Summary
The tri-methylation of histone H3 on K4 (H3K4me3) is a key epigenetic modification that is predominantly found at active gene promoters and is deposited by the complex of proteins associated with SET1 (COMPASS). CXXC zinc finger protein 1 (Cfp1) regulates this process by recruiting SET1 to chromatin and recognizing H3K4me3 via its plant homeodomain (Cfp1PHD). Here, we present a protocol for the purification and crystallization of the Drosophila melanogaster Cfp1PHD domain in complex with an H3K4me3 peptide (PDB: 9C0O). We describe steps for obtaining highly pure Cfp1PHD and diffraction-quality crystals. We then detail procedures for rapidly identifying crystals containing the H3K4me3-bound form of the Cfp1PHD domain.
For complete details on the use and execution of this protocol, please refer to Grégoire et al.1
Subject areas: Protein expression and purification, Structural Biology, X-ray Crystallography
Graphical abstract

Highlights
-
•
Production and purification of Cfp1PHD using affinity chromatography
-
•
Crystallization and structure determination of Cfp1PHD
-
•
Detailed troubleshooting to solve the H3K4me3-bound form of Cfp1PHD
-
•
Structural characterization of Cfp1PHD bound to H3K4me3
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The tri-methylation of histone H3 on K4 (H3K4me3) is a key epigenetic modification that is predominantly found at active gene promoters and is deposited by the complex of proteins associated with SET1 (COMPASS). CXXC zinc finger protein 1 (Cfp1) regulates this process by recruiting SET1 to chromatin and recognizing H3K4me3 via its plant homeodomain (Cfp1PHD). Here, we present a protocol for the purification and crystallization of the Drosophila melanogaster Cfp1PHD domain in complex with an H3K4me3 peptide (PDB: 9C0O). We describe steps for obtaining highly pure Cfp1PHD and diffraction-quality crystals. We then detail procedures for rapidly identifying crystals containing the H3K4me3-bound form of the Cfp1PHD domain.
Before you begin
Timing: 2 h
While this protocol has been optimized for the Drosophila melanogaster Cfp1PHD domain (DmCfp1PHD), it has been used for various GST-tagged proteins from other species by optimizing the buffer and crystallization conditions (e.g., crystallization method (sitting/hanging drop) and solutions). Here, we describe the protocol for obtaining crystals of Cfp1PHD bound to a peptide corresponding to the first 14 residues of histone H3 tri-methylated on lysine 4 (H3K4me31-14).2 Peptides spanning different regions neighboring H3K4 or harboring other post-translational modifications can be used. However, the crystallization conditions may vary.
Note: The H3K4me31-14 peptide must be at least 95% pure in a lyophilized form without TFA. Upon receipt, the peptide is stored at −20°C.
Note: When working with bacterial cultures, ensure sterile conditions. Use aseptic techniques (under flame) to avoid contamination. Clean everything (bench, pipettes, etc.) with 70% EtOH.
Note: A DNA plasmid (here, a pGEX-4T-1-based vector encoding DmCfp1PHD (56-119) in fusion with a Tobacco Etch Virus (TEV) cleavable glutathione-S-transferase needs to be available and kept at −20°C.
-
1.
Prepare all the buffers needed and cool them at 4°C.
-
2.
Prepare Luria-Bertani (LB) Broth/agar and autoclave at least one day before the experiment.
-
3.Prepare the gel filtration column (Superdex 75 prep column) for the step “protein purification of Cfp1PHD by affinity and size-exclusion chromatography steps”.
-
a.Wash and equilibrate with at least 2 column volumes using Buffer B at 4°C.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli: BL21-Rosetta | Novagen | Cat#70954-3 |
| Chemicals, peptides, and recombinant proteins | ||
| Isopropyl-beta-D-thiogalactoside (IPTG) | Fisher BioReagents | Cat#BP1755-100 |
| Ampicillin | Fisher BioReagents | Cat#BP1760-25 |
| Chloramphenicol | Fisher BioReagents | Cat#BP904-100 |
| Glutathione reduced | Fisher Scientific | Cat#J62166-14 |
| Sodium chloride (NaCl) | Fisher BioReagents | Cat#BP358-10 |
| 2-Mercaptoethanol (BME) | Fisher Scientific | Cat#125472500 |
| Glutathione agarose resin | Thermo Fisher Scientific | Cat#16101 |
| Tris-Base | Fisher BioReagents | Cat#BP152-5 |
| Polyethylene glycol 3350 | Sigma | Cat#88276 |
| Triton X-100 | Fisher BioReagents | Cat#BP151 |
| Glycerol | Fisher BioReagents | Cat#BP229-4 |
| Trichloroacetic acid (TCA) | Fisher Scientific | Cat#A322-500 |
| Zinc chloride | Fisher Scientific | Cat#A16281-22 |
| Methanol | Fisher Chemical | Cat#A412-4 |
| Bromophenol blue | Fisher Biotech | Cat#BP115-25 |
| Glacial acetic acid | Fisher Chemical | Cat#A38SI-212 |
| Dithiothreitol (DTT) | Fisher BioReagents | Cat#BP172-25 |
| Lysozyme | Fisher Scientific | Cat#BP535-10 |
| H3K4me3 (1–14) | GenScript | N/A |
| Mini-PROTEAN TGX gels (4%–20%) | Bio-Rad | Cat#4561094 |
| Recombinant DNA | ||
| pGEX-4T-1 based vector encoding DmCfp1PHD (56–119) | GenScript | N/A |
| Other | ||
| ӒKTA pure chromatography system | Cytiva | N/A |
| 33 mm syringe filter 0.45 μM | Fisherbrand | Cat#09-720-005 |
| Autoclave | N/A | N/A |
| 0.22 μM Nitrocellulose membrane filter | Millipore | SA1J789H5 |
| 24-well Cryschem M Sitting drop plate (round reagent reservoir) | Hampton Research | Cat#HR1-002 |
| Luer-Lok 50 mL syringe | BD | Cat#309653 |
| Heat block | Fisher Scientific | Cat#11-718 |
| HiLoad 16/600 Superdex 75 pg chromatography column | Cytiva | GE28-9893-33 |
| Low-speed centrifuge | Thermo Scientific | N/A |
| Eppendorf centrifuge 5424 R | Eppendorf | EP5404000138 |
| Avanti J-20 XP1 high-speed centrifuge | Beckman Coulter | N/A |
| Tube Rotator | Thermo Scientific | N/A |
| Stirrer | N/A | N/A |
| SolVac filter device | N/A | N/A |
| GENESYS 30 Visible spectrophotometer | Thermo Scientific | Cat# 840-277000 |
| SteREO Discovery.V8 microscope | Zeiss | N/A |
| CrystalCao PINE HT Cryo-loop 0.1–02 mm | Hampton Research | HR8-122 |
| Intelli-plate 96-3 LVR Protein crystallization plate | Hampton Research | HR3-185 |
| Centrifugal concentrator (3 kDa MWCO) | Millipore | UFC9003 |
| Crystal clear sealing tape | Hampton Research | HR4-511 |
| Erlenmeyer flasks, 4 L | N/A | N/A |
| Fraction collection tubes for FPLC | N/A | N/A |
| Probe sonicator | Misonix | N/A |
| Protein electrophoresis equipment | Bio-Rad | PowerPac Basic |
| Plasticware (measuring cylinders, beakers) | N/A | N/A |
| Shaker incubator | New Brunswick | Innova |
| ARI Crystal Gryphon dropsetter | Art Robbins Instruments | |
| ClearSeal Film | Hampton | Cat#HR4-521 |
| NanoDrop ONE | Thermo Scientific | N/A |
| Empty purification column | N/A | N/A |
Materials and equipment
Note: This protocol uses an AKTA Fast Protein Liquid Chromatography (FPLC) system from Cytiva. Other FPLC systems can be used, such as the model commercialized by Bio-Rad.
Note: This protocol uses a NanoDrop One (Thermo Scientific) spectrophotometer for protein quantification. Alternatives such as cuvette-based spectrophotometer or Qubit fluorometer can be used. The NanoDrop One allows you to quantify protein with small volumes (1μL) and the ability to measure the purity of the sample without the need for any reagents.
CRITICAL: When preparing buffers, read the Material Safety Data Sheet (MSDS) and ensure you understand the safety procedure when handling hazardous chemicals.
LB broth
| Reagent | Final concentration | Amount |
|---|---|---|
| Luria-Bertani (LB) broth | 2.5% (w/v) | 100 g |
| ddH2O | N/A | Up to 4 L |
| Total | N/A | 4 L |
Note: This recipe can make eight flasks containing 500 mL each.
Note: LB Broth needs to be autoclaved and then cooled down below 55°C before the addition of antibiotics (ampicillin (100 μg/mL) and chloramphenicol (50 μg/mL)).
Note: LB broth can be stored at room temperature for a few days, but it is best to prepare it fresh each time.
LB agar
| Reagent | Final concentration | Amount |
|---|---|---|
| Luria-Bertani (LB) broth | 2.5% (w/v) | 25 g |
| Agar | 1% | 10 g |
| ddH2O | N/A | Up to 1 L |
| Total | N/A | 1 L |
Note: LB agar needs to be autoclaved and cooled down below 55°C before adding the sterile filtered antibiotics of choice (ampicillin (100 μg/mL) and chloramphenicol (50 μg/mL)). LB agar plates can be stored at 4°C away from direct light.
Buffer A
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris pH 8 (1 M) | 50 mM | 50 mL |
| NaCl (5 M) | 150 mM | 30 mL |
| β-mercaptoethanol (14.3 M) | 5 mM | 350 μL |
| Glycerol (100%) | 10% | 100 mL |
| ddH2O | N/A | 820 mL |
| Total | N/A | 1000 mL |
Note: Store at 4°C for up to 1 month.
Note: The stock concentrations are indicated in parentheses.
Buffer B
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris pH 8 (1 M) | 20 mM | 20 mL |
| NaCl (5 M) | 150 mM | 30 mL |
| β-mercaptoethanol (14.3 M) | 5 mM | 350 μL |
| ddH2O | N/A | 950 mL |
| Total | N/A | 1000 mL |
Note: Prepare fresh buffer B each time and store at 4°C during the protocol.
Note: The stock concentrations are indicated in parentheses.
Note: To remove debris and avoid clogging the FPLC system, filter buffer B with a 0.22 μM filter.
Regeneration buffer (R buffer)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris pH 8 (1 M) | 50 mM | 2.5 mL |
| Glutathione reduced | 10 mM | 0.154 g |
| ddH2O | N/A | 47.5 mL |
| Total | N/A | 50 mL |
Note: Prepare fresh regeneration buffer each time and store at 4°C during the protocol.
Note: The stock concentrations are indicated in parentheses.
5X SDS loading buffer—Laemmli buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris pH 6.8 (1 M) | 250 mM | 12.5 mL |
| SDS | 10% | 5 g |
| β-mercaptoethanol (14.3 M) | 10% | Add before use |
| Glycerol (100%) | 25% | 12.5 mL |
| Bromophenol blue | 0.02% | 20 mg |
| ddH2O | N/A | 45 mL |
| Total | N/A | 50 mL |
Note: Transfer in 900 μL aliquots and store at room temperature. Add 100 μL BME/ tube before use.
Note: The stock concentrations are indicated in parentheses.
Step-by-step method details
Overexpression of Cfp1PHD in Escherichia coli
Timing: 4 days
This step allows the expression of Cfp1PHD in Escherichia coli.
Day 1
Timing: 2 h
-
1.Chemical transformation of competent Rosetta-DE3 Escherichia coli (E. coli) cells with the pGEX-4T-1 vector encoding DmCfp1PHD (amino acid 56-119) in fusion with a TEV cleavable glutathione-S-transferase.
-
a.Thaw competent cells on ice.
-
b.Add 1 μL (100 ng) of pGEX-4T-1_DmCfp1PHD to 50 μL of competent cells (Escherichia coli: Rosetta-DE3).
-
c.Incubate DNA-competent cells mixed on ice for 30 min.
-
d.Heat shock competent cells at 42°C in a heat block for 45 s.Note: A water bath can be used as an alternative approach to the heat block.
-
e.Incubate transformed competent cells on ice for 2 min.
-
f.Add 400 μL of LB broth to competent cells and incubate for 1 h at 37°C in a shaker incubator with constant shaking at 250 rpm.
-
g.Plate 50 μL of transformed cells onto LB/agar plate with ampicillin (100 μg/mL) and chloramphenicol (50 μg/mL) and place plates in a stationary incubator for 14–18 h at 37°C.Note: Place the LB/agar plates in a stationary incubator at 37°C 30 min before plating to allow the plates to warm up.Note: Competent cells are very sensitive to temperature. Make sure to thaw on ice and not warm up the cells.
-
a.
Day 2
Timing: 30 min
-
2.Inoculation of the starter culture.
-
a.Inoculate a liquid starter culture of 100 mL LB Broth containing 100 μg/mL Ampicillin and 50 μg/mL Chloramphenicol using a single colony from the LB agar plate.
-
b.Incubate the liquid starter culture in a shaker incubator at 250 rpm for 14–18 h at 37°C.
-
a.
Day 3
Timing: 4 h
-
3.Large scale overexpression.
-
a.Inoculate 8 x 500 mL of LB Broth supplemented with 100 μg/mL Ampicillin, 50 μg/mL Chloramphenicol and 100 μM ZnCl2 with ∼10 mL of starter culture and grow at 37°C in a shaker incubator at 250 rpm until the OD600 reaches 0.6 (approximately 2 h).Alternatives: Volumes can be adjusted according to flask size, while maintaining the same inoculation and shaking conditions as outlined in the main protocol.Note: Regularly check OD600 with a spectrophotometer to prevent the cells from reaching a density higher than OD600 0.6.
-
b.Lower the shaker incubator temperature to 18°C and induce protein expression by adding 500 μL of isopropyl-β-D-thiogalactopyranoside (IPTG) (100 mM) to each 500 mL culture.Note: Leave the flasks in the incubator at 18°C for at least 15 min to allow liquid culture to cool down before adding the IPTG.
-
c.Incubate in a shaker incubator at 250 rpm for 16 h.Note: Add 500 μL of 50% glycerol + 500 μL of overnight culture to produce glycerol stock. For subsequent overexpression of Cfp1PHD, a glycerol stock can be used to start on Day 2 of the protocol.Note: ZnCl2 is essential for the appropriate folding of Cfp1PHD. As such, ZnCl2 is unnecessary if working with other GST fusion proteins lacking zinc-binding sites.
-
a.
Day 4
Timing: 1 h
-
4.Harvesting cell pellets.
-
a.Add 1 L of culture to a centrifuge bottle and spin down at 4000 rpm for 30 min at 4°C.
-
b.Resuspend cell pellets in 30 mL of Buffer A and transfer each cell pellet to 50 mL tubes for storage.
-
c.Store cell pellets in a – 80°C freezer for long-term storage.
-
a.
Pause Point: You can proceed to protein purification or store the cell pellets.
Protein purification of Cfp1PHD by affinity and size-exclusion chromatography
Timing: 2 days
The purification step isolates the target protein, producing an ultra-pure and homogeneous sample essential for protein crystallization.
Note: Always keep cell lysate and purified protein sample on ice throughout all the purification steps.
Day 5
Timing: 1 h (for step 5)
Timing: 2 h (for step 6)
-
5.Cell lysis and isolation of soluble protein extract.
-
a.Thaw one bacterial cell pellet and complete the volume to 80 mL using Buffer A.
-
b.Add 2 mL of Triton X-100 and 2.4 mL of Lysozyme 10 mg/mL to cell lysate.
-
c.Maintaining the cell lysate on ice, lyse the cells using a probe sonicator. Sonicate the lysate three times for 1 min, with a 1-min rest period between each cycle.Note: To optimize cell lysis, ensure the sonicator tip is fully submerged in the cell lysate.Note: After sonication, transfer 2 x 30 μL of total cell lysate into 1.5 mL microcentrifuge tubes. Centrifuge one of the tubes at 15 000 rpm, transfer the supernatant to a new tube and resuspend the pellet in 30 μL of Buffer A. These three samples can be analyzed on SDS-PAGE gel electrophoresis to assess protein solubility.Alternatives: Other methods can also be used for bacterial cell lysis (e.g., French press).
-
d.Transfer cell lysate into high-speed centrifuge tubes and spin down at 16 000 rpm for 30 min at 4°C to remove cell debris and insoluble material.
-
e.Filter supernatant with a 0.45 μm filter using a syringe. Discard pellet.
-
a.
-
6.Cfp1PHD purification by affinity chromatography using glutathione Sepharose (GSH) beads.
-
a.Transfer 10 mL of GSH beads in an empty purification column and apply 50 mL of ddH2O followed by 100 mL of Buffer A to remove residual ethanol from the beads.
-
b.Apply the GSH beads to the filtered supernatant and incubate on a rotating wheel for 1 h at 4°C.
-
c.Thaw two aliquots of TEV (250 μg/aliquot) on ice.
-
d.Apply the supernatant and beads mixture to the purification column and let the supernatant flow through.Note: To make sure that the GSH beads have retained the GST-tagged protein, keep a 30 μL sample of the flow through to visualize by Coomassie-stained SDS-PAGE.
-
e.Wash glutathione Sepharose beads with 150 mL of Buffer A at 4°C.
-
f.Resuspend glutathione Sepharose beads in 30 mL of the Buffer B and transfer to a 50 mL conical tube.Note: Keep a 30 μL sample of the protein-bound beads to visualize by SDS-PAGE.
-
g.Add two aliquots of TEV protease to cleave the TEV site by leaving the tube on a rotating wheel overnight (16–18 h) at 4°C.Note: Add 5 μL of 5X SDS loading buffer to each sample and heat them at 95°C for 5 min and store them at – 20°C until further analysis by the SDS-PAGE.
-
a.
Day 6
Timing: 6 h
-
7.Size exclusion chromatography of Cfp1PHD using a Superdex 75 preparative column.
-
a.Equilibrate the Hi-Load 16/600 Superdex 75 column using 2 column volumes of Buffer B as described in the “before you begin” section.
-
b.Elute cleaved proteins by applying supernatant/beads on the empty purification column and collect flow through.
-
c.Wash beads with 10 mL of Buffer B to obtain residual cleaved proteins. The flow through corresponds to the purified cleaved protein of interest (Cfp1PHD).Note: Store the beads at 4°C in a 50 mL conical tube and cover with the Buffer B. When regenerated using the R Buffer, the beads are reusable.
-
d.Filter eluted protein with a 0.45 μm filter and syringe and transfer into two centrifugal concentrators (with a cut-off of 3 kDa).Note: Pre-equilibrate the 3 kDa MWCO centrifugal concentrators by adding the Buffer B in the concentrator and centrifuge for 10 min at 4000 rpm.Note: Take a 30 μL sample of the beads and eluted protein to visualize by SDS-PAGE and confirm that the GST tag was cleaved.
-
e.Centrifuge concentrators at 4000 rpm in rounds of 20 min (∼3-4 rounds) until the volume reached 2 mL combined (1 mL in each concentrator).
-
f.Transfer the concentrated protein to a 2 mL microcentrifuge tube and centrifuge at 15,000 rpm for 5 min to remove debris before applying it to the Superdex 75.Note: The FPLC system requires a 2 mL injection loop. However, the injection loop volume may be adjusted depending on the column volume or the type of chromatography steps needed.
-
g.Inject the 2 mL concentrated protein sample on the pre-equilibrated gel filtration column (S75prep) at a flow rate of 1 mL/min.
 CRITICAL: Ensure no air bubbles are in the injection syringe or the sample, as it can damage the FPLC.
CRITICAL: Ensure no air bubbles are in the injection syringe or the sample, as it can damage the FPLC. -
h.Collect the fractions of interest according to the purification profile. Cfp1PHD’s molecular weight is 7.7 kDa, so it should elute in the appropriate fractions according to the molecular weight standards.Note: Keep a 30 μL sample of each fraction to visualize on SDS-PAGE gel.
-
i.Pool the fractions of interest, concentrate the protein sample using a 3 kDa cut-off concentrator and measure the protein concentration using a nanodrop.Note: To proceed to crystallization, the protein sample must be concentrated to the desired concentration to set up crystal trays. Cfp1PHD was concentrated to 5 mg/mL.Note: Setting up crystal trays with freshly purified protein is recommended. However, the protein sample can be stored at 4°C or −80°C for short—or long-term storage. For long-term storage, add 20% glycerol to the protein sample and flash-freeze in liquid nitrogen before storing at – 80°C.
-
a.
Crystallization of Cfp1PHD in complex with an H3K4me3 peptide
Timing: time varies for crystals (hours to several weeks)
This step enables the crystallization of Cfp1PHD in a complex with an H3K4me3 peptide (Cfp1PHD-H3K4me3).
The optimal condition to obtain crystals is composed of 0.2 M Ammonium sulfate, 0.01 M Tris pH 8.5, 15% PEG 3350 by sitting drop at room temperature (22°C) with a protein to crystallization solution ratio of 1:1, at a protein concentration of 5 mg/mL in Buffer B.
-
8.Complex formation between Cfp1PHD and H3K4me3.
-
a.Prepare the synthetic peptide corresponding to the 14 residues of histone H3 tri-methylated on lysine 4 by resuspending the lyophilized peptide in ddH2O. A 95% pure (no TFA) H3K4me3 peptide (amino acid 1-14) was ordered from GenScript.
-
b.Transfer 100 μL of protein sample (per plate) of Cfp1PHD into a 1.5 mL microcentrifuge tube.
-
c.Dilute the H3K4me3 peptide to the appropriate concentration with Buffer B and add the peptide to the protein sample in a 1 protein: 3 peptide molar ratio. Incubate the protein-peptide sample for 1 h on ice.
-
a.
Note: The purity of the protein sample should be verified by SDS-PAGE followed by Coomassie-staining before proceeding to crystallization.
Note: Binding studies should be used to confirm the equilibrium dissociation constants between a reader and a histone peptide as it may influence the peptide:protein ratio used to carry out crystallization trials (e.g., Isothermal titration calorimetry, Surface plasmon resonance etc…).
Note: It is recommended not to freeze/thaw the protein sample before crystallization.
-
9.Set up sitting-drop 96-well plates using various sparse matrix screening screens. The screens used to crystallize Cfp1PHD in complex with H3K4me31-14 are the Proplex from Molecular Dimensions and Index from Hampton.
-
a.Add 100 μL of the appropriate crystallization solution to the wells of the sitting-drop plate.Alternatives: Hanging-drop plates can be used with glass cover slides.
-
b.Add 1 μL of crystallization solution from each deep well onto the shallow well, followed by 1 μL of protein-peptide complex to the shallow well for a 1:1 ratio between the protein sample and the crystallization solution.
-
c.Seal the plate with ClearSeal film (Hampton) (or similar sealing tape) and store the plate at room temperature.
-
d.Wait for crystals to appear. Crystals may appear within a day or weeks.Alternatives: Set up crystal plates using an ARI Gryphon drop setter.Note: This protocol can be applied to different protein/peptide complexes. However, the crystallization process can be impacted by many factors, such as protein concentration, purity of protein sample, crystallization condition, the use of seeds or additives and temperature. Please see troubleshooting problems 3 and 4 for potential solutions that may help facilitate the crystallization process.
-
a.
-
10.Optimization of crystals. Set up sitting-drop 24-well plates using the crystallization conditions that produced crystals.
-
a.Prepare optimized crystallization conditions. See Figure 4 for an example.
-
b.Add 500 μL of the appropriate crystallization solution to the 24-well reservoir tailored to enable crystallization using sitting-drop approaches.
-
c.Add 1 μL of crystallization solution from each deep well onto the shallow well, followed by 1 μL of protein-peptide complex.
-
d.Seal the plate with ClearSeal and store the plate at room temperature.
-
e.Wait for crystals to appear. Cfp1PHD crystals appear within 24–48 h.
-
a.
Figure 4.
Example of a crystal optimization plate setup used to obtain high quality crystals of Cfp1PHD (related to step 8)
A 24-well sitting drop plate optimizes the crystallization conditions that produce crystals using various sparse matrix screens (Step 9). From left to right, change the percentage of precipitant. From top to bottom, slightly change the concentration of salt. Here, 20% PEG 3350 (Precipitant) and 0.2 M Ammonium sulfate (Salt) were the initial targets. See Figure 3A for the image of a crystal obtained in well 1C.
Cryoprotection of crystals for X-ray crystallography
Timing: 1 h
-
11.Prepare Cryoprotectant Solution.
-
a.Mix 400 μL of crystallization solution with 100 μL of DMSO (Cryoprotection buffer) in a 1.5 mL tube.
-
b.On a slide, make 5 μL drops of cryoprotectant solution.
-
a.
-
12.Harvesting crystals.
-
a.Harvest the best crystals using the appropriate cryo-loop and soak them in the cryoprotectant solution.
-
b.Freeze and store in liquid nitrogen until X-ray diffraction data collection.
-
a.
Note: Multiple crystal morphologies may appear in a single crystallization condition. Initial screening of cryo-protectant conditions showed that glycerol can be used to cryo-protect a fraction of the Cfp1PHD crystals. However, none of these crystals showed detectable density for the H3K4me3 peptide. Given that some crystals dissolved upon incubation in glycerol, we tested other cryo-protectants (refer to problem 5), including DMSO. In that condition, (0.2 M Ammonium sulfate, 0.01 M Tris pH 8.5, 15% PEG 3350 and DMSO) nearly all the crystals could be harvested and cryo-protected. After data collection, we observed that some crystals showed different unit cell axis lengths than others (Refer to Expected outcomes). After molecular replacement using dataset collected on these crystals, a detectable positive Fourier map could be detected for the H3K4me3 peptide.
Expected outcomes
The protein yield obtained after purification typically ranges around 2.5 mg/L of bacterial culture. The purity of the Cfp1PHD is expected to be > 80% after affinity chromatography and > 90% after size exclusion chromatography, as shown in Figure 1. Cfp1PHD elutes at approximately 80 mL in a Hi-Load 16/600 Superdex 75 column (Figure 2), corresponding to its molecular weight of 7.7 kDa.
Figure 1.
Coomassie-stained SDS-PAGE showing the protein contents of the samples of various purification steps
Proteins were resolved on a 15% SDS-PAGE and stained with Coomassie. TCL: total cell lysis, P: pellet, SN: supernatant, FT: flow through, E: elution pellet, ESN: elution supernatant and Input, F28-33: Fractions collected of TEV-cleaved Cfp1PHD purified by size exclusion chromatography using a Superdex 75 preparative column.
Figure 2.
Fast protein liquid chromatography (FPLC) chromatogram showing the elution profile of Cfp1PHD input sample from Figure 1
Cfp1PHD sample colored in red overlayed with molecular weight standards (grey).
The condition used to obtain protein crystals is 0.2 M Ammonium sulfate, 0.01 M Tris pH 8.5, 15% PEG 3350 (Figure 3A), using sitting-drop vapor diffusion at room temperature (22°C) with a protein: crystallization solution ratio of 1:1, at a protein concentration of 5 mg/mL in Buffer B.1 Crystals appeared within 24–48 h. Optimization trials using varying concentrations of Ammonium sulfate and Tris pH 8.5 in a 24-well plate yielded bigger and better crystals, as shown in Figure 3A. The typical space group for any crystal is P212121. The expected unit cell dimensions (Å) for Cfp1PHD-H3K4me3 crystals are: 33.27 (a), 43.12 (b), 47.56 (c) and 32.4(a), 36.6(b), 55.7(c) for unbound Cfp1PHD crystals.
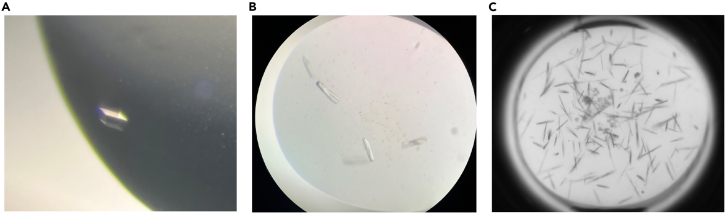
Figure 3.
Crystals of the Cfp1PHD-H3K4me3 complex
(A) Crystals of the Cfp1PHD-H3K4me3 complex grown in 0.2 M Ammonium sulfate, 0.01 M Tris pH 8.5, 15% PEG 3350 using sitting-drop vapor diffusion at room temperature (22°C) with a protein: crystallization solution ratio of 1:1, at a protein concentration of 5 mg/mL in Buffer B.
(B) Crystals grown using Index Hampton commercial screen after 5 days at room temperature (22°C).
(C) Poor crystals were obtained using the Index screen (Hampton) and a higher protein sample concentration. See potential solution 4 for troubleshooting.
Limitations
Limitations of the protocol are mentioned in the troubleshooting section below.
Troubleshooting
Problem 1
Protein yield is low after overexpression (related to step 3 in “overexpression of Cfp1PHD in Escherichia coli”.
Potential solution
First, never let the OD600 go over 0.5 when growing large-scale liquid cultures. Second, when adding the IPTG to induce protein expression, ensure that the incubator and liquid culture cool down to 18°C. If protein expression is still low, try inducing for a longer period of time (up to 24 h), using a different E. coli strain or employing a cDNA construct optimized for overexpression in bacteria.
Problem 2
The protein of interest is not soluble (related to steps 5-7 in “protein purification of Cfp1PHD by affinity and size-exclusion chromatography”).
Potential solution
First, ensure the protein sample remains on ice throughout the protocol. Second, use fresh buffers and test the pH. If the suggested buffer does not yield soluble protein, assess protein solubility in multiple buffers (Phosphate, Tris, HEPES, PBS-based buffers) supplemented with a high or low salt concentration, glycerol and detergents (e.g., Triton X-100, β-octylglucoside). Figure 1 illustrates high protein solubility in the supernatant fraction (SN) compared to the pellet (P).
Problem 3
Crystals of the protein of interest are not obtained (related to steps 8-10 in “crystallization of Cfp1PHD in complex with an H3K4me3 peptide”).
Potential solution
Several factors can impact the crystallization of the protein of interest, such as the solubility (refer to problem 2), the pH of the solution, choice of precipitant, and temperature. First, instead of using the recommended Index and Proplex screening kits, try other available sparse matrix available at Emerald Biostructures Inc.; Hampton Research; Molecular Dimensions; Qiagen-Nextal Biotechnology, Canada; Jena Bioscience, Germany; Axygen Biosciences. Crystal trays can be set up using various sparse matrix screens and an ARI Gryphon drop setter, allowing for accurate pipetting and fast screening. Second, try to set up your crystal plates at different temperatures (4°C or 16°C).
Problem 4
Crystals are not of diffraction quality (related to steps 8-10 in “crystallization of Cfp1PHD in complex with an H3K4me3 peptide”). See Figures 3B and 3C.
Potential solution
Figure 3B illustrates crystals obtained from the initial sparse matrix screening unsuitable for harvesting and collecting diffraction data. Therefore, once crystals are obtained, it’s crucial to optimize the crystallization solution (see Figure 4.) by slightly adjusting the concentration or pH of specific components based on the best initial conditions from the sparse matrix screens. This process will help you identify crystals of optimal diffraction quality, as shown in Figure 3A. If you obtain multiple tiny, thin crystals during the optimization phase, consider reducing the concentration of either the protein or the precipitant. Finally, additive screens (Hampton) or micro-seeding can be used to optimize crystals further.
Problem 5
Crystal structure does not contain peptides of interest (related to steps 11-12 in “cryoprotection of crystals for X-ray crystallography”).
Potential solution
During data refinement, no electronic density corresponding to the peptide may be observed (e.g., Cfp1PHD bound to H3K4me3). Observe how the crystals tolerate the cryoprotectant solution when harvesting and cryoprotecting crystals for X-ray crystallography. Different crystals (e.g., with varying morphologies) can respond differently to various cryoprotectants. Carefully monitor the crystals while soaking them in the cryoprotectant. If any deformation or cracks are observed, consider alternative cryoprotectants, such as polyethylene glycol and 2-propanol. A stepwise re-buffering approach may also be beneficial by gradually increasing the concentration of cryoprotectant in several steps to allow the crystals to adjust gradually. For Cfp1PHD bound to H3K4me3, we recommend using DMSO, as it helped cryoprotect all the crystals with varying morphologies and found that those who had a slightly longer axis contained the peptide.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jean-François Couture (jean-francois.couture@uottawa.ca).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Sabrina Grégoire (sgreg054@uottawa.ca).
Materials availability
The study did not generate other new or unique reagents.
Data and code availability
The study did not generate/analyze datasets.
Acknowledgments
S.G. acknowledges a scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Ontario Graduate Scholarship Program, as well as grants from NSERC and the Canadian Institutes of Health Research that support the J.-F.C. laboratory.
Author contributions
Conceptualization, J.G., M.J., S.G., and J.-F.C.; investigation, S.G., J.G., M.J., and S.C.; writing – original draft, S.G. and J.-F.C.; writing – review and editing, S.G., J.G., and J.-F.C.; funding acquisition, J.-F.C.; supervision, J.-F.C.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sabrina Grégoire, Email: sgreg054@uottawa.ca.
Jean-François Couture, Email: jean-francois.couture@uottawa.ca.
References
- 1.Grégoire S., Grégoire J., Yang Y., Capitani S., Joshi M., Sarvan S., Zaker A., Ning Z., Figeys D., Ulrich K., et al. Structural insights into an atypical histone binding mechanism by a PHD finger. Structure. 2024;32:1498–1506.e4. doi: 10.1016/j.str.2024.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Brown D.A., Cerbo V.D., Feldmann A., Ahn J., Ito S., Blackledge N.P., Nakayama M., McClellan M., Dimitrova E., Turberfield A.H., et al. The SET1 Complex Selects Actively Transcribed Target Genes via Multivalent Interaction with CpG Island Chromatin. Cell Rep. 2017;20:2313–2327. doi: 10.1016/j.celrep.2017.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study did not generate/analyze datasets.