Abstract
As promising noninvasive biomarkers, nucleic acids provide great potential to innovate cancer early detection methods and promote subsequent diagnosis to improve the survival rates of patient. Accurate, straightforward and sensitive detection of such nucleic acid-based cancer biomarkers in complex biological samples holds significant clinical importance. However, the low abundance creates huge challenges for their routine detection. As the next-generation diagnostic tool, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein (Cas) with their high programmability, sensitivity, fidelity, single-base resolution, and precise nucleic acid positioning capabilities are extremely attractive for trace nucleic acid-based cancer biomarkers (NABCBs), permitting rapid, ultra-sensitive and specific detection. More importantly, by combing with nanotechnology, it can solve the long-lasting problems of poor sensitivity, accuracy and simplicity, as well as to achieve integrated miniaturization and portable point-of-care testing (POCT) detection. However, existing literature lacks specific emphasis on this topic. Thus, we intend to propose a timely one for the readers. This review will bridge this gap by providing insights for CRISPR/Cas-based nano-biosensing development and highlighting the current state-of-art, challenges, and prospects. We expect that it can provide better understanding and valuable insights for trace NABCBs detection, thereby facilitating advancements in early cancer screening/detection/diagnostics and win practical applications in the foreseeable future.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02222-5.
Keywords: Cancer biomarkers, Nucleic acids, CRISPR/Cas, Nanotechnology, Ultra-sensitive detection
Introduction
Cancer is the leading cause of death worldwide. The World Health Organization (WHO) reports that cancer is responsible for nearly 10 million deaths annually, making up one-sixth of all deaths [1]. It is reported that most cancers are asymptomatic in the early stages, and 50% of them still only detected at an advanced stage [2], which seriously endanger human life and health. Thus, it is imperative to achieve their early detection [3]. As shown in Fig. 1, traditional protein biomarkers have been employed for early clinical cancer detection, typically through antigen-antibody interaction such as enzyme-linked immunosorbent assay (ELISA). Despite their advantages of non-invasion and portability, they are still not early enough, with low sensitivity and specificity. The emergence of nucleic acid-based cancer biomarkers (NABCBs) with advancements in molecular biology has introduced a promising new era in the realm of earlier detection. The methods based on NABCBs, at the gene level, enables a more precise and comprehensive guidelines for cancer earlier diagnosis/genotyping, individualized treatment, and postoperative monitoring to improve the survival rates of patients. Markedly, the low abundance and similar homology create huge challenge for PCR [4], next-generation sequencing [5], microarray [6] and other traditional methods in sensitivity, rapid, accurate of routine and portable point-of-care testing (POCT) detection. Thus, it is urgent to develop novel technologies to achieve more efficient detection of trace NABCBs in complex biological body fluids.
Fig. 1.
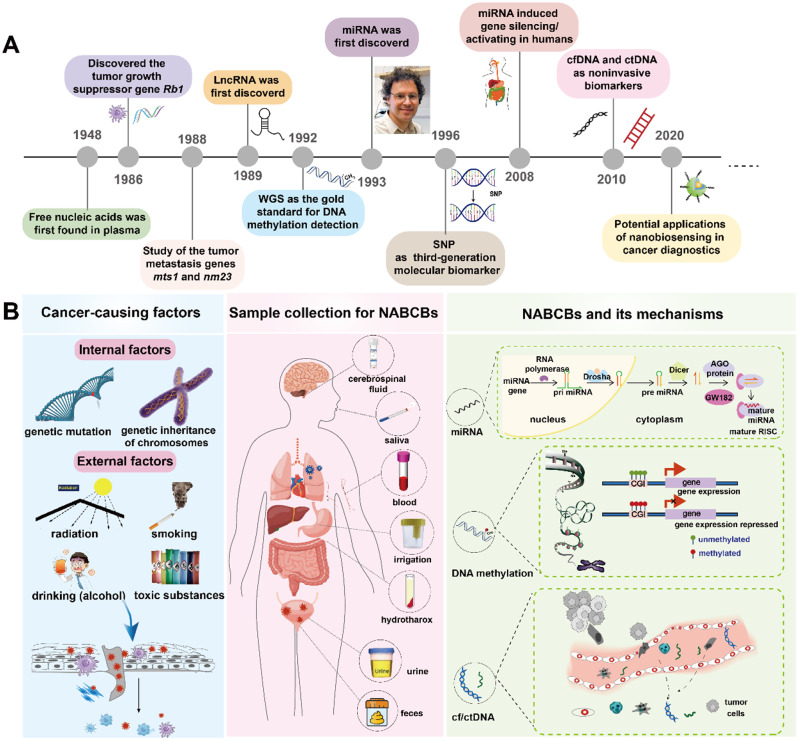
The characteristics of related biomarkers in different stages of cancer development and the application history of CRISPR/ nanotechnology-based biosensing in early detection of NABCBs. As the emerging gene biomarkers, NABCBs enable a more precise and comprehensive guidelines for cancer earlier diagnosis/genotyping compared with traditional protein biomarkers. The combined CRISPR-based nano-biosensing offers a promising strategy to the current dilemma of trace NABCBs detection
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) was a natural immune defense system discovered in E. coli in 1987 and was first named in 2002. Since 2012, the CRISPR/Cas9 system and gene editing technology have been revealed. As a promising nucleic acid detection tool, it has the significant advantages of nucleic acid targeting ability, programmability, sequence specificity, high fidelity/sensitivity, and high single-base resolution [7]. It means that CRISPR-based detection systems can be programmed to target specific genetic sequences, making it possible to customize detection for different cancer types or patient-specific mutations. Furthermore, CRISPR technology has emerged as a groundbreaking tool for nucleic acid analysis in molecular diagnostics, owing to the exceptional attributes like high sensitivity, specificity, rapidity, fast deployability and cost-effectiveness. In fact, it had been successfully applied to the detection of NABCBs several years ago. During 2015–2018, the advancements in the Cas9-based tumor models and the discovery of trans-cleavage of Cas12/13 have facilitated the direct detection of cancer-associated RNA and DNA biomarkers. By approximately 2020, the utilization of CRISPR technology was expected to expand to the detection of blood circulating cancer DNA (ctDNA), exosome RNA and other cancer biomarkers, demonstrating significant promise in the field of cancer liquid biopsy. At present, CRISPR-based technology has also been organically combined with fluorescence, colorimetry, electrochemistry, microfluidic and others methods to achieve early diagnosis, postoperative evaluation, genetic profiling/genotyping and monitoring of trace NABCBs that are challenging to detect.
Nanotechnology has undergone significant advancements since its inception in 1974. The introduction of the scanning tunneling microscope (STM) in 1981 marked a significant advancement in the analysis of microstructures, subsequently catalyzing the swift progression of nanotechnology. Then, the successfully preparation of pure carbon nanotubes, graphene opened a new stage of nano-material research. All these discoveries marked the promising beginning of an extensive experimental investigation in the realm of cancer diagnosis. Currently, nanomaterials have been widely explored for nanotechnology-based in vitro diagnostics, and several key aspects, such as signal generation, amplification, conversion, and collection are involved in the establishment of detection, which could be a versatile and effective paradigm for biochemical analyses. It has also shown great advantages in trace NABCBs detection, which promote technological innovation and improve the sensitivity, specificity and speed of detection. Specifically, due to its unique quantum size effect, nanomaterials are convenient for surface modification and signal amplification. In addition, nanomaterials can also be functionalized and modified by aptamers or oligonucleotides, which can achieve direct capture of specific DNA or RNA sequences [8]. More importantly, the small size of nanomaterials enables them to be integrated into microfluidic chips and handheld detection devices, which promotes the development of POCT detection and greatly shortens the detection time. These nano-biosensing can leverage CRISPR/Cas system for specific nucleic acid sequences identification with the concentrated NABCBs, enabling more precise targeting and ultra-sensitive detection (Fig. 1).
It can be witnessed that the CRISPR/Cas-based nano-biosensing holds significant potential for simple, accurate, and noninvasive detection of trace NABCBs, offering wide-ranging applications in cancer earlier detection and monitoring to complete cancer localization and individualized treatment. This approach offers advantages such as low risk, high survival rates, and noninvasiveness. In this paper, we are committed to presenting an overview of the icebreaking in trace NABCBs biosensing based on nanotechnology-leveraged CRISPR/Cas systems: from vital nucleic acid biomarkers, CRISPR/Cas toolbox, abundant nanomaterials, combined biosensing, advances to challenges and beyond. Specifically, the classification and pathogenesis of vital NABCBs are comprehensively reviewed, followed by the evolution of cancer detection methodologies. Next, the distinct operational principles of CRISPR/Cas toolbox and specialized function of nanotechnology were discussed by category, delineating their respective characteristics. Then, the latest practical implementation biosensing and their recent advances for NABCBs detection were emphatically summarized, with a particular emphasis on amplification-free techniques, isothermal amplification and nano-assisted methods. Finally, the existing challenges, potential countermeasures, and future prospects are presented. We expect that this review can provide valuable insights and further enhance the CRISPR-based nano-biosensing, facilitate its more effective utilization in early clinical diagnosis, and thus contributing to mitigate cancer mortality and safeguard human beings.
Cancer-related nucleic acids biomarkers
Traditional cancer biomarkers mainly exist in blood and tissues [9], such as proteins (cell surface antigens, enzymes and cytoplasmic proteins) or cells [10], and rely heavily on antibody-based immunoassays for detection. However, these methods are labor-intensive, low sensitivity/specificity, and may yield inconsistent outcomes. The urgent demand for accurate cancer earlier detection has prompted the discovery of NABCBs in blood, urine, saliva and other easily extractable body fluids. Compared with the traditional diagnosis technology, it provides an efficient, specific and noninvasive method, which can effectively reduce the potential health risks while improving the rate of earlier detection. Currently, the main types of NABCBs mainly include RNA, DNA and single nucleotide mutations (Fig. 2A). The following mainly focuses on the development process, different types, pathogenic mechanisms, specific characteristics and commercial products/clinical applications of NABCBs.
Fig. 2.
(A) A concise overview of the historical development and discovery of NABCBs. (B) Cancer-causing factors, samples collection and pathogenic mechanisms of NABCBs. Carcinogenic factors can be divided into external and internal factors, both of which contribute to the development of various types of cancers. Further, dysregulation of NABCBs can be observed during the initiation and progression of cancer, thereby facilitating the oncogenic process
The discovery of cancer-related nucleic acids biomarkers
Cancer is mainly caused by cell damage or gene mutations, and is influenced by environmental (radiation, smoking, drinking, toxic substances) or genetic (heredity, variation) factors. Among various cancers (such as lung cancer, breast cancer, gastric cancer, liver cancer, etc.), the internal changes are the dominant factors. They can lead to the occurrence and progress of cancer by the imbalance expression of suppressor/metastatic genes or pathogenic mechanisms of NABCBs. Furthermore, there is an observed up or down-regulation of NABCBs during the initial stages of tumor formation. As for the potential noninvasive biomarkers, NABCBs can be easily obtained from little saliva, blood, urine and other body fluids (Fig. 2B). It is well known that the NABCBs has experienced a long development process. The free nucleic acid in plasma was first discovered in 1948, and then the cancer-related genes (Rb1, mts1 and nm23) were identified in 1980. After that, microRNA (miRNA), single nucleotide polymorphism (SNP) and ctDNA/circulating free DNA (cfDNA) were gradually discovered and applied in the following 1930, 1996 and 2010 years. As shown in the following Table 1, we systematically summarized the characteristics of different NABCBs.
Table 1.
Characteristics and classification of common NABCBs
| NABCBs | Biomarkers | Expression | Samples | Cancer types | Ref. |
|---|---|---|---|---|---|
| RNA | miRNA-21 | Upregulated | - | BC | [11] |
| miR-1307 | Upregulated | - | OC | [12] | |
| miRNA-155 | Downregulated | - | PC | [13] | |
| lncRNA HOTAIR | Upregulated | - | GC/DC | [14] | |
| lncRNA LOC284454 | Upregulated | - | HNCs | [15] | |
| lncRNA BACE1AS | Upregulated | - | LiC | [16] | |
| DNA | ctDNA | - | Plasma; Tissue | PLC | [17] |
| ctDNA | - | Blood | LC | [18] | |
| cfDNA | - | Serum | CTCL | [19] | |
| cfDNA | - | Plasma; Cancer cell | GC | [20] | |
| DNA methylation | ZNF331 | - | Tissue; Cancer Cell | CRC | [21] |
|
SEPT9/ SDC2 |
- | Stool specimens | CRC | [22] | |
| SCARA5 | - | Tissue; Cell | NSCLC | [23] | |
| SNP/SNV | Myeloperoxidase MPO | - | - | LC | [24] |
| N-acetyltransferase1, NAT1 | - | - | BC | [25] | |
| CYP3A4 | - | - | PC | [26] |
Cancer-related RNA biomarkers
RNA plays a crucial role in gene expression, whether by regulating protein-encoded RNA or non-encoded RNA during transcription. Among them, miRNA and long-chain noncoding RNA (lncRNA) are extensively researched in the context of cancer and cancers.
miRNAs are non-coding single-stranded RNA molecules, typically 15–25 nucleotides in length, encoded by endogenous genes and are relatively conserved throughout evolution [27]. These molecules mediate post-transcriptional gene regulation by RNA-induced silencing complex (RISC) degrades mRNA or hinders its translation [28], playing a negative regulatory role by binding to the mRNA 3’-untranslated region. In 1993, the group of Ambros made the initial discovery of miRNAs during their investigation into the developmental processes of Caenorhabditis elegans [29]. Since this milestone over two decades ago, miRNA has been identified as a key regulator of complex biological processes associated with normal physiology and the pathogenesis of diseases, including cancers, muscle disorders and neurodegeneration diseases [30, 31]. Alterations in miRNA expression levels within serum and tissues can be used for the diagnosis, prognosis and curative effect evaluation of cancer. For instance, miRNA-21 is implicated in the initiation, progression, invasion, metastasis, and unfavorable prognoses of lung, breast, pancreatic, and colorectal cancers [32]. Studies have indicated that miRNA-155 is involved in the regulation of cancer progression in hematological cancers and solid cancers, such as breast cancer, gastric cancer and lung cancer [33]. The downregulation of let-7 family miRNAs in numerous cancer types is associated with cancer inhibition function, including lung cancer, pancreatic cancer, and colon cancer [34]. In addition, miR-17-92 cluster, miR-200 family, miR-34 family, miR-145 and various other miRNAs are also involved in diverse cancer pathophysiological processes.
lncRNA refers to non-coding RNA exceeding 200 nucleotides in length, which is involved in regulating gene expression, chromatin modification, transcription and splicing [35]. As the important cancer biomarker, lncRNA can play a role as oncogene or cancer suppressor in cervical cancer (CC). For example, the high expression of lncRNA HOXC13-AS is related to the poor prognosis of CC patients. Moreover, lncRNA has the capability to engage with miRNA in a manner akin to a “sponge”, thereby modulating the expression of miRNA target genes through binding and suppressing the activity of miRNA. This “competitive endogenous RNA” (Cernan) mechanism is particularly important in cervical cancer, which can regulate multiple cancer-related signaling pathways [36]. An example of this is the up-regulation of CTBP1-AS2 expression, where this lncRNA can be used as a decoy for miR-3163, leading to the subsequent occurrence and development of CC [37].
Cancer-related DNA biomarkers
Currently, within the realm of clinical medical testing, cancer-related DNA biomarkers predominantly encompass cfDNA and ctDNA. These biomarkers exist in cells, bodily fluids, or tissues, and alterations in their levels reflect the genetic characteristics and biological dynamics of cancers.
cfDNA was first described in plasma by Paul Mandel in 1958. It refers to endogenous double-stranded DNA (dsDNA) that is partially degraded in circulating blood, existing freely and extracellularly [38]. cfDNA covers a broader range, encompassing cancer-derived tuna, which means that the cellular DNA of cancer cells falls off or is released into the circulatory system after apoptosis. ctDNA is a noninvasive robust biomarker associated with cancer, characterized by a dsDNA containing specific mutations in the cancer-specific sequence. These mutations can be detected in blood cells [39] and various other bodily fluids such as serum, plasma, urine, cerebrospinal, saliva, semen, and exocrine secretions [40]. Notably, the remarkable features of cancer patients include the rapid proliferation and increased apoptosis of cancer cells, leading to a large amount of influx ctDNA into the bloodstream. This influx reflects the genomic characteristics of cancers, including point mutation, gene rearrangement and copy number variation. The existence and quantitative changes of ctDNA are closely related to cancer burden, invasiveness and therapeutic effect, rendering them frequently used as important biomarkers for early diagnosis, surveillance and prognosis evaluation of cancers.
Cancer-related single nucleotide mutation biomarkers
DNA methylation or single base mutation is crucial for identifying of NABCBs. These alterations can reflect the biological characteristics of cancers, aiding in the monitoring of cancer progression, diagnosis of cancer status, prediction of disease advancement, and assessment of therapeutic efficacy.
DNA methylation
DNA methylation is a form of chemical modification of DNA that has a huge impact on mammalian epigenetics, typically occurring at the C-5 position of CpG dinucleotide in vertebrate genomes [41, 42]. This process involves the addition of a methyl group to produce 5-methylcytosine (5mc), which can also be found in N-6 position of adenine and G-7 position of guanine [43]. At present, 5-hydroxymethylcytosine (5hmc), 5-formylcytosine (5fc) and 5-carboxycytosine (5cac) have also been identified. Numerous studies have demonstrated that abnormal DNA methylation at certain sites can silence cancer suppressor genes, thereby increasing the risk of cancer mutations. Specifically, in cancerigenesis, abnormal DNA methylation patterns, like hypermethylation in cancer suppressor gene promoters, can cause gene silencing, while demethylation in certain regions may activate proto-oncogenes. These methylation changes directly impact gene expression and contribute to cancer development. Therefore, DNA methylation is a crucial biomarker for early cancer diagnosis, and measuring its levels is vital for prognosis and diagnosis [44].
Point mutation
Single nucleotide mutation (SNV) and SNP both involve changes of single nucleotide in DNA sequence, such as substitution, insertion, or deletion [45]. The difference is that SNP usually refers to the single nucleotide variation with relatively high frequency (generally considered to be more than 1%) in the population, while the single nucleotide variation below 1% and association with individual rare diseases are usually called SNV. Both SNV and SNP are important biomarkers in genetic pathology, pharmacology and genetic diagnosis [46]. In addition, the presence of single-base mutations plays a crucial role in the onset and progression of significant human ailments, such as malignant cancers and autoimmune diseases. It can participate in disease progress by changing gene expression, affecting signal transduction pathway and interfering with DNA repair mechanism. This is coupled with the fact that conventional test is often difficult to achieve accurate detection due to the complexity and high cost of the process. Consequently, how to identify SNP/SNV efficiently and accurately through cost-effective and straightforward procedure has become a prominent research area and a notable challenge in recent years [47].
Commercial products and clinical application of NABCBs
In recent years, cancer biomarkers are increasingly employed in the diagnosis, treatment, and monitoring of cancer, leading to the rapid expansion of the market. By 2023, the early cancer screening sector has officially transitioned into the commercialization phase, but the market has not been well integrated, and the overall business model is still in the exploratory stage. Companies in the cancer gene detection industry, such as Ji Yinga, Maijing Gene, Youzhiyou, New Horizon Health, GNRGENE etc., are all committed to developing commercial products for cancer early detection. Currently, there are various NABCBs-related commercial products, which are based on different mainstream technical platforms (q-PCR and NGS) and detection indexes (miRNA, ctDNA, cfDNA and DNA methylation), providing strong support for early screening/detection of cancers (Table 2).
Table 2.
Commercialized products of NABCBs on the market
| Biomarkers | Company | Products | Year | Cancers | Methods | Sensitivity |
|---|---|---|---|---|---|---|
| miRNA | MicroMedMark | Yian™ | 2016 | PC | q-PCR | 76% |
| Mirxes | GASTRO Clear™ | 2019 | GC | q-PCR | 87.5% | |
| DunWILL | MiRNA7 TM | 2024 | CRC | q-PCR | 76.48% | |
| ctDNA | New Horizon Health | Glycoconjugate Clear™ | 2021 | LC | NGS | 97.9% |
| cfDNA | Huada Gene | Vaganin® | 2023 | LC | q-PCR | 76% |
| GENETRON | HCCscreen™ | 2021 | LC | CT Imaging | 92% | |
| Zhushi Biomedicine | Gan Xiaoning™ | 2023 | LC | NEEM-seq | 94.4% | |
| Herui Gene Technology | Lai Sining® | 2020 | LC | NGS | 95.7% | |
| DNA methylation | Nanjing Tengchen | Fei Changqing® | 2020 | GC | NGS | 72.32% |
| Guangzhou Jizhun Medicine | Fei Changan® | 2023 | NSCLC | CT Imaging -AI | 98.1% | |
| GNRGENE | Gananjian | 2020 | LC | q-PCR | 92.6% | |
| Beijing Eickelen | Aiweining | 2024 | GC | q-PCR | 92.07% | |
| SNP | Exact Science | Coloquard | 2019 | CRC | Immunochemistry | 94% |
Among them, DNA methylation detection kits are mature and have been widely used in the clinical diagnosis. For example, the Coloquard product approved by FDA, which is a colorectal cancer early screening product based on the combined testing of KRAS single base mutation and NDRG4/BMP3 methylation. In addition, Fei Changqing® product can achieve accurate screening of early non-small cell lung cancer (NSCLC) based on methylation detection with only 2 mL blood samples. As for cfDNA or ctDNA products, most of the detection kits (such as Glycoconjugate Clear™, HCCscreen™, Vaganin®, etc.) focus on liver cancer early detection. In fact, Mirxes developed GASTRO Clear™ product for early gastric cancer diagnosis based on miRNA in 2019, and obtained clinical recognition. Furthermore, there are several generic miRNA detection kits have been successfully introduced to the market. It can be witnessed that there is a noticeable shift in commercial products from imitation to technological innovation, with certain products being utilized in clinical and at-home testing. Overall, cfDNA, ctDNA and methylation have been recognized in clinical settings, whereas miRNA has not yet gained widespread adoption.
CRISPR/Cas toolbox
CRISPR/Cas system
CRISPR is a natural immune defense system present in bacteria and archaea, which was first discovered in E. coli by Japanese scientist Ishino et al. in 1987 [48]. Then it was officially named CRISPR by Mojica and Jansen officially in 2002 [49]. It forms a set of “molecular memory” by recording the genetic information fragments of viruses or plasmids that previously infected them, which can accurately identify and destroy these foreign genetic materials when encountering the same invaders again [50]. In short, the CRISPR/Cas system comprises two primary components: the genes responsible for encoding Cas proteins and the CRISPR array, which consists of repeat and spacer sequences. The mechanism of CRISPR/Cas action can be divided into three main parts: foreign DNA capture, crRNA synthesis, and targeted interference (Fig. 3A) [51].
Fig. 3.
(A) The immune defense system mechanism of CRISPR/Cas system. (B) The fundamental CRISPR-based molecular diagnosis strategies, the structure of Cas9/12a/13a/13b effectors and the schematic diagram of NASBACC/DETECTR/SHERLOCK/SHERLOCKv2 are summarized and compared
There are two mainly types of CRISPR/Cas systems. Class 1 (types I, III, IV) utilizes multiple Cas effecting proteins, and Class 2 (types II, V, VI) relys on a single Cas effecting protein. While, the most well-known and widely used ones in biotechnology and research are the toolkits Cas9, Cas12, Cas13, and Cas14, which all belong to the Class 2. Although they differ slightly in structural composition and working mechanism, they all play an important role in gene editing and molecular diagnosis (Table 3).
Table 3.
Classification of different effectors within the CRISPR/Cas system
| Cas protein | Cas9 | Cas12a | Cas13a | Cas14a |
|---|---|---|---|---|
| Type | II | V | VI | V |
| Guide RNA | sgRNA | crRNA | crRNA | crRNA |
| Target | dsDNA | ds/ssDNA | ssDNA | ssDNA |
| cis-cleavage | dsDNA | ds/ssDNA | ssRNA | ssDNA |
| Trans-cleavage | No | ssDNA | ssRNA | ssDNA |
| PAM/PFS | NGG | (T) TTN | Non-G | No |
| Components | RuvC, HNH | RuvC | Two HEPN | RuvC |
CRISPR/Cas molecular diagnosis technology
CRISPR technology offers significant advantages in molecular diagnosis. Cas9, a V-type CRISPR effector, was first discovered by Bolotin’s team in Streptococcus thermophilus in 2007 [52]. This RNA-directed endonuclease comprises two domains, HNH and RuvC. The target strand of protospacer adjacent motif (PAM) sequence containing 5-NGG-3 is guided by sgRNA (tracRNA and crRNA) to bind to Cas9 for cis-cleavage, which commonly used in gene editing and molecular diagnosis. The nucleic acid sequence-based amplification CRISPR/Cas 9 (NASBACC) technology has been developed to molecular diagnosis [53–56].
Cas12a, belongs to class 2 type V endonuclease in CRISPR/Cas system, typically ranges from 18 to 25 nucleotides in length. Cas13 in the type VI system, can degrade target RNA with the guidance of crRNA. The typical PAM sequence for Cas12a is TTTV, and in addition four of the preferred C’s sequences are TTTA, TCTA, TCCA and CCCA. But the PAM of Cas13a is preferred to non-G, and its length is from 22 to 30 nt. Different from Cas9, the trans-cleavage activity of Cas12a or Cas13 can be activated to achieve non-specific cleavage of surrounding single-stranded DNA (ssDNA). Combined with isothermal nucleic acid amplification methods, Cas12/13 have been developed for molecular diagnosis based on DETECTR, SHERLOCK and SHERLOCKv2 nucleic acid detection techniques (Fig. 3B) [57–60].
Cas14, a protein weighing only 40–70 KDa and consisting of 22–40 nucleotides [61], can even distinguish SNP with the elimination of PAM restriction [62–64]. However, there are few reports on the application of Cas14 in NABCBs detection at present, but with milder reaction conditions and higher specificity, it may have the potential of wider application in the future [65, 66].
Nanomaterial-integrated biosensing
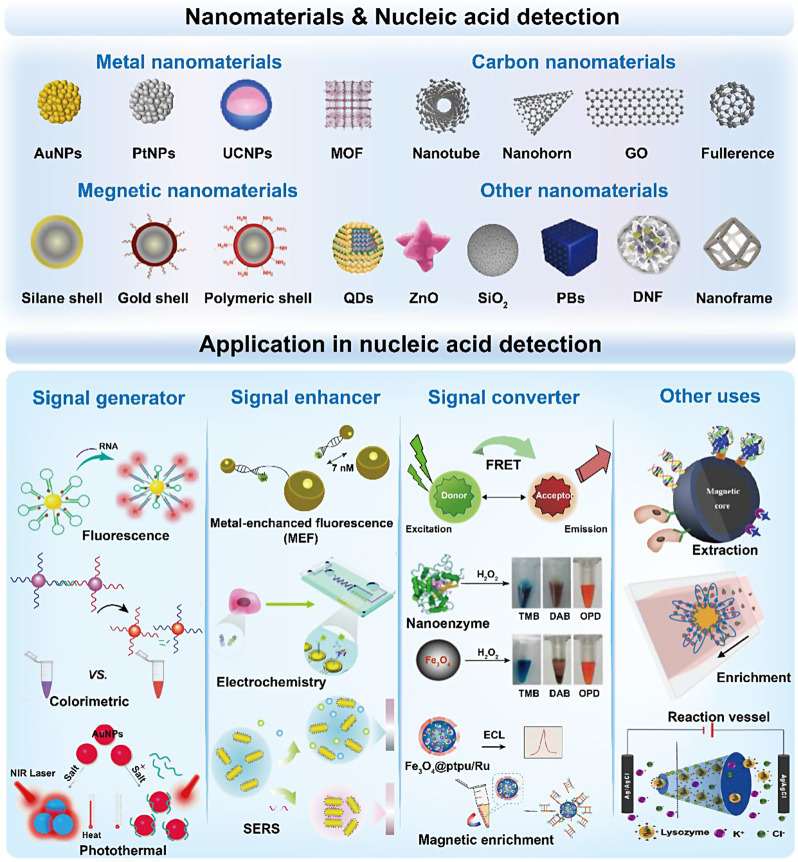
The rising prominence of nanotechnology has led to the integration of nanomaterials in biosensing analysis. In particular, these nanomaterials play a crucial role in simplifying sample extraction/separation, improving signal generation/transduction, accelerating molecular recognition/reaction, thereby enhancing overall detection performance. Specifically, these nanomaterials exhibit unique functions and advantages in trace NABCBs detection by acting as signal generators/visualizers, enhancers, converters and extraction/reaction vessels (Fig. 4) (Table 4).
Fig. 4.
The representative nanomaterials and their board application in nucleic acid biomarkers detection. Various metal, carbon, magnetic and other excellent nanomaterials have been widely used for nucleic acid detection due to their ability to serve as signal generators/enhancers/converters and sample extraction/reaction vessels
Table 4.
A summary of NABCBs detection based on nanomaterial-integrated biosensing
| Nanomaterials | Property | Function | Targets | LOD | Ref. |
|---|---|---|---|---|---|
| ANCs/MWCNT-NH2 | Nanotubes | Reaction vessel | lncRNA | 42.8 fM | [67] |
| AuNPs | Nanorods |
Optical probe |
cfDNA | 10 fM | [68] |
| QDs-DNA | Hydrogel | Signal generator | miR-17 | 182 fM | [69] |
| SiO2@DQD | Fluorescent microspheres | Fluorescence quencher | miR-133a | 0.3 fM | [70] |
| MoS2 nanosheets | Nanosheets | Reaction vessel | miR-499 | 381.78 fM | [71] |
| TC nanosheets | Nanosheets | Reaction vessel | miR-21 | 0.21 fM | [72] |
| S-BN/CN nanosheets | Nanosheets | Reaction vessel | miR-21 | 3.98 fM | [73] |
| [Ru(phen)2dppz]2+ | Metal nanomaterials | Optical signal switch | miR-17 | 1 fM | [74] |
| Fe3O4@PtPd/Ru(bpy)32+ | Magnetic-nano-luminescent material | Signal converter | miR-145 | 0.41 fM | [75] |
| Pt@Au |
Bimetallic nano enzyme |
Catalyst | miR-233/ lncRNA | - | [76] |
| rGO/GCE | Graphene oxide | Conduct electricity | miR-21 | 0.83 fM | [77] |
| MB/Fe3O4@COF/PD-AU | Nanocomposite | Sample extraction | ctDNA | 3.33 aM | [78] |
| Graphene microneedle patch | Graphene crystal | Sample extraction | cfDNA | - | [79] |
| gFET array | Graphene crystal | Reaction vessel | SNP | 1 aM | [80] |
Nanomaterials as a signal generator
As effective signal generator, nanomaterials can provide fluorescence, colorimetric, magnetic, photothermal and other signals in the process of nucleic acid detection. Fluorescent nanoparticles like quantum dots (QDs) [81], up-conversion nanoparticles (UCNPs) [82], long persistent nanomaterials and metal nanoclusters exhibit significant advantages over organic dyes and fluorescent proteins in terms of signal brightness [83, 84], photostability, and low background noise. As a result, they have been investigated for fluorescence sources in nucleic acid detection with enhanced precision and sensitivity. Moreover, gold nanoparticles (AuNPs), magnetic nanoparticles (Fe3O4), and photonic crystals can produce colorimetric signals through volume changes or aggregation in response to external stimuli, commonly utilized as visual detection signals in nucleic acid POCT detection. Based on the ability of magnetic relaxation switches, Fe3O4 can also directly serve as a signal visualizer for nucleic acid detection. In addition, AuNPs, gold nanorods (AuNRs), platinum nanoparticles (PtNPs), prussian blue nanoparticles (PBs) and carbon nanotubes can generate thermal signals [85] under near-infrared laser irradiation, enabling their application in vitro diagnostic scenarios when combined with thermal imaging technology [86, 87].
Nanomaterials as a signal enhancer
Nanomaterials have the capability to significantly augment and greatly amplify the faint signal of trace detection biomarkers. Due to their substantial specific surface area, nanomaterials facilitate the simultaneous labeling of active substances or signal molecules, such as enzymes and fluorescent materials, thereby enhancing the signal produced by a single biomarker. Among them, the utilization of AuNPs [88, 90], silica nanoparticles (SiO2NPs) [91, 92], polystyrene microspheres (PS), Fe3O4 [93, 94] and other nanoprobes, as composite carriers of enzymes and nucleic acids represents an effective method for weak signal enhancement detection. In addition, the sensitivity can be further enhanced by leveraging surface Raman enhancement, electrochemical enhancement, localized surface plasmon resonance (LSPR), and refractive index signals of photonic crystals. LSRP of inorganic metal nanomaterials can enhance the Raman signal for nucleic acid analysis [95]. AuNPs or carbon nanotubes can modulate the impedance of electrode materials to improve the sensitivity of electrochemical nucleic acid detection [96]. Photonic crystals possess excellent properties such as a stable reflection peak and a large specific surface area ratio (SVR) [97]. Suspension array technology based on photonic crystals is widely used in the detection of NABCBs [98].
Nanomaterials as a signal converter
Nanomaterials can also achieve signal conversion with unique properties, such as nanozyme catalysis (color change, chemiluminescence, kinetic energy) and fluorescence resonance energy transfer (FRET) [99]. Nanozymes refer to the enzyme mimics with catalytic activity of peroxidase or oxidase. Some metallic nanomaterials (PtNPs, Fe3O4, PBs) and metal-organic frameworks (Fe-MOF, Cu-MOF) can catalyze TMB and ABTS substrates to produce color changes [100]. In some cases, MOFs can also be used as signal converters to convert adsorbed molecular signals into detection signals. AuNPs, ZnO and other nanozyme can also catalyze luminol chemiluminescence to achieve the conversion of chemiluminescence signals. In addition, PtNPs can also be used as a nanomotor to achieve kinetic energy and distance conversion for motion-based detection platforms. Another more classical signal converter-FRET system, which uses nanomaterials with fluorescence properties (such as UCNPs/QDs/Go, etc.) as energy donors, transfer their energy to receptors or quenchers, and achieve the detection of various biomarkers [101, 102]. Furthermore, metal structures such as nanorods and nanowires with surface plasmon resonance effect can convert the absorbed optical signals into fluorescent signals with specific frequencies to enhance signal conversion.
Nanomaterials as sample extraction or reaction vessels
Nanomaterials have the potential to enhance sample extraction and separation processes by offering rapid and efficient methods. For instance, Fe3O4 can be used as a reaction container through modifying their surfaces, enabling rapid separation and enrichment of nucleic acid ability, which can be applied to nucleic acid extraction and enrichment process. Magnetic solid phase extraction (MSPE) represents a well-established technology for separation and extraction based on electrostatic adsorption, affinity, hydrogen bonding or ion exchange [103, 104]. In addition, specially functionalized nanoparticles (SFNPs) can selectively and rapidly capture targets (within 30 s) in automatic fine tuning (AFT) microfluidic systems through an acoustic vortex strategy. This SFNPS-assisted AFT approach has demonstrated the ability to increase target concentrations by nearly 200 times and has been successfully used for trace cancer biomarker detection. Some hollow (silica, metal, metal oxides, and MOFs) and metallic nanomaterials (nanorod, nanosphere, nanofilm, and nano bipyramid) can also be used as ideal reaction vessels. Due to the high catalytic performance and unique photothermal effect, it can significantly improve enzyme activity and catalytic efficiency, which is conducive to realizing ultra-high sensitivity detection of nucleic acid at the single-molecule level. For example, silica nanowires, with their high specific surface area and customizable surface chemistry, can act as reaction containers for immobilizing nucleic acid molecules and creating suitable reaction environments. AuNPs have excellent biocompatibility and surface plasmon resonance effect, which can be used to fix nucleic acid probes as reaction containers for nucleic acid detection. Furthermore, due to the ongoing advancements in nucleic acid nanotechnology, DNA nanoparticles, such as nanoflower (DNF), 3D nanoframes and others have emerged as crucial reaction carriers in cancer in vitro detection. In addition, nanopore reactors, which are user-friendly, have emerged as versatile tools for nucleic acid molecule detection, with third-generation nanopore sequencing technology showing potential to become a cost-effective sequencing method in the future [105]. Moreover, the unique photothermal effect of plasma nanomaterials lead to the efficient conversion of light into heat, making them advantageous vessels for nanoscale heating [106].
CRISPR-based biosensing for NABCBs detection
NABCBs play a crucial role in cancer by affecting the expression of proto-oncogenes and oncogenes, and thus contributing to epigenetic inheritance [107, 108]. However, due to the instability, dynamic expression changes and low abundance of most nucleic acids, it is difficult to achieve quickly, sensitively and specifically detection [109]. Based on the highly specific and sensitive of CRISPR-based diagnosis (CRISPR-Dx), the efficient enrichment, signal amplification and cancer identification of NABCBs can be realized, which can effectively enhance the early noninvasive tissue biopsy and precise therapy [110]. This process mainly involves extracting and sampling body fluids like blood, urine, cerebrospinal fluid, and saliva from patients. By incorporating CRISPR and nanotechnology, a dual signal amplification can be achieved, enabling sensitivity and precise detection of trace NABCBs in cancer early stage. Subsequently, the results of NABCBs can be used for identify cancer types and localize malignant tissues, guiding the medical committee to accomplish better clinical protocol and therapy (Fig. 5). The latest advancements have been made in the application of CRISPR-based biosensing in NABCBs detection, as shown in the following and summarized in Table 5.
Fig. 5.
The overall process of NABCBs liquid biopsy and the important significance for cancer early diagnosis and treatment. Noninvasive NABCBs biomarkers can be easily extracted from urine, saliva, blood, and other bodily fluids in the very early stage of cancers. After that, they can achieve rapid and accurate detection through dual signal amplification of CRISPR-based nano-biosensing. These detection results can assist in subsequent cancer diagnosis, location, guidelines for therapy and other clinical protocols
Table 5.
A summary of research conducted in the realm of different NABCBs based on CRISPR/Cas biosensing
| NABCBs | Cancer | Amplification | Readout | Sample | Time | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| let-7a | BC | CHA | Fluorescence | Cell | 120 min | 81.96 fM | [132] |
| miR-21 | BC | - | Fluorescence | Serum | 55 min | 10 fM | [68] |
| miR-17 | BC | TMSD | Fluorescence | Serum | 1 h | 182 aM | [69] |
| miR-31 | OSCC | ISAR | Colorimetry/Fluorescence | Salivary | 90 min | 384 aM | [133] |
| miR-17/miR-155/ miR-210 | NSCLC | - | Electrochemistry | Serum | 36 min | 50 aM | [125] |
| miR-27a/ let-7a/miR-141 | BC | EXPAR | Fluorescence | Cell lysates/ Serum | 60 min | 90 aM | [121] |
| miR-21/miR-17/miR-155 | CC | HCR | Fluorescence | Cell | 60 min | 1fM | [157] |
| miR-17 | BC/LC | - | Electrochemistry |
Cell extracts/ Lysates |
70 min | 1 fM | [74] |
| miR-21 | BC | RCA | Fluorescence | Exosomes /Serum | 3.5 h | 34.7 fM | [122] |
| miR-221 | BC/CC | LCR | Fluorescence | Cell | 183 min | 0.4 aM | [124] |
| miR-141 | - | - | Electrochemistry | - | 3 h | 0.331 fM | [137] |
| miR-10b | NSCLC/LC | RCA | Fluorescence/Colorimetry | Serum/Cell | 120 min | 1 fM | [126] |
| cfDNA-CTNNB1-S37F | NPC/ Sepsis | - | Electrochemistry | Interstitial fluid | 75 min | 1.2 fM | [79] |
| cfDNA-BRCA-1 | BC | - | Fluorescence/Colorimetry | Serum | 30 min | 0.34 fM | [145] |
| cfDNA-BRCA-1 | - | - | Glass nanopore | - | 1 h | 1 pM | [146] |
| ctDNA-EGFR L858R | NSCLC | HCR | Electrochemistry | Blood/ Serum | - | 3.3 aM | [78] |
| ctDNA | - | ESDR | Electrochemistry | Serum | - | 0.13 pM | [25] |
| ctDNA-PIK3CA | BC | - | - | Blood | 40 s | 0.65 nM | [157] |
| ctDNA-BRAF V600E | THCA | - | Fluorescence | Serum | 70 min | 0.37 fM | [137] |
| ctDNA-EGFR T790M | NSCLC | HCR | FRET | - | - | 316 fM | [135] |
| SNP/SNV | - | - | Fluorescence | Blood | 20 min | - | [116] |
| DNA methylation | - | - | Electrochemistry | - | - | 2 × 10− 4 U/mL | [138] |
| DNA methylation | - | RPA | - | - | - | 20 fM | [139] |
| SNP/SNV | - | - | Electrochemistry | - | 25 min | 15.7 aM | [141] |
| SNP/SNV | - | - | Colorimetry | Serum | - | 0.01% | [158] |
| SNP/SNV | - | - | Electrochemistry | - | - | 1 aM | [80] |
Amplification-free CRISPR for NABCBs detection
NASBACC, DETECTR and SHERLOCK are typical molecular diagnostic technologies based on CRISPR/Cas system, which can realize highly sensitive detection of specific nucleic acids based on their specific targeting and efficient cleavage activity. The CRISPR-based amplification-free methods do not require amplification and offer advantages such as shortened detection time, simplified operation, and reduced costs compared to traditional amplification-based detection methods.
Amplification-free CRISPR for RNA NABCBs detection
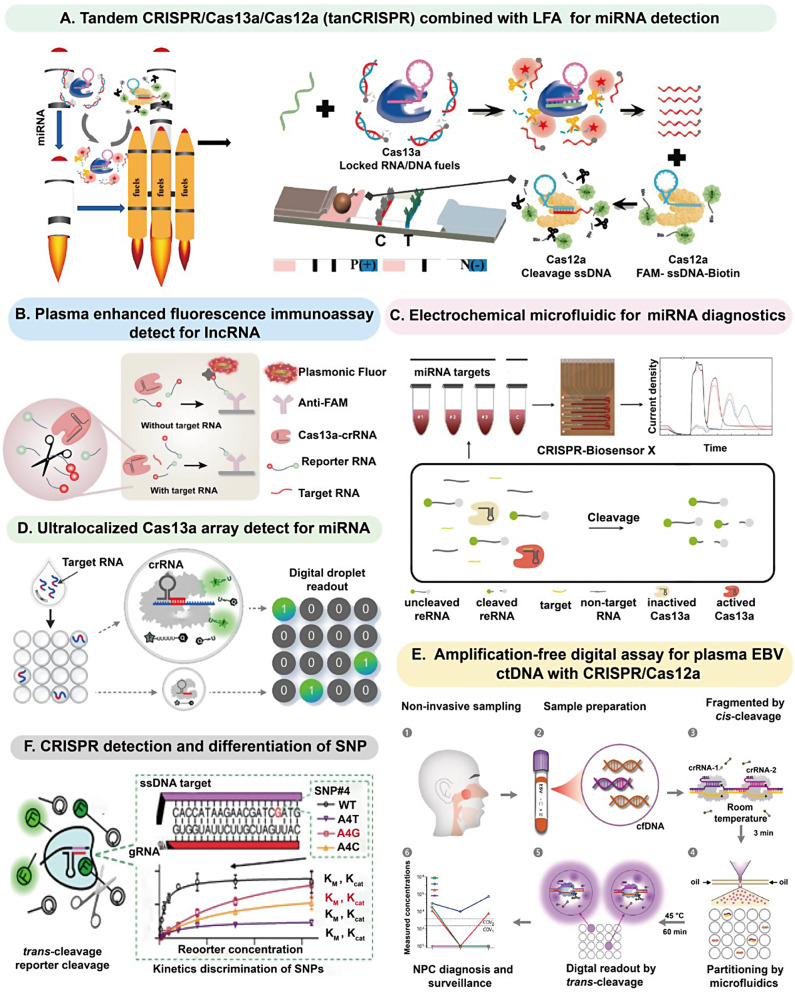
While obstacles persist in identifying non-coding RNA fragments especially short sequence, the integration of CRISPR technology offers enhanced prospects for the specific, efficient and sensitive detection of miRNAs and lncRNAs. CRISPR/Cas13a enables direct amplification-free detection of RNAs. For example, Tian et al. designed a tandem CRISPR/Cas13a/Cas12a direct biosensor by “locked RNA/DNA” probes, and achieved miRNA-21 detection via lateral flow assays (LFA) without amplification (Fig. 6A) [111]. Liu et al. proposed an amplification-free, ultrasensitive lncRNA H19 detection and quantification technique based on CRISPR/Cas13a combined with plasma enhanced fluorescence immunoassay (Fig. 6B) [112]. It is worth noting that short miRNAs can also be directly detected by Cas13, without requiring polymerase and ligase to convert them into dsDNA or ssDNA forms. For example, Bruch et al. achieved simultaneous detection of eight miRNAs by designing electrochemical microfluidic biosensing with four different chips, which take advantage of the trans-cleavage capability of CRISPR/Cas13a (Fig. 6C) [113]. Moreover, Tian et al. reported a supraliminal CRISPR/Cas13a-based amplification-free microfluidic platform for efficient and sensitive detection of miRNAs with the ability to absolutely and directly quantify them. In addition, this method exhibited a sensitivity that exceeds that of the single-Cas13 assay by over 10,000-fold (Fig. 6D) [114]. On this basis, Bruch et al. designed a low-cost, easy-to-operate microfluidic device based on CRISPR/Cas13a, which could be used to detect miRNA-19b in serum samples from pediatric brain cancer children without nucleic acid amplification. This innovative method greatly shortened the reading time to just 9 min, with the detection limit as low as 10 pM.
Fig. 6.
Amplification-free CRISPR methods for NABCBs detection. (A) Tandem CRISPR/Cas13a/Cas12a (tanCRlSPR) combined with LFA for miRNA detection. Reprinted with permission from Ref [111]. Copyright (2022). (B) CRISPR/Cas13a combined with plasma-enhanced fluorescence immunoassay for ultrasensitive lncRNA H19 detection. Reprinted with permission from Ref [112]. Copyright (2021). (C) Electrochemical microfluidic chips combined with CRISPR/Cas13a for miRNAs detection. Reprinted with permission from Ref [113]. Copyright (2021). (D) A supraliminal CRISPR/Cas13a-based amplification-free microfluidic platform for miRNA detection. Reprinted with permission from Ref [114]. Copyright (2021). (E) Micro-droplet digital assay with CRISPR/Cas12a for ctDNA detection. Reprinted with permission from Ref [115]. Copyright (2023). (F) CRISPR-based biosensing for SNP detection. Reprinted with permission from Ref [117]. Copyright (2022)
Amplification-free CRISPR for DNA NABCBs detection
CRISPR-based amplification-free DNA NABCBs detection technologies typically involve designing CRISPR/Cas complexes to recognize specific cancer-associated DNA sequences without pre-amplification of the target sequences. This approach does not require amplification commonly, but involves capturing, enriching, and identifying DNA, ultimately generating detectable signal outputs through cleavage or other biochemical reactions. Recently, Shu’s team developed a Cas12a-mediated unamplified digital DNA assay for cfDNA detection extracted from plasma, offering a useful diagnostic tool for patients with nasopharyngeal carcinoma (NPC). In this platform, double crRNA recognition was introduced to improve the detection accuracy. Notably, the assay demonstrated a reduced detection time of 60 min, as opposed to the approximately 90 min required by traditional qPCR and 120 min by droplet digital PCR (ddPCR). Furthermore, the method with absolute quantitative capabilities has been applied to clinical experiments, and achieving favorable results (Fig. 6E) [115].
Amplification-free CRISPR for Point mutation NABCBs detection
Amplification-free CRISPR technologies are also been widely used for the detection of point mutations, such as through methods like CRISPR-SNP microarrays or CRISPR-Chip, which typically involve the direct test of DNA or RNA samples. Chen et al. constructed a system that by introducing specific nucleotide mismatches as well as inserting PAM sequences, it was possible to increase the sensitivity and specificity of the reaction within 20 min. This system utilized a fluorescence biosensing that combined the kinetic rate of CRISPR/Cas12a targeting with the base and position characteristics of SNP associated with cancer variants [116]. Consequently, it could distinguish wide-type (WT) from mutants (Fig. 6F) [117].
Conventional CRISPR-assisted biosensing was not applicable for diagnosing methylation at individual CpG sites. Dongen et al. designed an in vitro diagnostic method based on CRISPR/Cas12a. By using methylation-sensitive restriction endonucleases (MSREs), the digestion of unmethylated fragments was facilitated, which had an impact on the formation of R-loops between the crRNA and target strand. Research findings indicated that factors such as fragment size, fragmentation position and number of fragments could influence the subsequent collateral trans-cleavage activity towards single-stranded DNA (ssDNA), enabling deducting the methylation position from the cleavage activity. Based on this, single CpG site methylation levels of a cancer gene were successfully determined [118].
Isothermal amplification integrated CRISPR for NABCBs detection
Isothermal amplification technology refers to a nucleic acid amplification method accomplished at a constant temperature, thereby simplifying equipment requirements and reducing operational complexity. Common isothermal amplification techniques mainly include loop-mediated isothermal amplification (LAMP), rolling circle amplification (RCA), strand displacement amplification (SDA), recombinase polymerase amplification (RPA), catalytic hairpin assembly (CHA), hybridization chain reaction (HCR), exponential amplification reaction (EXPAR) and others (Fig. 7). To further enhance the detection sensitivity and accuracy, CRISPR has been combined with isothermal amplification to enhance signals detection and achieve dual-layer signal amplification [119–124]. This integration significantly enhanced the detection of very low abundance nucleic acid targets, offering increased specificity and diverse signal outputs such as fluorescence and colorimetry.
Fig. 7.
Schematic diagram of different types of nucleic acid isothermal amplification methods. (A) LAMP, (B) RAP, (C) SDA, (D) RCA, (E) EXPAR, (F) CHA and (G) HCR
Isothermal amplification integrated CRISPR for RNA NABCBs detection
The present methods of RNA detection involve the utilization of CRISPR-based biosensing. These methods primarily entail the direct identification of target RNA with Cas13a, and subsequently converting the RNA target into the corresponding ssDNA or dsDNA sequences through the corresponding amplification or substitution reactions, which activate the trans-cleavage activity of the Cas in conjunction with the different signal output modes to detect the NABCBs.
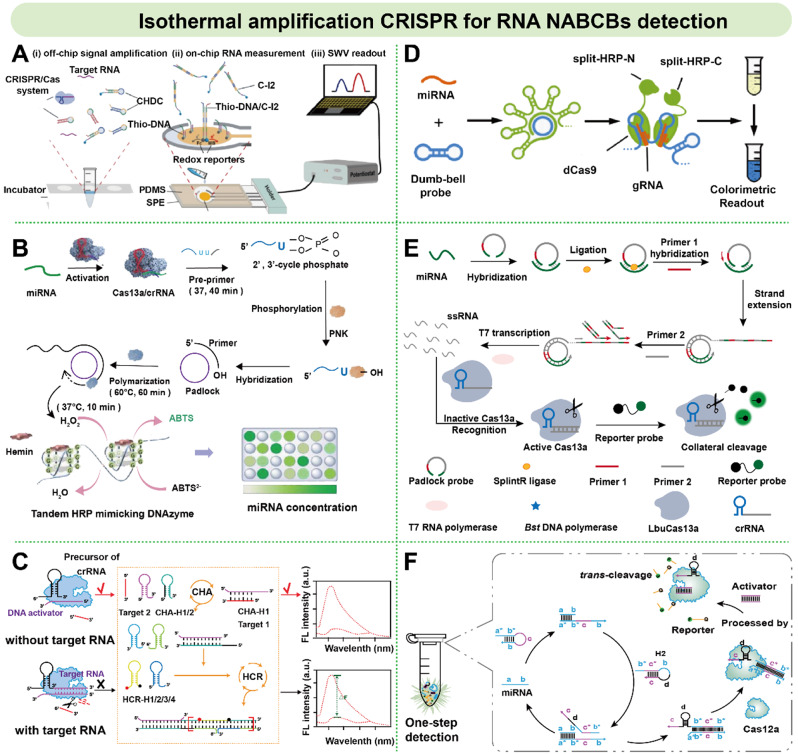
Although the direct activation of CRISPR/Cas system activity by miRNA is still in the innovative research stage, combined with isothermal amplification technology, miRNA signal transduction methods can detect NABCBs more sensitively and efficiently. Sheng et al. designed a catalytic hairpin DNA circuit (CHDC) combined with Cas13a to detect six types of RNA related to NSCLC. miRNA directly activates Cas13a, and then the intermediate product produced by displacement reaction is combined with surface thioDNA to form thioDNA/C-I2 complex, thus enhancing the electrochemical current between the electrode and redox reporter genes. This biosensing could reach the detection limit of 60 aM within 60 min, and exhibit high specificity (95.2%) and sensitivity (90%) (Fig. 8A) [125]. Zhou et al. developed a colorimetric biosensing detection system for directly activating Cas13a from miRNA-10b in serum and cell extracts. In this process, the cross-cutting substrate was used as the amplification primer to form a G tetrahedron for color response (Fig. 8B) [126].
Fig. 8.
Isothermal amplification CRISPR biosensing for RNA NABCBs detection. (A) A catalytic hairpin DNA circuit (CHDC) combined with Cas13a for six types of NSCLC related RNA detection. Reprinted with permission from Ref [125]. Copyright (2021). (B) A homogeneous Cas13a-based visual detection system (termed vCas) for specific and sensitive miRNA detection. Reprinted with permission from Ref [126]. Copyright (2021). (C) A miniature crRNA-mediated Cas12a system by designing CHA and HCR to specifically recognition lncRNA. Reprinted with permission from Ref [127]. Copyright (2024). (D) A hyper-branching rolling circle amplified Cas13a biosensing for miRNAs detection. Reprinted with permission from Ref [129]. Copyright (2018). (E) A fluorescent biosensing based on RCA-amplified Cas13a for miR-17 detection. Reprinted with permission from Ref [130]. Copyright (2020). (F) A crRNA-conjugated CHA circuit was designed for one-step detection of multiple miRNAs. Reprinted with permission from Ref [132]. Copyright (2022)
In addition, CRISPR combined with isothermal amplification for RNA NABCBs detection is mostly accomplished by designing DNA circuits and using single-stranded RNA targets as triggers to generate dsDNA to complete the amplification cycle process. There are fewer reports on lncRNA detection. Recently, Chen et al. proposed a miniature crRNA-mediated CRISPR/Cas12a system, which realized the specific detection of lncRNA pacer by designing modular components such as CHA and HCR (Fig. 8C) [127]. However, there are more reports on CRISPR combined with isothermal amplification for miRNA detection. For instance, Long et al. designed an HCR-based circuit system to initiate Cas12a and realize signal amplification for sensitive and specific detection of miRNA-21. The detection range was from 1 fM to 100 nM, and the calculated detection limit was 75.4 aM. This method avoided the high cost of conventional HCR and has the advantages of easy operation at room temperature [128]. In addition, other NABCBs detection platforms combined with isothermal amplification and CRISPR system are under constant investigation. For example, RCA is widely used because it requires only a small amount of cyclic DNA template to synthesize ultra-long DNA strands with efficient and stable synthesis efficiency. Qiu et al. used Cas9 and RCA for the first detection of circulating let-7a in non-small cell lung cancer patients by colorimetric signal output, and achieve effective detection of target miRNAs in clinical serum samples (Fig. 8D) [129]. Huang et al. proposed a fluorescent biosensing based on RCA-amplified Cas13a for detection of miR-17 in cancer cell extracts, achieving an ultra-low LOD of 200 aM (Fig. 8E) [130].
In addition, Peng et al. established a lock-Cas12a-based biosensing and combined it with SDA to detect target miRNA-21 in breast cancer cells, with a limit of 28.8 aM [131]. Yang et al. combined the isothermal EXPAR combined with Cas12a to realize the ultra-sensitive detection of miRNA-27a within 60 min with the double-layer signal amplification [121]. Chen et al. simplified the detection process and avoided the risk of cross-contamination by designing a crRNA-conjugated CHA circuit reaction and Cas-based end-point assay for one-step detection of multiple miRNAs under isothermal conditions (Fig. 8F) [132]. Other multiplexed analysis techniques for miRNA detection in combination with CRISPR technology are continuously being discovered and applied. For example, Chen et al. reported a sensitive detection of miRNA 31-5p (miR-31) associated with oral squamous cell carcinoma (OSCC) based on Cas12a-coupled IS primer amplification reaction (ISAR), which produced fluorescence/colorimetry signals for dual-mode paper-based strip detection [133].
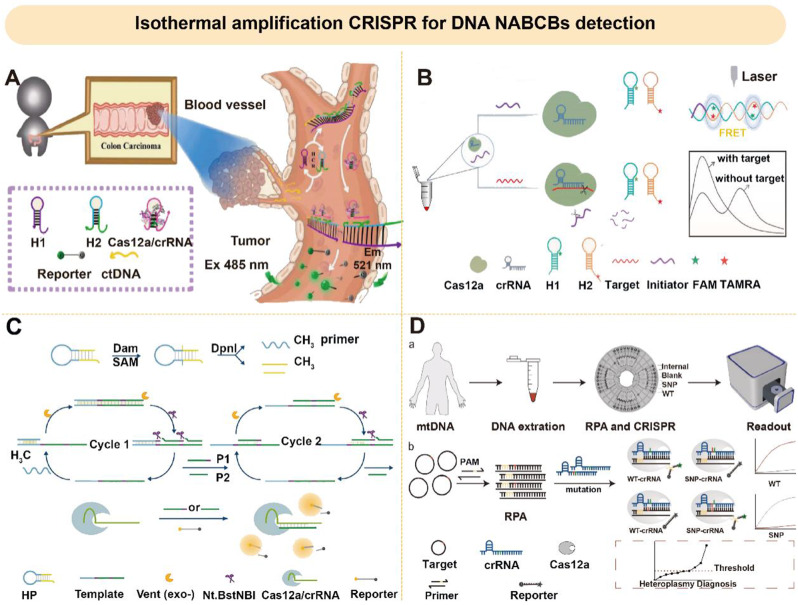
Isothermal amplification integrated CRISPR for DNA NABCBs detection
Combining the CRISPR system with isothermal amplification technology to detect DNA in cancer samples typically entails Cas-guided crRNA specifically recognizing PAM sequences or PFS sequences in dsDNA. The rapid amplification of specific DNA sequences by temperature cycling can generate millions copies of target DNA, thus significantly improving the detection sensitivity.
Usually, Cas12a-based fluorescent biosensing can detect trace ctDNA fragments due to its high sensitivity and selectivity. Li et al. developed a dual-signal sensing strategy combined with HCR. This strategy played an important role in the clinical diagnosis of cancer with the detection limit of 5.43 fM (Fig. 9A) [134]. Wu et al. also designed another fluorescent signaling system (HCR-FRET assay) with HCR and FRET for the diagnosis of T790M mutations in ctDNA in NSCLC (Fig. 9B) [135]. The main cause of papillary thyroid carcinoma (PTC) was the BRAF V600E mutation that leads to instability of the kinase encoded by this gene, which in turn promotes cancerigenesis through the mitogen-activated protein kinase (MAPK) pathway [136]. Zhang et al. further reported an amplification strategy combining 3D DNA Walker and Cas12a for rapid detection of BRAF V600E [137].
Fig. 9.
Isothermal amplification CRISPR for DNA/point mutation NABCBs detection. (A) A Cas12a-based fluorescent biosensing for ctDNA detection with HCR. Reprinted with permission from Ref [134]. Copyright (2023). (B) A hybridization chain reaction-fluorescence resonance energy transfer (HCR-FRET) system for biosensing assay of T790M of ctDNA Reprinted with permission from Ref [135]. Copyright (2023). (C) A fluorescent biosensing combining EXPAR and Cas12a for DNA methylation detection. Reprinted with permission from Ref [138]. Copyright (2018). (D) A proportional chip biosensing based on Cas12a for multiple heterogeneous SNPs detection. Reprinted with permission from Ref [141]. Copyright (2023)
Isothermal amplification integrated-CRISPR for point mutation NABCBs detection
In recent years, advancements have been made in the field of DNA methylation research, particularly in the development of fluorescent probes that offer high sensitivity, real-time monitoring, and dynamic tracking capabilities, demonstrating promising applications. On this basis, Sun et al. reported a fluorescent biosensing that combining EXPAR and Cas12a (EIC) capable of detecting DNA methyltransferase as low as 2 × 10− 4 U/mL, which characterized the process of DNA methylation on CpG islands (Fig. 9C) [138].
Moreover, paper-based biosensing has become a new type of biosensing in cancer diagnosis platform due to its dry storage characteristics and portability in DNA methylation detection. Zhou et al. reported a sensitive method to detect DNA methylation by lateral flow assays (LFA), and 20 fM methylated DNA could be identified by naked eyes. In this system, Lambda processing assisted RPA amplification, and then combined with Cas12a, thus it could minimize the background signal and realize rapid large-scale detection [139]. Wang et al. developed a dual methylation sensitive restriction endonuclease coupled with an RPA-assisted Cas13a system and integrated it into a lateral flow biosensing (LFB) for bedside detection of DNA methylation [140]. In addition, Huang et al. introduced Cas9 to generate a large number of DNA fragments as primers for EXPAR reaction. This fluorescence monitoring system exhibited high specificity for single base mismatch detection with the LOD of 0.82 aM [123]. Wu et al. designed a proportional chip biosensing based on Cas12a, which could detect multiple heterogeneous SNPs in mitochondrial DNA simultaneously after RPA amplification, and it could detect 8 samples within 25 min with the LOD as low as 15.7 aM (Fig. 9D) [141]. Recently, Zhu et al. further proposed a PAM-free Cas12a biosensing based on toe-dsDNA and improved RPA for SNP discrimination [142].
Nanomaterials-assisted CRISPR for NABCBs detection
Nanomaterials have demonstrated significant benefits in biomolecular diagnosis applications owing to their unique physicochemical properties. High specific surface area and exceptional characteristics of these multifunctional nanomaterials have the potential to significantly improve the detection performance of CRISPR/Cas-based molecular biosensing.
Nanomaterials-assisted CRISPR for RNA NABCBs detection
Although there are limited reports on the application of CRISPR-assisted nanotechnology for lncRNA detection, Chen et al. utilized a screen-printed carbon electrode (SPCE) decorated with gold nanocages and amidated multi-walled carbon nanotubes (AuNCs/MWCNT-NH2) to detect lncRNA MALAT1 in NSCLC [67]. The unique structural features of this electrode, including internal hollow and porous walls, a large surface area, superior electrical conductivity, and excellent biocompatibility, made it an innovative material in the realm of biosensing. Additionally, Broto et al. demonstrated that catalytic metal nanoparticles could detect lncRNA from acute myocardial infarction patients with Cas13a-mediated biosensing [76].
Presently, the AuNPS-based nanobeacon combined the advantages of magnetic beads (MBs) and the excellent performance of AuNPs in efficient fluorescence quenching. It could significantly shorten the detection time to 5 min and was able to detect target miRNAs as low as 10 fM (Fig. 10A) [68]. In this, Jay et al. utilized magnetic-gold nanoparticle (MGNPs) composite as signal probe for dark field imaging, which could observe miRNAs of five kinds of BC cells within 30 min [143]. Similarly, Yang et al. also developed a novel detection system using toehold-mediated strand displacement (TMSD) and quantum dots-DNA (QDs-DNA) hydrogel combined with Cas13a for the detection of miRNA in different cell systems. In this platform, the self-assembled QDs-DNA hydrogel has efficient and controllable optical properties, which could be used as a highly sensitive signal amplification element to effectively quench QDs (Fig. 10B) [69]. Zhao et al. developed a sensitive POCT miRNA detection method based on QDs fluorescent microspheres LFA, which utilized the high fluorescence signal of SiO2@DQD (Fig. 10C) [71]. In addition, nanosheets as reaction vessels or signal generators can greatly improve the catalytic efficiency of the reaction and enhance the sensitivity of the detection. For example, Peng et al. used MoS2 nanosheets as a significant fluorescence quenching agent to construct reporter, and it was coupled with Cas12a to detect miRNA-499 (Fig. 10D) [70]. Miao et al. combined TC nanosheets with Zn0.5Cd0.5S (ZCS) and Cas13a to construct a dual-mode biosensing platform to detect miRNA-21 [72]. Wang et al. prepared a new photoelectrochemical (PEC) biosensing for miRNA-21 detection. The heterojunction S-BN/CN nanosheets were prepared by compounding 2D g-C3N4, and it could provide binding sites for methylene blue (MB)-labeled hairpin DNA (MB-HP) signal probes and AuNPs [73].
Fig. 10.
Nanomaterials-assisted CRISPR for RNA NABCBs detection. (A) AuNPS-based nanobeacon fluorescent biosensing for miRNA detection. Reprinted with permission from Ref [68]. Copyright (2022). (B) A novel detection system based on CRISPR-Cas-catalyzed formation of QDs-DNA hydrogel for sensitive miRNA assay. Reprinted with permission from Ref [69]. Copyright (2023). (C) MoS2 nanosheets as a significant fluorescence quenching agent combined with Cas12a for miRNA-499 detection. Reprinted with permission from Ref [71]. Copyright (2024). (D) A QDs fluorescent microspheres LFA biosensing for miRNA detection. Reprinted with permission from Ref [70]. Copyright (2024). (E) A novel magnetic ECL biosensing by coupling Cas13a with Zn2+-dependent DNAzyme for miRNA-145 detection. Reprinted with permission from Ref [75]. Copyright (2024). (F) Detection of miRNA-21 by graphene oxide assisted RCA-Cas12a System. Reprinted with permission from Ref [77]. Copyright (2021)
Currently, metal nanomaterials have excellent electrochemical properties and have been widely used for electrochemical signal amplification and catalysts. Based on this, Zhou et al. adopted [Ru(phen)2dppz]2+ as an optical signaling switch to detect miRNA-17 based on Cas13a-powered electrogenerated chemiluminescence chip (PECL-CRSPR), with a detection limit as low as 1 fM [74]. Li et al. synthesized Fe3O4@PtPd/Ru(bpy)32+ magnetic nano-luminescent materials and combined them with Cas13a and Zn2+-dependent DNAzyme to design a novel magnetic ECL biosensing miRNA-21 detection, and the detection limit was reduced to 0.41 fM (Fig. 10E) [75]. Broto et al. used the bimetallic nanoenzyme (Pt@Au) composed of platinum and gold with high catalytic activity as the signal catalyst for RNA reporter cleavage. This experiment allowed quantitative and colorimetric detection of Cas13 mediated miRNA-223 and lncRNA-LIPCAR by through catalytic metal nanoparticles at room temperature [76]. Similarly, Luo et al. adopted Pt@Au as signal reporter for visual and colorimetric detection of Hepatocellular carcinoma (HCC)-related miRNAs [144]. Besides, the high specific surface area and adjustable electrical properties of graphene oxide make it an ideal material for constructing high-performance biosensing. Qing et al. designed a reduced graphene oxide-modified electrode (rGO/GCE)-based biosensing that combined RCA with Cas12a, providing a powerful tool for clinical diagnosis of cancers (Fig. 10F) [77].
Nanomaterials-assisted CRISPR for DNA NABCBs detection
Currently, the predominant techniques for cfDNA detection utilizing CRISPR primarily rely on fluorescence readouts, and the nanomaterials have been explored to enhance the fluorescence signals in these assays. For instance, Choi et al. designed a Cas12a-based metal-enhanced fluorescence (MEF) nucleic acid amplification biosensing for gene 1 (BRCF-1) detection in cfDNA from breast cancer patients, and the fluorescence color emitted by Cas12a upon cleaving FITC-AuNP-conjugated ssDNA shifted from violet to reddish purple (Fig. 11A) [145]. In addition, electrochemical biosensing based on nanomaterials played an increasingly important role in cfDNA detection. Yang et al. proposed a Cas9-based microneedle patch for trace cfDNA real-time detection. The electrochemical properties of graphene generated a stable and efficient electrical signal output, enabling diagnosis of Epstein-Barr virus (EBV), sepsis, and cfDNA in kidney transplantation (Fig. 11B) [79]. Furthermore, Wang et al. also developed a lead ion utilizing glass nanopores with Cas12a-bound DNA double-stranded blocks to complete the electrical signal output [146].
Fig. 11.
Nanoparticle-assisted CRISPR for DNA/point mutation NABCBs detection. (A) Cas12a-based metal-enhanced fluorescence (MEF) nucleic acid amplification biosensing for cfDNA detection. Reprinted with permission from Ref [145]. Copyright (2021). (B) Cas9-based microneedle patch for cfDNA real-time detection. Reprinted with permission from Ref [79]. Copyright (2022). (C) A novel 3D GR/Au Pt Pd nanoflower biosensing based on Cas9-triggered entropy-driven strand displacement reaction (ESDR) for ctDNA detection. Reprinted with permission from Ref [150]. Copyright (2021). (D) A novel MB/Fe3O4@COF/PD-AU based Cas12a biosensing for ctDNA detection. Reprinted with permission from Ref [78]. Copyright (2022). (E) Cas12a-mediated graphene field effect transistor (gFET) array for SNPs detection. Reprinted with permission from Ref [80]. Copyright (2023)
In recent years, some breakthroughs have also been made in the detection of ctDNA by electrochemical biosensing, and the nanomaterials have been extensively employed due to their exceptional electrocatalytic activity and stable chemical properties [147, 148]. Metal nanomaterials such as gold (Au), silver (Ag), platinum (Pt), silica, graphene, etc., have found wide applications in this regard [149]. Chen et al. innovatively designed a three-dimensional GR/Au Pt Pd nanoscale low biosensing platform for the detection of ctDNA. This method combined the advantages of site-specific cleavage of Cas9/sgRNA and the rapid amplification kinetics of entropy-driven strand displacement reaction (ESDR) (Fig. 11C) [67]. Similarly, Liu et al. designed a novel MB/Fe3O4@COF/PD-AU nanocomposite incorporating Cas12a for the detection of ctDNA epidermal growth factor receptor L858R mutation in non-small cell lung cancer. The nanomaterials were made of covalent organic skeletons (COFs) with a porous structure, which could produce high chemical stability through covalent bonding [150] and identify mutant targets using MB (Fig. 11D) [78]. Zhang et al. used CuCo-ONSs@AuNPs as an ultra-sensitive electrochemical signal and created a bimodal biosensing for EGFR L858R mutation in NSCLC patients [151]. In addition, Uygun et al. used inactivated Cas9 (dCas9) and sgRNA-modified graphene oxide screen-printed electrodes (GPHOXE) for label-free detection of ctDNA and cancer-associated mutations (PIK3CA exon 9 mutations) [152]. Moreover, other nanomaterials as carriers of electrical signal conversion, such as UiO-66-NH2 [153], gold nanosphere-like covalent organic framework (COFs-AuNPs) [154], complexes of gold and iron nanoparticles (AuNPs and FeNPs) were also used to diagnose ctDNA [155].
Nanomaterials-assisted CRISPR for Point mutation NABCBs detection
In the realm of SNP detection, nanomaterials like graphene are frequently employed due to its notable characteristics such as high specific surface area and excellent electrical conductivity. Balderston et al. reported a biosensing using graphene monolayer effector transistors, which could discriminate target genes with single nucleotide differences between wild-type and homozygous mutant types within 1 h by rapidly screening complexes of sgRNA and Cas9 [156]. In addition, Weng et al. reported an electrochemical biosensing based on a Cas12a-mediated graphene field effect transistor (gFET) array to detect SNPs in DNA without amplification, which has ultra-high sensitivity with the LOD of 1 aM (Fig. 11E) [80].
Challenges and prospects
The ongoing advancements in genomics have led to the identification of an increasing number of potential NABCBs, offering cancer patients a wider array of choices for early detection and tailored treatment. Known for the efficiency of CRISPR in molecular diagnosis and the unique advantages of nanotechnology in sample extraction/separation/detection, the CRISPR-based nano-biosensing has been increasingly utilized for trace NABCBs testing. These systems leverage the specific recognition and efficient signal amplification and by developing a series of innovative diagnostic tools to achieve integrated miniaturization and POCT detection. Although these technologies show great potential in early screening and monitoring of cancer, they still encounter some significant obstacles: (1) The intricate nature of multi-target detection and body fluid samples. How to effectively remove background interference, accurately capture trace NABCBs and ensure experimental specificity is a technical problem. (2) There is still a lack of standardized detection methods for NABCBs. In addition, in practical clinical application, there is also a lack of relevant guidelines for some NABCBs. (3) It is necessary to achieve the breakthrough of commercial products and realize the demand for timely diagnosis and continuous monitoring. It involves the interdisciplinary challenge of designing a more sensitive, portable, user-friendly and integrated biosensing. (4) Currently, some nanomaterials also exhibit some shortcomings, such as low activity/stability, poor dispersion/uniformity and potential environmental toxicity.
Based on the above existing challenges, we aim to provide some recommendations to solve the above problems: (1) Exploring more emerging biosensing based on novel programmable nuclease, nanoplatforms to enhance the multi-detection capability. Optimizing sample pretreatment steps, such as employing specific enrichment technology or high throughput screening methods to remove background interference. Developing a more designed algorithm of specific CRISPR guide RNA (gRNA) to reduce nonspecific binding. (2) Developing a more personalized detection plan by integrating genetics, epigenetics, and other omics data to achieve precise medical diagnostics. With the continuous development of molecular diagnostic technology, it is expected to establish cancer biomarker identification standards and improve relevant guidelines in the future. (3) Integrating immunological biosensing, biochip or nanopore sequencing and artificial intelligence-assisted image analysis into portable devices. This integration aims to promote continuous and real-time detection, simplify the detection process, create portable and cost-effective equipment, thus transforming molecules diagnosis and promoting commercialization. (4) Enhancing dispersion, biocompatibility and other properties by various physicochemical methods, such as coating additional surfaces layers, doping element lattices, changing electrical/optical/magnetic properties, or modulating mechanical strength/permeability/catalytic activity. In addition, adopting microbial/plant and more green synthesis methods to reduce the potential toxicity of nanomaterials (Fig. 12).
Fig. 12.
Challenges and prospects of NABCBs detection based on CRISPR-based nano-biosensing
Conclusion
In this paper, a comprehensive overview of the developmental process, various types, vital carcinogenic mechanisms and commercialized products of NABCBs are described in details. It delves into the specific functions of CRISPR/Cas toolbox and properties of diverse nanomaterials. Furthermore, the application of different NABCBs biosensing based on CRISPR and nanotechnology is further investigated. CRISPR-based platforms combined with isothermal amplification and nanomaterials exhibit high sensitivity and selectivity, rendering it extensively used in various domains such as biological analysis and disease diagnosis. There are still some persisting challenges in NABCBs detection, including the cross-interference/high background signal of multiplex detection, the lack of standardized methods/relevant guidelines of NABCBs, the urgent breakthrough of commercial products, and the long-standing drawbacks of nanomaterials. But we expect that these dilemmas can be addressed by exploring emerging biosensing, optimizing sample pretreatment steps, developing personalized strategies, integrating portable devices, adopting physicochemical modification/green synthesis methods. Looking ahead, with the ongoing promotion and optimization of CRISPR-based nano-biosensing, more potent programmable nuclease, ideal nanomaterials, cost-effective/portable devices, clinical products and non-nucleic acid targets detection platforms will be developed [159]. We firmly believe that this undoubtedly will provide a brighter future for cancer diagnosis, early diseases detection, and human life and health.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- AI
Artificial intelligence
- BC
Breast cancer
- BLCA
Bladder cancer
- CC
Cervical cancer
- CRC
Colorectal cancer
- CT
Computed tomography
- CTCL
Cutaneous T-cell lymphoma
- DC
Dendritic
- ELISA
Enzyme-linked immunosorbent assay
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- HNCs
Head and neck cancer
- LC
Liver cancer
- LiC
Leukaemia cell
- NEEM
seq-No End-repair Enzymatic Methyl-seq
- NGS
Next generation sequencing
- NPC
Nasal pharyngeal cancer
- NSCLC
Non-small cell lung cancer
- OC
Ovarian cancer
- OSCC
Oral squamous cell carcinoma
- PC
Prostatic cancer
- PLC
Primary liver cancer
- POCT
Portable point-of-care testing
- q-PCR
Real time fluorescence quantitative PCR
- Sepsis
Septicemia
- THCA
Thyroid carcinoma
- WGS
Whole genome sequencing
Author contributions
Weipan Peng: Writing-original draft, Draw figures, Investigation, Methodology, Data curation, Conceptualization. Mengting Shi: Writing-original draft, Draw figures, Investigation, Methodology, Data curation, Conceptualization. Bin Hu: Writing-original draft, Investigation, Methodology, Data curation. Jingyu Jia: Draw figures. Xinyue Li: Draw figures. Nan Wang: Writing-original draft, Draw figures. Shuli Man: Writing-review & editing, Supervision, Conceptualization, Project Administration. Shengying Ye: Supervision, Conceptualization, Project Administration. Long Ma: Writing-original draft, Writing-review & editing, Conceptualization, Investigation, Supervision, Project Administration, Funding acquisition.
Funding
This work was financially supported by National Natural Science Foundation of China (32072309, 32372415), Tianjin Municipal Science and Technology Committee (19JCYBJC27800, 22YFZCSN00070, 23JCJQJC00090).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Weipan Peng, Mengting Shi and Bin Hu are co-first authors.
Contributor Information
Shuli Man, Email: msl@tust.edu.cn.
Shengying Ye, Email: jbskxb@163.com.
Long Ma, Email: malong@tust.edu.cn, Email: woshimalong1983@163.com.
References
- 1.McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO Press, Adv. Nutr. 2016;7:418. [DOI] [PMC free article] [PubMed]
- 2.Pérez-Herrero E, Fernández-Medarde A. Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur J Pharm Biopharm. 2015;93:52–79. 10.1016/j.ejpb.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Sci U S A. 1995;92:5258–65. 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23:208–16. 10.1016/i.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Serratì S, Summa SD, Pilato B, Petriella D, Lacalamita R, Tommasi S, Pinto R. Next-generation sequencing: advances and applications in cancer diagnosis. Therapy. 2016;7355–65. 10.2147/OTT.S99807. [DOI] [PMC free article] [PubMed]
- 6.Mader A, Riehle U, Brandstetter T, Stickeler E, Hausen A, zur, Rühe J. Microarray-based amplification and detection of RNA by nucleic acid sequence-based amplification. Anal Bioanal Chem. 2010;397:3533–41. 10.1007/s00216-010-3892-4. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Qiao J, Zhao Z, Zhang Q, Zhang W, Man S, Ye S, Chen K, Ma L. A CRISPR/dCas9-enabled, on‐site, visual, and bimodal biosensing strategy for ultrasensitive and self‐validating detection of foodborne pathogenic bacteria. Food Front. 2023;4:2070–80. 10.1002/fft2.286. [Google Scholar]
- 8.Cheng Z, Li M, Dey R, Chen Y. Nanomaterials for cancer therapy: current progress and perspectives. J Hematol Oncol. 2021;14:1–27. 10.1186/s13045-021-01096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Chen W, Qi Y, Zhu Y. Evaluation of lncRNA FOXD3-AS1 as a biomarker for early-stage lung cancer diagnosis and subtype identification. J. Evidence-Based Complementary Altern. Med. 2022;2022. 10.1155/2022/5702014 [DOI] [PMC free article] [PubMed] [Retracted]
- 10.Sharma S. Tumor markers in clinical practice: General principles and guidelines. Indian J Med Paediatr Oncol. 2009;30:1–8. 10.4103/0971-5851.56328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talaat A, Helmy MA, Saadawy SF. Evaluation of miRNA-21 and CA-125 as a promising diagnostic biomarker in patients with ovarian cancer. Egypt J Med Hum Genet. 2022;23:123. 10.1186/s43042-022-00342-5. [Google Scholar]
- 12.Zhou Y, Wang M, Wu J, Jie Z, Chang S, Shuang T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J Ovarian Res. 2015;8:1–7. 10.1186/s13048-015-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng FR, Tang LJ, He Y, Garcia R. An update on the role of miRNA-155 in pathogenic microbial infections. Microbes Infect. 2015;17:613–21. 10.1016/j.micinf.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Elsayed ET, Salem P, Darwish E, Fayed AM. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int J Biol Markers. 2018;33:528–33. 10.1177/1724600818760244. [DOI] [PubMed] [Google Scholar]
- 15.Fan C, Wang J, Tang Y, Zhang S, Xiong F, Guo C, Zhou Y, Li Z, Li X, Li Y. Upregulation of long non-coding RNA LOC284454 may serve as a new serum diagnostic biomarker for head and neck cancers. BMC Cancer. 2020;20:1–9. 10.1186/s12885-020-07408-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie Y, Li Y, Xu Y, Jiao Y, Li W. Long noncoding RNA BACE1AS is an independent unfavorable prognostic factor in liver cancer. Oncol Lett. 2020;20:1–1. 10.3892/ol.2020.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W, Qiu L, Liu H, Xu Y, Zhan M, Zhang W, Xin Y, He X, Yang X, Bai J. Circulating tumor DNA as a potential prognostic and predictive biomarker during interventional therapy of unresectable primary liver cancer. J Gastrointest Oncol. 2020;11:1065. 10.21037/jgo-20-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Huang C, Zhou H, Liu D, Chen R, Li X, Cheng Y, Gao B, Chen J. Circulating tumor DNA predicts the outcome of chemotherapy in patients with lung cancer. Thorac Cancer. 2022;13:95–106. 10.1111/1759-7714.14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno Y, Shibata S, Miyagaki T, Ito Y, Taira H, Omori I, Hisamoto T, Oka K, Matsuda KM, Boki H. Serum cell-free DNA as a new biomarker in cutaneous T‐cell lymphoma. J Dermatol. 2022;49:1124–30. 10.1111/1346-8138.16520. [DOI] [PubMed] [Google Scholar]
- 20.Bu J, Lee TH, Jeong WJ, Poellmann MJ, Mudd K, Eun HS, Liu EW, Hong S, Hyun SH. Enhanced detection of cell-free DNA (cfDNA) enables its use as a reliable biomarker for diagnosis and prognosis of gastric cancer. PLoS ONE. 2020;15:0242145. 10.1371/journal.pone.0242145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, He T, Herman JG, Linghu E, Yang Y, Fuks F, Zhou F, Song L, Guo M. Methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin Epigenetics. 2017;9:1–12. 10.1186/s13148-017-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, Zhu Y, Miao J, Xiong S, Fei S. Aberrant DNA methylation of SEPT9 and SDC2 in stool specimens as an integrated biomarker for colorectal cancer early detection. Front Genet. 2020;11:643. 10.3389/fgene.2020.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Q, Liu Y, Kong X, Xian J, Ye L, Yang L, Guo S, Zhang Y, Zhou L, Xiang T. The novel methylation biomarker SCARA5 sensitizes cancer cells to DNA damage chemotherapy drugs in NSCLC. Front Oncol. 2021;11:666589. 10.3389/fonc.2021.666589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.London SJ, Lehman TA, Taylor JA. Myeloperoxidase genetic polymorphism and lung cancer risk. Cancer Res. 1997;57:5001–3. [PubMed] [Google Scholar]
- 25.Golka K, Prior V, Blaszkewicz M, Bolt HM. The enhanced bladder cancer susceptibility of NAT2 slow acetylators towards aromatic amines: a review considering ethnic differences. Toxicol Lett. 2002;128:229–41. 10.1016/s0378-4274(01)00544-6. [DOI] [PubMed] [Google Scholar]
- 26.Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90:1225–9. 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 27.Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater. 2019;31:1905311. 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 30.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–8. 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2014;8:706–13. 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng FR, Tang LJ, He Y, Garcia RC. An update on the role of miRNA-155 in pathogenic microbial infections. Microbes Infect. 2015;17:613–21. 10.1016/j.micinf.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. 2016;1402:271–86. 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 36.Aswathy R, Sumathi S. Defining new biomarkers for overcoming therapeutical resistance in cervical cancer using lncRNA. Mol Biol Rep. 2023;50:10445–60. 10.1007/s11033-023-08864-w. [DOI] [PubMed] [Google Scholar]
- 37.Yang S, Shi F, Du Y, Wang Z, Feng Y, Song J, Liu Y, Xiao M. Long non-coding RNA CTBP1-AS2 enhances cervical cancer progression via up-regulation of ZNF217 through sponging miR-3163. Cancer Cell Int. 2020;20:1–13. 10.1186/s12935-020-01430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel P. Les acides nucleiques du plasma sanguin chez 1 homme. C R Seances Soc Biol Fil. 1948;142:241–3. [PubMed] [Google Scholar]
- 39.Cheng X, Ma L. Enzymatic synthesis of fluorinated compounds. Appl Microbiol Biotechnol. 2021;105:8033–58. 10.1007/s00253-021-11608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R, Pulvirenti A, Ferro A. miRandola: extracellular circulating microRNAs database. PLoS ONE. 2012;7:47786. 10.1371/journal.pone.0047786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–64. 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 43.Murphy J, Mahony J, Ainsworth S, Nauta A, Sinderen DV. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol. 2013;79:7547–55. 10.1128/aem.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 45.Song Y, Zhang Y, Wang TH. Single quantum dot analysis enables multiplexed point mutation detection by gap ligase chain reaction. Small. 2013;9:1096–105. 10.1002/smll.201202242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe A, Huh YS, Strickland AD, Erickson D, Batt CA. Multiplex single nucleotide polymorphism genotyping utilizing ligase detection reaction coupled surface enhanced Raman spectroscopy. Anal Chem. 2010;82:5810–4. 10.1021/ac100921b. [DOI] [PubMed] [Google Scholar]
- 47.Wu K, Kong F, Zhang J, Tang Y, Chen Y, Chao L, Nie L, Huang Z. Recent progress in single-nucleotide polymorphism. Biosens Biosens. 2023;13:864. 10.3390/bios13090864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33. 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Zeng Z, She Q, Han W. The abortive infection functions of CRISPR-Cas and Argonaute. Trends Microbiol. 2023;31:405–18. 10.1016/j.tim.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 201;346:1258096. 10.1126/science.1258096 [DOI] [PubMed]
- 52.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 53.Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–29. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 54.Asmamaw M, Zawdie B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics. 2021;15:353–61. 10.2147/BTT.S326422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. J Annu Rev Biochem. 2016;85:227–64. [DOI] [PubMed] [Google Scholar]
- 56.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen JS, Ma E, Harrington LB, Costa MD, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9. 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhuang S, Hu T, Zho X, Zhou H, He S, Li J, Qiu L, Zhang Y, Xu Y, Pei H. meHOLMES: a CRISPR-cas12a-based method for rapid detection of DNA methylation in a sequence-independent manner. Heliyon. 2024;10:e24574. 10.1016/j.heliyon.2024.e24574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–44. 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–42. 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aquino-Jarquin G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine. 2019;18:428–31. 10.1016/j.nano.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA. Programmed DNA destruction by miniature CRIPR-Cas14 enzymes. Science. 2018;362:839–42. 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savage DF. Cas14: big advances from small CRISPR proteins. Biochemistry. 2019;58:1024–5. 10.1021/acs.biochem.9b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aman R, Mahas A, Mahfouz M. Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth Biol. 2020;9:1226–33. 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- 65.Hillary VE, Ceasar SA. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol Biotechnol. 2023;65:311–25. 10.1007/s12033-022-00567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou B, Yang R, Sohail M, Kong X, Zhang X, Fu N, Li B. CRISPR/Cas14 provides a promising platform in facile and versatile aptasensing with improved sensitivity. Talanta. 2023;254:124120. 10.1016/j.talanta.2022.124120. [DOI] [PubMed] [Google Scholar]
- 67.Chen M, Wu D, Tu S, Yang C, Chen D, Xu Y. A novel biosensor for the ultrasensitive detection of the lncRNA biomarker MALAT1 in non-small cell lung cancer. Sci Rep. 2021;11:3666. 10.1038/s41598-021-83244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo T, Li J, He Y, Liu H, Deng Z, Long X, Wan Q, Ding J, Gong Z, Yang Y. Designing a CRISPR/Cas12a-and Au-nanobeacon-based diagnostic biosensor enabling direct, rapid, and sensitive miRNA detection. Anal Chem. 2022;94:6566–73. 10.1021/acs.analchem.2c00401. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y, Yang J, Yang Y, Ji X, Li W, Wang F, Xie C, He Z. CRISPR/Cas13a catalyzed self-assembly of quantum dot-DNA hydrogel for microRNA assay. Sens Actuators B. 2023;393:134249. 10.1016/j.snb.2023.134249. [Google Scholar]
- 70.Zhao J, Han H, Liu Z, Chen J, Liu X, Sun Y, Wang B, Zhao B, Pang Y, Xiao R. Portable fluorescent lateral flow assay for ultrasensitive point-of-care analysis of acute myocardial infarction related microRNA. Anal Chim Acta. 2024;1295:342306. 10.1016/j.aca.2024.342306. [DOI] [PubMed] [Google Scholar]
- 71.Li P, Ye Y, Li Y, Xie Z, Ye L, Huang J. A MoS2 nanosheet-based CRISPR/Cas12a biosensor for efficient miRNA quantification for acute myocardial infarction. Biosens Bioelectron. 2024;251:116129. 10.1016/j.bios.2024.116129. [DOI] [PubMed] [Google Scholar]
- 72.Miao P, Wang B, Zheng G, Wang W, Lv Y, Zhang J, Yan M. CRISPR/Cas13a-mediated dual-modal biosensing platform based on the Zn0.5Cd0.5S/Ti3C2 Schottky heterojunction for the sensitive detection of miRNAs-21. Sens Actuators B. 2024;400:134829. 10.1016/j.snb.2023.134829. [Google Scholar]
- 73.Wang F, Zhang L, Liu H, Xie L, Ge S, Yu J. Bipedal DNA walker-CRISPR/Cas12a coupling with sulfur doped boron nitride–carbon nitride composite biosensing for the detection of miRNA-21. Sens Actuators B. 2024;403:135115. 10.1016/j.snb.2023.135115. [Google Scholar]
- 74.Zhou T, Huang R, Huang M, Shen J, Shan Y, Xing D. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific miRNA detection. Adv Sci. 2020;7:1903661. 10.1002/advs.201903661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Chen C, Luo F, Lin Z, Wang J, Huang A, Sun Y, Qiu B. Highly sensitive biosensor for specific miRNA detection based on cascade signal amplification and magnetic electrochemiluminescence nanoparticles. Anal Chim Acta. 2024;1288:342123. 10.1016/j.aca.2023.342123. [DOI] [PubMed] [Google Scholar]
- 76.Broto M, Kaminski MM, Adrianus C, Kim N, Greensmith R, Dissanayake-Perera S, Schubert AJ, Tan X, Kim H, Dighe AS. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs. Nat Nanotechnol. 2022;17:1120–6. 10.1038/s41565-022-01179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qing M, Chen SL, Sun Z, Fan Y, Luo HQ, Li NB. Universal and programmable rolling circle amplification-CRISPR/Cas12a-mediated immobilization-free electrochemical biosensor. Anal Chem. 2021;93:7499–507. 10.1021/acs.analchem.1c00805. [DOI] [PubMed] [Google Scholar]
- 78.Liu F, Peng J, Lei YM, Liu RS, Jin L, Liang H, Liu HF, Ma SY, Zhang XH, Zhang YP. Electrochemical detection of ctDNA mutation in non-small cell lung cancer based on CRISPR/Cas12a system. Sens Actuators B. 2022;362:131807. 10.1016/j.snb.2022.131807. [Google Scholar]
- 79.Yang B, Kong J, Fang X, Programmable. Nat Commun. 2022;13:3999. 10.1038/s41467-022-31740-3. CRISPR-Cas9 microneedle patch for long-term capture and real-time monitoring of universal cell-free DNA. [DOI] [PMC free article] [PubMed]
- 80.Weng Z, You Z, Li H, Wu G, Song Y, Sun H, Fradlin A, Neal-Harris C, Lin M, Gao X. CRISPR-Cas12a Biosensor array for ultrasensitive detection of unamplified DNA with single-nucleotide polymorphic discrimination. ACS sens. 2023;8:1489–99. 10.1021/acssensors.2c02495. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Liu J, Wang M, Su X. Determination of ascorbic acid and ascorbate oxidase based on quaternary CuInZnS QDs/thiochrome ratiometric fluorescence sensing system. Talanta. 2020;214:120814. 10.1016/j.talanta.2020.120814. [DOI] [PubMed] [Google Scholar]
- 82.Jalani G, Naccache R, Rosenzweig DH, Lerouge S, Haglund L, Vetrone F, Cerruti M. Real-time, non-invasive monitoring of hydrogel degradation using LiYF4:Yb3+/Tm3+ NIR-to-NIR upconverting nanoparticles. Nanoscale. 2015;7:11255–62. 10.1039/c5nr02482j. [DOI] [PubMed] [Google Scholar]
- 83.Gupta BK, Kumar A, Kumar P, Dwivedi J, Pandey GN, Kedawat G. Probing on green long persistent Eu2+/Dy3+ doped Sr3SiAl4O11 emerging phosphor for security applications. J Appl Phys. 2015;117. 10.1063/1.4922983.
- 84.Shang L, Dong S, Nienhau GU. Ultra-small fluorescent metal nanoclusters: synthesis and biological applications. Nano Today. 2011;6:401–18. 10.1016/j.nantod.2011.06.004. [Google Scholar]
- 85.Nie L, Ou Z, Yang, Xing D. Thermoacoustic molecular tomography with magnetic nanoparticle contrast agents for targeted tumor detection. Med Phys. 2010;37:4193–200. 10.1118/1.3466696. [DOI] [PubMed] [Google Scholar]
- 86.Marzouk SAM, Al-Marzouqi MH. Analyzer for continuous monitoring of H2S in gas streams based on a novel thermometric detection. Sens Actuators B. 2012;162:377–83. 10.1016/j.snb.2011.12.107. [Google Scholar]
- 87.Lu W, Chen Y, Liu Z, Tang W, Feng Q, Sun J, Jiang X. Quantitative detection of microRNA in one step via next generation magnetic relaxation switch sensing. ACS Nano. 2016;10:6685–92. 10.1021/acsnano.6b01903. [DOI] [PubMed] [Google Scholar]
- 88.Wu Y, Guo W, Peng W, Zhao Q, Piao J, Zhang B, Wu X, Wang H, Gong X, Chang J. Enhanced fluorescence ELISA based on HAT triggering fluorescence turn-on with enzyme-antibody dual labeled AuNP probes for utrasensitive detection of AFP and HBsAg. ACS Appl Mater Interfaces. 2017;9:9369–77. 10.1021/acsami.6b16236. [DOI] [PubMed] [Google Scholar]
- 89.Li YS, Zhou Y, Meng XY, Zhang YY, Liu JQ, Zhang Y, Wang NN, Hu P, Lu SY, Ren HL, Liu ZS. Enzyme-antibody dual labeled gold nanoparticles probe for ultrasensitive detection of κ-casein in bovine milk samples. Biosens Bioelectron. 2014;61:241–4. 10.1016/j.bios.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 90.Ma L, Yin LJ, Li XY, Chen S, Peng L, Liu GZ, Ye SY, Zhang WL. S L Man Biosens Bioelectron. 2022;195:113646. 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y, Tian XL, Li YS, Pan FG, Zhang YY, Zhang JH, Yang L, Wang XR, Ren HL, Lu SY, Li ZH, Chen QJ, Liu ZS, Liu JQ. An enhanced ELISA based on modified colloidal gold nanoparticles for the detection of pb (II). Biosens Bioelectron. 2011;26:3700–4. 10.1016/j.bios.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 92.Jia CP, Zhong XQ, Hua B, Liu MY, Jing FX, Lou XH, Yao SH, Xiang JQ, Jin QH, Zhao JL. Nano-ELISA for highly sensitive protein detection. Biosens Bioelectron. 2009;24:2836–41. 10.1016/j.bios.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 93.Liu D, Yang J, Wang HF, Wang Z, Huang X, Wang Z, Niu G, Walker ARH, Chen X. Glucose oxidase-catalyzed growth of gold nanoparticles enables quantitative detection of attomolar cancer biomarkers. Anal Chem. 2014;86:5800–6. 10.1021/ac500478g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang J, Yao C, Li X, Wu Z, Huang C, Fu Q, Lan C, Cao D, Tang Y. Silver nanoprism etching-based plasmonic ELISA for the high sensitive detection of prostate-specific antigen. Biosens Bioelectron. 2015;69:128–34. 10.1016/j.bios.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 95.Guo R, Yin F, Sun Y, Mi L, Shi L, Tian Z, Li T. Ultrasensitive simultaneous detection of multiplex disease-related nucleic acids using double-enhanced surface-enhanced raman scattering nanosensors. ACS Appl Mater Interfaces. 2018;10:25770–8. 10.1021/acsami.8b06757. [DOI] [PubMed] [Google Scholar]
- 96.Yang C, Dou B, Shi K, Chai Y, Xiang Y, Yuan R. Multiplexed and amplified electronic sensor for the detection of microRNAs from cancer cells. Anal Chem. 2014;86:11913–8. 10.1021/ac503860d. [DOI] [PubMed] [Google Scholar]
- 97.Toader O, John S. Square spiral photonic crystals: robust architecture for microfabrication of materials with large three-dimensional photonic band gaps. Phys Rev E. 2002;66. 10.1103/PhysRevE.66.016610. [DOI] [PubMed]
- 98.Zhao Y, Zhao X, Pei X, Hu J, Zhao W, Chen B, Gu Z. Multiplex detection of tumor markers with photonic suspension array. Anal Chim Acta. 2009;633:103–8. 10.1016/j.aca.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 99.Zeng GL, Yun XY. Discovery and current application of nanozyme. Prog Biochem Biophys. 2013;40:892–902. [Google Scholar]
- 100.Zhang Z, Zhang X, Liu B, Liu J. Molecular imprinting on inorganic nanozymes for hundred-fold enzyme specificity. J Am Chem Soc. 2017;139:5412–9. 10.1021/jacs.7b00601. [DOI] [PubMed] [Google Scholar]
- 101.Cui F, Qiu Q, Peng G, Li X, Liu X, Chen X. Core-shell CdSeTe/ZnS quantum dots for the detection of microRNA-155 based on the fluorescence resonance energy transfer technique via the formation of a network structure. Anal Methods. 2019;11:4137–45. 10.1039/C9AY00783K. [Google Scholar]
- 102.Kuningas K, Ukonaho T, Päkkilä H, Rantanen T, Rosenberg J, Lövgren T, Soukka T. Upconversion fluorescence resonance energy transfer in a homogeneous immunoassay for estradiol. Anal Chem. 2006;78:4690–6. 10.1021/ac0603983. [DOI] [PubMed] [Google Scholar]
- 103.He J, Huang M, Wang D, Zhang Z, Li G. Magnetic separation techniques in sample preparation for biological analysis: a review. J Pharm Biomed Anal. 2014;101:84–101. 10.1016/j.jpba.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 104.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry: J Int Soc Anal Cytol. 1990;11:231–8. 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 105.Hu J, Chen M, Fang X, Wu L. Fabrication and application of inorganic hollow spheres. Chem Soc Rev. 2011;40:5472–91. 10.1039/c1cs15103g. [DOI] [PubMed] [Google Scholar]
- 106.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Noble metals on the nanoscale: optical and photothermal properties and some applications in imaging, sensing, biology, and medicine. Acc Chem Res. 2008;41:1578–86. 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 107.Ju X, Tang Y, Qu R, Hao S. The emerging role of Circ-SHPRH in cancer. Onco Targets Ther. 2021;14:4177–88. 10.2147/OTT.S317403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao N, Guo M, Zhang C, Wang C, Wang K. Pan-cancer methylated dysregulation of long non-coding RNAs reveals epigenetic biomarkers. Front Cell Dev Biol. 2022;10:882698. 10.3389/fcell.2022.882698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao J, Xia K, He P, Wei G, Zhou X, Zhang X. Recent advances of nucleic acid-based cancer biomarkers and biosensors. Coord Chem Rev. 2023;497. 10.1016/j.ccr.2023.215456.
- 110.Gong S, Zhang S, Lu F, Pan W, Li N, Tang B. CRISPR/Cas-Based in vitro diagnostic platforms for cancer biomarker detection. Anal Chem. 2021;93:11899–909. 10.1021/acs.analchem.1c02533. [DOI] [PubMed] [Google Scholar]
- 111.Tian B, Wang Y, Tang W, Chen J, Zhang J, Xue S, Zheng S, Cheng G, Gu B, Chen M. Tandem CRISPR nucleases-based lateral flow assay for amplification-free miRNA detection via the designed locked RNA/DNA as fuels. Talanta. 2024;266:124995. 10.1016/j.talanta.2023.124995. [DOI] [PubMed] [Google Scholar]
- 112.Liu L, Wang Z, Wang Y, Luan J, Morrissey JJ, Naik RR, Singamaneni S. Plasmonically enhanced CRISPR/Cas13a-based bioassay for amplification‐free detection of cancer‐associated RNA. Adv Healthc Mater. 2021;10:2100956. 10.1002/adhm.202100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bruch R, Johnston M, Kling A, Mattmüller T, Baaske J, Partel S, Madlener S, Weber W, Urban GA, Dincer C. CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens Bioelectron. 2021;177. 10.1016/j.bios.2020.112887. [DOI] [PubMed]
- 114.Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo Z, Han Z, Wang Z, Zhou X. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2021;15:1167–78. 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- 115.Jiang C, Zheng X, Lin L, Li X, Liao Y, Jia W, Shu B. CRISPR Cas12a-mediated amplification-free digital DNA assay improves the diagnosis and surveillance of nasopharyngeal carcinoma. Biosens Bioelectron. 2023;237:115546. 10.1016/j.bios.2023.115546. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y, Mei Y, Jiang X. Universal and high-fidelity DNA single nucleotide polymorphism detection based on a CRISPR/Cas12a biochip. Chem Sci. 2021;12:4455–62. 10.1039/d0sc05717g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blanluet C, Huyke DA, Ramachandran A, Avaro AS, Santiago JG. Detection and discrimination of single nucleotide polymorphisms by quantification of CRISPR-Cas catalytic efficiency. Anal Chem. 2022;94:15117–23. 10.1021/acs.analchem.2c03338. [DOI] [PubMed] [Google Scholar]
- 118.Dongen JEV, Berendsen JTW, Eijkel JCT, Segerink LI. A CRISPR/Cas12a-assisted in vitro diagnostic tool for identification and quantification of single CpG methylation sites. Biosens Bioelectron. 2021;194:113624. 10.1016/j.bios.2021.113624. [DOI] [PubMed] [Google Scholar]
- 119.Chi Z, Wu Y, Chen L, Yang H, Khan MR, Busquets R, Huang N, Lin X, Deng R, Yang W, Huang J. CRISPR-Cas14a-integrated strand displacement amplification for rapid and isothermal detection of cholangiocarcinoma associated circulating microRNAs. Anal Chim Acta. 2022;1205. 10.1016/j.aca.2022.339763. [DOI] [PubMed]
- 120.Wang R, Zhao X, Chen X, Qiu X, Qing G, Zhang H, Zhang L, Hu X, He Z, Zhong D, Wang Y, Luo Y. Rolling circular amplification (RCA)-assisted CRISPR/Cas9 cleavage (RACE) for highly specific detection of multiple extracellular vesicle microRNAs. Anal Chem. 2019;92:2176–85. 10.1021/acs.analchem.9b04814. [DOI] [PubMed] [Google Scholar]
- 121.Yang Y, Yang J, Gong F, Zuo P, Tan Z, Li J, Xie C, Ji X, Li W, He Z. Combining CRISPR/Cas12a with isothermal exponential amplification as an ultrasensitive sensing platform for microRNA detection. Sens Actuators B. 2022;367. 10.1016/j.snb.2022.132158.
- 122.Zhang G, Zhang L, Tong J, Zhao X, Ren J. CRISPR-Cas12a enhanced rolling circle amplification method for ultrasensitive miRNA detection. Microchem J. 2020;158. 10.1016/j.microc.2020.105239.
- 123.Huang M, Zhou X, Wang H, Xing D. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic ncid detection. Anal Chem. 2018;90:2193–200. 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 124.Yan X, Zhang J, Jiang Q, Jiao D, Cheng Y. Integration of the ligase chain reaction with the CRISPR-Cas12a system for homogeneous, ultrasensitive, and visual detection of microRNA. Anal Chem. 2022;94:4119–25. 10.1021/acs.analchem.2c00294. [DOI] [PubMed] [Google Scholar]
- 125.Sheng Y, Zhang T, Zhang S, Johnston M, Zheng X, Shan Y, Liu T, Huang Z, Qian F, Xie Z. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens Bioelectron. 2021;178:113027. 10.1016/j.bios.2021.113027. [DOI] [PubMed] [Google Scholar]
- 126.Zhou T, Huang M, Lin J, Huang R, Xing D. High-fidelity CRISPR/Cas13a trans-cleavage-triggered rolling circle amplified DNAzyme for visual profiling of microRNA. Anal Chem. 2021;93:2038–44. 10.1021/acs.analchem.0c03708. [DOI] [PubMed] [Google Scholar]
- 127.Chen X, Huang C, Zhang J, Hu Q, Wang D, You Q, Guo Y, Chen H, Xu J, Hu M. Mini crRNA-mediated CRISPR/Cas12a system (MCM-CRISPR/Cas12a) and its application in RNA detection. Talanta. 2024;268:125350. 10.1016/j.talanta.2023.125350. [DOI] [PubMed] [Google Scholar]
- 128.Long K, Cao G, Qiu Y, Yang N, Chen J, Yang M, Hou C, Huo D. Hybridization chain reaction circuit controller: CRISPR/Cas12a conversion amplifier for miRNA-21 sensitive detection. Talanta. 2024;266:125130. 10.1016/j.talanta.2023.125130. [DOI] [PubMed] [Google Scholar]
- 129.Qiu XY, Zhu LY, Zhu CS, Ma JX, Hou T, Wu XM, Xie SS, Min L, Tan DA, Zhang DY, Zhu L. Highly effective and low-cost microRNA detection with CRISPR-Cas9. ACS Synth Biol. 2018;7:807–13. 10.1021/acssynbio.7b00446. [DOI] [PubMed] [Google Scholar]
- 130.Huang M, Huang R, Yue H, Shan Y, Xing D. Ultrasensitive and high-specific microRNA detection using hyper-branching rolling circle amplified CRISPR/Cas13a biosensor. Sens Actuators B. 2020;325. 10.1016/j.snb.2020.128799.
- 131.Peng J, Liu T, Guan L, Xu Z, Xiong T, Zhang Y, Song J, Liu X, Yang Y, Hao X. A highly sensitive Lock-Cas12a biosensor for detection and imaging of miRNA-21 in breast cancer cells. Talanta. 2024;273:125938. 10.1016/j.talanta.2024.125938. [DOI] [PubMed] [Google Scholar]
- 132.Chen P, Wang L, Qin P, Yin BC, Ye BC. An RNA-based catalytic hairpin assembly circuit coupled with CRISPR-Cas12a for one-step detection of microRNAs. Biosens Bioelectron. 2022;207. 10.1016/j.bios.2022.114152. [DOI] [PubMed]
- 133.Chen M, Luo R, Li S, Li H, Qin Y, Zhou D, Liu H, Gong X, Chang J. Paper-based strip for ultrasensitive detection of OSCC-associated salivary microRNA via CRISPR/Cas12a coupling with IS-Primer amplification reaction. Anal Chem. 2020;92:13336–42. 10.1021/acs.analchem.0c02642. [DOI] [PubMed] [Google Scholar]
- 134.Li M, Luo N, Liao X, Zou L. Proximity hybridization-regulated CRISPR/Cas12a-based dual signal amplification strategy for sensitive detection of circulating tumor DNA. Talanta. 2023;257:124395. 10.1016/j.talanta.2023.124395. [DOI] [PubMed] [Google Scholar]
- 135.Wu X, Ju T, Li Z, Li J, Zhai X, Han K. Target-independent hybridization chain reaction-fluorescence resonance energy transfer for sensitive assay of ctDNA based on Cas12a. Anal Chim Acta. 2023;1261. 10.1016/j.aca.2023.341170. [DOI] [PubMed]
- 136.Elisei R, Ugolini C, Viola D, Lupi C, Biagini A, Giannini R, Romei C, Miccoli P, Pinchera A, Basolo F. BRAFV600E mutation and outcome of patients with papillary thyroid carcinoma: a 15-year median follow-up study. J Clin Endocrinol Metab. 2008;93:3943–9. 10.1210/jc.2008-0607. [DOI] [PubMed] [Google Scholar]
- 137.Zhang W, Zhao S, Xie Z, Chen S, Huang Y, Zhao Z, Yi G. The fluorescence amplification strategy based on 3D DNA walker and CRISPR/Cas12a for the rapid detection of BRAF V600E. Anal Sci. 2022;38:1057–66. 10.1007/s44211-022-00131-5. [DOI] [PubMed] [Google Scholar]
- 138.Sun H, Zhou S, Liu Y, Lu P, Qi N, Wang G, Yang M, Huo D, Hou C. A fluorescent biosensor based on exponential amplification reaction-initiated CRISPR/Cas12a (EIC) strategy for ultrasensitive DNA methyltransferase detection. Anal Chim Acta. 2023;1239:340732. 10.1016/j.aca.2022.340732. [DOI] [PubMed] [Google Scholar]
- 139.Zhou S, Dong J, Deng L, Wang G, Yang M, Wang Y, Huo D, Hou C. Endonuclease-assisted PAM-free recombinase polymerase amplification coupling with CRISPR/Cas12a (E-PfRPA/Cas) for sensitive detection of DNA methylation. ACS Sens. 2022;7:3032–40. 10.1021/acssensors.2c01330. [DOI] [PubMed] [Google Scholar]
- 140.Wang X, Zhou S, Chu C, Yang M, Huo D, Hou C. Dual methylation-sensitive restriction endonucleases coupling with an RPA-assisted CRISPR/Cas13a system (DESCS) for highly sensitive analysis of DNA methylation and its application for point-of-care detection. ACS Sens. 2021;6:2419–28. 10.1021/acssensors.1c00674. [DOI] [PubMed] [Google Scholar]
- 141.Wu X, Zhao Y, Guo C, Liu C, Zhang Q, Chen Y, Liu Y, Zhang X, Ratio CRISPR. A ratiometric biochip based on CRISPR/Cas12a for automated and multiplexed detection of heteroplasmic SNPs in mitochondrial DNA. Biosens Bioelectron. 2023;241:115676. 10.1016/j.bios.2023.115676. [DOI] [PubMed] [Google Scholar]
- 142.Zhu Y, Lin Y, Gong B, Zhang Y, Su G, Yu Y. Dual toeholds regulated CRISPR-Cas12a sensing platform for ApoE single nucleotide polymorphisms genotyping. Biosens Bioelectron. 2024;255:116255. 10.1016/j.bios.2024.116255. [DOI] [PubMed] [Google Scholar]
- 143.Kim JJ, Hong JS, Kim H, Choi M, Winter U, Lee H, Im H. CRISPR/Cas13a-assisted amplification-free miRNA biosensor via dark-field imaging and magnetic gold nanoparticles. Sens Diagn. 2024;3:1310–8. 10.1039/d4sd00081a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luo B, Zhou J, Zhan X, Ying B, Lan F, Wu Y. Visual and colorimetric detection of microRNA in clinical samples based on strand displacement amplification and nanozyme-mediated CRISPR-Cas12a system. Talanta. 2024;277:126310. 10.1016/j.talanta.2024.126310. [DOI] [PubMed] [Google Scholar]
- 145.Choi JH, Lim J, Shin M, Paek SH, Choi JW. CRISPR-Cas12a-based nucleic acid amplification-free DNA biosensor via au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett. 2021;21:693–9. 10.1021/acs.nanolett.0c04303. [DOI] [PubMed] [Google Scholar]
- 146.Wang J, Gui C, Zhu J, Zhu B, Zhu Z, Jiang X, Chen D. A novel design of DNA duplex containing programmable sensing sites for nanopore-based length-resolution reading and applications for Pb2+ and cfDNA analysis. Analyst. 2023;148:4346–55. 10.1039/d3an01126g. [DOI] [PubMed] [Google Scholar]
- 147.Cho IH, Kim DH, Park S. Electrochemical biosensors: perspective on functional nanomaterials for on-site analysis. Biomater Res. 2020;24:1–12. 10.1186/s40824-019-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang J. Nanomaterial-based electrochemical biosensors. Analyst. 2005;130:421–6. 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 149.Zhu C, Yang G, Li H, Du D, Lin Y. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal Chem. 2015;87:230–49. 10.1021/ac5039863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao X, Pachfule P, Thomas A. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Chem Soc Rev. 2021;50:6871–913. 10.1039/d0cs01569e. [DOI] [PubMed] [Google Scholar]
- 151.Zhang H, Gao H, Mu W, Que L, Gu X, Rong S, Ma H, Miao M, Qi X, Chang D, Pan H. Electrochemical-fluorescent bimodal biosensor based on dual CRISPR-Cas12a multiple cascade amplification for ctDNA detection. Anal Chem. 2024. 10.1021/acs.analchem.4c03012. [DOI] [PubMed] [Google Scholar]
- 152.Uygun ZO, Yeniay L, Gi̇rgi̇n Sağın F. CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs. Anal Chim Acta. 2020;1121:35–41. 10.1016/j.aca.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 153.Dong J, Li X, Hou C, Hou J, Huo D. A novel CRISPR/Cas12a-mediated ratiometric dual-signal electrochemical biosensor for ultrasensitive and reliable detection of circulating tumor deoxyribonucleic acid. Anal Chem. 2024;96:6930–9. 10.1021/acs.analchem.3c05700. [DOI] [PubMed] [Google Scholar]
- 154.Wei Y, Fu Y, Li C, Chen S, Xie L, Chen M. Ultrasensitive detection of circulating tumor DNA using a CRISPR/Cas9 nickase-driven 3D DNA walker based on a COF-AuNPs sensing platform. Mikrochim Acta. 2024;191:671. 10.1007/s00604-024-06749-8. [DOI] [PubMed] [Google Scholar]
- 155.Zhao JJ, Zhu JF, Wang WQ, Qian ZF, Fan ST. CRISPR/Cas12a cleavage triggered nanoflower for fluorescence-free and target amplification-free biosensing of ctDNA in the terahertz frequencies. Biomed Opt Express. 2024;15:5400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Balderston S, Taulbee JJ, Celaya E, Fung K, Jiao A, Smith K, Hajian R, Gasiunas G, Kutanovas S, Kim D. Discrimination of single-point mutations in unamplified genomic DNA via Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Eng. 2021;5:713–25. 10.1038/s41551-021-00706-z. [DOI] [PubMed] [Google Scholar]
- 157.Jia HY, Zhao HL, Wang T, Chen PR, Yin BC, Ye BC. A programmable and sensitive CRISPR/Cas12a-based microRNA detection platform combined with hybridization chain reaction. Biosens Bioelectron. 2022;211:114382. 10.1016/j.bios.2022.114382. [DOI] [PubMed] [Google Scholar]
- 158.Shao N, Han X, Song Y, Zhang P, Qin L. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal Chem. 2019;91:12384–91. 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- 159.Li YR, Zhao ZZ, Liu YJ, Wang N, Man SL, Ma L, Wang S. CRISPR/Cas system: the accelerator for the development of non-nucleic acid target detection in food safety. J Agric Food Chem. 2023;71:13577–94. 10.1021/acs.jafc.3c03619. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yang B, Kong J, Fang X, Programmable. Nat Commun. 2022;13:3999. 10.1038/s41467-022-31740-3. CRISPR-Cas9 microneedle patch for long-term capture and real-time monitoring of universal cell-free DNA. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.