base pairing with adenine (A · 8oxoG) (33). While Fpg excises 8oxoG and MutY excises adenine from the A · 8oxoG mispair in DNA, MutT sanitizes the nucleotide precursor pool of 8oxodGTP by hydrolyzing it to 8oxodGMP and pyrophosphate, thereby preventing the incorporation of the oxidized nucleotide into DNA during replication (12, 32). These three enzymes are also conserved in mammalian cells, thus underscoring the importance of preventing an 8oxoG mutagenic effect (1). However, in the budding yeast Saccharomyces cerevisiae, only a functional homologue of Fpg, Ogg1 (S. cerevisiae 8-oxoguanine-DNA glycosylase), has been isolated and characterized (49). Neither sequence homology searches nor screening for yeast genes that would complement the mutator phenotype of the fpg mutY double mutant of E. coli permitted the identification of the MutY and MutT homologues. This raises the possibility that S. cerevisiae may have evolved alternative strategies to combat the mutagenic effects of 8oxodGTP precursor. Indeed, in S. cerevisiae the mismatch repair (MMR) system can act at A · 8oxoG mispairs, suggesting that this pathway may substitute for both MutY and MutT in this organism (35).
We have previously demonstrated that the major apurinic/apyrimidinic (AP) endonuclease S. cerevisiae AP endonuclease 1 (Apn1) and major human AP endonuclease 1 (Ape1) are involved in the NIR pathway and that these enzymes shared common substrate specificity towards various oxidatively damaged bases (13, 18). These enzymes nick AP sites on the 5′ side, as well as directly incise the DNA containing oxidatively damaged bases to create 3′ hydroxyl group for DNA repair synthesis (18). The latter observation led to the suggestion that the Apn1-initiated pathway, as well as the Ape1-initiated, NIR pathway is intimately connected with the BER pathway to cleanse genomic DNA of potentially mutagenic and cytotoxic lesions (13). Apn1, which is localized to the nucleus and mitochondria (39, 51), also performs additional functions in DNA repair (37). It possesses a 3′ diesterase activity, which removes a multitude of 3′-blocking groups at single-strand breaks in DNA (e.g., 3′-phosphoglycolate and 3′ phosphate) that are induced by oxidative agents (21, 22). Yeast mutants (apn1Δ) lacking Apn1 are hypersensitive to several DNA-damaging agents (e.g., methyl- and ethylmethane sulfonate) that create AP sites, as well as to agents (H2O2 and tert-butyl hydroperoxide) that create oxidative DNA lesions (40). These apn1Δ mutants also exhibit a mutator phenotype mainly characterized by a 60-fold increase rate of A · T to C · G transversion, predicted to arise as a result of insertion of dGMP on opposite AP sites during translesion synthesis (27).
S. cerevisiae also possesses a second AP endonuclease (Apn2), and loss of this enzyme dramatically increases the sensitivity to genotoxic agents, as well as to the spontaneous mutation rate of apn1Δ mutants (2). Apn2 possesses a 3′ diesterase and a 3′→5′ exonuclease; both of these have levels of activity that are 30- to 40-fold higher than its AP endonuclease activity (47). To date, no biological function has been assigned to the 3′→5′ exonuclease activity of Apn2. Interestingly, the major human AP endonuclease, Ape1, also possesses 3′→5′ exonuclease activity, which has been documented to remove mismatch deoxyribonucleoside monophosphate at the 3′ termini of gapped or nicked DNA (7). As for Apn1, earlier studies showed that the highly purified protein lacks 3′→5′ exonuclease activity on double- and single-stranded DNA (21). Later, using duplex oligonucleotides, Vance and Wilson (48) demonstrated that a glutathione S-transferase (GST)-tagged form of Apn1 can excise a single nucleotide at the 3′ side of a nick, thereby generating a one-nucleotide gap. More recent studies revealed that Apn1 displayed the ability to remove more than one nucleotide following incision of an AP site (20). In fact, E. coli endonuclease IV (Nfo), a counterpart of Apn1, also possesses an intrinsic 3′→5′ exonuclease activity, which is very sensitive to ionic strength, metal ions, EDTA, and reducing conditions (25). These observations suggest that Apn1 may indeed possess a 3′→5′ exonuclease activity.
In this study, we attempt to elucidate the role of the 3′→5′ exonuclease activity of Apn1 in preventing 8oxoG-induced mutagenesis in S. cerevisiae. We show that Apn1 contains robust 3′→5′ exonuclease activity, which can remove regular and oxidized nucleotides at 3′ termini of a gap or a nick. In parallel, the Nfo and Ape1 exonuclease activities were also characterized using the same DNA substrates. Consistent with the biochemical studies, we demonstrate that Apn1 strongly suppresses the rate of G · C to T · A transversions in the ogg1 mutants. Our model suggests that Apn1-dependent 3′→5′ exonuclease is a major repair pathway for cleansing 8oxodGMP that is misincorporated opposite adenine and cytosine during DNA replication in S. cerevisiae.
MATERIALS AND METHODS
Oligonucleotides.
All other oligodeoxyribonucleotides were purchased from Eurogentec (Seraing, Belgium), including regular oligonucleotides and those containing 8oxoG, 5,6-dihydrouridine (DHU), uridine (dU), tetrahydrofuranyl (THF), and inosine (dI). The oligonucleotide sequences for the recessed strand referred to as Exo-X were d(GTGGCGCGGAGACTTAGAGAX), where X is 8oxoG, DHU, dU, THF, dI, or a regular nucleotide; for the 3′ part of the nicked strand, the sequence was pd(ATTTGGCGCGGGGAATTCC); and for the complementary strands, the sequences were d(GGAATTCCCCGCGCCAAATNTCTCTAAGTCTCCGCGCCAC) and d(NTCTCTAAGTCTCCGCGCCAC), whereN is dA, dG, dC, or T. The oligonucleotide d(GTGGCGCGGAGACTTAGAGAXATTTGGCGCGGGGAATTCC), where X is THF, was hybridized to a complementary 40-mer strand. As shown in Fig. 1A, the resulting fully recessed and nicked duplexes are referred to as X · N, X · Nrecs, and X · Nnick, respectively, where X is either a regular or modified nucleotide and N is dA, dG, dC, or T. Oligonucleotides were 5′ end labeled by T4 polynucleotide kinase (New England Biolabs, OZYME, Saint Quentin Yvelines, France) in the presence of [γ-32P]ATP (4,500 Ci/mmol; ICN Biomedicals, SARL, Orsay, France) as recommended by the manufacturers. The labeled oligonucleotides were annealed to their appropriate complementary oligonucleotides in a buffer containing 50 mM NaCl and 10 mM HEPES/KOH (pH 7.5) at 65°C for 3 min.
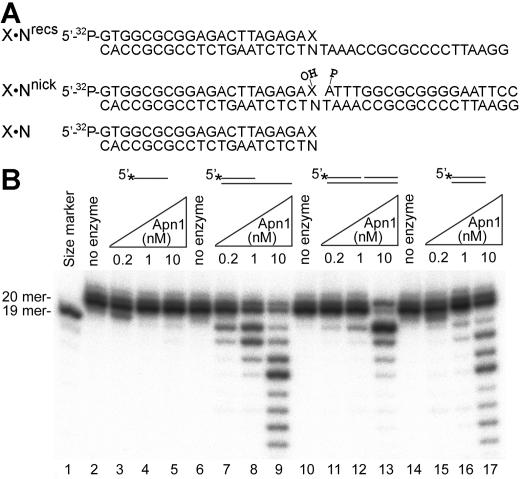
FIG. 1.
The 3′→5′ exonuclease activity of Apn1 on different DNA substrates. (A) DNA substrates used in this study. See the text for details. (B) Increasing concentration of Apn1 (0 to 10 nM) were incubated for 5 min at 30°C with 5 nM of 5′ 32P-labeled 20-mer oligonucleotides. Lane 1, 19-mer size marker; lanes 2 to 5, ExoG single-stranded oligonucleotide; lanes 6 to 9, G · Crecs recessed duplex; lanes 10 to 13, G · Cnick nicked duplex; lanes 14 to 17, G · C fully duplex. For details, see Materials and Methods.
Yeast strains and plasmids.
The strains used in this study were derived from the parent strain FF18733 (MATa his7 leu2 lys1 ura3 trpΔ). The parent strain and CD138 (ogg1Δ::TRP1) were kindly provided by Serge Boiteux (CEA, Fontenay aux Roses, France). DRY139, DRY140, and DRY142 were created by one-step gene targeting as previously described (30).
Construction of GST fusion protein. The plasmid pTW340 (URA3 LEU2; Ampr) was kindly provided by Tom Wilson (Ann Arbor, Mich.); it contains the OGG1 gene fused to GST under the control of the copper-inducible promoter CUP1 in the dual-host expression vector pYEX-Phiz (Eric Phizicky via Tom Wilson). The OGG1 gene was removed by restriction digestion using EcoRI and SmaI, and the linearized vector was purified. A PCR fragment bearing the entire coding sequence of the mutT gene was amplified from the E. coli genome by using Pfu polymerase and the primers mutT-YEX-F1 and mutT-YEX-R1. To generate the GST-MutT fusion by GAP repair, 100 ng of the linearized vector and 0 or 300 to 1,800 ng of the MutT-PCR fragment were cotransformed into S. cerevisiae strain YW607. Transformants were selected for on uracil-free minimal medium agar plates. The pGst-MutT plasmid was isolated from yeast and amplified in bacteria, and the DNA sequence was verified. The vector pYES and pGst-MutT were introduced into the yeast strains, and MutT protein expression was verified by Western blotting with anti-GST antibodies as previously described (20).
Enzymes.
The E. coli exonuclease III (Xth) protein was purchased from Roche Diagnostics (Meylan, France). The Klenow fragments of E. coli DNA polymerase I deficient in exonuclease activity (FKexo−) was purchased from New England Biolabs (OZYME, Saint Quentin Yvelines, France). The E. coli endonuclease VIII protein (Nei) was generously provided by Dmitry Zharkov (Novosibirsk, Russia). Purifications of the Nfo, Apn1, and Ape1 proteins were performed as previously described (19).
Exonuclease assay.
The standard reaction mixture (20 μl) contained 3 nM 5′ 32P-labeled oligonucleotide duplex, 100 mM KCl, 20 mM HEPES/KOH, pH 7.6, 0.1 mg/ml bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), 5 mM MgCl2 and either 0.5 nM of Apn1 or 25 pM Xth, unless otherwise stated. For Ape1, the reaction buffer was 20 mM HEPES/KOH, pH 7.4, 0.1 mg/ml BSA, 30 mM KCl, 1 mM DTT, 1 mM MgCl2, and 1 nM enzyme (6). For Nfo, the reaction buffer was 25 mM HEPES/KOH, pH 8.4, 0.1 mg/ml BSA, and 1 nM enzyme (25). Reactions were performed at 30°C (37°C for Ape1, Nfo, and Xth) for 5 min and stopped by the addition of 10 μl of 1.5% sodium dodecyl sulfate-0.3 mg/ml proteinase K, followed by incubation for 10 min at 50°C. The mixture was desalted by hand-made spin-down columns filled with Sephadex G25 (Amersham Biosciences) equilibrated in 5 M urea. Purified reaction products were heated at 65°C for 2 min and separated by electrophoresis in denaturing 20% (wt/vol) polyacrylamide gels (20:1; 7.5 M urea-0.5× Tris-borate-EDTA). The gels were exposed to a Fuji FLA-3000 Phosphor Screen and analyzed using with Jauge V3.12 software.
To measure linear velocity, 1 to 200 nM duplex oligonucleotide substrate was incubated under standard reaction conditions. The Km and kcat values were determined from Lineweaver-Burk plots.
3′→5′ exonuclease activity in whole-cell extract.
S. cerevisiae whole-cell extracts were prepared as previously described (37), except that lysis buffer contained 40 mM HEPES, pH 7.6, 0.3 M KCl, 1 mM DTT, 0.1% NP-40, and protease inhibitor cocktail (Roche, Switzerland). Reactions were carried in the standard reaction buffer supplied with 20 μM dNTPs, 0.5 mM rATP, and either 3 μg of extract or 5 mU of FKexo−. Reactions were performed at 30°C for 1 h and stopped by incubation at 65°C for 10 min in the presence of 3.5 mM EDTA. When required, reaction products were reannealed by slow cooling at room temperature and incubated for 30 min at 37°C with 500 nM Fpg and 500 nM Nei. Reaction products were analyzed as described above.
Spontaneous mutation rates.
Spontaneous mutation analysis was performed using the fluctuation test method previously described (50). At least, 7 to 11 independent colonies of the indicated yeast strains were grown in 1 ml of minimal synthetic (SD) medium lacking arginine for 2 days at 30°C (45). For each culture, cells were diluted to an initial titer of ∼4,000 to 10, 000 cells in SD medium lacking arginine, and 24 1-ml cultures were distributed in 24-multiwell non-tissue-culture-treated plates (Falcon, Becton Dickinson). This therefore created 7 to 11 24-well plates for the analysis. The plates were placed in an incubator at 30°C for ∼24 h, followed by the addition of canavanine (50 μg/ml) to each well, and immediately returned to the incubator for 5 to 7 days to select for the canavanine forward mutants. Cell density (∼1.0 × 106) just before the addition of canavanine was measured by plating dilutions on yeast extract-peptone-glucose (YPD) agar plates and counting the colonies after 3 days at 30°C. Each experiment was carried out independently four times. Mutation rates were calculated from the cell density determined before the addition of canavanine and the number of wells with no mutations by using the Poisson distribution (31).
Spontaneous mutation spectra.
For each strain tested, at least 25 independent colonies were inoculated to an initial density of ∼102 cells per ml and grown at 30°C in YPD for 2 days; ∼1.0 × 106 or 107 cells spread onto SD solid medium lacking arginine but containing 50 μg/ml of canavanine. From each culture, a single mutant colony was selected, streaked onto selective plates, and grown for 3 days at 30°C. Another 15 to 20 independent canavanine-resistant mutants were directly obtained from three of the independent mutation rate assays. These mutants were purified, and a single colony was kept for DNA extraction. The canavanine-resistant mutants were grown in 5-ml YPD cultures to saturation, followed by chromosomal DNA extraction as previously described (41). The CAN1 gene was amplified by PCR using Pfx (Invitrogen Canada, Inc., Burlington, Ontario) and 20 ng of genomic DNA with the primers CAN1-FF [d(GTTGGATCCAGTTTTTAATCTGTCG)] and CAN1-R3 [d(CGGTGTATGACTTATGAGGGTGAG)]. The PCR product (i.e., the CAN1 gene; 1,773 bp) was purified by using a ManuPCR 030 plate (Millipore Canada, Ltd., Etobicoke, Ontario) and prepared for automated sequencing (3100 Genetic Analyzer; AB Applied Biosystems/Hitachi) using the following three primers: CAN1-F2 [d(GGGAGGTTCTTAGGTTGGGTTTCCT)], CAN1-F3 [d(GGCATATTCTGTCACGCAGTCCTTG)], and CAN1-R2 [d(GTACCTTGAAATGTGAAGGCAGCGT)]. Sequence alignments and analyses were performed using the software programs Vector NTI Suite 8 and Chromas 2.
RESULTS
Apn1 has a progressive 3′→5′ exonuclease activity that eliminates mispairs and modified nucleotides at 3′ termini.
Previously, we observed nonspecific exonuclease activity when incubating 5′-labeled regular duplex oligonucleotide with an excess amount of Apn1 (data not shown). To investigate the ability of Apn1 to degrade regular DNA, we constructed different substrates including single-stranded, recessed duplex, nicked duplex, and fully duplex oligonucleotides. As shown in Fig. 1A, X · Nrecs is a standard recessed duplex containing a 20-nucleotide-long, 5′ single-stranded tail, where X is a nucleotide at the 3′ termini of the 5′ 32P-labeled strand and N is a complementary nucleotide in the lower nonlabeled strand. The substrates X · Nnick and X · N are nicked and fully duplex oligonculeotides, respectively. Using these substrates, we found that increasing concentrations of the purified Apn1 protein degraded the duplex DNAs (Fig. 1B, lanes 7 to 17) but not the single-stranded DNA (lanes 3 to 5). Moreover, Apn1 exonuclease activity was more efficient on the recessed duplex (lanes 7 to 9) than on either the nicked duplex (lanes 11 to 13) or the entire duplex (lanes 15 to 17). Interestingly, at high protein concentrations, Apn1 appeared to remove an unlimited number of nucleotides from either the recessed or fully duplex DNA in a progressive manner (lanes 9 and 17). However, in the case of the nicked duplex and at high protein concentrations, Apn1 seemed to preferentially remove only the first nucleotide, followed by the very slow removal of the second and third ones (lane 13).
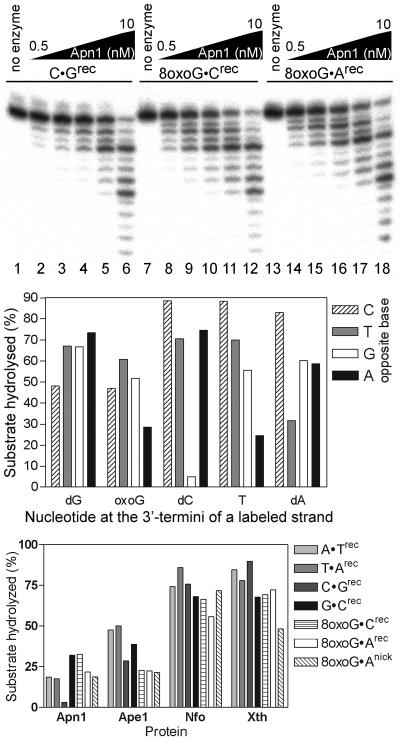
We next examined the 3′→5′ degradation of recessed duplex oligonucleotides containing either a regular or a modified nucleotide at the 3′ termini of a gap. As shown in Fig. 2, top, Apn1 exhibited a robust 3′→5′ exonuclease activity on C · Grecs, 8oxoG · Crecs, and 8oxoG · Arecs oligonucleotides, indicating that the enzyme can remove both regular and oxidized nucleotides. Interestingly, Apn1 excised 8oxoG from 8oxoG · Crecs and 8oxoG · Arecs (Fig. 2, top, lanes 7 to 11 and 14 to 17) more efficiently than C from C · Grecs (lanes 2 to 5), suggesting that the enzyme may display preference for the type of nucleotide at the 3′ side of a gap and a nick.
FIG. 2.
Substrate specificity of Apn1 exonuclease. (Top) Apn1 exonuclease activity towards 8oxoG residues. Recessed duplex (5 nM) was generated by annealing a 5′-end 32P-labeled 20-mer fragment containing either regular nucleotide or 8oxoG at 3′ termini (X) and a 40-mer template fragment containing one of the four natural nucleotides at position 20 (N). (Middle) The 3′→5′ degradation of recessed duplex containing a 3′ nucleotide paired to one of the four natural nucleotides. The axis of abscissas shows the nature of a nucleotide at 3′ termini (X) of the labeled strand. (Bottom) Comparison of Apn1, Ape1, Nfo, and Xth exonuclease activities towards recessed and nicked duplexes. The reaction mixture contained 0.5 nM Apn1 and 1.0 nM Ape1, Nfo, or Xth. For details, see Materials and Methods.
To check whether the 3′→5′ exonuclease activity of Apn1 could discriminate mismatched and modified nucleotides, the substrate specificity of the enzyme was investigated using recessed duplex oligonucleotides containing either regular or modified 3′-terminal nucleotides such as 8oxoG, DHU, dU, THF, and dI. As shown in Fig. 2, middle, Apn1 excised the mismatched regular nucleotides with higher efficiency than the corresponding matched nucleotides. Quite surprisingly, Apn1 degraded C · Grecs much less efficiently than all other matched nucleotide pairs (Fig. 2, middle). In addition, Apn1 processed 8oxoG · Arecs less efficiently than 8oxoG · Crecs, 8oxoG · Trecs, and 8oxoG · Grecs (Fig. 2, middle). The 3′-terminal DHU and dU were excised less efficiently than mismatched dC and T, whereas THF and dI residues were the most preferred substrates among other modifications; their removal was independent of the opposite base (data not shown).
Since bacterial (Nfo and Xth) and human (Ape1) AP endonucleases also have 3′→5′ exonuclease activity, we investigated whether these enzymes would remove 3′-terminal regular and oxidized nucleotides under reaction conditions optimal for their exonuclease activities. As shown in Fig. 2, bottom, the AP endonucleases were capable of acting on each type of recessed duplex oligonucleotides tested and with good efficiency. No significant differences were observed between G · Crecs and 8oxoG · Crecs for Apn1, Nfo, and Xth. Overall, the efficiencies of the removal of a 3′-terminal nucleotide by Ape1, Nfo, and Xth varied less than twofold, depending on the nucleotide pairs tested. Furthermore, recessed duplexes were preferred substrates for all AP endonucleases tested (data not shown).
Characterization of the 3′→5′ exonuclease activity of Apn1.
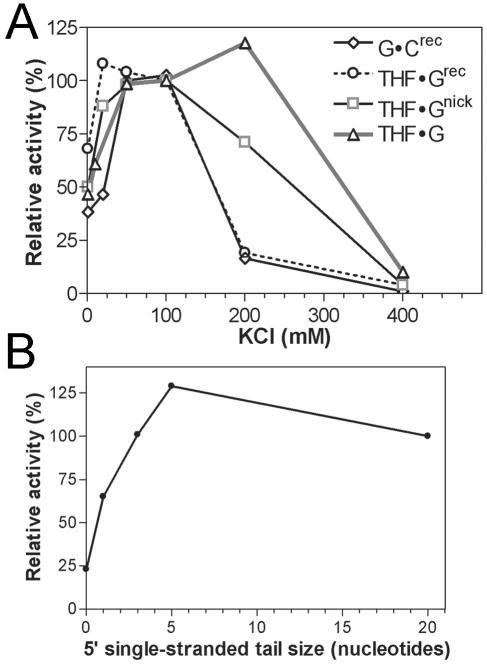
Previously, it has been shown that for Nfo and Ape1 the reaction conditions for the AP endonuclease and exonuclease activities are different. Therefore, we investigated the effects of pH, ionic strength, and magnesium concentration on Apn1 exonuclease activity. We found that the maximal exonuclease activity was observed at MgCl2 concentrations above 2.5 mM and at pH 7.2 to 7.6 and that these conditions were also optimal for the AP endonuclease activity (data not shown). However, as shown in Fig. 3A, the maximal AP endonuclease activity on THF · G was observed at 200 mM KCl, whereas the exonuclease activity on recessed duplexes (G · Crecs and THF · Grecs) was strongly inhibited. Interestingly, the exonuclease activity on a nicked duplex (THF · Gnick) was less sensitive to salt than on a recessed duplex (THF · Grecs). To determine the type of duplex structure required for the optimal exonuclease activity, we used G · Crecs oligonucleotides with 5′ single-stranded tails of various lengths. As shown in Fig. 3B, maximal 3′→5′ exonuclease activity was observed on the recessed duplexes when the tail length was ≥5 nucleotides. In the absence of a recessed end, as in the case of a fully duplex oligonucleotide, Apn1 degraded this substrate with three- to sixfold-lower efficiency than the recessed duplexes (Fig. 3B).
FIG. 3.
Activity profiles of Apn1. (A) Exonuclease and AP endonuclease activities depending on ionic strength. Oligonucleotide duplex containing a single THF residue was used to measure AP endonuclease activity. The reaction contained 0.5 nM Apn1. (B) Exonuclease activity on duplexes with varied sizes of a 5′ single-stranded tail. For comparison, the activity towards a standard recessed duplex with a 20-mer tail was assumed to be 100%.
To further characterize the substrate specificities of Apn1, the kinetic parameters for the exonuclease activity towards recessed and nicked duplex DNAs were measured. A comparison of the kinetic constants showed that the THF residue at the 3′ side of a nick was the preferred substrate, compared to the AP site and the 3′-terminal nucleotides (Table 1). Unexpectedly, the apparent kcat/Km values for G · Crecs were about 20-fold-higher than for C · Grecs (Table 1). Importantly, exonuclease degradation of the mismatched G · Arecs was 2.3-fold higher cthan for the matched G · Crecs (Table 1), suggesting that Apn1 has a slight preference for the mismatched nucleotides. Remarkably, 8oxoG · Crecs was repaired somewhat more efficiently than G · Crecs, whereas 8oxoG · Arecs was degraded less efficiently than G · Crecs and G · Arecs (Table 1), suggesting that excision of 3′-terminal oxidized nucleotides depends on the opposite nucleotide. It is noteworthy that the kcat/Km ratio for exonuclease degradation of G · Arecs (0.43 nM−1 · min−1) was similar to that of AP endonuclease activity (0.48 nM−1 · min−1), suggesting that Apn1 is a very efficient exonuclease.
TABLE 1.
Steady-state kinetic parameters of Apn1 exonuclease activity towards recessed and nicked duplex oligonucleotides
| Base paira | Parameter
|
||
|---|---|---|---|
| Km (nM)c | kcat (min−1) | kcat/Km (nM−1 · min−1) | |
| 8oxoG · Crecs | 13 ± 3 | 4.0 ± 0.3 | 0.31 |
| 8oxoG · Arecs | 18 ± 4 | 2.0 ± 0.2 | 0.11 |
| G · Crecs | 36 ± 3 | 6.9 ± 0.2 | 0.19 |
| G · Arecs | 18 ± 4 | 7.7 ± 0.7 | 0.43 |
| C · Grecs | 47 ± 9 | 0.53 ± 0.03 | 0.01 |
| C · Arecs | 21 ± 5 | 7.2 ± 0.6 | 0.34 |
| 8oxoG · Anick | 3.8 ± 1 | 0.4 ± 0.02 | 0.10 |
| THF · Gnick | 6.0 ± 0.5 | 4.1 ± 0.1 | 0.68 |
| AP siteb | 24 ± 4.4 | 11 ± 1.0 | 0.48 |
The first letter in the nucleotide pairs is a residue at the 3′ termini in the recessed (recs) or nicked (nick) strand. The second letter is the corresponding opposite nucleotide in the complementary strand.
Duplex oligonucleotide containing a single THF · G base pair for AP endonuclease activity.
Kinetic constant values are means ± standard deviation.
Comparison of the efficiency of wild-type (WT) and mutant cell extracts to elongate DNA substrates containing either mismatch or THF residue at 3′ termini of a nick.
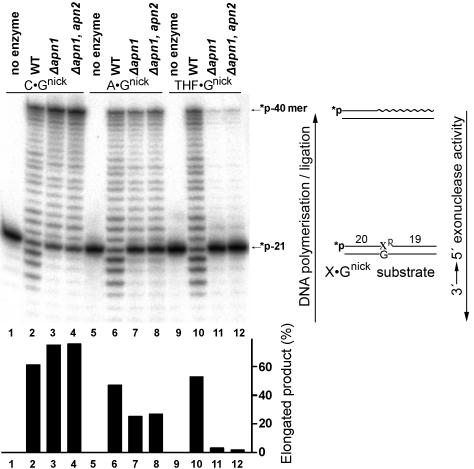
Analysis of the kinetic parameters of 3′→5′ exonuclease activity suggests that Apn1 might interfere with DNA polymerase and DNA ligase activities. To examine this, we reconstituted the repair of 5′-labeled C · Gnick, A · Gnick, and THF · Gnick oligonucleotides in yeast cell extracts of WT, apn1Δ, and apn1Δ apn2Δ in the presence of dNTPs and rATP. As shown in Fig. 4, incubation of C · Gnick generated elongated products up to the full-length 40-mer product with 60 to 80% efficiency in all extracts (lanes 2 to 4). The appearance of DNA fragments with intermediate sizes (21 to 39 mer) during repair of the C · Gnick, A · Gnick, and THF · Gnick duplexes indicates that restoration of the 40-mer fragment was dependent on a DNA polymerase, which catalyzed the strand displacement (lanes 2 to 4, 6 to 8, and 10). Interestingly, when dNTPs were omitted, DNA ligase activity in the extracts efficiently sealed the C · Gnick, 8oxoG · Anick, and 8oxoG · Cnick duplexes without generating intervening products (data not shown). The conversion of C · Gnick to a 40-mer product in WT extracts (lane 2) was slightly less efficient than in the mutants (lanes 3 and 4), due to the partial degradation of a 5′-labeled 21-mer fragment by 3′→5′ exonuclease activity in WT extracts (lanes 2, 6, and 10) but not in the apn1Δ mutants. In contrast to C · Gnick, conversion of the A · Gnick and THF · Gnick duplexes to the full-sized 40-mer products was 2- and 20-fold-less efficient, respectively, in apn1Δ mutant extracts (lanes 7,8,11 and 12) than in WT extract (lanes 6 and 10). This result suggests that efficient elongation of the primers containing mismatches and 3′-blocking groups depends on Apn1 exonuclease function.
FIG. 4.
Repair of a mismatch and THF residue at 3′ termini of a nick in yeast cell extracts. The 5′ 32P-labeled nicked duplex was incubated with crude extract. Reaction products were separated in the gel, and the amount of elongated products was quantified. For details, see Materials and Methods.
Apn1-dependent repair of nicked and recessed DNA containing 3′-terminal 8oxoG residue.
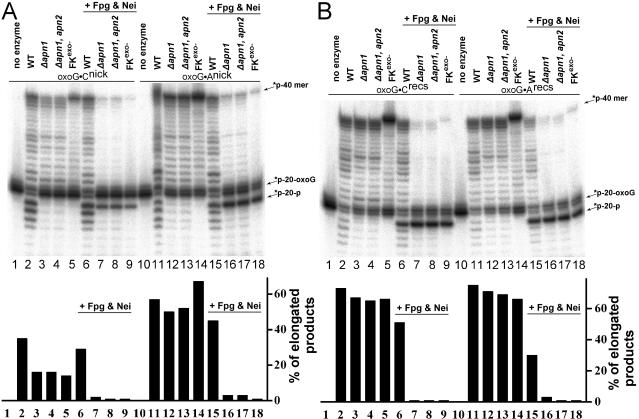
To examine whether Apn1 eliminates 8oxoG residues incorporated during DNA synthesis at 3′ termini, we reconstituted the repair of 5′-labeled 8oxoG · Cnick, 8oxoG · Anick, 8oxoG · Crecs,and 8oxoG · Arecs duplexes in yeast cell extracts of WT, apn1Δ, and apn1Δ apn2Δ strains (Fig. 5). In these experiments, the FKexo− DNA polymerase lacking proofreading function was used as a control for blunt DNA synthesis. Following incubation of the oligonucleotides with extracts, we checked the newly synthesized DNA for 8oxoG residues by treating it with the Fpg and Nei proteins that are known to excise the oxidized base (16, 46). The appearance of the 3′-phosphorylated 20-mer fragment lacking an 8oxoG residue after DNA glycosylase treatments will indicate the presence of 8oxoG in the elongated fragments. It should be noted that Nei and excess of Fpg can also excise 8oxoG when it is opposite to A (4, 16). As shown in Fig. 5A, the 5′-labeled 21-mer fragment in 8oxoG · Cnick and 8oxoG · Anick was efficiently elongated in WT extracts generating full-sized 40-mer product (lanes 2 and 11). Fpg/Nei treatment of the elongated products generated a 20-mer fragment and slightly reduced the amount of 40-mer fragments, indicating that nearly 20% of the newly synthesized DNA in WT extracts contained 8oxoG residues (Fig. 5A, lanes 6 and 15). This result suggests that 80% of the 8oxoG residues are efficiently eliminated during DNA synthesis in WT extracts. The elongation of the 8oxoG · Cnick oligonucleotide in apn1Δ and apn1Δ apn2Δ extracts (Fig. 5A, lanes 3 and 4) was less efficient (15%) than WT (34%). Furthermore, >90% of the newly synthesized DNA fragments in apn1Δ mutants contained 8oxoG residue (Fig. 5A, lanes 7 and 8), suggesting that Apn1 is required to eliminate the majority of 8oxoG residues during DNA synthesis. Similarly, after incubation of the 5′-labeled 8oxoG · Anick in either apn1Δ or apn1Δ apn2Δ extracts, >95% of the newly synthesized DNA fragments (Fig. 5A, lanes 12 and 13, respectively) contained 8oxoG residue (Fig. 5A, lanes 16 and 17, respectively), suggesting that Apn1 can eliminate 8oxoG when it is opposite to A. As expected, incubation of the 8oxoG · Cnick and 8oxoG · Anick duplexes with FKexo− produced elongated fragments containing 8oxoG residues (Fig. 5A, lanes 5 and 14, respectively), and the Fpg/Nei treatment cleaved >95% of the newly synthesized products (Fig. 5A, lanes 9 and 18, respectively).
FIG. 5.
Repair of 3′-terminal 8oxoG in yeast cell extracts. (A) Nicked duplex oligonucleotides. (B) Recessed duplex oligonucleotides. The 5′ 32P-labeled duplex was incubated with either crude extract or FKexo−. The presence of 8oxoG in the elongated products was revealed by subsequent Fpg/Nei treatment. Reaction products were separated in the gel, and the amount of elongated products was quantified. *p-20-oxoG is a 21-mer oligonucleotide containing 8oxoG at the 3′ end; *p-20-p is a Fpg/Nei incision product lacking the 8oxoG base and containing 3′ phosphate. For details, see Materials and Methods.
Similar results were obtained with 5′-labeled 8oxoG · Crecs and 8oxoG · Arecs duplexes, suggesting that Apn1 may eliminate 8oxoG residues during DNA replication (Fig. 5B). However, DNA synthesis from the recessed duplexes was about two times more efficient than the nicked substrates. Furthermore, in WT extracts, 30% and 60% of newly synthesized DNA product generated from the 8oxoG · Crecs and 8oxoG · Arecs substrates, respectively, contained 8oxoG residues (Fig. 5B, lanes 6 and 15), suggesting that DNA polymerases compete with Apn1 and prevent removal of mutagenic adducts from the 3′ termini. Interestingly, the higher level of 8oxoG present in the synthesized DNA from the 8oxoG · Arecs substrate was consistent with a recent report demonstrating that 8oxoG · A resembles normal base pairing without distortion and thus evades detection, whereas 8oxoG · C causes localized distortion, which is recognized as a mismatch (17).
Apn1 and Ogg1 represent overlapping pathways for repair of misincorporated 8oxoG residues in DNA.
Based on the above biochemical data, we hypothesized that Apn1 3′→5′ exonuclease activity would clean up newly synthesized DNA from misincorporated 8oxoG residues and thus perform a function similar to that of the MutT and MTH1 proteins in E. coli and human, respectively. Furthermore, if Apn1 reduces the level of 8oxoG residues in the genome, it may also serve as a back up function for Ogg1. Therefore, to investigate whether Apn1 can initiate an alternative repair pathway to 8oxoG-DNA glycosylase and 8oxodGTP phosphotase, we constructed apn1Δ ogg1Δ double mutant and a plasmid expressing the E. coli MutT protein in S. cerevisiae. As shown in Table 2, apn1Δ and ogg1Δ single mutants displayed 4- and 10-fold increases in the rate of spontaneous forward can1r mutations and that expression of GST-tagged MutT in these strains did not significantly suppress the spontaneous mutator phenotype. Remarkably, the apn1Δ ogg1Δ double mutant exhibited a synergistic 45-fold increase in the rate of spontaneous can1r mutations (Table 2). Furthermore, expression of GST-MutT reduced the mutation rate of the apn1Δ ogg1Δ double mutant by nearly twofold (Table 2). Importantly, the MutT effect was small but statistically significant.
TABLE 2.
Spontaneous rates and spectrum of canr mutations in yeast DNA repair deficient mutants
| Straina | Expt | Mutation rate canr/cell generation (10−8) | Fold increase |
|---|---|---|---|
| FF18733/pYES | 1 | 1.9 ± 0.8 | 1 |
| 2 | 2.1 ± 0.6 | 1 | |
| FF18733/pGst-MutTb | 1 | 1.7 ± 0.7 | |
| 2 | 1.8 ± 0.5 | ||
| DRY139 (apn1Δ::LEU2)/pYES | 1 | 7.7 ± 1.3 | 4 |
| 2 | 8.3 ± 1.8 | 3.9 | |
| DRY139 (apn1Δ::LEU2)/pGst-MutT | 1 | 6.8 ± 1.2 | 3.2 |
| 2 | 5.7 ± 1.4 | 3.2 | |
| CD138 (ogg Δ::TRP1)/pYES | 1 | 20.3 ± 3.1 | 10.7 |
| 2 | 23.7 ± 2.8 | 11.2 | |
| CD138 (ogg1 Δ::TRP1)/pGst-MutT | 1 | 16.5 ± 2.6 | 9.7 |
| 2 | 15.3 ± 2.9 | 8.5 | |
| DRY140 (apn1Δ::LEU2 | 1 | 89.0 ± 9.3 | 46.7 |
| ogg1Δ::TRP1)/pYES | 2 | 93.0 ± 11.3 | 44.3 |
| DRY140 (apn1Δ::LEU2 | 1 | 55.0 ± 6.4 | 26.2 |
| ogg1Δ::TRP1)/pGst-MutT | 2 | 49.0 ± 5.8 | 27.2 |
| DRY142 (apn1Δ::LEU2 | 1 | 105.0 ± 12.2 | 55.3 |
| ogg1Δ::TRP1 rad30::KAN)/pYES | 2 | 113.0 ± 9.7 | 53.8 |
All strains were isogenic to the parent strain FF18733 (kindly provided by Serge Boiteux).
The entire coding sequence of the E. coli mutT gene was cloned by gap repair into pYEX-Phiz, which contains the gst gene under the control of a copper-inducible promoter, to produce plasmid pGst-MutT. pGst-MutT expresses a 47-kDa protein.
It is well documented that the translesion synthesis DNA polymerase η (Rad30) is required to bypass a 8oxoG lesion on a template DNA by the insertion of dCMP, thereby restoring the correct genetic information for the next round of DNA replication (54, 55). The ogg1Δ rad30Δ double mutant displays a higher rate of can1r forward mutations than the ogg1Δ or rad30Δ single mutants, indicating that the DNA polymerase η prevents mutagenic replication of 8oxoG in DNA (15). To examine whether Apn1 mutation might be synergistic with deficient translesion synthesis (TLS) we constructed the apn1Δ ogg1Δ rad30Δ triple mutant. As shown in Table 2, the triple mutant displayed an only slightly enhanced spontaneous mutation rate compared to the apn1Δ ogg1Δ double mutant. This result may suggest that in the absence of Ogg1, Apn1 and Rad30 are involved in the same repair pathway to prevent 8oxoG-induced mutagenesis. Taken together, these genetic data are consistent with the notion that Apn1 3′→5′ exonuclease eliminates 8oxoG after incorporation by a DNA polymerase, thereby preventing accumulation of this oxidized base in DNA.
The apn1Δ ogg1Δ double mutant shows a strong preference for G · C to T · A and A · T to C · G transversion mutations.
If 8oxoG residues are at the origin of the spontaneous mutator phenotype of an apn1Δ ogg1Δ double mutant, the spectrum of spontaneous can1r mutations in this strain should be strongly biased in favor of the G→T and A→C transversions. To test this hypothesis, sequence analyses of a number of independent can1r mutations were performed with the WT and mutant yeast strains. As shown in Table 3, the spectrum of can1r mutations in the WT strain was composed of G→T transversions (28.9%), −1 and −2 deletions (23.7%), insertions (13.2%), A→C transversions (10.5%), and other base pair substitutions (23.7%). An excess of G · C to T · A transversions in the WT strain used in the present work was similar to that obtained in a previous study (27). Remarkably, can1r mutation spectra in the apn1Δ ogg1Δ double mutant exhibited a strong bias in favor of G · C to T · A (50%) and T · A to G · C (25%) transversion events that represented 75% of all mutations (Table 3). Indeed, G→T and A→C events were 70- and 108-fold higher in the double mutant than in the WT strain. Interestingly, the mutant also exhibited a 65-fold-higher increase in G→C and A base substitutions than the WT strain. The high levels of spontaneous G→T and A→C tranversions in apn1Δ ogg1Δ strain suggests that misincorporated 8oxoG residues serve as the origin of the mutations.
TABLE 3.
Spectrum of canr mutations
| Mutation type |
APN1 OGG1
|
apn1Δ ogg1Δ
|
Fold increase | ||||
|---|---|---|---|---|---|---|---|
| No. | % | Rate (10−8) | No. | % | Rate (10−8) | ||
| G · C to T · A | 11 | 28.9 | 0.58 | 20 | 50 | 40.5 | 70 |
| A · T to C · G | 4 | 10.5 | 0.21 | 10 | 25 | 22.75 | 108 |
| A · T to G · C | 3 | 7.9 | 0.16 | ||||
| A · T to T · A | 3 | 5.3 | 0.11 | ||||
| G · C to A · T | 2 | 5.3 | 0.11 | 3 | 7.5 | 6.8 | 65 |
| G · C to C · G | 2 | 5.3 | 0.11 | 3 | 7.5 | 6.8 | 65 |
| Deletions | 9 | 23.7 | 0.47 | 2 | 5 | 4.55 | |
| Insertions | 5 | 13.2 | 0.26 | 1 | 2.5 | 2.28 | |
| Complexa | 1 | 2.5 | 2.28 | ||||
| Total | 38 | 40 | |||||
Complex mutation: C→G and insertion (+1 bp).
DISCUSSION
The misincorporation of regular and/or damaged nucleotides during DNA replication and repair processes results in mutations. For example, misincorporation of 8oxodGMP by DNA polymerases using 8oxodGTP as a precursor for DNA synthesis generates either 8oxoG · A or 8oxoG · C base pairs, yielding A · T to C · G and G · C to T · A transversion mutations, respectively, after the second round of DNA replication (5, 32). In E. coli and mammalians, the MutT and MTH1 proteins, respectively, sanitize the nucleotide precursor pool of 8oxodGTP by hydrolyzing it to 8oxodGMP and pyrophosphate, thereby preventing its misincorporation into the genome (44). The physiological relevance of this function is supported by the strong A · T to C · G mutator phenotype of MutT-deficient strains of E. coli (53). In S. cerevisiae, there is no evidence for a MutT-related protein; as such, one might expect this organism to display an increased level of A · T to C · G transversions. However, this is not the case, raising the possibility that a molecular mechanism(s) preventing the mutagenic action of 8oxodGTP exists in yeast (3, 35). One may suggest that this mechanism involves the function of Apn1, as the apn1Δ mutant exhibits a spectrum of mutations (T to G transversion) similar to that of E. coli mutT strains (27). While it was previously proposed that the increased T-to-G transversions in apn1Δ mutants are due primarily to unrepaired AP sites (27), here we demonstrate that these mutations might occur as a result of the mutant inability to remove misincorporated 8oxoG. Furthermore, the fact that Apn1 also plays a key role in the NIR pathway for oxidatively damaged bases implies that unrepaired oxidized bases may also contribute to the mutator phenotype (18).
In this work, we demonstrated that Apn1 has an efficient exonuclease activity towards duplex DNA under reaction conditions that are also optimal for its AP endonuclease activity. The enzyme exhibits progressive exonuclease activity and prefers a 5′-tailed recessed duplex as a substrate (Fig. 1 and 3B). Apn1 can also process fully blunt-ended duplex oligonucleotide, albeit at lower efficiency. We show that the 3′-terminal THF residue is a preferred substrate for Apn1 (kcat/Km = 0.68), which suggests that the 3′→5′ exonuclease serves to eliminate 3′-blocking groups (Table 1). Comparison of Apn1 kinetic parameters for AP endonuclease (THF · G; kcat/Km = 0.48) and 3′→5′ exonuclease (G · Crecs; kcat/Km = 0.19) activities shows that relative exonuclease activity is high. Previously, it was shown that with a 39-mer nicked duplex oligonucleotide containing four sequential C · G pairs at the 3′ side of the nick, Apn1 excises only a single nucleotide without proceeding further (48). Here, we show that the kcat/Km value for the Apn1 exonuclease with the C · Grecs substrate (kcat/Km = 0.01) was 20-fold lower than that for G · Crecs (kcat/Km = 0.19), indicating that the exonuclease activity strongly depends on the DNA sequence context.
Recently, Cheng and colleagues (6) have shown that recombinant histidine-tagged Ape1 protein removes 3′-terminal nucleotides from mismatched T · G, A · G and G · G pairs much more efficiently (∼35 to 50 fold) than from the matched C · G pair. The authors proposed that Ape1 may ensure fidelity of the BER pathway in vivo, since DNA polymerase β lacks a proofreading exonuclease activity (7). In this work, we demonstrate that Apn1, Nfo, Xth, and Ape1 eliminate mismatched and matched nucleotides, as well as 8oxoG that is present at the 3′ termini of nick or gap sites, with good efficiency (Fig. 2). Although Apn1 degraded the G · Arecs and A · Grecs substrates more efficiently than the corresponding matched T · Arecs and A · Trecs duplexes, overall we did not observe strong preference for mismatched and oxidized nucleotides. For example, using a nontagged Ape1, we found only a two- to threefold difference between matched and mismatched pairs. In fact, similar results were obtained by other groups (29, 52). We suggest that two factors may account for the differences in substrate specificity reported for Ape1: the N-terminal histidine tag and the use of C · G pair as a matched control instead of a G · C pair.
To substantiate the role of Apn1 exonuclease, we reconstituted the repair of nicked and recessed duplex oligonucleotides in cell extracts of S. cerevisiae WT, apn1Δ, and apn1Δ apn2Δ strains. We demonstrated that Apn1-dependent removal of THF is indispensable for the repair of the THF · Gnick duplex (Fig. 4). Furthermore, elimination of the 3′-terminal mismatched regular nucleotide (A · Gnick) by Apn1 also stimulates the restoration of the nicked duplex. In contrast, DNA synthesis from the matched base pair was slightly less efficient in WT and was perhaps due to the competing action of an active 3′→5′ exonuclease, which is not present in either the apn1Δ or apn1Δ apn2Δ extracts (Fig. 4). Results obtained with the cell extracts support the idea that in vivo Apn1 exonuclease might be involved in the repair of single-strand breaks containing 3′-blocking groups and mismatched nucleotides.
Since purified Apn1 degrades 8oxoG · Crecs more efficiently than corresponding matched G · Crecs duplex, we speculated that Apn1 might provide the damage-cleansing function to replicative DNA polymerases. Therefore, we examined whether Apn1 might stimulate DNA synthesis in cell extracts by excision of the 3′-terminal 8oxoG residues in 8oxoG · C and 8oxoG · A duplexes (Fig. 5). As expected, elimination of the oxidized nucleotides present at the 3′ end of the nick or the gap strongly depends on Apn1, indicating the involvement of 3′→5′ exonuclease in the specific cleansing of newly synthesized DNA. Interestingly, DNA polymerase-catalyzed extension beyond 8oxoG can prevent Apn1-dependent removal of the oxidized nucleotides in the recessed duplexes and (to a lesser extent) in the nicked duplexes. We note that although Apn2 contains strong 3′→5′ exonuclease activity, we did not observe any significant contribution by this enzyme to the repair of 3′-terminal residues in cell extracts, suggesting that Apn2-catalyzing activities are minor to that of Apn1. On the basis of our findings, it appears that Apn1-dependent excision of misincorporated 8oxoG may constitute a repair pathway specific to the newly synthesized DNA. Consistent with the above results, it was recently demonstrated that Ape1 is the major activity involved in the repair of 3′-terminal 8oxoG residues in the reconstituted system and in the human cell extracts (G. Dianov, personal communication).
In S. cerevisiae, three repair networks prevent the mutagenic action of 8oxoG residues occurring in DNA template: the 8oxoG-DNA glycosylase Ogg1 (49), the MMR system (35), and the postreplication repair system consisting of DNA polymerase η and the Rad6-Rad18 complex (9, 15). At present, little is known about the mechanisms for sanitization of oxidatively damaged precursors during DNA synthesis in budding yeast. It has been proposed that the MMR system might act on mispairs resulting from misincorporation of 8oxodGTP opposite A (35). Indeed, Colussi and et al. (8) showed that mammalian MMR could selectively eliminate 8oxoG incorporated from the oxidized dNTP pool. However, S. cerevisiae MMR-deficient mutants do not display an increased rate of A→C transversions, and the human MMR pathway does not discriminate misincorporated 8oxoG, suggesting the existence of an alternative repair pathway (28, 35). Intriguingly, data obtained with the yeast extracts suggest that Apn1 reduces 8oxoG incorporation opposite C and A; therefore, this enzyme might prevent G→T and A→C transversions. This would imply that Apn1 could provide a backup mechanism for the 8-oxoguanine-DNA glycosylase and the MMR system in S. cerevisiae. To support this notion, we compared the spontaneous mutation rates in apn1Δ single, ogg1Δ single, and apn1Δ ogg1Δ double mutants and the strains harboring a plasmid construct that expresses the E. coli MutT protein. Consistent with the biochemical data, the apn1Δ ogg1Δ mutant exhibited a synergistic (more than additive) 4- to 10-fold increase in can1r mutation compared to the single mutants (Table 2). Although deletion of the major AP endonuclease activity in cells severely disabled the BER pathway, the synergistic interaction between APN1and OGG1 indicated that Apn1 is involved in the DNA glycosylase-independent repair pathway for 8oxoG residues. Furthermore, the expression of MutT in apn1Δ ogg1Δ double mutant suppressed by a factor of two the spontaneous mutation rate, suggesting that Apn1 reduces the misincorporation of oxidized nucleotides during DNA replication. It is noteworthy that MutT was unable to restore the normal spontaneous mutation frequency to the apn1Δ, ogg1Δ, and apn1Δ ogg1Δ mutants. The absence of a MutT homologue in S. cerevisiae suggests that MMR and Apn1 may be functional analogues of MutT in yeast. Therefore, we propose that these alternative repair pathways render the heterologous 8oxodGTP triphosphatase less important for sanitization of the nucleotide pool in yeast.
The work of several groups has established that in S. cerevisiae, MMR system and error-free translesion DNA synthesis cooperate with Ogg1 to prevent the mutagenic effect of 8oxoG residues (9, 15, 35). In the proposed model, the DNA polymerase η coded by gene RAD30 promotes error-free bypass of 8oxoG, following the MMR-dependent removal of A mispaired with 8oxoG. Consistent with this, the mutation rate was about the same in the msh2Δ and msh2Δ rad30Δ strains and in the ogg1Δ msh2Δ and ogg1Δ msh2Δ rad30Δ strains, indicating a role for RAD30 in the postreplication repair pathway for 8oxoG residues. Here, we examined whether apn1Δ ogg1Δ double mutation is synergistic with RAD30. We show that the ogg1Δ apn1Δ rad30Δ strain has only slight increase in can1r mutability compared to ogg1Δ apn1Δ strain suggesting an epistatic interaction of Apn1 with postreplication repair pathways. Since DNA polymerase η prevents mutation when 8oxoG occurs in the template strand, it is tempting to speculate that the increased mutation rates in the ogg1Δ apn1Δ rad30Δ compared to the ogg1Δ apn1Δ mutant are mainly due to the direct oxidation of DNA bases. It should be noted that elevated mutation rates exhibited by the apn1Δ ogg1Δ rad30Δ triple mutant cannot be due to unrepaired AP sites accumulating in this mutant, as these lesions are mutagenic in the presence of Rad30 and not in its absence (56).
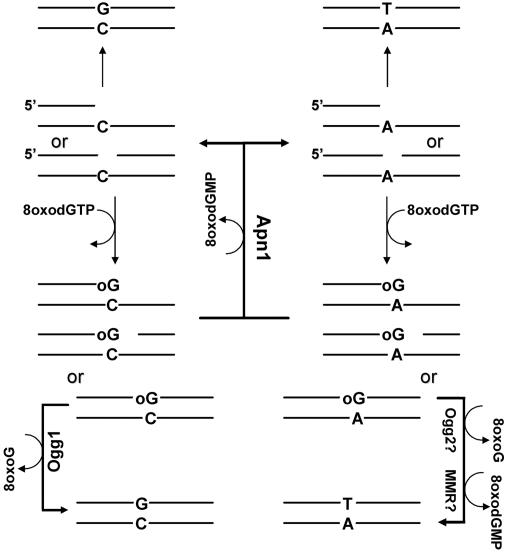
The absence of MutY- and MutT-related proteins may suggest that proofreading activities of the yeast replicative DNA polymerases might decrease the 8oxodGMP misincorporation. However, recent structural studies of high-fidelity DNA polymerases revealed that 8oxoG can form mismatches that evade polymerase error detection mechanisms, potentially leading to the stable incorporation of lethal mutations (11, 17). Indeed, genetic studies of yeast and the present study indicate that DNA replication machinery promotes the mutagenic incorporation of dAMP opposite to 8oxodGMP (15, 35). Furthermore, it has been demonstrated that the oxidized dNTP pool makes a significant contribution to the steady-state level of 8oxoG in DNA of mismatch repair-deficient cells, indicating that DNA replication machinery is prone to misincorporate 8oxodGMP (42). On the basis of our biochemical and genetic results, we propose a model (Fig. 6) where the 3′→5′ exonuclease activity of Apn1 prevents the mutagenic effect of 8oxoG. Although the level of specificity of Apn1 for a modified 3′ nucleotide is not high, we propose that the stalled DNA polymerase might direct Apn1 to 3′ blocking termini in vivo. Indeed, the misincorporation of 8oxodGMP during DNA synthesis results in very low nucleotide extension beyond 8oxoG (10). In addition, the polymerase switches from high-fidelity replicase to low-fidelity lesion bypass polymerases, once it encounters the damaged DNA (36, 38). Therefore, in our current model, repair of the newly synthesized DNA take place when the stalled DNA polymerase switches to accommodate the 3′→5′ exonuclease activity of Apn1, resulting in the exonucleolytic removal of the mutagenic adduct, e.g., 8oxoG.
FIG. 6.
Role of Apn1 in the prevention of 8oxoG-induced mutagenesis in S. cerevisiae. Misincorporation of 8oxodGMP by DNA polymerases with 8oxodGTP as a precursor for DNA synthesis generates either oG · A and oG · C base pairs at 3′ termini of nick and gap sites. Apn1 eliminates 8oxoG at the 3′ termini, restoring efficient DNA synthesis. In the absence of Apn1, DNA polymerase and DNA ligase generate fully duplex DNA containing 8oxoG opposite to C or A. 8oxoG in fully duplex DNA is removed either by the Ogg1 glycosylase or probably by MMR and Ogg2.
The fact that the 3′→5′ exonuclease function is evolutionarily conserved among the major AP endonucleases of bacterial, yeast, and mammalian origins underscores the importance of this activity in performing a damage-cleansing function. In the case of Apn1, we demonstrate that it contains progressive exonuclease activity, which is as efficient as its AP endonuclease. We therefore propose that Apn1 exonuclease serves as an alternative backup function to the BER pathway and to the sanitization of the nucleotide precursor pool of 8oxodGTP in S. cerevisiae. Observations that ogg1−/− null mice have no marked tumor predisposition (26, 34) indicate redundancy in the repair mechanisms and perhaps a possible role of AP endonucleases in preventing the accumulation of 8oxoG residues in the genome. In conclusion, an alternative DNA glycosylase-independent repair pathway for highly mutagenic lesions initiated by AP endonucleases provides new insights into a molecular mechanism assuring the stability of the genome and cancer prevention.
Acknowledgments
We thank Jacques Laval and Pat Foster for discussions, Dmitry Zharkov for the E. coli Nei protein, and Grigory Dianov for sharing results prior to publication.
This work was supported by grants from the European Community (RISC-RAD FI6R-CT-2003-508842), the Association pour la Recherche sur le Cancer, and Electricité de France Contrat Radioprotection (to M.S.), and the Natural Sciences and Engineering Council of Canada (to D.R.). A.A.I. was supported by a postdoctoral fellowships from the Association pour la Recherche sur le Cancer. D.R. is the recipient of a senior scholarship from the Fonds de la Recherche en Sante du Quebec.
REFERENCES
- 1.Barnes, D. E., and T. Lindahl. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38:445-476. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, R. A. 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 19:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boiteux, S., L. Gellon, and N. Guibourt. 2002. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 32:1244-1253. [DOI] [PubMed] [Google Scholar]
- 4.Castaing, B., A. Geiger, H. Seliger, P. Nehls, J. Laval, C. Zelwer, and S. Boiteux. 1993. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 21:2899-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, K. C., D. S. Cahill, H. Kasai, S. Nishimura, and L. A. Loeb. 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G → T and A → C substitutions. J. Biol. Chem. 267:166-172. [PubMed] [Google Scholar]
- 6.Chou, K. M., and Y. C. Cheng. 2003. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J. Biol. Chem. 278:18289-18296. [DOI] [PubMed] [Google Scholar]
- 7.Chou, K. M., and Y. C. Cheng. 2002. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature 415:655-659. [DOI] [PubMed] [Google Scholar]
- 8.Colussi, C., E. Parlanti, P. Degan, G. Aquilina, D. Barnes, P. Macpherson, P. Karran, M. Crescenzi, E. Dogliotti, and M. Bignami. 2002. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12:912-918. [DOI] [PubMed] [Google Scholar]
- 9.de Padula, M., G. Slezak, P. Auffret van Der Kemp, and S. Boiteux. 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 32:5003-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einolf, H. J., and F. P. Guengerich. 2001. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 276:3764-3771. [DOI] [PubMed] [Google Scholar]
- 11.Freisinger, E., A. P. Grollman, H. Miller, and C. Kisker. 2004. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 23:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grollman, A. P., and M. Moriya. 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9:246-249. [DOI] [PubMed] [Google Scholar]
- 13.Gros, L., A. A. Ishchenko, H. Ide, R. H. Elder, and M. K. Saparbaev. 2004. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 32:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gros, L., M. K. Saparbaev, and J. Laval. 2002. Enzymology of the repair of free radicals-induced DNA damage. Oncogene 21:8905-8925. [DOI] [PubMed] [Google Scholar]
- 15.Haracska, L., S. L. Yu, R. E. Johnson, L. Prakash, and S. Prakash. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 25:458-461. [DOI] [PubMed] [Google Scholar]
- 16.Hazra, T. K., T. Izumi, R. Venkataraman, Y. W. Kow, M. Dizdaroglu, and S. Mitra. 2000. Characterization of a novel 8-oxoguanine-DNA glycosylase activity in Escherichia coli and identification of the enzyme as endonuclease VIII. J. Biol. Chem. 275:27762-27767. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, G. W., M. Ober, T. Carell, and L. S. Beese. 2004. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 431:217-221. [DOI] [PubMed] [Google Scholar]
- 18.Ischenko, A. A., and M. K. Saparbaev. 2002. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415:183-187. [DOI] [PubMed] [Google Scholar]
- 19.Ishchenko, A. A., G. Sanz, C. V. Privezentzev, A. V. Maksimenko, and M. Saparbaev. 2003. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 31:6344-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jilani, A., R. Vongsamphanh, A. Leduc, L. Gros, M. Saparbaev, and D. Ramotar. 2003. Characterization of two independent amino acid substitutions that disrupt the DNA repair functions of the yeast Apn1. Biochemistry 42:6436-6445. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, A. W., and B. Demple. 1988. Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: substrate specificity and kinetics. J. Biol. Chem. 263:18017-18022. [PubMed] [Google Scholar]
- 22.Johnson, A. W., and B. Demple. 1988. Yeast DNA diesterase for 3′-fragments of deoxyribose: purification and physical properties of a repair enzyme for oxidative DNA damage. J. Biol. Chem. 263:18009-18016. [PubMed] [Google Scholar]
- 23.Kamath-Loeb, A. S., A. Hizi, H. Kasai, and L. A. Loeb. 1997. Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J. Biol. Chem. 272:5892-5898. [DOI] [PubMed] [Google Scholar]
- 24.Kasai, H., P. F. Crain, Y. Kuchino, S. Nishimura, A. Ootsuyama, and H. Tanooka. 1986. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 7:1849-1851. [DOI] [PubMed] [Google Scholar]
- 25.Kerins, S. M., R. Collins, and T. V. McCarthy. 2003. Characterization of an endonuclease IV 3′-5′ exonuclease activity. J. Biol. Chem. 278:3048-3054. [DOI] [PubMed] [Google Scholar]
- 26.Klungland, A., I. Rosewell, S. Hollenbach, E. Larsen, G. Daly, B. Epe, E. Seeberg, T. Lindahl, and D. E. Barnes. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96:13300-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunz, B. A., E. S. Henson, H. Roche, D. Ramotar, T. Nunoshiba, and B. Demple. 1994. Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc. Natl. Acad. Sci. USA 91:8165-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson, E. D., K. Iams, and J. T. Drummond. 2003. Strand-specific processing of 8-oxoguanine by the human mismatch repair pathway: inefficient removal of 8-oxoguanine paired with adenine or cytosine. DNA Repair (Amsterdam) 2:1199-1210. [DOI] [PubMed] [Google Scholar]
- 29.Lebedeva, N. A., S. N. Khodyreva, A. Favre, and O. I. Lavrik. 2003. AP endonuclease 1 has no biologically significant 3′ → 5′-exonuclease activity. Biochem. Biophys. Res. Commun. 300:182-187. [DOI] [PubMed] [Google Scholar]
- 30.Leduc, A., C. H. He, and D. Ramotar. 2003. Disruption of the Saccharomyces cerevisiae cell-wall pathway gene SLG1 causes hypersensitivity to the antitumor drug bleomycin. Mol. Genet. Genomics 269:78-89. [DOI] [PubMed] [Google Scholar]
- 31.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistane. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 33.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minowa, O., T. Arai, M. Hirano, Y. Monden, S. Nakai, M. Fukuda, M. Itoh, H. Takano, Y. Hippou, H. Aburatani, K. Masumura, T. Nohmi, S. Nishimura, and T. Noda. 2000. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA 97:4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni, T. T., G. T. Marsischky, and R. D. Kolodner. 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell 4:439-444. [DOI] [PubMed] [Google Scholar]
- 36.Plosky, B. S., and R. Woodgate. 2004. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr. Opin. Genet. Dev. 14:113-119. [DOI] [PubMed] [Google Scholar]
- 37.Popoff, S. C., A. I. Spira, A. W. Johnson, and B. Demple. 1990. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA 87:4193-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 39.Ramotar, D. 1997. The apurinic-apyrimidinic endonuclease IV family of DNA repair enzymes. Biochem. Cell. Biol. 75:327-336. [PubMed] [Google Scholar]
- 40.Ramotar, D., S. C. Popoff, E. B. Gralla, and B. Demple. 1991. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 11:4537-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose, M. D. 1987. Isolation of genes by complementation in yeast. Methods Enzymol. 152:481-504. [DOI] [PubMed] [Google Scholar]
- 42.Russo, M. T., M. F. Blasi, F. Chiera, P. Fortini, P. Degan, P. Macpherson, M. Furuichi, Y. Nakabeppu, P. Karran, G. Aquilina, and M. Bignami. 2004. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell. Biol. 24:465-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 44.Sekiguchi, M., and T. Tsuzuki. 2002. Oxidative nucleotide damage: consequences and prevention. Oncogene 21:8895-8904. [DOI] [PubMed] [Google Scholar]
- 45.Sherman, F., G. R. Fink, and J. B. Hicks. 1982. Methods in yeast genetics: laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Tchou, J., H. Kasai, S. Shibutani, M. H. Chung, J. Laval, A. P. Grollman, and S. Nishimura. 1991. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. USA 88:4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unk, I., L. Haracska, S. Prakash, and L. Prakash. 2001. 3′-Phosphodiesterase and 3′→5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 21:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vance, J. R., and T. E. Wilson. 2001. Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3′ phosphatases. Mol. Cell. Biol. 21:7191-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Kemp, P. A., D. Thomas, R. Barbey, R. de Oliveira, and S. Boiteux. 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93:5197-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Von Borstel, R. C. 1978. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 20:1-24. [DOI] [PubMed] [Google Scholar]
- 51.Vongsamphanh, R., P. K. Fortier, and D. Ramotar. 2001. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol. 21:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, D. M., IIII. 2003. Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J. Mol. Biol. 330:1027-1037. [DOI] [PubMed] [Google Scholar]
- 53.Yanofsky, C., E. C. Cox, and V. Horn. 1966. The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl. Acad. Sci. USA 55:274-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, F., Y. Zhang, D. K. Rajpal, X. Wu, D. Guo, M. Wang, J. S. Taylor, and Z. Wang. 2000. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J. Biol. Chem. 275:8233-8239. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., F. Yuan, X. Wu, O. Rechkoblit, J. S. Taylor, N. E. Geacintov, and Z. Wang. 2000. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 28:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, B., Z. Xie, H. Shen, and Z. Wang. 2004. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 32:3984-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]