Abstract
Here we describe 2 mutations in growth and differentiation factor 5 (GDF5) that alter receptor-binding affinities. They cause brachydactyly type A2 (L441P) and symphalangism (R438L), conditions previously associated with mutations in the GDF5 receptor bone morphogenetic protein receptor type 1b (BMPR1B) and the BMP antagonist NOGGIN, respectively. We expressed the mutant proteins in limb bud micromass culture and treated ATDC5 and C2C12 cells with recombinant GDF5. Our results indicated that the L441P mutant is almost inactive. The R438L mutant, in contrast, showed increased biological activity when compared with WT GDF5. Biosensor interaction analyses revealed loss of binding to BMPR1A and BMPR1B ectodomains for the L441P mutant, whereas the R438L mutant showed normal binding to BMPR1B but increased binding to BMPR1A, the receptor normally activated by BMP2. The binding to NOGGIN was normal for both mutants. Thus, the brachydactyly type A2 phenotype (L441P) is caused by inhibition of the ligand-receptor interaction, whereas the symphalangism phenotype (R438L) is caused by a loss of receptor-binding specificity, resulting in a gain of function by the acquisition of BMP2-like properties. The presented experiments have identified some of the main determinants of GDF5 receptor-binding specificity in vivo and open new prospects for generating antagonists and superagonists of GDF5.
Introduction
The development of the appendicular skeleton follows a sequence of highly regulated events beginning with the condensation of skeletal precursor cells and their subsequent differentiation into chondrocytes (1). This process is controlled by a complex molecular network of signaling pathways, one of the most prominent being the signaling cascade of the TGF-β superfamily. To date, at least 25 different bone morphogenetic proteins (BMPs) and growth and differentiation factors (GDFs) that belong to this superfamily have been described (2). Like all BMPs, GDF5 is synthesized as a larger precursor molecule that is subsequently processed to mature proteins that form homo- and heterodimers with other BMPs. The mature domain is highly conserved between BMPs and GDFs, which contain 7 cysteine residues; 6 of these form intrachain disulfide bonds, creating a rigid 3D cysteine-knot structure, whereas the fourth of the 7 cysteines is needed for dimerization (3). Part of the necessary specificity of BMP action is ensured by differential affinities of distinct ligands to their receptors. Signaling of the TGF-β superfamily members requires the binding of the ligand to cell surface receptors consisting of 2 types of transmembrane serine/threonine kinase receptors classified as type 1 and type 2. For example, BMP2 binds with high affinity to the BMP receptors type 1A (BMPR1A) and type 1B (BMPR1B) whereas GDF5 preferentially binds to BMPR1B (4, 5). Intracellular effectors of the activated type 1 receptors are the SMADs that, when phosphorylated by the receptors, translocate to the nucleus, where they participate in the transcriptional regulation of genes involved in cartilage and bone formation (6–8).
Analysis of the brachypod (bp) mouse mutant showed that Gdf5 has a key role in chondrocyte differentiation and joint formation (9). Gdf5bp-J mice are characterized by a severe limb reduction phenotype consisting of shortening of the humerus and femur, hypoplasia of all phalanges and metacarpals, and lack of interphalangeal joints. This and the expression of Gdf5 in the areas of the joint interzone suggest a specific role in joint formation. However, Gdf5 is initially expressed in cells surrounding the condensations, and overexpression results in broader and longer cartilage but not in the formation of additional joints (10, 11). Furthermore, the abnormalities observed in Gdf5bp-J mice appear to be due to a defect in early chondrogenesis that results in smaller condensations (12). Thus, it is unlikely that the major role of Gdf5 is to specify joints. Rather, the molecule elicits its function by increasing the proliferation rate and recruitment of mesenchymal precursor cells (13). The chondrogenic potential of GDF5 is further supported by the finding that GDF5 administered subcutaneously or intramuscularly on carrier matrices induces cartilage and bone or dense connective tissue formation reminiscent of that in ectopic tendon (14, 15). Consequently, GDF5 has been considered for use as a pharmaceutical agent to induce cartilage and bone in, for example, a spinal fusion model (16).
Mutations in human GDF5 result in skeletal malformation syndromes including brachydactyly type C (BDC) (OMIM 113100) (17), a condition characterized by shortening of digits and hypersegmentation of phalanges, and the recessive acromesomelic dysplasias of the Hunter-Thompson, Grebe, and DuPan types, which are characterized by short stature, severe limb shortening, and profound brachydactyly (18–20). The great majority of mutations in GDF5 described so far are nonsense and frame shift mutations that presumably result in a complete loss of function. Dominant and recessive mutations in the prodomain of GDF5 have been described, but their role in the pathogenesis of BDC remains to be shown (21). Mutations in BMPR1B, the high-affinity receptor for GDF5, cause brachydactyly type A2 (BDA2) (OMIM 112600), a condition characterized by shortening of the index finger due to hypoplasia/aplasia of the middle phalanx (22). As shown by in vitro and in vivo overexpression studies, BDA2 mutations act in a dominant-negative manner by interfering with type 2 receptor–mediated transphosphorylation. In contrast, BMPR1B loss-of-function mutations are recessive and phenotypically more similar to homozygous GDF5 mutations (23). Inactivation of Bmpr1b in the mouse results in a comparable phenotype (24, 25).
The biological availability and thus activity of GDFs and BMPs is in part regulated by binding to strong inhibitors such as NOGGIN (NOG), CHORDIN, or GREMLIN (26). The inactivation of Nog in the mouse results in a massive overproduction of cartilage and a complete loss of joint formation (27). Heterozygous loss-of-function mutations in human NOG have a similar albeit less severe effect that causes proximal symphalangism (SYM1) (OMIM 185800), carpal-tarsal coalition syndrome (TCC) (OMIM 186570), and multiple synostosis syndrome (SYNS1) (OMIM 186500) (28–31). All 3 conditions are characterized by the absence of joint cartilage and bony fusions of the affected elements. Thus, mutations interfering with the stochiometry of the BMP–BMP-inhibitor complex result in brachydactyly and abnormal joint formation.
Here we present the molecular analysis of 2 mutations in GDF5 that give rise to opposing phenotypes, brachydactyly with shortening or loss of phalanges and SYM1 with joint fusions. The 2 mutations are located in close proximity to each other within the receptor-interaction interface. The brachydactyly mutation (L441P) results in a loss of function through a reduced binding affinity to the BMPR1B receptor, whereas the SYM1 mutation (R438L) causes increased activity in all functional tests, a gain of function likely mediated through higher binding affinity to the BMPR1A receptor.
Results
Point mutations in GDF5 result in BDA2 and proximal SYM1.
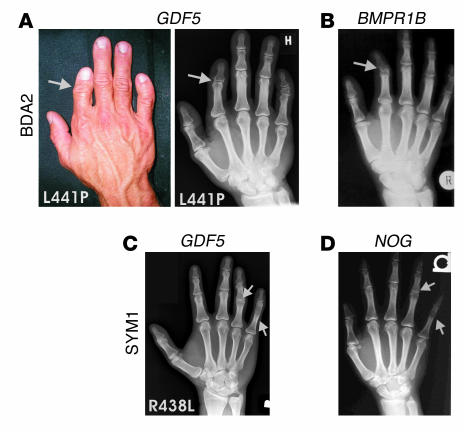
BDA2 was clinically diagnosed in patients from a large pedigree who showed short index fingers and variable clinodactyly. X-rays showed hypoplasia or aplasia of the second phalanx of digit II and, to a variable extent, shortening and shape abnormalities of the middle phalanx of digit V. The phenotype was very similar to that in previously published cases of BDA2, but mutations in BMPR1B could not be identified. By screening for candidate genes, we identified a heterozygous T1322C mutation in GDF5 in all affected members of the family, resulting in the exchange of leucine at position 441 to proline (L441P). The phenotype is shown in Figure 1A. For comparison, the phenotype of an individual with a mutation in BMPR1B, as previously described (22), is also shown.
Figure 1.
Phenotypes of the right hands of patients carrying L441P and R438L mutations. (A) The L441P mutation is associated with brachydactyly, characterized by a short index finger and bending of finger V (clinodactyly). X-rays show missing middle phalanges in finger II and hypoplasia of the middle phalanges in finger V. (B) The phenotype is very similar to that of BDA2, which is caused by mutations in BMPR1B. (C) The R438L mutation results in SYM1, characterized by bony fusion of the proximal interphalangeal joint of finger V and an abnormal interphalangeal joint in finger IV. (D) The phenotype is very similar to that of SYM1, which is caused by mutations in NOG.
In a second family, SYM1 was diagnosed on the basis of patients’ inability to bend fingers IV and V in their interphalangeal joints and loss of flexion creases at the corresponding sites. X-rays of the hands showed a complete bony fusion between the proximal and middle phalanges of digit V and, to a lesser extent, digit IV. Screening for mutations in NOG, the gene known to be mutated in SYM1, showed no mutation. However, sequencing GDF5 revealed a change of G to T at position 1632 in all affected members of the family, resulting in the exchange of arginine at position 438 to leucine (R438L). The phenotype is shown in Figure 1C. For comparison, the phenotype of an individual with SYM1 and a mutation in NOG (28) is also shown.
R438L and L441P mutations are located within the receptor interaction site.
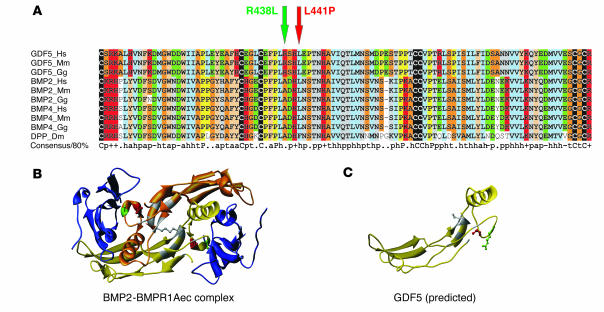
To study evolutionary conservation and to predict specific functions for the amino acid residues at the mutated sites, a multiple alignment between the highly conserved signaling domains of GDF5 and BMPs of different species was constructed. As shown in Figure 2A, both sites are completely conserved in GDF5 proteins of distantly related species. L441 is conserved in other members of the BMP family such as BMP2 or BMP4. In contrast, R438 is only present in members of the GDF family, whereas all BMPs including the drosophila BMP homolog decapentaplegic (DPP) have an alanine at this position. We used the BMP2-BMPR1A structure (Protein Data Bank [PDB] entry 1REW; http://www.rcsb.org/pdb/) as a model to simulate GDF5-BMPR1B interaction (Figure 2B). The alignment allowed us to locate homologous residues of GDF5 in the BMP2 structure. The mutated residues R438 and L441 are located within the receptor interaction interface of GDF5 (Figure 2C).
Figure 2.
Protein sequence alignment of GDF5 homologs and 3D models for GDF5 receptor binding. (A) Primary sequence alignment of GDF5, BMP2, and BMP4 from different species, including drosophila DPP. The positions of R438L and L441P mutations in GDF5 are indicated by arrows. Note the widespread conservation of residue L441 in BMPs and the specificity of R438 conservation for the GDF5 subfamily (replaced by A in the BMPs). (B) 3D model of a BMP2 dimer (yellow and gold) linked via a disulfide bridge and bound to the ectodomain of BMPR1A (BMPR1Aec). The amino acids mutated in GDF5 are indicated in red (L441) and green (R438). Note their position within the receptor interaction site. (C) Predicted structure of a GDF5 monomer. The positions of the mutated amino acids are indicated in red (L441) and green (R438).
Functional analysis of GDF5 mutants in micromass culture.
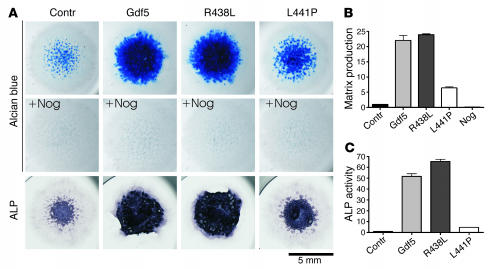
To analyze the functional consequences of the GDF5 mutations, we infected chicken micromass cultures with replication-competent avian sarcoma (RCAS) viruses expressing WT Gdf5 and mutated Gdf5 and measured cell differentiation and cartilaginous matrix production by alkaline phosphatase (ALP) and Alcian blue. As expected, infection of micromass cells with WT GDF5–expressing virus resulted in a massive induction of cartilage production as indicated by the increase in Alcian blue and ALP staining (Figure 3). In contrast, infection with the L441P mutant construct resulted in a moderate increase in staining and very little induction of ALP, which might be a secondary effect of increased proliferation. Infection with the R438L mutant resulted in strong induction of Alcian blue and ALP at levels similar to those found in WT Gdf5. Coinfection of cultures with Gdf5 (WT and mutants) and Nog completely inhibited cartilage formation.
Figure 3.
Functional analysis of Gdf5 mutants in micromass culture. (A) Chicken micromass cultures were assayed after 4 days for extracellular matrix production and analyzed after 7 days for ALP activity. Cells were infected with virus containing WT Gdf5 or mutant sequences and coinfected with Nog or not coinfected. The mutants differed drastically in their biological activity. Coinfection with Nog completely represses chondrogenesis irrespective of the Gdf5 variant expressed. Magnification, ×1 objective (AxioCam HRc camera; Zeiss). (B) Alcian blue incorporation into the extracellular matrix of micromass cultures reflecting the production of proteoglycan-rich cartilaginous matrix measured at day 4 was quantified after extraction. Biological activity of R438L mutant was comparable to that of Gdf5, whereas the L441P mutant displayed only small effects on matrix production. (C) ALP activity of micromass cultures at day 7 was quantified by a specific enzymatic assay. While Gdf5 and R438L induced ALP activity effectively, L441P caused only a small amount of induction, slightly above control activity. Contr, control.
Analysis of receptor specificity of GDF5 mutants in cell culture.
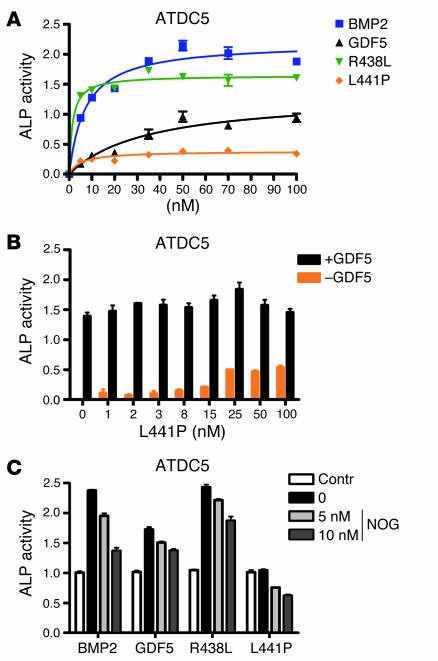
To further analyze the specific interaction of the mutants with the 2 different type 1 receptors, we constructed expression vectors encoding the GDF5 mutants and produced human recombinant mature GDF5. The protein was shown to be pure by silver-stained PAGE analysis. To identify functional differences among the group of mutants and between the mutants and WT Gdf5, we treated a chondrogenic progenitor cell line (ATDC5) and a premyoblastic cell line (C2C12) with the respective recombinant proteins. As shown in Figure 4A, treatment of ATDC5 cells with WT GDF5 protein resulted, as previously described, in an induction of ALP activity. In this system, BMP2 was even more potent when compared with GDF5. Stimulation of cells with the R438L mutant resulted in a strong ALP induction at levels similar to those in BMP2, indicating a gain of function for this mutant. In contrast, treatment with the L441P mutant caused only a low degree of induction, significantly less than with WT GDF5. Thus the L441P mutant has almost no activation capacity. To test the effect of the L441P mutant on WT GDF5, we stimulated cells with 10 nM WT GDF5 and added increasing amounts of L441P mutant (Figure 4B). The results showed neither an increase nor a decrease in ALP activity, with rising concentrations of the mutant indicating that L441P in this setting has no influence on WT GDF5 signaling. The induction of ALP activity by BMP2 was suppressed by the addition of recombinant NOG. Suppression was also obtained with WT GDF5 and the R438L mutant, suggesting that NOG was still able to effectively bind and inactivate the mutant (Figure 4C). However, GDF5 appeared to be inhibited less efficiently than BMP2.
Figure 4.
Characterization of ALP induction by GDF5 mutants in chondrogenic ATDC5 cells. ALP activity was measured after stimulation of ATDC5 cells with increasing amounts of recombinant proteins after 3 days. (A) Recombinant BMP2, GDF5, and R438L display characteristic dose-dependent inductions of ALP activity in ATDC5 cells. The R438L mutant induces significantly higher ALP activity than WT GDF5, whereas L441P is almost inactive. (B) Stimulation of ATDC5 cells with L441P or a combination of GDF5 and L441P. Cells were incubated with increasing amounts of L441P with or without 10 nM GDF5. No stimulating or suppressing effect of increasing amounts of L441P on GDF5-dependent induction of ALP activity was observed. (C) Inhibition of ALP induction by recombinant NOG in stimulated ATDC5 cells. ALP was determined after incubation with 5 nM recombinant BMP2, GDF5, R438L, or L441P, and cotreatment with increasing amounts of NOG. GDF5, R438L, and L441P display similar inhibition profiles by NOG.
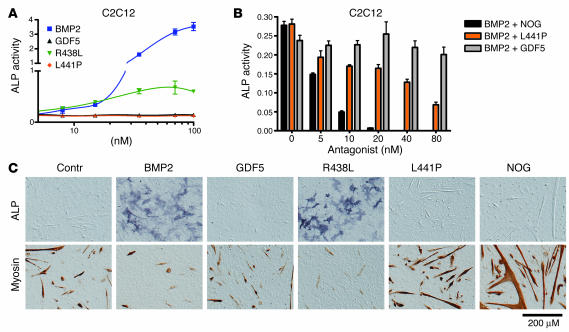
C2C12 cells showed strong induction after treatment with BMP2 but practically no response when treated with WT GDF5 (Figure 5A). The R438L mutant resulted in ALP induction significantly higher than that achieved with WT GDF5. C2C12 cells are mesenchymal progenitor cells that have the potential to differentiate into muscle cells, osteoblasts, or adipocytes. We made use of this quality by staining for osteogenesis (ALP activity) and myogenesis (myosin expression) to test the ability of the GDF5 mutants to direct cell differentiation (Figure 5C). Untreated cells differentiated into myocytes, but treatment with BMP2 resulted in strong induction of ALP, inducing osteogenesis. Treatment with GDF5 had no major effect, but the myotubes appeared smaller than in untreated cells. Inhibition of endogenous Bmp activity by NOG strongly increased the amount of myosin-positive cells. Similar results were obtained for the L441P mutant. Treatment of C2C12 cells with the R438L mutant, however, resulted in strong induction of ALP and suppression of myoblast formation, indicating that this mutant has a BMP2-like effect. We quantified ALP activity photometrically and showed that WT GDF5 and the L441P mutant had no effect whereas the R438L mutant resulted in ALP induction, albeit less than with WT BMP2. To test a dominant effect of the L441P mutant on BMP2, we treated cells with 5 nM BMP2 and increasing amounts of mutant protein. For comparison, we performed the same experiment for NOG and WT GDF5 (Figure 5B). The inductive activity of BMP2 was substantially suppressed by adding increasing concentrations of NOG. Likewise, the addition of L441P mutant resulted in BMP2 suppression, indicating that the mutant interfered with BMP2 signaling. The addition of WT GDF5 resulted in a small reduction of ALP activity, but overall there was no detectable major effect. Thus in this setting the L441P mutant inhibited BMP2-induced ALP activity with higher efficiency than WT GDF5.
Figure 5.
Effects of GDF5 mutants on the differentiation of premyoblastic C2C12 cells. ALP activity was analyzed after stimulation of C2C12 cells with the recombinant proteins for 3 days. Differentiation markers ALP and myosin were assayed 5 days after addition of the proteins. (A) WT GDF5 and the L441P mutant did not induce significant ALP activity in C2C12 cells. In contrast, treatment with BMP2 resulted in a strong induction, and the R438L mutant induced significant ALP activity, albeit to a lower degree than BMP2. (B) Addition of NOG inhibited BMP2-induced ALP activity in a dose-dependent fashion. A similar antagonism was observed with the addition of L441P mutant. The addition of WT GDF5 had no major effect. (C) Differentiation of C2C12 cells was determined using ALP staining (osteoblastic lineage) or immunohistochemical analysis of myosin expression (muscle lineage). Treatment of C2C12 cells with WT GDF5 had a slightly negative effect on their spontaneous differentiation along the muscle lineage, as indicated by the reduced size of myoblasts. L441P treatment increased muscle formation with results similar to the effects of the BMP inhibitor NOG. Incubation with BMP2, in contrast, resulted in strong induction of ALP activity and suppression of muscle differentiation. Similar results were obtained for the R438L mutant, indicating that this mutant displayed BMP2-like activity in this assay. Magnification, ×10 objective (AxioCam HRc camera; Zeiss).
Measurements of protein-protein binding.
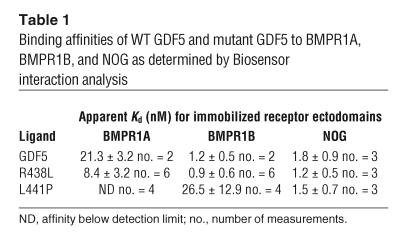
To further analyze ligand-receptor binding, we performed binding studies of the WT GDF5 and mutant GDF5 proteins to immobilized receptor ectodomains using the BIA2000 system. As shown in Table 1, WT GDF5 demonstrated low binding affinity to BMPR1A but high affinity to BMPR1B and NOG. The L441P mutant showed a strong reduction in binding affinity to BMPR1B, and its binding affinity to BMPR1A was below detection level. Normal affinity was found for NOG. In contrast, the R438L mutant showed strong binding to all 3 components tested, i.e., BMPR1A, BMPR1B, and NOG. These results further support our previous findings in cell culture and demonstrate that the R438L mutant has lost its specificity for BMPR1B and is able to signal through the BMPR1A receptor as well.
Table 1.
Binding affinities of WT GDF5 and mutant GDF5 to BMPR1A, BMPR1B, and NOG as determined by Biosensor interaction analysis
Expression analysis of Gdf5 pathway components and overexpression of Gdf5 mutants in vivo.
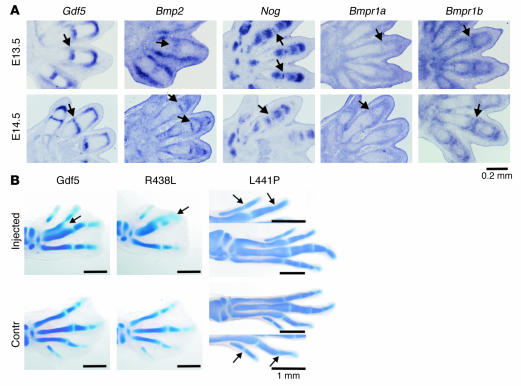
To localize the expression of ligands and receptors during joint development, we performed in situ hybridization using probes specific for Gdf5, Bmp2, Bmpr1a, Bmpr1b, and Nog on mouse limbs at stages E13.5 and E14.5 (Figure 6A). We found Bmpr1a to be ubiquitously expressed with higher levels in the developing joint, as previously reported (32). In contrast, Bmp2 expression was absent from the joint region at E13.5 and did not appear until E14.5. As shown in Figure 6A, expression was observed around the joint area in 2 stripes. However, this expression domain was shown to correspond to developing tendons and was thus oriented along the longitudinal axis of the digit and not the transverse axis, as expected for a gene expressed in the joint. At E14.5, we observed Bmp2 expression in a small band of cells in the middle of the joint cavity that is known to subsequently undergo apoptosis. At E13.5, Nog was expressed in the ends of the phalangeal anlagen, sparing the future joint, with the exception of a small band of cells in the middle of the joint interzone. Nog expression in the joint interzone disappears at E14.5 and is then replaced by expression of Bmp2. Furthermore, this is the only domain where the expression patterns of Nog and Gdf5 overlap. The inhibition of Gdf5 in these cells is likely to pave the way for interzone formation.
Figure 6.
Expression analysis during joint development and overexpression of Gdf5 in vivo. (A) In situ hybridization on mouse limb sections at E13.5 and E14.5 with probes specific for Gdf5, Bmp2, Nog, Bmpr1a, and Bmpr1b. The area of joint formation is indicated by arrows. Note strong expression of Gdf5 at both stages but Bmp2 expression in the joint only at E14.5. The longitudinal stripes of Bmp2 expression in the joint area at E13.5 correspond to expression in developing ligaments and not the joint interzone. Nog was expressed in a small band of cells in the joint interzone at E13.5 but not at E14.5. Bmpr1b was expressed in chondrocytes flanking the joint interzone. Bmpr1a shows ubiquitous expression with higher levels in the perichondrium and the developing joints. (B) Overexpression of WT Gdf5 as well as the Gdf5 mutants R438L and L441P in chick embryos using RCAS retroviral system. Alcian blue staining was used to visualize cartilage. For comparison, the injected left limb is shown next to the uninfected right limb, which serves as the control. Note enlargement of skeletal elements, joint fusions, and fusions between digits in WT Gdf5 and R438L mutant infected limbs at stage HH32. Changes in the L441P-injected limb were less severe and were only observed at a later stage (HH34–35). A higher magnification (arrows in B) shows joint fusions. Magnification, ×10 objective (A); ×1.6 objective (WT Gdf5 and R438L in B); ×1.25 objective (L441P in B) (AxioCam HRc camera; Zeiss).
To analyze the effect of the Gdf5 mutants in vivo, we overexpressed WT Gdf5 and mutant Gdf5 in chick embryos using the RCAS retroviral system (Figure 6B). Overexpression of WT Gdf5 as well as the R438L mutant resulted in thickening of skeletal anlagen as well as in the fusion of joints as demonstrated by Alcian blue–stained skeletal preparations of Hamburger Hamilton stage 32 (HH32) embryos. As expected from our in vitro results, overexpression of the L441P mutant had very little effect, and no significant phenotype was observed at this stage. However, single joint fusions and a moderate enlargement of distal phalanges was observed after longer incubation times (a day later at HH34–35), indicating some residual activity of the L441P mutant.
Discussion
The present work describes 2 mutations in the BMP-like signaling molecule GDF5 that give rise to the human limb malformation syndromes BDA2 and SYM1, previously shown to be associated with mutations in BMPR1B and NOG, respectively. Prediction of the GDF5 structure based on the known structure of the BMP2–BMPR1A complex implied that both mutations are located within the interaction interface of ligand and receptor. We therefore hypothesized that the GDF5 mutants display altered receptor-binding affinities and by this mechanism interfere with regular GDF5 signaling. We performed binding studies with the GDF5 mutants and showed that the L441P mutant has a dramatically reduced affinity to BMPR1B. In contrast, no difference in binding to NOG was observed, confirming that the gross tertiary structure of the mutant was not significantly altered. In accordance with these findings, we observed a severe reduction of chondrocyte differentiation and cartilage formation by the L441P mutant in our cell assays, suggesting lack of signaling through BMPR1B. However, the mutation is unlikely to result in complete inactivation because loss-of-function mutations in GDF5 cause BDC, a condition characterized by a distinct and more severe phenotype (33).
Ligand cross-linking experiments showed that GDF5 binds efficiently to singly expressed type 2 and type 1b receptors, but positive signaling activity was only detected when both receptors were present (4). More recent studies have shown that BMP receptors (types 1a, 1b, and 2) form homomeric and heteromeric complexes even in the absence of ligand and that both type 1 receptors have similar affinities to their ligands (34, 35). Since the L441P mutation affects only the type 1 receptor-binding site, the mutant should still be able to bind to the BMPR2 receptor and may consequently elicit a negative effect on the entire ligand-receptor complex. This effect is likely to be intensified by the fact that 75% of the GDF5 dimer molecules will contain at least 1 mutated molecule. The L441P mutation reported here to cause BDA2 was previously described in a cosanguineous Pakistani family with recessive DuPan syndrome (20). This condition also belongs to the group of acromesomelic dysplasias but is less severe than the Grebe and Hunter-Thompson dysplasias. The DuPan phenotype resembles a homozygous loss-of-function mutation in BMPR1B, as recently described (23). Thus, the similarities between the L441P-associated phenotypes and those associated with BMPR1B mutations are striking and strongly support our conclusion that the L441P mutation results in a selective loss of GDF5 signaling through the BMPR1B receptor. Differences between BDC and the L441P phenotype on one hand and DuPan and BMPR1B-associated phenotype on the other are likely to be due to the binding of GDF5 to other receptors and/or negative effects of the L441P mutant on the entire signaling cascade.
The R438L mutation described here causes a phenotype indistinguishable from that observed in individuals with NOG mutations (28). However, as shown in cell culture and by our in vitro binding assays, binding of the R438L mutant to NOG was unaltered in spite of the fact that R438 lies within the NOG interaction region, making it unlikely that the SYM1 phenotype described here is caused by an abnormal interaction of the mutant protein with NOG. Structural and mutational analysis of the BMP2 and BMPR1A binding sites revealed a specific type of protein-protein interface consisting of a large hydrophobic contact area (36). The residues L51 and D53 (corresponding to L437 and S439 in GDF5, respectively) are particularly exposed and essential for binding to BMPR1A. Interestingly, R438 resides between these 2 residues within this receptor interaction site. R438 is conserved throughout species and within different GDFs but differs markedly in other BMPs, including the drosophila BMP analog DPP, in which the positively charged amino acid arginine is replaced by the nonpolar and hydrophobic alanine. In the mutant, R438 is replaced by the hydrophobic residue leucine, resulting in a GDF5 with BMP2-like properties. Mutagenesis of these sites in BMP2 has shown altered binding affinities for BMPR1A (5). For example, converting L51 into proline resulted in a complete loss of binding to the type 1 receptor but normal binding to NOG and the type 2 receptor (36). Our binding studies show that the R438L mutant is still able to bind to BMPR1B with high affinity. However, in contrast to WT GDF5, the mutant is also able to bind to BMPR1A, albeit with lower affinity than BMP2. Thus the R438L mutation converts GDF5 into a molecule with BMP2-like properties.
This hypothesis is supported by our cell assays. The R438L mutant is highly active in all tests and appears to be even more potent than WT GDF5. The major difference between the mutant and the WT protein, however, becomes apparent through testing the differentiation of C2C12 cells into osteoblasts versus myoblasts. C2C12 cells are mesenchymal progenitor cells that spontaneously differentiate into muscle cells when reaching confluence. Treatment with BMPs resulted in differentiation along the osteoblastic lineage, as shown by a drastic increase in ALP activity. GDF5 has no effect on this phenomenon, presumably because Bmpr1b is, in contrast to Bmpr1a, nearly absent in C2C12 cells (2). In this assay, the R438L mutant behaves like BMP2, thus inducing osteoblastic differentiation and inhibiting muscle differentiation. We conclude that the R438L mutant GDF5 binds to both BMPR1A and BMPR1B, loses its preferential binding to BMPR1B, and thus takes on receptor affinities similar to those of BMP2.
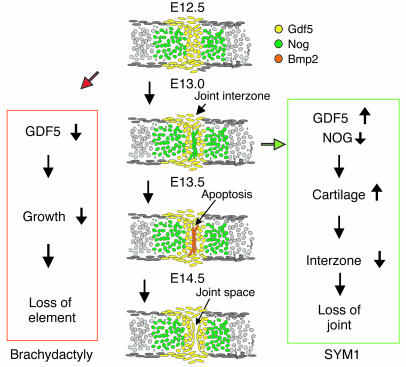
The consequences of this double signaling activity of the R438L mutant can be 2-fold. First, as shown in our in vitro assays, the mutant is likely to be more active and will thus enhance the natural function of WT GDF5. Second, the activation of the BMP2/BMPR1A pathway may result in aberrant signaling interfering with the normal function of GDF5. In situ hybridization experiments on developing joints in the mouse indicated that signaling through the 1a receptor is possible within the Gdf5 expression domain. Bmp2-induced signaling was tightly regulated on the level of expression and the local release of inhibitors such as Chordin and Nog, indicating that the activation of aberrant Bmp2-like signaling is likely to result in a disturbance of joint formation. To study a gain of function in vivo, we overexpressed WT Gdf5 and mutant Gdf5 in chicken limb buds. We observed joint fusions and overall enlargements of the cartilaginous anlagen similar to that found in previous studies (10) consistent with the proposed function of Gdf5 as a cartilage inducer. Similar results were obtained for WT Gdf5 and mutant Gdf5, indicating that the mutant proteins retained WT activity. Treatment of limb buds by implantation of BMP2-soaked beads resulted in the induction of apoptosis similar to that in vivo, in which BMPs are thought to be involved in the regulation of apoptosis in the interdigital mesenchyme (11, 37). In particular, Bmp2 is able to induce apoptosis through a Smad-independent, PKC-dependent signaling pathway (38). We did not observe an increase in apoptosis when expressing WT Gdf5 and mutant Gdf5 in chick limbs, which shows that the increased activity of the R438L mutant had no major effect on this pathway. These results indicate that the R438L mutant elicits its pathology through a gain of function, probably by recruiting additional type 1a receptors expressed in the region at the critical time of joint formation. Thus, as shown schematically in Figure 7, the lack of joint formation observed in SYM1 was induced either by overactive GDF5 or by downregulated NOG. Both result in an imbalance of signal versus inhibitor, leading to the persistence of cartilage in the future joint interzone. In this model, proliferation and recruitment of mesenchymal cells caused by GDF5 is inhibited by NOG in the center of the joint interzone, resulting in dedifferentiation of interzone cells. The subsequent downregulation of NOG expression in these cells and the upregulation of BMP2 expression finally permit apoptosis and joint formation. In contrast, selective inhibition of GDF5 as in the L441P mutant primarily affects longitudinal growth and thus causes shortening or loss of individual skeletal elements.
Figure 7.
Schematic drawing of proposed mechanism of metacarpophalangeal joint development and expression of Gdf5, Nog, and Bmp2 in mouse limbs corresponding to E12.5, E13.0, E13.5, and E14.5. Gdf5 induces longitudinal growth at the distal end of each element. Bmp inhibitor Nog is expressed in the cartilage anlagen and the joint interzone first, while Bmp2 is expressed later. Apoptosis, mechanical forces, and other factors lead to formation of a joint space. Boxes on left and right show proposed molecular pathogenesis for brachydactyly and SYM1, respectively. Reduction of GDF5 signal, as in the L441P mutant, results in reduced growth of phalanges and thus shortening of elements. Increased GDF5 signaling, as in the R438L mutant, or mutations in NOG, both result in the persistence of cartilage in the joint region and a deficiency in joint interzone formation. Clinically this results in lack of joint formation and thus SYM1.
The study of the mutations described here extends our knowledge of human GDF5-related phenotypes. Furthermore, the functional analysis showed that mutations in different genes of the same pathway can result in identical phenotypes, provided the mutations have similar effects on the affected signals. Inactivation of the ligand-receptor complex as in BMPR1B receptor mutants causes a BDA2 phenotype identical to the L441P mutation in GDF5 described here. Likewise, the phenotype caused by inactivation of the BMP-antagonist NOG can be mimicked by a gain-of-function mutation in GDF5 that results from a loss of signaling specificity. The present experiments localize some of the main determinants of GDF5-binding specificity, suggesting that GDF5 with altered binding affinities may function as an inhibitor of BMP signaling. The study of such mutated GDF5 peptides may improve our ability to make use of the pharmacological properties of GDF5 in the clinical applications of fracture healing and tendon and nerve regeneration.
Methods
Patients.
One family from Norway and 1 family from the US, whose members experienced BDA2 and SYM1, respectively, were investigated. DNA was extracted with standard methods from peripheral blood or buccal swabs. Sequencing of GDF5, NOG, and BMPR1B was performed as described previously (17, 21, 22, 28). All participants gave informed consent.
Alignment and 3D structure prediction of GDF5.
Protein sequence alignments comprising the highly conserved cysteine knot domains of 10 members of the GDF5, BMP2, and BMP4 family and from DPP (drosophila DPP) were aligned using CLUSTAL X (http://bio.ifom-firc.it/docs/clustal/clustalx.html) (39) and colored using CHROMA (http://www.lg.ndirect.co.uk/chroma/) (40). The 3D structure prediction of GDF5 was generated by the 3D-PSSM server (http://www.sbg.bio.ic.ac.uk/∼3dpssm/index2.html) (41) using the BMP2 structure (PDB entry 3BMP) as a template. Images of the molecular structure were produced using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera/) (42).
In situ hybridization.
In situ hybridization was performed on 7-μm sections of paraffin-embedded mouse limbs, stage E13.5 and E14.5, by using digoxygenin-labeled riboprobes as described (43). The following previously described riboprobes were used: Bmp2 (32), Bmpr1a (44), Bmpr1b (24), Gdf5 (43), and Nog (45). All animal experiments were approved by the ethics committee of the Landesamt für Arbeitsschutz, Gesundheitsschutz, und technische Sicherheit (Berlin, Germany).
Viral constructs.
The coding sequence of chicken Gdf5 was cloned into p–SLAX-13 and used as a template for generating the R438L and L441P mutations corresponding to the human mutations. In vitro mutagenesis was performed with the Quickchange Kit (Stratagene) according to the manufacturer’s recommendations. Cloning into the RCAS vector was performed as described previously (46). RCAS-Nog was a kind gift from A. Vortkamp (Max-Planck-Institut für Molekulare Genetik).
Micromass cultures.
Micromass cultures were prepared as described previously (22) with minor modifications. Briefly, fertilized chicken eggs were obtained from Tierzucht Lohmann and incubated at 37.5°C in a humidified egg incubator for about 4.5 days. Ectoderm was removed, and cells were isolated from the limb buds at stage HH23–24 by digestion with 0.1% collagenase type Ia and 0.1% trypsine. Micromass cultures were plated at a density of 2 × 105 cells/10-μl drop. Infection was performed with 1 μl of 2 concentrated viral supernatants: RCASBP-A, containing the cDNA encoding WT–chicken Gdf5 (WT-chGdf5), R438L-chGdf5, or L441P-chGdf5; and RCASBP-B, containing the cDNA encoding WT-chNog. Culture medium (DMEM-F12, 2% chicken serum, 4 mM L-glutamine, 1000 U/ml penicillin, and 100 μg/ml streptomycin) was replaced every 2 days. For each condition, 4 replicates were performed in parallel.
Cell assays.
Quantitative ALP assays were performed as previously described (47) with minor modifications: ATDC5 and C2C12 cells were seeded at a density of 1 × 104 cells/96-well plates in growth media (for ATDC5: DMEM/Ham’s F12, 5% FCS, 10 μg/ml transferrin, 30 nM sodium selenite, and 2 mM L-glutamine in 5% CO2; for C2C12: high-glucose DMEM, 10% FCS, and 2 mM L-glutamine in 10% CO2). After 24 hours, cells were starved for 5 hours in media with reduced FCS (for ATDC5: DMEM/Ham’s F12, 0.5% FCS, and 2 mM L-glutamine in 5% CO2; for C2C12: high-glucose DMEM, 2% FCS, and 2 mM L-glutamine in 10% CO2). Stimulation with recombinant proteins was performed in starving media for 3 days. ALP activity was measured in triplicate for ATDC5 and C2C12 cells in 96-well plates by lysing cells in ALP-buffer1 (0.1 M glycine, pH 9.6, 1% Nonidet P-40 [Sigma-Aldrich], 1 mM MgCl2, and 1 mM ZnCl2). ALP activity of micromass cultures was determined by homogenizing 4 replicates separately in ALP-buffer1; after centrifugation only supernatant was used for further proceedings. After the addition of 1 volume of ALP-buffer2 (5 mM p-nitrophenyl phosphate [p-NPP], 0.1 M glycine, pH 9.6, 1 mM MgCl2, and 1 mM ZnCl2), ALP activity was determined spectrophotometrically. The amount of p-NP released from the substrate p-NPP was recorded at 405 nm and used to calculate ALP activity.
Alcian blue staining was performed by fixing micromass cultures at day 4, then incubating with 0.1% Alcian blue, pH 1, overnight. Quantification of the staining was achieved after extensive washings with water by extraction with 6 M guanidine-HCl for 8 hours at room temperature. Dye concentration was determined spectrophotometrically at A650.
For differentiation of C2C12, cells were grown on coverslips in high-glucose DMEM supplemented with 10% FCS, 2 mM L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Before reaching confluence, cells were stimulated with 10 nM recombinant proteins in high-glucose DMEM supplemented with only 2% FCS, 2 mM L-glutamine, and penicillin/streptomycin over 5 days. ALP staining was performed to visualize osteoblastic differentiation. Myoblastic differentiation was analyzed by immunocytochemistry using an antibody against myosin heavy chain (anti-myosin-IgG mouse, 1:300; Sigma-Aldrich). Detection was achieved after extensive washings by Vectastain Kit (Vector Laboratories) and diaminobenzidine tetrahydrochloride (DAB) staining (Vector Laboratories) according to the manufacturer’s recommendations.
Preparation of recombinant proteins.
For the expression of mutant GDF5, patient DNA carrying the R438L or L441P mutants was amplified by PCR with specific GDF5 primer pair F_MP52_Afl II (GCAAGAACCTTAAGGCTCGC) and R_MP52_Nco I (CGGGGTCCATGGAGTTCATC) with Pfx DNA Polymerase (Invitrogen Corp.). The amplified DNA was ligated into the protein expression vector pKOT277 and confirmed by DNA sequencing.
The proteins were expressed in inclusion bodies using the bacterial Escherichia coli strain W3110BP and isolated using homogenization buffer (25 mM Tris HCl, pH 7.3, and 10 mM EDTA, pH 8.0) and wash buffer (1 M urea, 20 mM Tris HCl, and 5 mM EDTA, pH 8.3). After centrifugation, the inclusion bodies were dissolved in 6 M guanidine HCl, 50 mM Tris, 150 mM NaCl, and 3 mM DTT, pH 8.0. Further purification was carried out on a reversed phase column Aquapore Octyl (100 × 10 mm, 20 μm, no. 186470; Applied Biosystems) with a gradient from 100% eluent A (0.1% trifluoroacetic acid [TFA]) to 100% eluent B (0.1% TFA, 90% CH3N) in 104 minutes (flow rate, 3 ml/min).
Mutant proteins were dissolved in buffer (6 M guanidine HCl, 50 mM Tris, 150 mM NaCl, and 3 mM DTT, pH 8.0), the protein concentration adjusted to 2.6 mg/ml, and the pH adjusted to between 8 and 9. After 2 hours incubation at room temperature, refolding buffer [1 M NaCl, 50 mM Tris, 5 mM EDTA, 1 mM oxidized glutathione (GSSG), 2 mM glutathione (GSH), and 33 mM 3-(3-cholamidopropyl) dimethylammonio-1-propanesulfonate, pH 9.5] were added under gentle agitation so that a final protein concentration of 0.16 mg/ml was reached. The solution was then incubated for 48 hours at 22°C; refolding was stopped by changing the pH to 3–4 by adding HCl to 18% (vol/vol). After centrifugation, the nonrefolded monomer was separated from the dimer form through a second RP-HPLC under the same conditions. Purity was determined by a Western blot and SDS-PAGE silver stain, and the fractions containing the dimerized protein were pooled and lyophilized. Subsequently, the proteins were dissolved in 10 mM HCl and stored at –80°C. BMP2 was prepared as previously described (48). Human recombinant NOG was a kind gift from A. Economides (Regeneron Pharmaceuticals Inc.).
Biosensor interaction analysis.
The BIA2000 system (Biacore) was used to record the binding of GDF5 proteins to immobilized NOG (R&D Systems) and receptor ectodomains as described (5, 36). In brief, the biotinylated proteins were fixed to streptavidin-coated matrix of biosensor CM5 at a density of 200 resonance units. WT GDF5 and mutant proteins L441P or R338L at 25, 50, and 75 nM in HBS buffer (10 mM HEPES, pH 7.4, 500 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P20) were perfused at a flow rate of 10 μl/min at 25°C. The association period was 5 minutes, and the dissociation period was set to 3 minutes. Free receptors and NOG were regenerated on the biosensor by perfusion with 4 M MgCl2. We calculated apparent Kd from the kinetic rate constants kon for association and koff for dissociation (Kd = koff/kon).
Retroviral infection of chicken limbs.
Production of concentrated viral supernatant and injection into the limb field of HH10 chicken embryos was performed as described previously (43). The same virus preparations were used as for the micromass cultures. Embryos were harvested between stages HH32–35 and stained with Alcian blue to visualize cartilage.
Statistical analyses and graphics.
Nonlinear regression and graphical view of enzymatic assays were determined using GraphPad Prism version 4.00 for Windows (GraphPad Software).
Acknowledgments
We thank A. Economides for the generous supply of recombinant human NOGGIN. We acknowledge Britta Hoffmann for technical support and Lutz Schomburg for critical remarks on the manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to S. Mundlos and by NIH grants HD22567 and M01-RR88435 to D. Krakow. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at UCSF (supported by NIH grant P41 RR-01081).
Footnotes
Nonstandard abbreviations used: ALP, alkaline phosphatase; BDA2, brachydactyly type A2; BMP, bone morphogenetic protein; BMPR1A, BMP receptor type 1a; DPP, decapentaplegic; GDF, growth and differentiation factor; HH, Hamburger Hamilton stage; RCAS, replication-competent avian sarcoma; SYM1, symphalangism.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Kornak U, Mundlos S. Genetic disorders of the skeleton: a developmental approach [review] Am. J. Hum. Genet. 2003;73:447–474. doi: 10.1086/377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebald W, Nickel J, Zhang JL, Mueller TD. Molecular recognition in bone morphogenetic protein (BMP)/receptor interaction. Biol. Chem. 2004;385:697–710. doi: 10.1515/BC.2004.086. [DOI] [PubMed] [Google Scholar]
- 3.Luyten FP. Cartilage-derived morphogenetic protein-1. Int. J. Biochem. Cell Biol. 1997;29:1241–1244. doi: 10.1016/s1357-2725(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 4.Nishitoh H, et al. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J. Biol. Chem. 1996;271:21345–21352. doi: 10.1074/jbc.271.35.21345. [DOI] [PubMed] [Google Scholar]
- 5.Kirsch T, Nickel J, Sebald W. BMP-2 antagonists emerge from alterations in the low-affinity binding epitope for receptor BMPR-II. EMBO J. 2000;19:3314–3324. doi: 10.1093/emboj/19.13.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 7.Miyazono K, Kusanagi K, Inoue H. Divergence and convergence of TGF-beta/BMP signaling. J. Cell. Physiol. 2001;187:265–276. doi: 10.1002/jcp.1080. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 9.Storm EE, et al. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 10.Francis-West PH, et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 11.Merino R, et al. Expression and function of Gdf-5 during digit skeletogenesis in the embryonic chick leg bud. Dev. Biol. 1999;206:33–45. doi: 10.1006/dbio.1998.9129. [DOI] [PubMed] [Google Scholar]
- 12.Takahara M, et al. Developmental failure of phalanges in the absence of growth/differentiation factor 5. Bone. 2004;35:1069–1076. doi: 10.1016/j.bone.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Archer CW, Dowthwaite GP, Francis-West P. Development of synovial joints. Birth Defects Res. C. Embryo Today. 2003;69:144–155. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- 14.Erlacher L, et al. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J. Bone Miner. Res. 1998;13:383–392. doi: 10.1359/jbmr.1998.13.3.383. [DOI] [PubMed] [Google Scholar]
- 15.Wolfman NM, et al. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-beta gene family. J. Clin. Invest. 1997;100:321–330. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiro RC, Thompson AY, Poser JW. Spinal fusion with recombinant human growth and differentiation factor-5 combined with a mineralized collagen matrix. Anat. Rec. 2001;263:388–395. doi: 10.1002/ar.1119. [DOI] [PubMed] [Google Scholar]
- 17.Polinkovsky A, et al. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat. Genet. 1997;17:18–19. doi: 10.1038/ng0997-18. [DOI] [PubMed] [Google Scholar]
- 18.Thomas JT, et al. Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat. Genet. 1997;17:58–64. doi: 10.1038/ng0997-58. [DOI] [PubMed] [Google Scholar]
- 19.Thomas JT, et al. A human chondrodysplasia due to a mutation in a TGF-beta superfamily member. Nat. Genet. 1996;12:315–317. doi: 10.1038/ng0396-315. [DOI] [PubMed] [Google Scholar]
- 20.Faiyaz-Ul-Haque M, et al. Mutation in the cartilage-derived morphogenetic protein-1 (CDMP1) gene in a kindred affected with fibular hypoplasia and complex brachydactyly (DuPan syndrome) Clin. Genet. 2002;61:454–458. doi: 10.1034/j.1399-0004.2002.610610.x. [DOI] [PubMed] [Google Scholar]
- 21.Schwabe GC, et al. Brachydactyly type C caused by a homozygous missense mutation in the prodomain of CDMP1. Am. J. Med. Genet. 2004;124A:356–363. doi: 10.1002/ajmg.a.20349. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann K, et al. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demirhan O, et al. A homozygous BMPR1B mutation causes a new subtype of acromesomelic chondrodysplasia with genital anomalies. J. Med. Genet. 2004;42:314–317. doi: 10.1136/jmg.2004.023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127:605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- 25.Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development. 2000;127:621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- 26.Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators [review] Dev. Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- 27.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y, et al. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet. 1999;21:302–304. doi: 10.1038/6821. [DOI] [PubMed] [Google Scholar]
- 29.Brown DJ, et al. Autosomal dominant stapes ankylosis with broad thumbs and toes, hyperopia, and skeletal anomalies is caused by heterozygous nonsense and frameshift mutations in NOG, the gene encoding noggin. Am. J. Hum. Genet. 2002;71:618–624. doi: 10.1086/342067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi T, et al. Mutations of the NOG gene in individuals with proximal symphalangism and multiple synostosis syndrome. Clin. Genet. 2001;60:447–451. doi: 10.1034/j.1399-0004.2001.600607.x. [DOI] [PubMed] [Google Scholar]
- 31.Dixon ME, Armstrong P, Stevens DB, Bamshad M. Identical mutations in NOG can cause either tarsal/carpal coalition syndrome or proximal symphalangism. Genet. Med. 2001;3:349–353. doi: 10.1097/00125817-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Zou H, Wieser R, Massague J, Niswander L. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev. 1997;11:2191–2203. doi: 10.1101/gad.11.17.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savarirayan R, et al. Broad phenotypic spectrum caused by an identical heterozygous CDMP-1 mutation in three unrelated families. Am. J. Med. Genet. 2003;117A:136–142. doi: 10.1002/ajmg.a.10924. [DOI] [PubMed] [Google Scholar]
- 34.Gilboa L, et al. Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol. Biol. Cell. 2000;11:1023–1035. doi: 10.1091/mbc.11.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nohe A, et al. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 2002;277:5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- 36.Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat. Struct. Mol. Biol. 2004;11:481–488. doi: 10.1038/nsmb756. [DOI] [PubMed] [Google Scholar]
- 37.Yokouchi Y, et al. BMP-2/-4 mediate programmed cell death in chicken limb buds. Development. 1996;122:3725–3734. doi: 10.1242/dev.122.12.3725. [DOI] [PubMed] [Google Scholar]
- 38.Hay E, Lemonnier J, Fromigue O, Marie PJ. Bone morphogenetic protein-2 promotes osteoblast apoptosis through a Smad-independent, protein kinase C-dependent signaling pathway. J. Biol. Chem. 2001;276:29028–29036. doi: 10.1074/jbc.M011265200. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodstadt L, Ponting CP. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–846. doi: 10.1093/bioinformatics/17.9.845. [DOI] [PubMed] [Google Scholar]
- 41.Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 44.Morgan EA, Nguyen SB, Scott V, Stadler HS. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development. 2003;130:3095–3109. doi: 10.1242/dev.00530. [DOI] [PubMed] [Google Scholar]
- 45.Albrecht AN, et al. The synpolydactyly homolog (spdh) mutation in the mouse – a defect in patterning and growth of limb cartilage elements. Mech. Dev. 2002;112:53–67. doi: 10.1016/s0925-4773(01)00639-6. [DOI] [PubMed] [Google Scholar]
- 46.Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knaus P, Sebald W. Cooperativity of binding epitopes and receptor chains in the BMP/TGFbeta superfamily. Biol. Chem. 2001;382:1189–1195. doi: 10.1515/BC.2001.149. [DOI] [PubMed] [Google Scholar]
- 48.Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur. J. Biochem. 1996;237:295–302. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]