Abstract
Objectives: Hepatocellular carcinoma (HCC) mostly developsfrom cirrhosis, so we compared the miRNA profiles of patients with cirrhosis who developed HCC with those who did not, and screened for miR-145-5p, which may be involved in the progression of liver cancer. The study’s purpose was to explore the mechanism of miR-145-5p in cirrhosis that becomes HCC. Methods: Cell counting kit 8 (CCK-8) and clone formation assays were employed to calculate cell proliferative ability. Quantitative real-time PCR (qRT-PCR) was conducted to measure the mRNA levels of miR-145-5p, plasminogen activator inhibitor 1 (PAI-1), and Circular RNA PVT1 (circPTV1). Results: MiR-145-5p was downregulated in HCC, and miR-145-5p inhibited Huh-7 cell viability and clone formation. The Area Under the Curve (AUC) of miR-145-5p for distinguishing between the HCC and Tumor-free groups was 0.900, indicating a high diagnostic accuracy. PAI-1 was identified as a downstream target of miR-145-5p. Silencing PAI-1 decreased the viability and clone formation of Huh-7 cells. circPVT1 directly binding to miR-145-5p in HuH-7 cells. circPVT1 regulates cell viability and clone formation through miR-145-5p. In summary, we identified miRNA miR-145-5p, which was lowly expressed in HCC, and inhibited the viability and clone formation of HCC cells. Conclusion: Our research elucidates the regulatory role of the circPVT1/miR-145-5p/PAI-1 axis in HCC, suggesting its potential as a novel diagnostic biomarker or therapeutic target for HCC.
Keywords: miR-145-5p, PAI-1, hepatocellular carcinoma, viability, clone formation
Introduction
Hepatocellular carcinoma (HCC) is a highly aggressive invasive malignancy that has a poor prognosis, and rapid progression. In China, HCC has a high prevalence and ranks third in mortality among malignant tumors. As an inflammation-associated neoplasm, over 80% of HCC cases are linked to liver cirrhosis and hepatitis, with HCC developing in 3%-5% of cirrhotic patients annually [1]. Nevertheless, not all cases of hepatitis and liver fibrosis progress to HCC, as only one-third of cirrhotic individuals ultimately develop this malignancy [2]. The precise mechanisms responsible for the progression of liver cirrhosis to hepatocellular carcinoma (HCC) remain unclear. Thus, there is a pressing need for further investigation into the etiology and pathogenesis linking liver cirrhosis and HCC, as well as the implementation of appropriate preventive and therapeutic interventions to impede the transition from liver cirrhosis to HCC.
MicroRNAs, which are small non-coding RNA molecules consisting of 18-25 nucleotides, play a crucial role in gene expression regulation by modulating protein translation and mRNA cleavage [3]. Due to their ability to meet multiple criteria for ideal biomarkers, such as ease of use and sensitivity, microRNAs have been identified as promising biomarkers for a variety of human diseases. MicroRNAs may function as tumor suppressors or oncogenes, involved in regulating tumorigenesis and the development of human cancers [4]. Additionally, microRNAs exhibit a high level of specificity to tissue or cell type, suggesting the potential for using specific microRNAs to elucidate disease onset and progression [5]. The demonstrated sensitivity of microRNAs and their ability to fluctuate in response to disease progression or therapeutic intervention underscores the potential for non-invasive for accurate disease diagnosis and monitoring. Previous research has demonstrated miR-145-5p has tumor suppressor roles in multiple malignancies, including ovarian cancer, colorectal cancer, breast cancer, prostate cancer, and cervical cancer [6-10].
Plasminogen activator inhibitor-1 (PAI-1 or SERPIN E1) is a fast and specific inhibitor of the serine proteases urokinase-type (uPA) and tissue-type plasminogen activator (tPA). Both uPA and tPA convert the inactive zymogen plasminogen into the active serine protease plasmin, which degrades extracellular proteins such as fibrin and laminin. Accordingly, plasminogen activation plays important physiologic roles in fibrinolysis and tissue remodeling; PAI-1 is one of the most useful biochemical prognostic markers in several types of cancers [11]. In a multicentre prospective randomized trial of node-negative breast cancer, patients with low levels of uPA and PAI-1 in their primary tumor had a very good prognosis, thus uPA and PAI-1 may be candidates for adjuvant chemotherapy. In contrast, the risk of disease recurrence for node-negative patients with high levels of uPA and PAI-1 is similar to the risk for patients with three or more tumour cell positive axillary lymph nodes [12]. The malignant cells’ production of uPA aligns with the traditional model of uPA bound to uPAR on their surface, facilitating pericellular proteolysis and promoting invasion. High PAI-1 levels are associated with poor prognoses in oral cancer, which is consistent with the hypothesis that malignant cells express PAI-1 to promote tumor growth, invasion, and metastasis by protecting the malignant cells against uPA-mediated proteolysis [13,14].
The purpose of this study was to evaluate the effects of miR-145-5p on the viability and clone formation of HCC cells. We further experimentally identified a novel mechanism through which miR-145-5p suppresses clone formation by directly targeting PAI-1 based on the bioinformatic analysis of its putative target genes. According to our research, the miR-145-5p/PAI-1 pair may serve as targets for slowing HCC proliferation.
Materials and methods
Cell culture
The human HCC cells HCC-LM3, Bel-7402, and Huh7, and normal hepatic cell line L-02, were obtained from the Shanghai Institute of Cell Biology. L-02 cells were grown in RPMI-1640 medium (Gibco) with 10% Fetal Bovine Serum (FBS) (Gibco), while HCC-LM3, Bel-7402, and Huh7 cells were grown in DMEM (Gibco) with 10% FBS. All cells were incubated at 37°C with 5% CO2. Authentication of the cells was performed using short tandem repeats (STR) profiling, and they were confirmed to be free of mycoplasma contamination.
RNA interference
The RiboBio Company (Guangzhou, China) synthesized siRNAs and mimics of indicated miRNAs. A riboFECTTM CP (RiboBio) was used for transfecting siRNAs and miRNAs.
Cell counting kit 8 (CCK-8) assay
Following transfection, Huh-7 cells were distributed into 96-well plates at a density of 1.0 × 103 cells per well and incubated in DMEM medium supplemented with 10% FBS for time intervals of 12, 24, 48, and 72 hours. Cell proliferation was assessed using the CCK8 detection kit (Engreen Biosystem Co., Ltd., China), with absorbance readings taken at 450 nm using a microplate reader (Molecular Devices, USA).
Clone formation assay
In six-well plates, 600 cells were seeded per well and maintained in DMEM medium supplemented with 10% FBS at 37°C in an incubator with 5% CO2 for a duration of 2 weeks, with medium replacement every 3 days. Following two washes with PBS, crystal violet was used to stain the fixed cells after they were fixed in methanol for 15 minutes at 25°C. Clone formation was then quantified manually.
RNA extraction and qRT-PCR
Total RNA samples were extracted using TRIzolTM reagent (Life Technologies, USA). PrimeScriptTM RT Master Mix (Takara, Japan) was used to carry out the reverse transcription, and TB GreenTM Premix Ex TaqTM (Takara, Japan) was used to perform quantitative real-time PCR (qRT-PCR) on an ABI PRISM 7500 real-time PCR System (Applied Biosystems, USA). Using the 2-ΔΔCt method and GAPDH as a housekeeping gene, relative expression levels were determined. The extracted total RNA was stored at -80°C for future experiments.
The miRNeasy Mini Kit (Qiagen) was employed for miRNA extraction. The miRNA RT-PCR Quantitation Kit (Qiagen) was used to perform subsequent reverse transcription and qRT-PCR analyses on miRNAs, including the internal reference U6. The amplification process involved an initial denaturation step at 95°C for 3 minutes, followed by 40 cycles of a melting temperature of 95°C for 15 seconds and an annealing temperature of 62°C for 34 seconds.
Luciferase reporter assay
Online software Targetscan website (http://www.targetscan.org/vert_72/) predicted the putative binding sites between PAI-1 and miR-145-5p, while Circular RNA Interactome (https://circinteractome.nia.nih.gov/) identified the binding sites between circPVT1 and miR-145-5p. To produce the pmir-GLO-PAI-1 (PAI-1-WT) or pmir-GLO-circPVT1 (LYAR-WT) reporter vectors, wild-type PAI-1 and circPVT1 containing predicted miR-145-5p binding sites were constructed and cloned into the pmir-GLO Dual-luciferase vector (Promega, USA). To create PAI-1-MUT and circPVT1-MUT reporter vectors, by using a GeneTailor Site-Directed Mutagenesis System (Invitrogen), we synthesized mutated sequences for integration into the dual-luciferase pmir-GLO vector. With Lipofectamine 3000, PAI-1-WT, PAI-1-MUT, or circPVT1-WT reporter vectors were co-transfected with miR-145-5p mimic or negative control (NC mimic) into Huh-7 cells. Next, a 48-h luciferase assay kit (Promega) was used to measure luciferase activity, and the results were normalized to Renilla luciferase activity.
Statistical analysis
Data analysis was conducted using SPSS software version 16. Each experiment was replicated at least three times, and results were reported as mean ± standard deviation. Statistical significance was assessed using the χ2-Test and Student’s t-test. A p-value of less than 0.05 was deemed significant.
Results
MiR-145-5p was downregulated in HCC and may act as a marker for the diagnosis of HCC
In our prior investigation, we conducted a miRNA data analysis of cirrhosis and liver cancer utilizing the GEO dataset GSE116054. The study encompassed 49 patients with liver fibrosis and cirrhosis. Through long-term follow-up, 25 patients who eventually evolved into HCC were designated as the HCC group, alongside an additional 24 patients who did not progress to HCC, designated as the Tumor-free group. Through examination of miR-145-5p levels in both groups, a notable downregulation of miR-145-5p was observed in the majority of HCC tissues in comparison to the Tumor-free group (P<0.05) (Figure 1A). The study used Receiver Operating Characteristic (ROC) analysis to determine that the Area Under the Curve (AUC) of miR-145-5p in distinguishing between the HCC group and the Tumor-free group was 0.900, suggesting the potential diagnostic utility of miR-145-5p for HCC (Figure 1B). Additionally, the study demonstrated downregulation of miR-145-5p in HCC cell lines HCC-LM3, Bel-7402, and Huh7 compared to a normal hepatic cell line L-02, as illustrated in Figure 1C. Subsequently, to investigate the involvement of miR-145-5p in the development of liver cirrhosis and HCC, Huh-7 cells exhibiting the lowest expression levels of miR-145-5p were chosen for further analysis. miR-145-5p was either upregulated or downregulated using mimic and inhibitor techniques. MiR-145-5p was markedly increased and silenced by mimic and inhibitor, as illustrated in Figure 1D and 1E.
Figure 1.
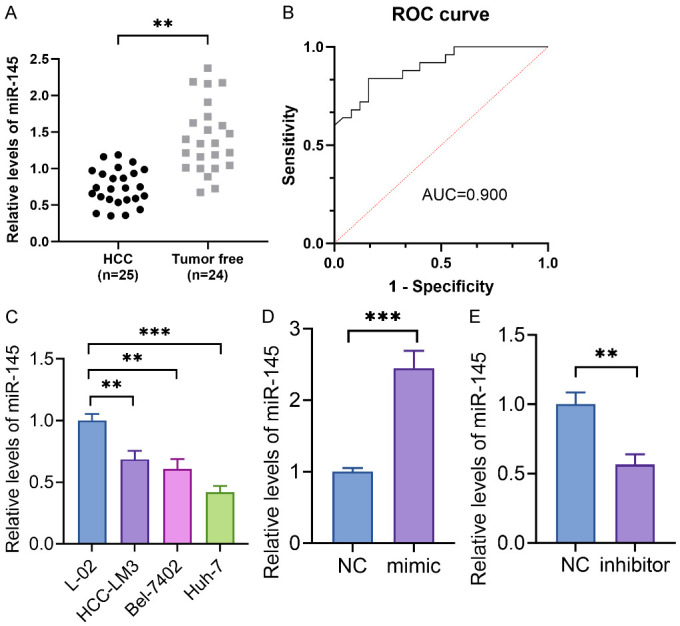
MiR-145-5p was downregulated in HCC and may act as a marker for the diagnosis of HCC. A: The miR-145-5p levels in HCC and tumor-free groups. B: The ROC curve of miR-145-5p in diagnosing HCC and liver cirrhosis. C: miR-145-5p was downregulated in HCC cell lines HCC-LM3, Bel-7402, Huh7, and a normal hepatic cell line L-02. D and E: miR-145-5p was markedly increased and silenced by mimic and inhibitor. **P<0.01; ***P<0.001.
MiR-145-5p suppressed Huh-7 cell proliferation
To explore the function of miR-145-5p in HCC, function experiments in vitro were performed in Huh-7 cells. We designed specific mimics and inhibitors that targeted miR-145-5p and transfected them into Huh-7 cells. Next, we applied CCK8 and clone formation assays to determine the effect of miR-145-5p on Huh-7 cell proliferation. As the representative photos show in Figure 2A and 2C, miR-145-5p mimic significantly inhibited Huh-7 cell viability (P<0.05); meanwhile, the cloned cell number was also remarkably reduced (P<0.05). On the contrary, silencing of miR-145-5p obviously improved Huh-7 cell viability (P<0.05) (Figure 2B) and cloned cell number (P<0.05) (Figure 2D).
Figure 2.

MiR-145-5p suppressed Huh-7 cell proliferation. A: miR-145-5p mimic significantly inhibited Huh-7 cell viability. B: miR-145-5p inhibitor enhanced the viability of Huh-7 cells. C: miR-145-5p mimic significantly inhibited the cloned cell number of Huh-7 cells. D: miR-145-5p inhibitor enhanced the cloned cell number of Huh-7 cells. *P<0.05; **P<0.01.
PAI-1 was upregulated in HCC tissues and cell lines
To deepen our understanding of the role of miR-145-5p in Huh-7 cells, we conducted a further analysis of the GSE116054 dataset. Our findings indicate a significant upregulation of PAI-1 mRNA levels in tissues of patients with HCC (n=25) compared to patients in tumor-free groups (n=24) (Figure 3A), suggesting a role for PAI-1 in promoting the progression of liver cirrhosis to HCC by exacerbating liver fibrosis. Subsequent examination of PAI-1 levels in HCC cell lines revealed a similar pattern, with markedly elevated levels observed in HCC cells compared to normal cells, particularly in Huh-7 cells (Figure 3B). Inhibition of PAI-1 expression using siRNA (si-PAI-1) resulted in decreased viability and clone formation of Huh-7 cells (Figure 3C, 3D).
Figure 3.

Silencing PAI-1 decreased the viability and clone formation. A: PAI-1 was upregulated in HCC tissues of patients with HCC. B: PAI-1 markedly elevated in HCC cells compared to normal cells, particularly in Huh-7 cells. C: Inhibition of PAI-1 expression using siRNA (si-PAI-1) decreased the viability of Huh-7 cells. D: Silencing of PAI-1 decreased the clone formation. *P<0.05; **P<0.01; ***P<0.001.
PAI-1 was involved in miR-145-5p-mediated proliferation in Huh-7 cells
To further investigate the relationship between PAI-1 and miR-145-5p, bioinformatic analysis revealed complementary binding sites between miR-145-5p and the 3’-UTR of PAI-1 (Figure 4A). It is hypothesized that miR-145-5p may modulate liver fibrosis by targeting PAI-1, thereby influencing the progression from liver cirrhosis to liver cancer. Through a luciferase assay, we demonstrated direct targeting of miR-145-5p by PAI-1. Additionally, only the wild-type PAI-1 plasmids, pmir-GLO-PAI-1 (WT), resulted in reduced luciferase activity. When mutations were introduced to the theoretical binding sites provided by PAI-1 for miR-145-5p, there was no change in luciferase activity (Figure 4B). A correlation analysis of mRNA levels in 25 HCC patients revealed a negative correlation between miR-145-5p and PAI-1 (Figure 4C). Additionally, transfection with a miR-145-5p mimic reduced the mRNA levels of PAI-1 (Figure 4D), while transfection with a miR-145-5p inhibitor increased PAI-1 mRNA levels (Figure 4E), indicating a possible regulatory relationship between miR-145-5p and PAI-1.
Figure 4.
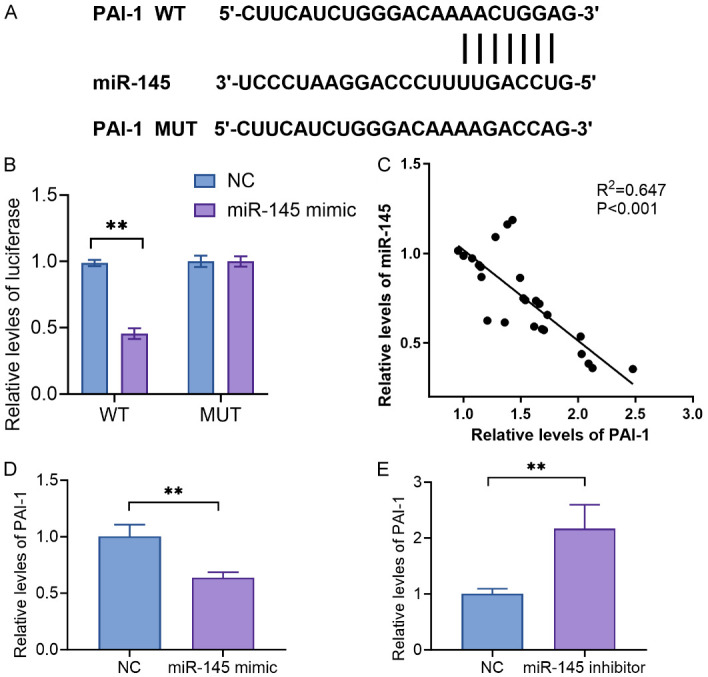
PAI-1 is involved in miR-145-5p-mediated viability and clone formation in Huh-7 cells. A: Bioinformatic analysis revealed complementary binding sites between miR-145-5p and the 3’-UTR of PAI-1. B: miR-145-5p directly targeted PAI-1 based on a luciferase assay. C: miR-145-5p negatively correlated with PAI-1 in 25 HCC patients. D: Transfection with a miR-145-5p mimic reduced the mRNA levels of PAI-1. E: Transfection with a miR-145-5p inhibitor increased PAI-1 mRNA levels. **P<0.01.
circPVT1 directly targets miR-145-5p in HuH-7 cells
Circular RNA interactome (https://circinteractome.nia.nih.gov/) showed circPVT1 binds directly to miR-145-5p using online bioinformatics analysis software. The miR-145-5p binding sequences on circPV-T1 were mutated from 5’-…ACUGGA…-3’ to 5’-…ACACCUA…-3’ (Figure 5A). Next, we co-transfected two plasmid vectors with WT or MUT circPVT1 and miR-145-5p mimic in HuH-7 cells, and then detected the luciferase activity. We found that the luciferase activity was reduced when co-transfected miR-145-5p mimic and circPVT1-WT 3’UTR. On the contrary, in HuH-7 cells co-transfected with miR-145-5p mimic and circPVT1-MUT 3’UTR, luciferase activity remained unchanged (Figure 5B). The levels of circPVT1 were also found to be upregulated in the HCC group compared to the tumor-free group (Figure 5C). The correlation analysis between circPVT1 and miR-145-5p was performed in the HCC group, and we found circPVT1 had a miR-145-5p negative correlation (Figure 5D). Also, circPVT1 had a positive correlation with PAI-1 in HCC group (Figure 5E).
Figure 5.

circPVT1 directly targeted miR-145-5p in HuH-7 cells. A: circPVT1 directly binds to miR-145-5p. B: Co-transfected WT or MUT of circPVT1 3’UTR and miR-145-5p mimic in HuH-7 cells to confirm that miR-145-5p binding to circPVT1. C: circPVT1 was found to be upregulated in the HCC group compared to tumor-free group. D: circPVT1 had a negative correlation with miR-145-5p in the HCC group. E: circPVT1 had a positive correlation with PAI-1 in the HCC group. F: circPVT1 was upregulated in HCC cells versus normal cells. G: Silencing circPVT1 could increase the mRNA level of miR-145-5p, and overexpression of circPVT1 could inhibit the mRNA level of miR-145-5p. H: Downregulating circPVT1 reduced PAI-1, while upregulating circPVT1 enhanced PAI-1. *P<0.05; **P<0.01; ***P<0.001.
The levels of circPVT1 in cell lines were measured. Similar to that of tissues, circPVT1 was upregulated in HCC cells versus normal cells (Figure 5F). Moreover, silencing circPVT1 could increase the mRNA level of miR-145-5p, and conversely, circPVT1 overexpression could inhibit the mRNA level of miR-145-5p (Figure 5G). On the contrary, downregulating circPVT1 reduced PAI-1, while upregulating circPVT1 enhanced PAI-1 (Figure 5H).
circPVT1 regulates cell proliferation through miR-145-5p
To verify the effect of circPVT1 on regulating cell viability and clone formation by targeting miR-145-5p, pcDNA3.1-circPVT1 was transfected into HuH-7 cells that were transfected with miR-145-5p mimic (Figure 6A). CCK-8 assay demonstrated that co-transfecting pcDNA3.1-circPVT1 increased the proliferative ability in HuH-7 cells transfecting with miR-145-5p mimic (Figure 6B). In addition, the cloned number was also improved by co-transfecting pcDNA3.1-circPVT1 and miR-145-5p mimic compared to only transfected miR-145-5p mimic (Figure 6C). Thus, all the results showed that circPVT1 regulated miR-145-5p-mediated regulation of HCC cells.
Figure 6.

circPVT1 regulates cell viability and clone formation through miR-145-5p. A: pcDNA3.1-circPVT1 was transfected into HuH-7 cells that were transfected with miR-145-5p mimic. B: Co-transfecting pcDNA3.1-circPVT1 increased the proliferative ability in HuH-7 cells transfected with miR-145-5p mimic. C: The cloned number was also improved by co-transfecting pcDNA3.1-circPVT1 and miR-145-5p mimic compared to only transfected miR-145-5p mimic. *P<0.05; **P<0.01.
Discussion
In our prior investigation, we analyzed miRNA data from patients with cirrhosis-liver cancer using the GEO dataset, incorporating a cohort of 49 individuals with liver fibrosis and cirrhosis. Following an extended period of monitoring disease advancement, the patients were stratified into two categories: the tumor-free group, comprising patients diagnosed with cirrhosis who did not develop HCC, and the HCC group, consisting of patients diagnosed with cirrhosis who subsequently progressed to HCC. Through a comparative analysis of miRNA expression in liver tissues from two distinct groups, it was observed that miR-145-5p exhibited a significant increase as the disease progressed within the Tumor free group. This suggests a potential correlation between the up-regulation of miR-145-5p levels and the reduced risk of hepatocellular carcinoma in liver cirrhosis within the tumor-free group. Previous research indicated that miR-145-5p played an important role in tumor growth and aggressiveness, possibly serving as biomarkers and therapeutic targets [15-17]. Consistent with the findings, we found that miR-145-5p was downregulated in HCC tissues and cell lines. By ROC curves, miR-145-5p was discovered to be a diagnostic biomarker for HCC, with an AUC of 0.900. In Huh-7 cells, upregulating miR-145-5p inhibited cell viability and clone formation, while downregulating miR-145-5p enhanced them.
In order to deepen our understanding of the role of miR-145-5p in Huh-7 cells, we conducted additional analysis of the mRNA expression profile within the GSE116054 dataset. Our findings indicate that miR-145-5p directly targets the downstream gene PAI-1, as evidenced by the decreased mRNA levels of PAI-1 following transfection with a miR-145-5p mimic and increased levels following transfection with a miR-145-5p inhibitor. Furthermore, knockdown of PAI-1 resulted in a reduction in cell viability and clone formation. Plasminogen Activator (PA) has been identified as a significant gene implicated in the progression of liver cirrhosis to hepatocellular carcinoma (HCC). Research indicates that the plasminogen activator (PA)/plasmin system plays a crucial role in regulating the activity of matrix metalloproteinases (MMPs) and extracellular matrix (ECM) degradation. PA has the ability to activate plasmin, which in turn activates MMPs, whereas plasminogen activator inhibitor-1 (PAI-1) can inhibit MMP activation [18,19]. Hepatic stellate cells (HSC) play a crucial role in the regulation of hepatic extracellular matrix (ECM) deposition through the expression of plasminogen activator (PA) and plasminogen activator inhibitor-1 (PAI-1). The activation of HSCs serves as a pivotal factor in the pathogenesis of liver fibrosis. As liver fibrosis advances, the upregulation of PAI-1 outpaces that of PA, resulting in impaired ECM degradation and exacerbation of liver fibrosis progression [20,21]. Prior studies have demonstrated that enhancing PA expression and modulating PAI-1 levels can effectively ameliorate fatty liver and liver necrosis [22].
Circular RNAs (circRNAs) are a new subtype of ncRNAs that lack both polyadenylation and 5’-3’ polarity. According to the ceRNA hypothesis, circRNAs may act as miRNA sponges, regulating downstream target gene transcription. CircPVT1 enhanced ccRCC proliferative and invasive phenotype by sponging miR-145-5p [23]. Similarly, in CRC and endometrial cancer, circPVT1 promoted tumor progression by sponging miR-145-5p [24,25]. In our study, using dual-luciferase reporter assays, wt and miR-145 mimics significantly reduced luciferase intensity, whereas mut or miRNA mimics did not reduce it significantly, demonstrating that circPVT1 binds to miR-145-5p in HCC cells. CircPVT1 is predominantly located in the cytoplasm of cells. On the other hand, PAI-1 is primarily found in the extracellular environment, where it plays a significant role in blood circulation by interacting with the fibrinolytic system and regulating fibrin degradation. Rescue experiments revealed that the overexpression of circPVT1 promoted cell viability and clone formation that was dependent on miR-145.
In summary, we identified a miRNA miR-145-5p, that was lowly expressed in HCC, and inhibited the viability and clone formation of HCC cells. As a result of being competitively sponged by circPVT1, miR-145-5p inhibits the progression of HCC by regulating the expression of its target gene PAI-1. Our research demonstrates that the circPVT1/miR-145-5p/PAI-1 axis operates as a regulator and contributes to HCC development, possibly providing a new biomarker for diagnosis, and a therapeutic target.
Acknowledgements
This work was supported by Shanxi Province Natural Science Research Facing Project (Project Number: 202103021224421).
Disclosure of conflict of interest
None.
References
- 1.Dopazo C, Søreide K, Rangelova E, Mieog S, Carrion-Alvarez L, Diaz-Nieto R, Primavesi F, Stättner S. Hepatocellular carcinoma. Eur J Surg Oncol. 2024;50:107313. doi: 10.1016/j.ejso.2023.107313. [DOI] [PubMed] [Google Scholar]
- 2.Baraka K, Abozahra RR, Badr E, Abdelhamid SM. Study of some potential biomarkers in Egyptian hepatitis C virus patients in relation to liver disease progression and HCC. BMC Cancer. 2023;23:938. doi: 10.1186/s12885-023-11420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kar S, Mukherjee R, Guha S, Talukdar D, Das G, Murmu N. Modulating the acetylation of α-tubulin by LncRNAs and microRNAs helps in the progression of cancer. Cell Biochem Funct. 2024;42:e3953. doi: 10.1002/cbf.3953. [DOI] [PubMed] [Google Scholar]
- 4.Fanoodi A, Maharati A, Akhlaghipour I, Rahimi HR, Moghbeli M. MicroRNAs as the critical regulators of tumor angiogenesis in liver cancer. Pathol Res Pract. 2023;251:154913. doi: 10.1016/j.prp.2023.154913. [DOI] [PubMed] [Google Scholar]
- 5.Kim T, Croce CM. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023;55:1314–1321. doi: 10.1038/s12276-023-01050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao S, Zhang Y, Pei M, Wu L, Li J. miR-145 inhibits mitochondrial function of ovarian cancer by targeting ARL5B. J Ovarian Res. 2021;14:8. doi: 10.1186/s13048-020-00762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mozammel N, Amini M, Baradaran B, Mahdavi SZB, Hosseini SS, Mokhtarzadeh A. The function of miR-145 in colorectal cancer progression; an updated review on related signaling pathways. Pathol Res Pract. 2023;242:154290. doi: 10.1016/j.prp.2022.154290. [DOI] [PubMed] [Google Scholar]
- 8.Qu H, Li X, Chen F, Zhang M, Lu X, Gu Y, Lv M, Lu C. LncRNA PVT1 influences breast cancer cells glycolysis through sponging miR-145-5p. Genes Genomics. 2023;45:581–592. doi: 10.1007/s13258-023-01368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng H, Huang Y, Liu Q, Liu H, Long T, Zhu C, Wu X. MiR-145 suppresses the motility of prostate cancer cells by targeting cadherin-2. Mol Cell Biochem. 2021;476:3635–3646. doi: 10.1007/s11010-021-04188-0. [DOI] [PubMed] [Google Scholar]
- 10.Hu C, Liu T, Zhang W, Sun Y, Jiang D, Zhang X, Liu Y, Mao S, Xu Y, Pan J, Wang J, Huang Y, Yang S, Yang K. miR-145 inhibits aerobic glycolysis and cell proliferation of cervical cancer by acting on MYC. FASEB J. 2023;37:e22839. doi: 10.1096/fj.202201189RR. [DOI] [PubMed] [Google Scholar]
- 11.Thu YM, Suzawa K, Tomida S, Ochi K, Tsudaka S, Takatsu F, Date K, Matsuda N, Iwata K, Nakata K, Shien K, Yamamoto H, Okazaki M, Sugimoto S, Toyooka S. PAI-1 mediates acquired resistance to MET-targeted therapy in non-small cell lung cancer. PLoS One. 2024;19:e0300644. doi: 10.1371/journal.pone.0300644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbeck N, Kates RE, Gauger K, Willems A, Kiechle M, Magdolen V, Schmitt M. Urokinase-type plasminogen activator (uPA) and its inhibitor PAI-I: novel tumor-derived factors with a high prognostic and predictive impact in breast cancer. Thromb Haemost. 2004;91:450–456. doi: 10.1160/TH03-12-0798. [DOI] [PubMed] [Google Scholar]
- 13.Chin D, Boyle GM, Williams RM, Ferguson K, Pandeya N, Pedley J, Campbell CM, Theile DR, Parsons PG, Coman WB. Novel markers for poor prognosis in head and neck cancer. Int J Cancer. 2005;113:789–797. doi: 10.1002/ijc.20608. [DOI] [PubMed] [Google Scholar]
- 14.Hundsdorfer B, Zeilhofer HF, Bock KP, Dettmar P, Schmitt M, Kolk A, Pautke C, Horch HH. Tumour-associated urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in normal and neoplastic tissues of patients with squamous cell cancer of the oral cavity - clinical relevance and prognostic value. J Craniomaxillofac Surg. 2005;33:191–196. doi: 10.1016/j.jcms.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Yadav AK, Singh N, Yadav SK, Bhatt MLB, Pandey A, Yadav DK, Yadav S. Expression of miR-145 and miR-18b in peripheral blood samples of head and neck cancer patients. Indian J Clin Biochem. 2023;38:528–535. doi: 10.1007/s12291-023-01119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong L, Willoughby CE, McKenna DJ. The suppression of the epithelial to mesenchymal transition in prostate cancer through the targeting of MYO6 using MiR-145-5p. Int J Mol Sci. 2024;25:4301. doi: 10.3390/ijms25084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou W, Yang Y, Wang W, Yang C, Cao Z, Lin X, Zhang H, Xiao Y, Zhang X. Pseudogene OCT4-pg5 upregulates OCT4B expression to promote bladder cancer progression by competing with miR-145-5p. Cell Cycle. 2024;23:645–661. doi: 10.1080/15384101.2024.2353554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhar D, Baglieri J, Kisseleva T, Brenner DA. Mechanisms of liver fibrosis and its role in liver cancer. Exp Biol Med (Maywood) 2020;245:96–108. doi: 10.1177/1535370219898141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nahm JH, Lee HS, Kim H, Yim SY, Shin JH, Yoo JE, Ahn SH, Choi JS, Lee JS, Park YN. Pathological predictive factors for late recurrence of hepatocellular carcinoma in chronic liver disease. Liver Int. 2021;41:1662–1674. doi: 10.1111/liv.14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewidar B, Meyer C, Dooley S, Meindl-Beinker AN. TGF-β in hepatic stellate cell activation and liver fibrogenesis-updated 2019. Cells. 2019;8:1419. doi: 10.3390/cells8111419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwabe RF, Tabas I, Pajvani UB. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology. 2020;158:1913–1928. doi: 10.1053/j.gastro.2019.11.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EH, Park KI, Kim KY, Lee JH, Jang EJ, Ku SK, Kim SC, Suk HY, Park JY, Baek SY, Kim YW. Liquiritigenin inhibits hepatic fibrogenesis and TGF-β1/Smad with Hippo/YAP signal. Phytomedicine. 2019;62:152780. doi: 10.1016/j.phymed.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Z, Chen Z, Zhong Q, Zhu D, Xie Y, Shangguan W, Xie W. CircPVT1 promotes progression in clear cell renal cell carcinoma by sponging miR-145-5p and regulating TBX15 expression. Cancer Sci. 2021;112:1443–1456. doi: 10.1111/cas.14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Su M, Xiang B, Zhao K, Qin B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem Biophys Res Commun. 2019;512:716–722. doi: 10.1016/j.bbrc.2019.03.121. [DOI] [PubMed] [Google Scholar]
- 25.Wang YY, Duan H, Wang S, Quan YJ, Huang JH, Guo ZC. Elevated circular RNA PVT1 promotes eutopic endometrial cell proliferation and invasion of adenomyosis via miR-145/Talin1 axis. Biomed Res Int. 2021;2021:8868700. doi: 10.1155/2021/8868700. [DOI] [PMC free article] [PubMed] [Google Scholar]


