Abstract
Mature osteoclasts, multinucleated giant cells responsible for bone resorption, are terminally differentiated cells with a short life span. Recently, we have demonstrated that osteoclast apoptosis is regulated by ERK activity and Bcl-2 family member Bim. In this paper, we summarize the methods we used to study osteoclast apoptosis in vitro and in vivo. Using adenovirus and retrovirus vectors, we were able to introduce foreign genes into osteoclasts and examine their effects on osteoclast survival in vitro. In addition, we established the modified methods for in situ hybridization and BrdU labeling of bone sections from mice to study osteoclast survival in vivo. The detailed methods described here could be useful for studying the biological process in bone.
Keywords: Apoptosis, Macrophage Colony-Stimulating Factor, Osteoclasts
Introduction
Osteoclasts, terminally differentiated cells with a short life span, are multinucleated giant cells primarily responsible for bone resorption (1). They undergo rapid apoptosis in the absence of trophic factors such as macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) (2). However, the molecular events implicated in these processes still remain elusive.
Apoptosis is the genetically programmed cell death to remove the unwanted cells (3). The abnormalities of apoptosis regulation induce various sicknesses such as cancer, autoimmune diseases and degenerative disorders (4). There are two distinct apoptosis signal pathways in mammals. One pathway is initiated by death receptors, members of tumor necrosis factor receptor (TNF-R) family. The other pathway is regulated by pro- and anti-apoptotic Bcl-2 family member via mitochondrial release of cytochrome c and caspase-9 activation (5, 6). The anti-apoptotic Bcl-2 family members include mammalian Bcl-2 and Bcl-xL and they share similarity within three or four Bcl-2 homology (BH) domains. So far, more than 20 pro-apoptotic Bcl-2 family proteins have been identified in mammals. They can be further divided into 2 groups: multi-domain members possess homology in two or three BH regions, such as Bax and Bak, whereas the BH3 domain-only proteins, such as Bad, Bid and Bim/Bod, share only the short BH3 region (7). The pro-apoptotic activity of BH3-only proteins is strictly regulated at both the transcriptional and post-translational level to prevent inappropriate cell death (7).
The BH3-only protein Bim was first identified as a Bcl-2-interacting protein by screening a λ phage expression library constructed from mouse thymic lymphoma cells (8). Bim is expressed in hematopoietic, epithelial, neuronal and germ cells (9), and alternative splicing generates various Bim isoforms, including Bims, BimL and BimEL. Experiments with knock-out mice have shown that Bim is essential for apoptosis of T lymphocytes, B lymphocytes, myeloid cells and neurons (10-14). Pro-apoptotic activity of Bim is regulated both transcriptionally and post-transcriptionally (7). We recently showed that ubiquitin-dependent regulation of Bim levels is critical for controlling osteoclast apoptosis (15).
The purpose of this paper is to summarize approaches recently developed and optimized to study molecular mechanism of osteoclast apoptosis in vitro and in vivo. We developed protocols of adenovirus- and retrovirus-mediated gene transduction systems for mature osteoclasts and osteoclast precursors, respectively. Furthermore, we established the optimal protocols for in situ hybridization and BrdU labeling of bone sections from mice to study osteoclast life span in vivo. These assays can be used to understand how bone homeostasis is maintained.
Materials and Methods
Antibodies and chemicals
Antibodies to Bid and Bax were purchased from Cell Signaling Technology. (Beverly, MA). We used three different antibodies to Bim in this study, i.e. M-20 (Sant Cruz Technology, Santa Cruz, CA) for immunoprecipitation, anti-Bim monoclonal antibody (BD Bioscience Pharmingen, San Jose, CA) for Western blotting, and anti-Bim polyclonal antibody (Oncogene Research Products Cambridge, MA) for immunocytochemistry. Anti-Bcl-xL and anti-ubiquitin were from Santa Cruz Technology. Recombinant mouse M-CSF was obtained from TECHNE Co. (Minneapolis, MN) and soluble RANKL was from Wako Pure Chemical Co. (Osaka, Japan). Alpha modified-minimum essential medium (αMEM) was purchased from GIBCO BRL, Life Technologies Inc. (Rockville, MD), and fetal bovine serum (FBS) was from Sigma (St. Louis, MO). Bacterial collagenase was purchased from Wako Pure Chemical Co. (Osaka, Japan), 1α,25(OH)2D3 from Calbiochem(La Jolla, CA) and dispase from Godoshusei (Tokyo, Japan). The broad-spectrum caspase inhibitor zVAD-FMK was from Calbiochem (La Jolla, CA). Alexa 488 and 568 labeled antibodies to rabbit IgG, Mitotracker® Green, MitoTracker® Red and Hoechst 33342 were purchased from MolecularProbes (Eugene, OR). ApoAlert® Cell Fraction Kit was purchased from BD Biosciences Clontech (Palo Alto, CA). Other chemicals and reagents used in this study were of analytical grade.
Construction and gene transduction of adenovirus vectors
The recombinant adenovirus vectors carrying either constitutively active MEK1 (ser218 and Ser222 to Glu, AxMEK1CA), dominant negative Ras (Ser17 to Asn, AxRasDN), myristoylated Akt (The Src myristoylation sequence was added to Akt Δ4-129, myr-Akt) or enhanced green fluorescent protein (EGFP) gene under the control of the CAG (cytomegalovirus IE enhancer + chicken β-actin promoter + rabbit β-globin poly (A)+ signal promoter) was constructed by homologous recombination between expression cosmid cassette and the parental virus genome in 293 cells (16). Adenovirus vector expressing BimL under the control of the CMV promoter was constructed using Adeno-X™ Adenoviral Expression Systems from BD Biosciences Clontech (Palo Alto, CA). Titers of virus stocks were determined by endpoint cytopathic effect assay with the following modifications (17). 50 μl of DMEM/10%FBS was dispensed into each well of a 96-well tissue culture plate, and then eight rows of three fold serial dilutions of the virus starting from 10-4 dilutions were prepared. 3 x 105 293 cells in 50 μl of DMEM/10%FBS was added to each well every 3d. 12 days later, the endpoint of the cytopathic effect was determined by microscopy, and the 50% tissue culture infectious dose (TCID50) was calculated. One TCID50/ml approximately corresponds to one plaque forming unit (PFU)/ml (17). The efficiency of infection is affected not only by the concentration but also by the ratio of viruses to cells, the multiplicity of infection (MOI). In this study, MOI is expressed as a measure of titer how many PFUs of viruses were added to every cell. Mouse cocultures on day 5, when OCLs began to appear, were incubated with a small amount of αMEM containing the recombinant adenoviruses for 1 h at 37°C at an indicated MOI. The cells were washed twice with PBS and further incubated with αMEM/10%FBS at 37°C. Experiments were performed 24 h after the infection.
Construction and gene transduction of retroviral vectors
Retroviral vectors, pMx-BimEL-IRES-EGFP and pMx-mtBimEL-IRES-EGFP were constructed by inserting full-length mouse cDNA of bim EL and mutated bim EL , in which the two ubiquitin acceptor lysine residues (Lys3 and Lys108) were mutated to arginine, into pMx-IRES-EGFP vector (provided by Dr. Kitamura). Retrovirus packaging was performed by transfection of the pMx vectors into packaging cell line BOSC23 (2 x 106) cells. Virus stocks were prepared by collecting the media from cultures 48 h after transfection. Mouse bone marrow macrophage cells (3 x 105) were incubated with 3 ml of virus stock for 4 h in the presence of mouse recombinant M-CSF (50 ng/ml) and polybrene (8 μg/ml). Cells were washed twice with PBS and further incubated in the presence of 100 ng/ml M-CSF. Experiments were performed 7 days after the infection.
Animals, cells and cultures
Newborn ddY mice and 8-week-old male ddY mice were purchased from Shizuoka Laboratories Animal Center (Shizuoka, Japan). The breeding and genotyping of bim-/- mice (Chimeric mouse were bred to C57BL/6 mice. Heterozygous mice were interbred more than 12 times. The genetic background was same as C57BL/6 mice) was performed as previously described (10). The transgenic mice in which a reporter lacZ gene was introduced into the bim locus was generated by homologous recombination. To obtain large numbers of cells for biochemical analyses, we utilized the co-culture system established by Takahashi et al. (18). Mouse primary osteoblastic cells were obtained from 1-day-old mouse calvaria by enzymatic digestion and bone marrow cells were from tibiae of adult mice. Osteoblastic cells (5 x 105 cells/dish) were co-cultured with bone marrow cells (1 x 107 cells/dish) on 10 cm dishes or collagen gel-coated dishes in the presence of 10 nM 1α,25(OH)2D3 and 1 μM PGE2. For protein analysis, OCLs were purified following a modification of the method originally reported by Tezuka et al. (19). The crude OCL preparation placed on plastic dishes was washed with αMEM, and treated with 5 ml of αMEM containing 0.1% collagenase and 0.2% dispase for 10 min to remove osteoblastic cells. The purity of OCLs and their precursors was more than 90% at the final preparation.
Survival of OCLs
OCLs were purified 24 h after the infection and some of the cultures were subjected to tartrate-resistant acid phosphatase (TRAP) staining. Osteoclasts adhere to the bone surface through specialized discrete structures called “podosomes” in the clear zone, which consist mainly of dots containing F-actin. The rounded and spreaded appearance with ringed structure of podosomes (actin ring) is a characteristic of polarized osteoclasts. Cell viability/survival is expressed as morphologically intact TRAP-positive multinucleated cells. Other cultures were further incubated for the indicated times, and then the number of living OCLs was counted. The number of viable cells remaining at the different time points is shown as a percentage of the cells at time zero (20).
DNA extraction and electrophoretic analysis
Purified OCLs were lysed by incubating at 60°C overnight in a digestion buffer containing 150 mM NaCl, 25 mM EDTA, 100 μg/ml proteinase K, and 0.2% SDS. The DNA was extracted twice with phenol/chloroform/isoamylalcohol and once with chloroform, and precipitated in ethanol with 150 mM CH3COONa, pH 5.2. The DNA was dissolved in TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) and treated with 20 mg/ml RNase A. The procedure for DNA extraction and precipitation were repeated. 2 μg of DNA was separated by electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining with UV light illumination (21).
TUNEL assay
Cells undergoing apoptosis were identified by means of the TdT-mediated dUTP-dioxigenin nick-end labeling (TUNEL) method, which specifically labels the 3’-hydroxyl terminal of DNA strand breaks. For the TUNEL procedure, all agents, including buffers, were part of a kit (Apoptosis in situ Detection KIT, Wako Pure Chemical Co.); the staining procedure was carried out according to the manufacturer’s recommendation. Negative controls included omission of TdT. Positive controls included treatment of the samples with DNase I. Apoptotic cells were recognized by their dark nuclear staining (TUNEL-positive) and nuclei of non-apoptotic cells were visualized by staining with methyl green.
Western blotting
Cells were washed twice with ice-cold PBS and lysed in lysis buffer [20 mM Tris-HCl (pH7.5), 150 mM NaCl, 1% TritonX-100, 12 mM β-glycerophosphate, 5 mM EGTA, 0.5% deoxycholate, 3 mM DTT, 10 mM NaF, 1 mM Na3VO4, 2 μM leupeptin, 20 μg/ml aprotinin, and 1 mM PMSF]. After 30 min incubation on ice, cell lysates were cleared by centrifugation at 12,000 × g for 20 min. An equal amount (20 μg per lane) of protein was fractionated on 4-20% gradient SDS-PAGE gels under reducing conditions, transferred electrophoretically onto PVDF membranes, and probed sequentially with an appropriate primary antibody followed by secondary antibodies coupled to horseradish peroxidase (Promega Co., Madison, WI). Immunoreactive proteins were visualized using ECL Western blotting detection reagents (Amersham Co., Arlington Heights, IL) following the procedures recommended by the supplier. The blots were stripped by incubating for 20 min in stripping buffer [2% SDS, 100 mM 2-mercaptoethanol, 62.5 mM Tris-HCl (pH 6.7)] at 50°C and reprobed by other antibodies.
Ubiquitylation assay
2 × 106 Cells were washed three times with PBS and the medium was changed to fresh αMEM/10% FBS with 50 ng/ml M-CSF, 50 μM MG132, or 20 μM lactacystin. Twelve hours after changing medium, cells were washed three times with PBS and lysed in lysis buffer [20 mM Hepes-NaOH (pH 7.4), 150 mM NaCl, 10 mM EDTA, 1 mM Na3VO4, and 1 mM PMSF, 0.05% NP-40, 10% glycerol] cell lysates were homogenized by pipetting with 1 ml silinge and 26G needle more than 50 times. After homogenization, cellular debris was pelleted by centrifugation at 12,000 × g for 20 min. Supernatants (cell lysates) were precleared by adding 20 μl of protein-A agarose. Immunoprecipitation was performed by incubating 500 μl of cell lysate with 2 μg of anti-Bim antibody for 2 h, then adding 20 μl of protein-A agarose. After incubation for 1 h at 4°C with end-over-end mixing, the immune complexes were recovered by centrifugation and washed twice with washing buffer [20 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM EGTA, 2 mM DTT, and 1 mM PMSF]. The immunoprecipitates were then subjected to SDS-PAGE and immunoblotted with anti-ubiquitin antibody.
Immunofluorescence
For immunofluorescence analysis, cells were plated on sterile FBS-coated glass coverslips and purified by treatment with αMEM containing 0.1% collagenase and 0.2% dispase. After purification, OCLs were fixed in 3.7% (vol/vol) formaldehyde in PBS for 10 min, and then washed three times in PBS. Cells were permeabilized in 0.05% saponin for 30 min, and blocked in 5% normal goat serum (Boehringer) for 30 min and incubated in appropriate primary antibodies, washed in PBS, incubated with fluorescein-conjugated secondary antibody, and finally washed in PBS and mounted in FluorSave. Cells were examined using a confocal imaging system (MRC-600; Bio-Rad Laboratories).
RT-PCR and real time PCR
mRNAs was isolated from OCLs, and reverse-transcribed by SuperScript III First-Strand Synthesis system for RT-PCR (Invitrogen), according to the manufacturer’s protocol. The primers we utilized to detect bim and gapdh were as follows: bim: 5’-ATGGCCAAGCAACC-TTCTGA-3’ (sense) and 5’-GACCATACCTCTTCCGTAACT -3’ (antisense). gapdh: 5’-GTATGTCGTGGAGTCTACTGGTGT-3’ (sense) and 5’- CCGGATGTACCGGAGGTTCCTCAT-3’ (antisense).
The primers utilized to detect, bax, bcl-xL were as previously reported by Okahashi et al. (22). Reverse transcribed mRNA were analyzed by ABI Prism® 7000 Sequence Detection System (Applied Biosystems, CA). The primers we utilized in real time PCR to detect common form of all bim splice variants and bim EL specific form were as follows: bim common form, 5’-CTTCCATACGACAGTCTC-3’ (antisense). and 5’- CTTCTGGTGGGAGTTTACCAA-3’(antisense). bim EL specific form, 5’-GTCCTCCAGTGGGTATTTCT (sense) and 5’-TCCTCCTTGGACTTCTAGAC-3’ (antisense).
In situ hybridization
Five-week-old male wild-type mice were sacrificed and the tibiae were fixed in 4% paraformaldehyde/PBS overnight at 4°C, processed, embedded in paraffin. 3-μm-thick paraffin sections were cut, deparaffinized with Xylene and rehydrated with decreased concentrations of ethanol. The sections were then incubated overnight with complementary digoxigenin-labeled riboprobes for mouse bim L , procollagen type IA, and procollagen type IIA, followed by extensive washes to remove excess probe. Fluorescent-linked anti-digoxigenin antibodies are used to visualize the hybridized probe.
In vivo BrdU labeling
Five-week-old bim+/+ and bim-/- mice were fed with water containing 1 mg/ml of 5’-bromo-2’-deoxyuridine (BrdU) for 1 week (labeling period). Mice were then sacrificed either on the next day (group A) or after 6 weeks (group B) of the labeling period. The tibiae were removed and immersed in 4% buffered paraformaldehyde for paraffin sections. The sections were washed with PBS three times, treated with 2.5% hyaluronidase and blocked with 3% FBS in TBST for 30 min. The sections were then incubated with the fluorescein-conjugated anti-BrdU antibody at a dilution of 1:50 overnight at 4°C. After washing with PBS, they were mounted in FluorSave. Sections were examined using a confocal imaging system (MRC-600; Bio-Rad Laboratories).
Statistical analysis
Each series of experiments was repeated at least three times. The results obtained from a typical experiment were expressed as the means ± SD (standard deviation). Significant differences were determined using factorial analysis of variance (ANOVA).
Results and Discussion
Osteoclasts undergo spontaneous apoptosis without any cytokines or supporting cells
To study the survival of osteoclasts in vitro, we used osteoclast-like cells (OCLs) purified from co-cultures by removing osteoblastic cells (18). The transduction of osteoclasts had been extremely difficult before we demonstrated that adenovirus vector-mediated gene delivery system can be successfully used to transduce foreign genes into osteoclasts both in vitro and in vivo (23, 24). We also found that ERK activity is important for osteoclast survival using adenovirus system. Downregulation of ERK activity by dominant negative Ras (RasDN) overexpression inhibited the survival of OCLs, whereas ERK activation after the introduction of constitutively active MEK (MEKCA) remarkably lengthened their survival (Fig. 1A). As shown in Fig. 1B, DNA fragmentation was observed in OCLs 24 h after purification by removing osteoblastic cells, indicating that OCLs undergo rapid apoptosis without any supporting cells or cytokines (20, 25, 26). We further determined that ~80% of the nuclei expressing RasDN underwent apoptosis after 12 h of the purification as shown in Fig. 1C by TUNEL staining. In contrast, positive TUNEL staining was hardly observed in MEKCA-expressed OCLs even 24 h after the purification, indicating that Ras/ERK pathways greatly contribute to preventing apoptosis of osteoclasts.
Fig. 1. Osteoclasts undergo spontaneous apoptosis.
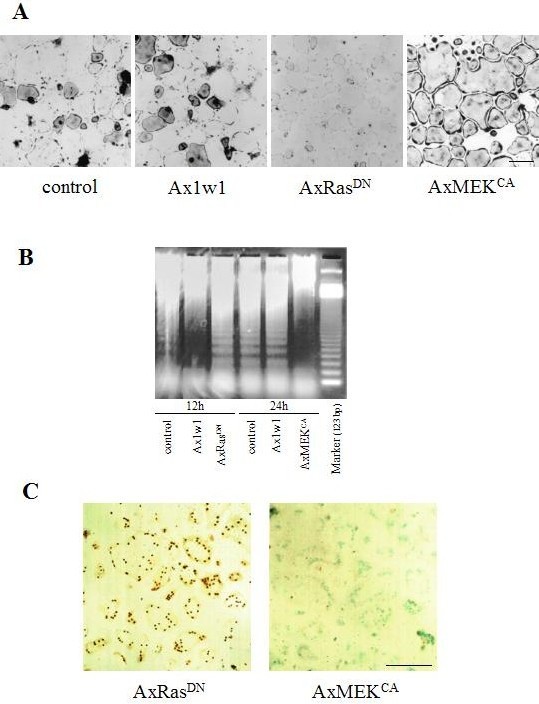
(A) After purification, non-infected OCLs or OCLs infected with Ax1w1, AxRasDN, or AxMEKCA were incubated with αMEM/10%FBS. The cultures remaining 18 h after purification were subjected to TRAP staining. (B) DNA ladder assay for apoptosis. Fragmentation of DNA in uninfected OCLs and OCLs infected with Ax1w1 was not detectable within 12 h, but observed after culturing for 24 h. DNA fragmentation was promoted in RasDN-expressed OCLs, and clear fragmentation was observed within 12 h. In contrast, DNA ladder formation, i.e. apoptosis, was strongly suppressed by expressing MEKCA. The 123-bp ladder DNA was used as a marker. (C) The TUNEL staining of MEKCA- or RasDN-expressed OCLs was conducted 24 or 12 h after the purification, respectively. Apoptotic cells were recognized by their dark nuclear staining (TUNEL-positive) and nuclei of nonapoptotic cells were visualized by staining with methyl green. Bar: 50 μm. (Ref. 20; Copyright 2000. The Rockfeller University Press).
Rapid induction of Bim protein in purified OCLs
Because osteoclasts contain abundant mitochondria that are involved in cytochrome c-mediated caspase-9 activation, we investigated whether mitochondrial pathways are involved in osteoclast apoptosis. As shown in Fig. 2A and B, depolarization of mitochondrial transmembrane potential, chromatin condensation and cytochrome c release from mitochondria into cytoplasm were observed in OCLs after purification. To further investigate the role of mitochondrial pathways in osteoclast survival, we examined the expression levels of pro- or anti-apoptotic Bcl-2 family members in purified OCLs. Immunoblot analysis revealed a significant increase in Bim expression levels in a time-dependent manner after removal of M-CSF, while those of Bid, Bax or Bcl-xL remained unchanged (15). Induction of Bim was reversed by M-CSF or, albeit less efficiently, by RANKL treatment (15). However, stimulation of M-CSF did not change bim mRNA expression in OCLs (15), demonstrating that the changes in Bim protein levels are due to post-transcriptional mechanism.
Fig. 2. Mitochondrial pathway is involved in osteoclast apoptosis.
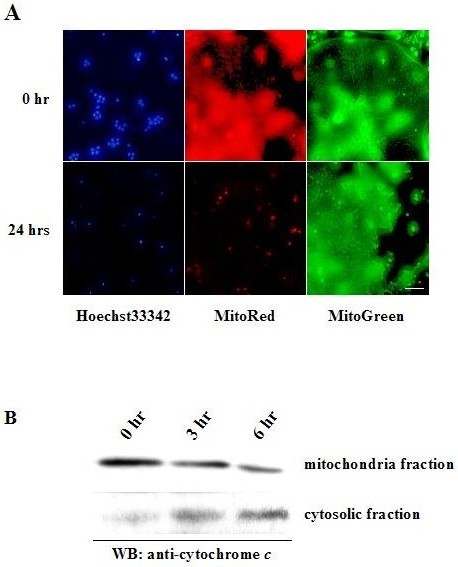
(A) The nuclei of purified OCLs at time zero were stained uniformly with Hoechst 33258, whereas fragmentation and condensation of nuclei were observed 24 h after the purification. MitoRed staining, an indicator of mitochondrial membrane potential, was markedly decreased in OCLs 24 h after purification, as compared to the staining at time zero. MitoGreen, which preferentially accumulates in mitochondria regardless of mitochondrial membrane potential, was used to detect the localization of mitochondria. Bar: 10 μm. (B) Subcellular fractionation of purified OCLs. Cell membranes were fractionated by ApoAlert Cell Fractionation Kit and the resulting fractions were immunoblotted with anti-cytochrome c antibody. Release of cytochrome c from mitochondria to cytosol was increased in a time-dependent manner.
Ras/ERK pathway is involved in regulation of Bim in OCLs
We found that the ERK pathway promotes survival of OCLs (20), while others found that the PI3-kinase/Akt pathway has an effect on this process (27, 28). To analyze whether these pathways are involved in downregulation of Bim, we used adenovirus vectors encoding constitutively active MEK1 (MEKCA) or Akt (myr-Akt), which contains a Src myristoylation signal that promotes association with the plasma membrane, causing constitutive activation. Enforced expression of MEKCA downregulated the expression level of Bim, while that of myr-Akt had less effect. These results indicate that Ras/ERK signaling is an important pathway for regulation of Bim expression level in OCLs (15).
Bim deficiency promotes osteoclast survival in vitro
To investigate the role of Bim in osteoclast apoptosis in vitro, we performed the OCL survival assay using bone marrow cells from bim-/- and bim+/+ mice. After purification by removing osteoblasts, bim-/- and bim+/+ OCLs were incubate with αMEM containing 10% FBS. Almost all bim+/+ OCLs disappeared within 24 hours, whereas more than 90% of bim-/- OCLs remained alive. The elongated survival of bim-/- OCLs was abrogated by the overexpression of Bim with adenovirus vector (15). These results indicate that the expression level of Bim play a critical role in spontaneous apoptosis of osteoclasts.
Expression of Bim in skeletal tissues
To analyze the expression of bim in skeletal tissues in vivo, we performed in situ hybridization analysis using a bimL antisense probe which detects bimS , bimL and bimEL . Bim mRNA was strongly expressed in the bone trabeculae of 5-week-old male mouse metatarsal bone, which was co-localized with tartrate-resistant acid phosphatase (TRAP) enzymatic staining, i.e. with osteoclasts (Fig. 3). We used bone sections from bim-deficient mice as negative controls (data not shown). On the other hand, bim transcripts were hardly detectable in osteoblasts or chondrocytes, whose localization was determined by procollagen type IA and type IIA expression, respectively (Fig. 3). The expression pattern of bim in the skeletal tissues was confirmed further by X-gal staining of mutant mice, in which a lacZ reporter gene was knocked into the bim locus by homologous recombination (Fig. 3).
Fig. 3. In situ hybridization of the section of the metatarsal bones from 5-week-old male wild type mouse using digoxigenin-labeled mouse bimL (B), procollagen type IA (C) and type II A (D) riboprobes, and TRAP enzymatic staining (A).
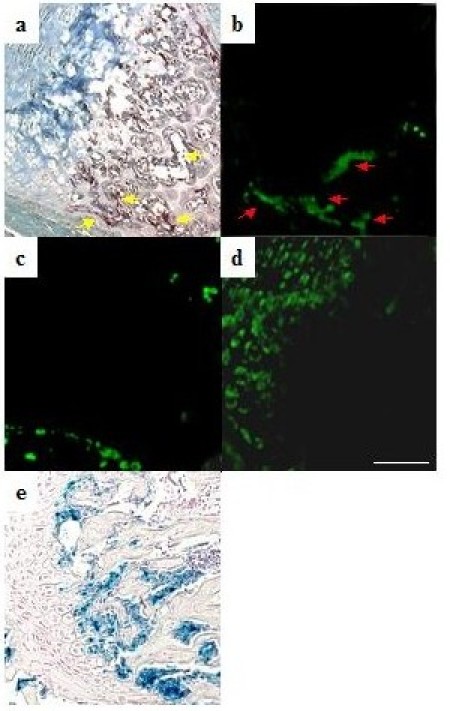
The labeling was detected by anti-digoxigenin antibody and Alexa 488-labeled anti-rabbit IgG antibody. Note the colocalization of bim transcripts with TRAP staining (osteoclasts) (A & B). No positive bimL staining was colocalized with procollagen type IA staining (osteoblasts) or type IIB staining (chondrocytes). X-gal staining of the tibiae from 5-week-old transgenic mice in which lacZ gene was introduced into the bim locus by homologous recombination also showed the clear positive staining in osteoclasts but not in chondrocytes or osteoblasts (E). Bar: 100 μm. (Ref. 15; Copyright 2003. The European Molecular Biology Organization).
Elongated life span of bim-/- osteoclasts in vivo
We next examined whether bim-/- osteoclasts have a longer life span in vivo. Five-week-old bim+/+ and bim-/- mice were fed with water containing 1 mg/ml of BrdU for 1 week (labeling period). Mice were then sacrificed either on the next day (group A) or after 6 weeks (group B) of the labeling period. Immunostaining with anti-BrdU antibody demonstrated that almost similar proportions of osteoclasts were positively stained in group A bim+/+ and bim-/- mice (50 and 48%, respectively). However, the proportion of BrdU-positive osteoclasts was markedly decreased to < 5% in group B bim+/+ mice, while that in group B bim-/- mice was maintained at 33% (Fig. 4). This suggests that bim-/- osteoclasts have a longer life span than bim+/+ osteoclasts in vivo.
Fig. 4. bim-/- osteoclasts have longer life span in vivo.
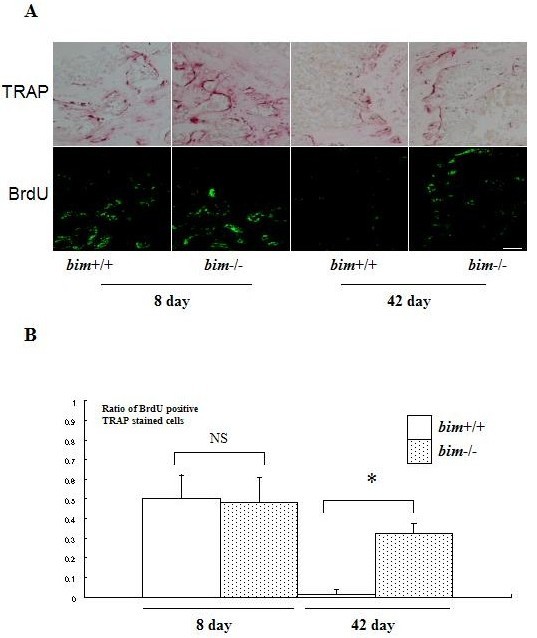
(A) Five-week-old bim+/+ and bim-/- mice (n=4) were fed with water containing 1 mg/ml BrdU for 1 week (labeling period). Mice were then sacrificed either on the next day (group A) or after 6 weeks (group B) of the labeling period, and their tibia were examined by anti-BrdU immunohistochemistry. Bar: 100 μm. (B) More than 100 osteoclasts were examined by BrdU immunostaining in the serial sections of tibia, and the number of BrdU-positive osteoclasts was counted. 50% of bim+/+ osteoclasts and 48% of bim-/- osteoclasts in group A were positively stained by BrdU. However, the proportion of BrdU-positive osteoclasts was markedly reduced to less than 5% in group B bim+/+ mice, due to the apoptotic cell death, while more than 30% of group B bim-/- osteoclasts still exhibited BrdU labeling. NS = not significantly different. *Significantly different, P < 0.005. (Ref. 15; Copyright 2003. The European Molecular Biology Organization).
M-CSF stimulation promotes ubiquitylation-dependent degradation of Bim in OCLs
Induction of Bim protein without changes in mRNA expression levels suggested that Bim is regulated post-transcriptionally and rapidly. To elucidate the detail of Bim regulation, we investigated the effect of proteasome inhibitors, such as lactacystin or MG132, on Bim expression level in OCLs. Interestingly, treatment with these inhibitors greatly enhanced the expression of Bim both in OCLs and their precursors. This indicates that the ubiquitin/proteasome degradation system is involved in the regulation of Bim expression in these cells. In fact, immunoprecipitation of Bim followed by immunoblot analysis using anti-ubiquitin antibody demonstrated a high level of Bim poly-ubiquitylation in OCLs cultured in M-CSF, which was reduced after M-CSF removal (15). Furthermore, we found that Bim is associated with c-Cbl and that overexpression of c-Cbl suppressed the expression of Bim by promoting its ubiquitylation, while that of v-Cbl, which can act in a dominant-negative fashion, increased Bim level even in the presence of M-CSF, indicating the important role of c-Cbl in Bim ubiquitylation (15).
Retrovirus-mediated Bim mutant expression in osteoclast precursors
One of the first steps in the ubiquitin/proteasome degradation system includes selective modification of ε-NH2 groups of lysine residues by ubiquitylation. We constructed retroviral vectors encoding wild type (wt) or mutated (mt) BimEL, which lacks all the ubiquitin acceptor lysine residues (Lys3 and Lys108). The both lysine residues are mutated to arginine. These vectors contain EGFP tracer that enables to detect the infected cells. bim-/- osteoclast precursors were infected either with control virus (pMX-IRES-EGFP), pMxBimEL-IRES-EGFP or pMxmtBimEL-IRES-EGFP in the presence of M-CSF and a pan-caspase inhibitor zVAD-FMK. Removal of zVAD-FMK induced rapid apoptosis in the cells expressing mtBimEL within 18 h even in the presence of M-CSF, while almost all control virus- or 77% of pMx-IRES-BimEL-infected cells survived for at least 24 h. Removal of both zVAD-FMK and M-CSF caused apoptosis in BimEL and mtBimEL-expressing cells (Fig. 5A). MG132 treatment clearly upregulated the expression of wtBimEL, while it did not affect mtBim expression (Fig. 5C). Immunoprecipitated mtBim was not ubiquitinated even in the presence of M-CSF and MG132, as compared to wtBim (Fig. 5D). These results indicate that ubiquitylation and proteasome-mediated degradation are critical modulators of the pro-apoptotic cascades regulated by Bim, at least in osteoclasts and their precursors.
Fig. 5. Effect of mutations in Bim that prevent ubiquitylation.
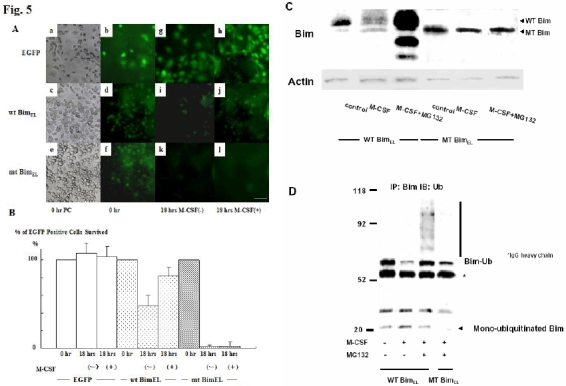
(A) Phase contrast (a, c, & e) and immunofluorescence microscopy.: bim-/- bone marrow cells cultured in the presence of M-CSF were infected with either pMx-IRES-EGFP, pMxBimEL-IRES-EGFP or pMxmtBimEL-IRES-EGFP: zVAD-FMK was added just after the retroviral infection. After 7 days of the retrovirus infection, when gene expression was confirmed by EGFP fluorescence (b, d, & f), cultures were deprived of zVAD-FMK. Eighteen hours after zVAD-FMK removal, more than 70% of pMx-IRES-EGFP- and pMxBimEL-IRES-EGFP-infected cells survived as identified by EGFP fluorescence (h and j), compared to the survival rate of 5% in pMxmtBimEL-IRES-EGFP virus-infected cells (l). Both pMxBimEL-IRES-EGFP- or pMxmtBimEL-IRES-EGFP-infected cells died when M-CSF was removed from the cultures (i & k). Bar: 50 μm. (B) The survival ratio of EGFP positive cells. Almost 100% of the control cells survived even 18h after z-VAD-FMK removal in the presence of M-CSF. 18 hours after zVAD-FMK removal, more than 70% of pMx-IRES-EGFP- and pMxBimEL-IRES-EGFP-infected cells survived as identified by EGFP fluorescence (EGFP and wtBimEL), compared to the survival rate of 5% in pMxmtBimEL-IRES-EGFP virus-infected cells (mtBimEL). (C) The proteasome inhibitors MG132 enhanced the expression level of wtBim in pMxBimEL-IRES-EGFP-infected OC precursors even in the presence of M-CSF, while no obvious upregulation of mtBim was observed in pMxmtBimEL-IRES-EGFP infected cells. (D) WtBim or mtBim was immunoprecipitated from cell lysates of pMxBimEL-IRES-EGFP-infected or pMxmtBimEL-IRES-EGFP-infected cells using anti-Bim polyclonal antibody, and the immunoprecipitates were immunoblotted with anti-ubiquitin antibody. Treating the cells with the proteasome inhibitor MG132 strongly increased the ubiquitination of Bim, while no ubiquitination of mtBim was observed. (Ref. 15; Copyright 2003. The European Molecular Biology Organization).
In summary, our experiments highlight that a combination of adenovirus and retrovirus-mediated gene transfer into osteoclasts and their precursors can be powerful tools for studying osteoclast function such as their survival and bone resorbing activity. Adenovirus vectors are also useful for in vitro gene transfer to cultured osteoblasts, chondrocytes and synoviocytes, and moreover, they are one of the most efficient vectors for in vivo gene delivery into intra-articular tissues including synovium (24). We also established the modified protocols for in situ hybridization and BrdU labeling of bone sections from mice. These methods enabled us to obtain a new insight into the molecular mechanism and dynamics of bone homeostasis and inflammatory diseases leading to progressive bone and joint destruction.
Acknowledgments
The authors thank R. Yamaguchi and M. Ikeuchi (Department of Orthopaedic Surgery, The University of Tokyo), who provided expert technical assistance. We also thank H. Katagiri (Tohoku University) and T. Asano (The University of Tokyo) for MEKCA and myrAKT adenoviruses, T. Kitamura (Institute of Medial Science, The University of Tokyo) for pMx vectors, J Adams and S Cory (WEHI) for bim-/- mice. This work was supported by fellowships and Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, the Health Science Research Grants from Ministry of Health and Welfare of Japan, Uehara Memorial Award, Nakatomi Health Science Foundation Award, Grants-in-Aid from the Research Society for Metabolic Bone Diseases to S.T, and by the NHMRC (Canberra), the Leukemia and Lymphoma Society of America and the Dr. Josef Steiner Cancer Research Foundation (Bern).
Appendix
Protocols
Adenovirus infection
Day 0: Seed 1.5 × 106 osteoblastic cells in 10 cm tissue culture dishes in 10 ml αMEM/10%FBS containing 10 nM 1α,25(OH)2D3 and 1 μM PGE2.
Day 1: Aspirate off media and start coculture by adding 1.5 × 107 bone marrow cells in 10 ml fresh media with 1α,25(OH)2D3 and PGE2.
Day 3: Add only 5 ml fresh media containing 1α,25(OH)2D3 and PGE2 without aspirating medium.
Day 5: Aspirate media and add 1ml of αMEM containing the recombinant adenovirus at an indicated MOI. Then incubate at 37°C for 1 h with intermittent shaking. Wash twice with PBS and further incubated at 37°C in αMEM/10%FBS containing 10 nM 1α,25(OH)2D3 and 1 μM PGE2.
Day 6: Aspirate medium and wash twice with PBS. Add 5 ml of αMEM containing 0.1% collagenase and 0.2% dispase and incubate for 10 min at 37°C. Tap the dishes to remove osteoblastic cells. The purity of OCLs was > 90% at the final preparation. Carefully wash twice with αMEM without injuring purified OCLs. Then start the experiments using the purified OCLs.
Retrovirus infection
Day 0: Seed 1.0 × 107 bone marrow cells in 6 cm tissue culture dishes in 10 ml αMEM/10%FBS with mouse recombinant M-CSF (50ng/ml).
Day 2: Aspirate off media and incubate with 1 ml of αMEM containing the retrovirus at an indicated MOI for 4 h in the presence of mouse recombinant M-CSF (50 ng/ml) and polybrene (8 μg/ml). Wash twice with PBS and further incubated at 37°C in αMEM/10%FBS containing 100 ng/ml M-CSF.
Day 4, 6, 8: Aspirate off media and replace with fresh complete media.
Day 9: Start the experiments using retrovirus-infected bone marrow macrophage.
In situ hybridization
Place slides in RNase-free glass coplin jars.
Deparaffinize and rehydrate in:
xylene for 3 X 2 minutes
100% EtOH for 3 X 1 minutes
95% EtOH for 1 X1 minutes
70% EtOH for 1 X 1 minutes
1XPBS for 1 X 1 minutes.
Fix sections with 4% paraformaldehyde buffered with PBS (PFA-PBS) for 15 min at room temperature.
Wash in 1X PBS for 5 minutes.
Digest slides with 10 μg/ml proteinase K for 15 min in PBS.
Treat again with 4% PFA-PBS for 10 min to inactivate proteinase K
Wash in 1X PBS for 3 minutes.
Incubate with 0.2 N HCl for 10 minutes
Wash in 1X PBS for 5 minutes.
Acetylate slides with 0.3% acetic anhydride in the presence of 0.1 M triethanolamine for 10 min.
Treat with 3% hydrogen peroxide/methanol for 30 min. Dehydrate with:
1XPBS for 1 X 5 minutes
70% EtOH for 1 X 3 minutes
95% EtOH for 1 X 3 minutes and air dried.
Add 28.5 μl hybridization buffer to 1.5 μl probe solution and mix thoroughly.
Boil hybridization solution for 5 minutes, place on ice.
Pipette hybridization solution onto section.
Place in a prewarmed humidity chamber and incubate for 18 hours at 52°C.
Preheat 200 ml 2XSSC to 52°C.
Remove slides from the humidity chamber and place in Coplin jars.
Wash slides in 2XSSC for 2 X 1 minutes.
Treat tissues with RNase A 30 μg / ml / 2XSSC for 30 minutes at 37°C.
Wash slides in 2XSSC for 2 X 1 min.
Incubate in 50% formamide / 2XSSC for 5 minutes at 52°C.
Wash slides in 3 X TBS-T for 3 min.
Block slides by PBS containing 1% bovine serum albumin for 10 minutes.
Incubate with HRP-conjugated anti-DIG rabbit polyclonal antibody at a dilution of 1:100 for 24 h at 4°C.
Return slides to a Coplin jar and wash in DIG I buffer for 10 minutes. Repeat.
Wash slides in 3 X TBS-T for 3 min.
Immersed slides in a diaminobenzidine solution for 5-10 min at room temperature to visualize immunoreactivity.
Wash slides in water for 3 min
Counterstain in methylgreen for 1 to 2 minutes
Rinse thoroughly in running water.
Coverslip slides with permanent mounting media.
In vivo BrdU labeling
Feed 5-week-old bim+/+ and bim-/- mice with water containing 1 mg/ml of 5’-bromo-2’-deoxyuridine (BrdU) for 1 week (labeling period). Replace with fresh BrdU water every other day.
Remove the tibiae and fix in 4% paraformaldehyde/PBS at 4°C for 8 hours.
Embed the tibiae in paraffin and section paraffin-embedded samples 3-mm-thick.
Place slides in glass coplin jars.
Deparaffinize and rehydrate in:
xylene for 3 X 2 minutes
100% EtOH for 3 X 1 minutes
95% EtOH for 1 X 1 minutes
70% EtOH for 1 X 1 minutes
1XPBS for 3 X 1 minutes.
Treat with 2.5% hyaluronidase and block with 3% FBS in TBST for 30 min at room temperature.
Incubate sections with the fluorescein-conjugated anti BrdU antibody at a dilution of 1:50 overnight at 4°C.
Wash with PBS for 5min three times.
Coverslip slides with FluorSave.
Examine the sections using a confocal imaging system (MRC-600; Bio-Rad Laboratories).
References
- Baron R. Molecular mechanisms of bone resorption by the osteoclast. Anat Rec. 1989;224(2):317–324. doi: 10.1002/ar.1092240220. [DOI] [PubMed] [Google Scholar]
- Hughes DE, Wright KR, Uy HL, Sasaki A, Yoneda T, Roodman GD, Mundy GR, Boyce BF. Bisphosphonates promote apoptosis in murine osteoclasts in vitro and in vivo. J Bone Miner Res. 1995;10(10):1478–1487. doi: 10.1002/jbmr.5650101008. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13(15):1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- Strasser A, O'Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103(6):839–842. doi: 10.1016/S0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17(2):384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly LA, Cullen L, Visvader J, Lindeman GJ, Print C, Bath ML, Huang DC, Strasser A. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am J Pathol. 2000;157(2):449–461. doi: 10.1016/S0002-9440(10)64557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286(5445):1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415(6874):922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, Johnson EM. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29(3):615–628. doi: 10.1016/S0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29(3):629–643. doi: 10.1016/S0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Villunger A, Scott C, Bouillet P, Strasser A. Essential role for the BH3-only protein Bim but redundant roles for Bax, Bcl-2, and Bcl-w in the control of granulocyte survival. Blood. 2003;101(6):2393–2400. doi: 10.1182/blood-2002-07-2132. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, Fukuda A, Hikita A, Seto H, Okada T et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. Embo J. 2003;22(24):6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc Natl Acad Sci USA. 1996;93(3):1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae Y, Makimura M, Saito I. A simple and efficient method for purification of infectious recombinant adenovirus. Jpn J Med Sci Biol. 1994;47(3):157–166. doi: 10.7883/yoken1952.47.157. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, Moseley JM, Martin TJ, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123(5):2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- Tezuka K, Sato T, Kamioka H, Nijweide PJ, Tanaka K, Matsuo T, Ohta M, Kurihara N, Hakeda Y, Kumegawa M. Identification of osteopontin in isolated rabbit osteoclasts. Biochem Biophys Res Commun. 1992;186(2):911–917. doi: 10.1016/0006-291x(92)90832-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Katagiri H, Kanegae Y, Takayanagi H, Sawada Y, Yamamoto A, Pando MP, Asano T, Verma IM, Oda H et al. Reciprocal role of ERK and NF-kappaB pathways in survival and activation of osteoclasts. J Cell Biol. 2000;148(2):333–342. doi: 10.1083/jcb.148.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi E, Nakamura I, Ikebe T, Akiyama S, Takahashi N, Suda T. Activation of NF-kappaB is involved in the survival of osteoclasts promoted by interleukin-1. J Biol Chem. 1998;273(15):8799–8805. doi: 10.1074/jbc.273.15.8799. [DOI] [PubMed] [Google Scholar]
- Okahashi N, Koide M, Jimi E, Suda T, Nishihara T. Caspases (interleukin-1beta-converting enzyme family proteases) are involved in the regulation of the survival of osteoclasts. Bone. 1998;23(1):33–41. doi: 10.1016/S8756-3282(98)00069-6. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Takahashi T, Takayanagi H, Miyazaki T, Oda H, Nakamura K, Hirai H, Kurokawa T. Modulation of osteoclast function by adenovirus vector-induced epidermal growth factor receptor. J Bone Miner Res. 1998;13(11):1714–1720. doi: 10.1359/jbmr.1998.13.11.1714. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Juji T, Miyazaki T, Iizuka H, Takahashi T, Isshiki M, Okada M, Tanaka Y, Koshihara Y, Oda H et al. Suppression of arthritic bone destruction by adenovirus-mediated csk gene transfer to synoviocytes and osteoclasts. J Clin Invest. 1999;104(2):137–146. doi: 10.1172/JCI6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne WC, Baron R. Src kinase activity is essential for osteoclast function. J Biol Chem. 2004;279(17):17660–17666. doi: 10.1074/jbc.M311032200. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol. 2003;160(5):709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong BR, Besser D, Kim N, Arron JR, Vologodskaia M, Hanafusa H, Choi Y. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell. 1999;4(6):1041–1049. doi: 10.1016/S1097-2765(00)80232-4. [DOI] [PubMed] [Google Scholar]
- Glantschnig H, Fisher JE, Wesolowski G, Rodan GA, Reszka AA. M-CSF, TNFalpha and RANK ligand promote osteoclast survival by signaling through mTOR/S6 kinase. Cell Death Differ. 2003;10(10):1165–1177. doi: 10.1038/sj.cdd.4401285. [DOI] [PubMed] [Google Scholar]


