Abstract
The genus Preussia is widely distributed and includes species with ecological and biotechnological importance. In this study, morphological and phylogenetic analyses of the ITS and LSU rDNA sequences revealed two novel species, P. jejuensis sp. nov. and P. koreensis sp. nov., as well as one previously unrecorded species in Korea, P. isomera, from horse dung collected in Seopjikoji, Jeju Island, Republic of Korea. P. jejuensis sp. nov. is unique in producing conidia from conidiomata instead of teleomorphic structures, a feature not observed in any other known species of the Preussia genus. P. koreensis sp. nov. is morphologically distinguished by the absence of a neck, a smaller ascocarp diameter, smaller asci, and larger ascospores. In the phylogenetic analysis, P. jejuensis sp. nov. was closely related to P. isomera and S. minimoides, while P. koreensis sp. nov. was closely related to P. arizonica, P. persica, and S. minima. However, P. jejuensis sp. nov. and P. koreensis sp. nov. were clearly distinguished from their related species. This study expands the understanding of the biodiversity of coprophilous fungi and its distribution in Jeju Island, a region where horse breeding has been practiced for centuries, emphasizing the ecological importance of dung as a fungal habitat.
Keywords: Horse dung, Preussia jejuensis sp. nov., P. koreensis sp. nov., P. isomera, Seopjikoji
1. Introduction
Coprophilous fungi are microorganisms that inhabit animal feces or feces-contaminated soil [1]. They are commonly found in the feces of livestock, such as cattle, horses, and sheep, as well as in the feces of wild mammals and birds [2]. Coprophilous fungi are directly involved in the decomposition of feces and participate in carbon, nitrogen, and energy cycling [3, 4]. They play a crucial role in the mineralization of herbivore feces [5]. In addition, coprophilous fungi interact with insects such as dung beetles and flies, contributing to the composition and stability of the ecosystem within dung habitats [6]. Coprophilous fungi also play an important role in nutrient decomposition and maintaining ecological balance through succession processes over time in dung habitats [7].
Sporomiaceae is a family that includes 25 genera, including Preussia, Sporormiella, Westerdykella, and Niesslella, and is widely distributed worldwide. Sporomiaceae are mainly known as saprophytes in various substrates such as decaying plant debris, soil, feces, etc. [8]. Sporomiaceae have been reported not only as saprophytes but also as plant endophytes existing inside various plants [9–11]. The genus Preussia, reported by Fuckel in 1867 [12], has been isolated from various sources, mainly wood, soil, dead plants, or feces [13–17]. The genus Preussia has been described as having the following morphological characteristics: bitunicate ascomycetes with non-ostiolate, globose to subglobose ascomata, 8-spored, broadly clavate or subglobose asci, and ascospores with germ slits that are mostly surrounded by a gelatinous sheath [12]. The genus Preussia was reexamined by Cain, and as a result, coprophilous species without ostioles were included in the genus Preussia, giving it a broader ecological concept [18]. Preussia is morphologically very similar to the Sporormiella genus, which has the morphological characteristics of coprophilous bitunicate ascomycetes with ostiolate perithecioid ascomata and cylindrical to cylindric claviform asci [19]. Morphological differences between the genera Preussia and Sporomiella include the presence or absence of ostioles in the ascocarps. However, in 1973, von Arx demonstrated that the formation of ostioles can vary depending on growth conditions, proving that the presence of ostioles in ascocarps is an incorrect classification criterion [20]. Later, von Arx and van der Aa incorporated the coprophilous species with ampulliform and ostiolate ascomata into the genus Sporormiella, and defined the species with turbinate, ostiolate or non-ostiolate ascomata occurring on plant debris, wood or soil as Preussia in 1987 [21]. However, regarding the classification criteria of von Arx and van der Aa for Preussia and Sporormiella, in 1997 Guarro et al. reported that P. aquilirostrata and P. intermedia, isolated from both soil and feces, showed both non-ostiolate and ostiolate ascomata under the same culture conditions [22]. Therefore, the classification criteria of von Arx and van der Aa were no longer clear classification criteria [17, 22]. However, in 2010, analysis of Asa Kruys and Mats Wedin, based on sequence analysis of the ITS-LSU, mtSSU, and β-tubulin genes made it possible to distinguish between the genera Preussia and Sporormiella [23]. Various species belonging to the genus Preussia are known to produce secondary metabolites that exhibit antibacterial activity, such as preussomerins [13, 14, 24–26].
Some species of the genus Preussia have been studied for their ability of producing secondary metabolites. Preeuciafurans dibenzofurans including A and B, spirobinaphthalene, spiropreussiones A and B and spiropreussomerin A were isolated from Preussia sp. [27, 28]. Preussiadins A, and B, A1 to A3, and leptosins A and C were isolated from P. typharum [29]. Ethanol extract of Preussia species showed antibacterial and antiplasmodial activity including cytotoxicity [27–29]. Recently, benzyl 2,4-di(benzyloxy)benzoate and six known compounds (2‒7) were isolated from Preussia sp. found in frozen soil of the Himalaya Mountain [30]
In the Republic of Korea, Preussia typharum (NNIBRFG4211) is the only species reported so far among the genus Preussia [31]. The strain isolated from freshwater environment in Jecheon showed laccase activity. We have been studying biodiversity of fungi in Jeju Island in the Republic of Kore, an UNESCO World Natural Heritage [32]. Historically, Jeju Island has also been famous as the place for horse raising including breeding [33]. Thus, to search for culturable fungi associated with horses living in Southeastern side of Jeju Island, we collected horse dung and investigated coprophilous fungi. In this study, we report two novel and one unrecorded Preussia species with their morphological features and the results of molecular genetic identification.
2. Materials and methods
2.1. Collection of dung samples
Horse dung samples were collected from horses living in Seopjikoji (33°25′48.5″N 126°56′03.5″E), Southeast side of Jeju Island, the Republic of Korea. The collected dung samples were put into clean plastic bags and stored in an icebox and transported to the laboratory. The transported dungs were stored in a refrigerator at 4 °C until the fungal isolation experiment.
2.2. Fungal isolation
The inside part of the dung sample (3 g) was added to 50 mL sterile plastic tube containing 10 mL of sterilized distilled water and the dung sample tube was vortexed for 30 min. After vortex, the suspended dung solution was diluted from 10−1 to 10−2. The diluted dung solution (100 µL) was spread on several Dichloran Glycerol 18% (DG18; BD, Franklin Lakes, NJ) agar using a sterile plastic rod. DG18 agar was chosen for its low water activity, which inhibits the rapid growth of fast-growing molds, thereby facilitating colony enumeration and isolation. The DG18 agar with the suspended dung sample was cultured in a 25 °C incubator for 1 month without light. After incubation, the grown fungal colonies were selected and sing spore culture was purely isolated to Potato Dextrose Agar (PDA; BD, Franklin Lakes, NJ). Purely isolated fungal strains were cultured in an incubator at 25 °C for 2 weeks to confirm whether pure separation was achieved. Six fungal strains coded as DUCC15667, DUCC15684, DUCC15669, DUCC15670, DUCC15668, and DUCC15730 were selected for detailed morphological and molecular phylogenetic analysis. Stock cultures of strains DUCC15667 (NIBRFGC000512613), DUCC15668 (NIBRFGC000512614), and DUCC15669 (NIBRFGC000512615) with NIBRFGC numbers were deposited in the National Institute of Biological Resources (NIBR) located in Incheon in the Republic of Korea.
2.3. Morphology analysis
The colony morphology of the selected six DUCC fungal strains was examined on PDA, Malt Extract Agar (MEA; BD, Franklin Lakes, NJ), Czapek Yeast Extract Agar (CYA; BD, Franklin Lakes, NJ), and Oatmeal Agar (OA; BD, Franklin Lakes, NJ). Agar plugs from fungi grown on PDA were transferred to each medium, and colony morphology was assessed after 14 days of incubation at 25 °C in darkness. The microstructures of the fungal isolates, cultivated for a designated period, were observed under an optical microscope (BX53, OLYMPUS, Tokyo, Japan). Measurements of microstructures were conducted by measuring the length and thickness of ascocarp, ascus, and ascospore with 50 measurements.
2.4. Molecular analysis and phylogenetic analysis
Approximately 100 mg of fungal hyphae were collected after growing on cellophane-layered PDA for seven days and DNA extraction was carried out following the method described by Kim [34]. The extracted DNA was used as a template for PCR amplification of the Internal Transcribed Spacer region (ITS), large subunit rDNA region (LSU rDNA)) regions by T100 Thermal Cycler (BIO-RAD, Hercules, USA) using the primer sets listed in Supplementary Table S1. The size of the amplified PCR products was checked on a 1% (w/v) agarose gel through electrophoresis. The size confirmed PCR products were purified using the High Pure PCR Product Purification Kit (Roche, Indianapolis, USA). The nucleotide sequence analysis of the PCR products was outsourced to BIONICS Co. Ltd. (Seoul, Korea). The determined nucleotide sequences were edited with Chromas Software (https://technelysium.com.au/wp/chromas). The edited ITS and LSU rDNA sequences were aligned and trimmed using the ClustalW alignment tool in MEGA X to extract complete ITS and LSU rDNA sequences [35]. For the homologous sequence search, the complete sequences were blasted in the Basic Local Alignment Search Tool (BLAST) in NCBI database (https://www.ncbi.nlm.nih.gov). For phylogenetic analysis, closely related fungal strains were selected based on the results of BLAST homologous sequence search. Reference nucleotide sequences of the ITS and LSU rDNA in species of Preussia and Sporomiella were downloaded from the GenBank DNA database (https://www.ncbi.nlm.nih.gov/genbank/) and are provided in Table 1. The downloaded reference sequences datasets were aligned using the ClustalW alignment tool in MEGA X [35] to eliminate redundancy (the 18S rDNA region, the overlapping ITS-LSU region, and the LSU end) and improved manually where necessary. These ITS and LSU sequences of reference species were used for similarity analysis using the distance analysis with pairwise deletion option. To construct the phylogenetic tree, the maximum likelihood method was used based on the aligned sequences [36]. The Kimura 2-parameter model was used to estimate genetic differences [37]. The reliability of the nodes in the phylogenetic tree was confirmed by bootstrap analysis of 1,000 replicates. Together with the ITS and LSU rDNA sequences of the 6 DUCC fungal strains, these complete sequences were concatenated into a single FASTA file and realigned. Phylogenetic trees were constructed based on the concatenated sequences using the maximum likelihood method with the Kimura 2-parameter model to estimate genetic differences [35, 37]. Gaps and ambiguous bases were eliminated using the partial deletion method with a 70% of site coverage cutoff, there were a total of 875 positions in the final dataset. Node reliability was assessed through bootstrap analysis of 1,000 replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 45 nucleotide sequences. All positions with less than 70% site coverage were eliminated, i.e., fewer than 30% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There was a total of 875 positions in the final dataset.
Table 1.
Information on the nucleotide sequences used in phylogenetic analysis.
| Species name | Strain | Substrate | Origin | GenBank accession number* |
|
|---|---|---|---|---|---|
| ITS | LSU | ||||
| Preussia isomera | DUCC15667, NIBRFGC000512613 | Horse dung | Republic of Korea | OR835259 | OR835275 |
| P. isomera | DUCC15684 | Horse dung | Republic of Korea | OR835263 | OR835279 |
| P. jejuensis | DUCC15669, NIBRFGC000512615 | Horse dung | Republic of Korea | OR835261 | OR835277 |
| P. jejuensis | DUCC15670 | Horse dung | Republic of Korea | OR835262 | OR835278 |
| P. koreensis | DUCC15668, NIBRFGC000512614 | Horse dung | Republic of Korea | OR835260 | OR835276 |
| P. koreensis | DUCC15730 | Horse dung | Republic of Korea | PQ520562 | PQ520563 |
| P. aemulans | CBS 28767 | Greenhouse soil | Netherlands | DQ468017 | DQ468037 |
| P. africana | s15 | Zebra dung | South Africa | AY510421 | AY510385 |
| P. africana | s17 | Viburnum tinus leaves | Spain | AY510418 | AY510383 |
| P. alloiomera | S:Lundqvist 21345-p | Goat dung | Norway | GQ203771 | GQ203731 |
| P. antarctica | CBS 222.89 | Soil | Norway | KX710224 | KX710224 |
| P. arizonica | DS0008 | Ephedra trifurca | USA | OP876770 | OP876770 |
| P. bipartis | S:Lundqvist 17250-a | Ptarmigan dung | Sweden | GQ203774 | GQ203733 |
| P. borealis | S:Lundqvist 16745-c | Horse dung | Romania | GQ203775 | GQ203734 |
| P. dubia | S:Strid 19562-G | Horse dung | Iceland | GQ203777 | GQ203736 |
| P. flanaganii | CBS 112.73 (T) | – | Mexico | NR_077168 | NG_064098 |
| P. fleischhakii | CBS 36149 | – | Mexico | NR_077168 | NG_064098 |
| P. fleischhakii | CBS 56563 | Wheat field soil | Germany | DQ468019 | DQ468039 |
| P. funiculata | F:Huhndorf et al. 2577 (T) | Porcupine dung | USA | GQ203762 | GQ203722 |
| P. isomera | CBS 318.65 (T) | – | USA | NR_103588 | NG_064045 |
| P. lignicola | 18ALIC002 | Leaves | France | MT520567 | MT472604 |
| P. longisporopsis | S:Lundqvist 16551-g | Rabbit dung | Hungary | GQ203784 | GQ203742 |
| P. mediterranea | S34 | Daphne gnidium leaves | Spain | DQ468025 | DQ468045 |
| P. mediterranea | S22 | Quercus ilex leaves | Spain | DQ468021 | DQ468041 |
| P. minipascua | UPS:Kruys 306 | Cow dung | Sweden | GQ203787 | GQ203745 |
| P. persica | GLMC 447 | Prunus domestica | Germany | MT153731 | MT156301 |
| P. persica | CBS 117680 (T) | Dead leaf | Iran | NR_137730 | GQ292752 |
| P. polymorpha | CBS 117679 (T) | Dead leaf | Iran | NR_137729 | GQ292751 |
| P. procaviae | CBS 146827 (T) | Procavia capensis dung | Namibia | NR_171769 | NG_074498 |
| P. procaviicola | CBS 146981(T) | Dung of Procavia sp. | Namibia | NR_173052 | NG_076741 |
| P. pseudominima | s25 | Leaf litter | Puerto Rico | AY510424 | AY510389 |
| P. terricola | CBS 317.65 (T) | Musasa pientum | Honduras | NR_156524 | NG_064044 |
| P. typharum | CBS 107.69 | Deer dung | Japan | MH859270 | MH871002 |
| P. vulgaris | S:Strid 18884 | Hare dung | Sweden | GQ203767 | GQ203727 |
| Sporormiella australis | s6 | Gazelle dung | Namibia | AY510412 | AY510377 |
| S. australis | s7 | Zebra dung | South Africa | AY510413 | AY510378 |
| S. intermedia | UAMH 7460 | – | – | DQ468020 | DQ468040 |
| S. intermedia | s4 | Goat dung | Cefalonia | AY510416 | AY510381 |
| S. minima | s13 | Gazelle dung | Namibia | AY510426 | AY510391 |
| S. minima | s21 | Rhinoceros dung | South Africa | AY510425 | AY510390 |
| S. minimoides | s10 | Pig dung | Argentina | AY510423 | AY510388 |
| S. minimoides | s18 | Prunus lusitanica | Canary Islands | AY510422 | AY510387 |
| S. similis | s19 | Dung | Arizona | AY510419 | AY510386 |
| S. similis | CBS 80473 | Saline dessert soil | Kuwait | DQ468028 | DQ468048 |
| Alternaria cerealis | CBS 119544 (T) | Avena sativa | New Zealand | NR_136117 | NG_069254 |
* Accession numbers of sequences newly generated in this study are indicated in bold. 28S large subunit of the nrDNA; ITS internal transcribed spacer regions of the nrDNA and intervening 5.8S nrDNA. (T): type stain.
Bayesian Inference analysis was conducted based on DNA sequences aligned with ClustalW [38], applying the Markov Chain Monte Carlo (MCMC) method in MrBayes v. 3.2.7 [39] using the GTR+I + G model. A total of four chains were used in the MCMC analysis, with tree sampling conducted over 500,000 generations. Trees were sampled every 100 generations. Convergence between the chains was assessed by monitoring the average standard deviation (SD) of split frequencies, with convergence assumed when the SD was 0.05 or lower. Trees sampled before convergence were discarded with a burn-in of 2,500 generations, and the remaining trees were used to construct a 50% majority-rule consensus tree and estimate Bayesian posterior probabilities (BPP). BPP values below 0.95 were not considered significant. The results obtained from MCMC analysis were visualized with FigTree v1.4.4 (tree.bio.ed.ac.uk/software/figtree/). The ITS and LSU rDNA sequences of 6 DUCC fungal strains were registered in the GenBank database with or without the 18S rDNA region and the overlapping ITS-LSU region and their accession numbers are given in Table 1. All the primer set and amplification conditions used in this study [40–42] are given in Supplement Table S1.
3. Results
3.1. Molecular phylogenetic analysis
Preussia is very similar, morphologically and genetically, to the genus Sporormiella, so reference sequences for molecular analysis included sequences from species belonging to these two genera. The determined ITS region sequences from the PCR amplicon of NIBRFGC000512613 (DUCC15667) and DUCC15684 were 520 bp and 466 bp. GenBank blastn search results showed that both sequences were the closest to the ITS sequence of type strain Preussia isomera CBS 318.56. After trimming, the complete ITS of 443 bp was obtained from strains NIBRFGC000512613 and DUCC15684. The pairwise comparison of this sequence with the trimmed ITS sequences of all strains listed in Table 1 showed that P. isomera CBS 318.65 had 100% sequence similarity only to strains NIBRFGC000512613 and DUCC15684. As a result of constructing a ML phylogenetic tree based on the ITS sequences, strains NIBRFGC000512613 and DUCC15684 were most closely related to P. isomera CBS 318.56 (Figure 1). Both the determined LSU sequences of strains NIBRFGC000512613 and DUCC15684 were 904 bp and 860 bp, respectively. GenBank blastn search results showed that these sequences were the most like the LSU sequence of P. isomera CBS 318.56. Approximately half of the species registered in the genera Preussia and Sporormiella in the GenBank DNA database have LSU sequence lengths shorter than 500 bp. Therefore, to include more sequences in the comparison, the trimmed LSU sequences of all strains listed in Table 1 along with the trimmed 439 bp of strains NIBRFGC000512613 and DUCC15684 were used for pairwise comparison analysis and phylogenetic tree construction. The results of pairwise comparison revealed that P. isomera CBS 318.65 showed 100% sequence similarity to strains NIBRFGC000512613 and DUCC15684. A ML phylogenetic tree based on the LSU sequences also showed that strain NIBRFGC000512613 and DUCC15684 were grouped with P. isomera CBS 318.65 (Figure 2). To perform a better resolved phylogenetic analysis, the concatenated ITS (443 bp) and LSU (439 bp) sequences were used for phylogenetic analysis using the strains listed in Table 1. As a result of constructing a ML phylogenetic tree based on the concatenated ITS and LSU rDNA sequences, strains NIBRFGC000512613 and DUCC15684 were the most closely related to P. isomera CBS 318.56 (Figure 3). The Bayesian inferred phylogenetic tree generated based on the concatenated sequences of ITS and LSU rDNA also showed that strains NIBRFGC000512613 and DUCC15684 clustered well with P. isomera CBS 318.65 with high bootstrap values (>99) and were clearly separated from other species (Figure 4).
Figure 1.
Maximum-likelihood phylogenetic tree based on the ITS sequences of selected Preussia species and related genera. The tree was generated using the Kimura 2-parameter model and the ML method. Strains identified in this study are indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values which node reliability was less than 50 were removed. The reference sequences used to construct the ML phylogenetic tree are shown in Table 1. Alternaria cerealis CBS 119544 was used as an outgroup. (T): type strain.
Figure 2.
Maximum-likelihood phylogenetic tree based on the LSU rDNA region sequences of selected Preussia species and related genera. The tree was generated using the Kimura 2-parameter model and the ML method. Strains identified in this study are indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values which node reliability was less than 50 were removed. The reference sequences used to construct the ML phylogenetic tree are shown in Table 1. Alternaria cerealis CBS 119544 was used as an outgroup. (T): type strain.
Figure 3.
Maximum-likelihood phylogenetic tree based on concatenated the ITS and LSU rDNA sequences of selected Preussia species and related genera. The tree was generated using the Kimura 2-parameter model and the ML method. Strains identified in this study are indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values which node reliability was less than 50 were removed. The reference sequences used to construct the ML phylogenetic tree are shown in Table 1. Alternaria cerealis CBS 119544 was used as an outgroup. (T): type strain.
Figure 4.
Bayesian inference phylogenetic tree based on concatenated the ITS and LSU rDNA sequences of selected Preussia species and related genera. The tree was generated using the GTR+I + G model and the MCMC method. Strains identified in this study are indicated in bold. The number of nodes represents the Bayesian posterior probabilities through 500,000 generations. The reference sequences used to construct this phylogenetic tree are shown in Table 1. Alternaria cerealis CBS 119544 was used as an outgroup. (T): type strain.
The determined ITS region sequences from the PCR amplicon of stains NIBRFGC000512615 (DUCC15669) and DUCC15670 were 514 bp and 514 bp. GenBank blastn search results showed that these sequences were the most similar to the ITS sequence of S. minimoides s10. After trimming, the complete ITS of 443 bp was obtained from strains NIBRFGC000512615 and DUCC15670. The pairwise comparison of this sequence with the trimmed ITS sequences of all strains listed in Table 1 showed S. minimoides s10 possessed 94.28% sequence similarity to strains NIBRFGC000512615 and DUCC15670. As a result of constructing the ML phylogenetic tree based on the ITS sequence, strains NIBRFGC000512615 and DUCC15670 formed a separate clade and were adjacent to the clade with P. isomera CBS 318.56, NIBRFGC000512613 and DUCC15684 (Figure 1). The determined LSU sequences of strains NIBRFGC000512615 and DUCC15670 were 897 bp and 872 bp, respectively. GenBank blastn search results showed that they were close to the LSU sequences of P. isomera CBS 318.56, S. minimoides s10, and S. minimoides s18. Again, the trimmed LSU sequences of all strains listed in Table 1 were compared with the trimmed 439 bp of strains NIBRFGC000512615 and DUCC15670. The results of pairwise comparison revealed that strains NIBRFGC000512615 and DUCC15670 had 97.85% sequence similarity to P. isomera CBS 318.65 and S. minimoides s10, and 98.18% to S. minimoides s18. A ML phylogenetic tree based on the LSU sequences also showed that strains NIBRFGC000512615 and DUCC15670 were separated from other clades (Figure 2). P. isomera CBS 318.65, S. minimoides s10, and S. minimoides s18 are located in neighboring clades. Concatenated ITS (413 bp) and LSU (439) sequences were used for phylogenetic analysis. In the ML phylogenetic tree, strains NIBRFGC000512615 and DUCC15670 formed their own clade and were clearly separated from other species with high bootstrap values (>99). In the Bayesian inference phylogenetic tree based on the concatenated sequence of ITS and LSU rDNA, strains NIBRFGC000512615 and DUCC15670 were also clearly separated from other Preussia and Sporormiella species, similar to the ML phylogenetic tree, and were well supported with a high bootstrap value (>99) (Figure 4). These molecular results suggest that P. jejuensis strains NIBRFGC000512615 and DUCC15670 are the same species and could be a novel species (Table 2).
Table 2.
Representative morphological characteristics between Preussia species that are phylogenetically close to P. jejuensis and P. koreensis.
| Scientific name | Ascomata | Asci | Ascospores | Conidiomata | Conidia | Reference |
|---|---|---|---|---|---|---|
|
Preussia jejuensis NIBRFGC000512615 |
Absent | Absent | Absent | Hyaline to brown, circular or oval, 4–8 µm of peridium, smooth, 67–158 × 64–152 µm | Oval, with oil droplets present at both ends or at one end, hyaline, 2.5–3.7 × 1.7–2.4 µm | In this study |
|
P. koreensis NIBRFGC000512614 |
Black, shiny, smooth, not ostiolated, 6.1–8.5 µm of peridium, 65–140 µm diam. Aggregated ascomata were frequently observed | Octospores, cylindrical to clavate, elongate and often curved, (50-)60–82(–100) × (10.9-)11.5–15.3 µm | 4-celled, cylindrical to oval, straight or slightly curved, hyaline first soon turn in pale green, dark brown when mature, 26.3–36.2 × 4.2–5.8 µm | Absent | Absent | In this study |
|
P. isomera NIBRFGC000512613 |
Black, shiny, smooth, not ostiolated, 5.3–9.4 µm of peridium, 100–180 µm diam | Octospores, clavate or sub-ellipsoidal, thick walled, 42.8–59 × 16.5–19.3 µm | Cylindrical to oval, broadly rounded at both ends, 8.0–9.2 × 4.1–5.1 µm | Absent | Absent | In this study |
|
P. isomera CBS 318.65 |
Black, shiny, smooth, not ostiolated, 5–9 µm of peridium, 100–350 µm diam | Octospores, clavate or sub-ellipsoidal, thick walled, 43–55 × 15–18 µm | Cylindric-oval, broadly rounded at both ends, 7.0–9.0 × 4.0–4.5 µm | Absent | Absent | [18] |
|
P. arizonica DS0008 |
Dark brown to black, glabrous, ostioles not evident, subglobose to globose | Octospores, cylindrical to clavate, elongate and often curved, (98-)100–180(–185) × (16-)18–22(–24) µm | 4-celled, uniseriate or biseriate, cylindrical, elongate, hyaline to olivaceous when young, dark brown when mature, 18–24(–25) × 4–8(–9) µm | Absent | Absent | [9] |
|
S. minima s13 |
Dark-brown, glabrous, subglobose to nearly pyriform, 55–60 × 35–45 µm with neck, 9–13 × 7–11 µm of peridium, 150–190 × 75–100 µm | 8-spored, fissitunicate, broadly cylindrical, 80–140 × 15–18 µm | 4-celled, cylindrical, straight or curved, hyaline when young, becoming yellowish-brown, dark brown when mature, 26–34 × 4–6 µm | Absent | Absent | [20] |
|
S. minimoides s10 |
Dark-brown, subglobose to pyriform, 50–80 × 40–50 µm with neck, 180–240 × 100–150 µm | Octospores, subcylindrical, stipe about 4–6 µm long, 90–105(–110) × 16–19(–20) µm | 4-celled, cylindrical, straight or curved, dull-brown and opaque, transversely septate, 28–36 × 6–7 µm | Absent | Absent | [44] |
|
P. persica CBS 117680 |
Single neck, 120–160 µm diam. with 8.5–13 µm of peridium; more than one neck, 95–105 × 80–90 µm with 9.5–12 µm of peridium | Cylindrical, 8-spored, 75–90 × 13.5–15.5 µm | 4-celled, straight or slightly curved, hyaline at first, soon turning pale green to olivaceous-brown, yellowish brown and finally dark brown at maturity, 28–31 × 5–6 µm | Absent | Absent | [15] |
The determined ITS region sequences from the PCR amplicon of stains NIBRFGC000512614 (DUCC15668) and DUCC15730 were 520 bp and 521 bp, respectively. GenBank blastn search results showed that these sequences were closely related to the ITS sequence of Preussia arizonica DS0008, P. persica CBS 117680, and Sporormiella minima s13. After trimming, the complete ITS of 438 bp was obtained from strains NIBRFGC000512614 and DUCC15730. Pairwise comparison of these sequences with the trimmed ITS sequences of all strains listed in Table 1 showed that strains NIBRFGC000512614 and DUCC15730 had 98.17% sequence similarity to P. arizonica DS0008 and S. minima s13 and 98.63% to P. persica CBS 117680. As a result of constructing the ML phylogenetic tree based on the ITS sequence, strains NIBRFGC000512614 and DUCC15730 formed a separate clade and were adjacent to the clade with P. persica CBS 117680 and GLMC 447 (Figure 1). The determined LSU sequences of strains NIBRFGC000512614 and DUCC15730 were 848 bp and 849 bp, respectively. GenBank blastn search results showed that they were close to the LSU sequences of Preussia arizonica DS0008, P. persica CBS 117680, and Sporormiella minima s13. Again, the trimmed LSU sequences of all strains listed in Table 1 were compared with the trimmed 439 bp of strains NIBRFGC000512614 and DUCC15730. The results of pairwise comparison showed that strains NIBRFGC000512614 and DUCC15730 had 99.77% sequence similarity to Preussia arizonica DS0008, P. persica CBS 117680, and Sporormiella minima s13.
The ML phylogenetic tree based on the LSU sequences also showed that strains NIBRFGC000512614 and DUCC15730 were distantly related to other lineages (Figure 2). Concatenated ITS (438 bp) and LSU (439) sequences were used for phylogenetic analysis. In the ML phylogenetic tree, strains NIBRFGC000512614 and DUCC15730 formed their own clade and were separated from other species with high bootstrap values (>100). In the Bayesian inference phylogenetic tree based on the concatenated sequence of ITS and LSU rDNA, strains NIBRFGC000512614 and DUCC15730 were also clearly separated from other Preussia and Sporormiella species, similar to the ML phylogenetic tree, and were well supported with a high bootstrap value (>100) (Figure 4). These molecular results suggest that P. koreeensis strains NIBRFGC000512615 and DUCC15670 are the same species and could be a novel species.
3.2. Taxonomy
Preussia isomera Cain, Canad. J. Bot. 39 (7): 1643 (1961) [MB#337564] (Figure 5)
Figure 5.
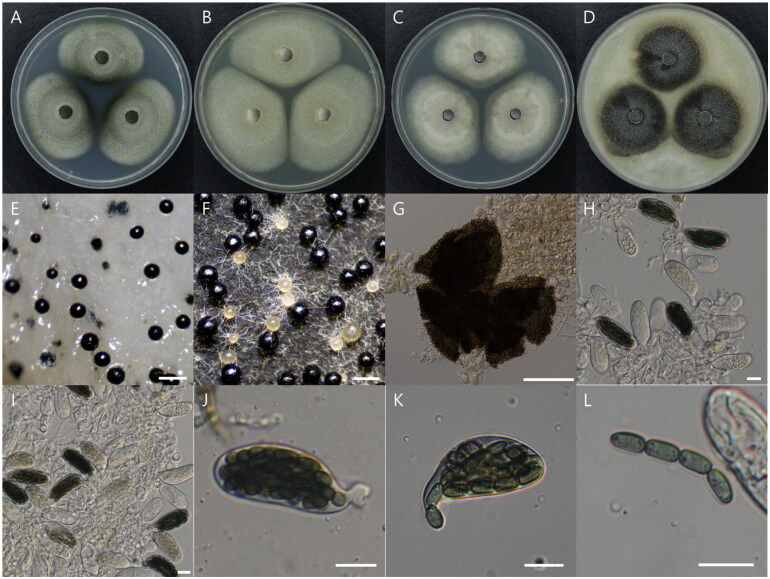
Morphological characteristics of Preussia isomera (NIBRFGC000512613) after 14 days of incubation at 25 °C on PDA (a), MEA (B), CYA (C), and OA (D). E-G: ascocarp. H-K: asci. L: 4 celled ascospore. Scale bar, E-F = 200 μm, g = 100 μm, H-L = 10 μm.
Typification
Jeju Island, the Republic of Korea, dung of horses living in Seopjikoji (33°25′48.5″N 126°56′03.5″E), collected on October 18, 2023. NIBRFGC000512613.
Description
Colony round, with thread-like margins, flat, gray-green in color, 40–50 mm in diameter on PDA and CYA, 54–55 mm in diameter on MEA. On OA, colony 36–38.5 mm in diameter, radial, a slightly fluffy texture, dark olive green. Ascocarps tightly packed, often gregarious, superficial or submerged in agar, globose shaped, black, shiny, smooth, not ostiolated, 100–180 µm in diameter. Peridium thin, textura angularis, 5.3–9.4 µm in diameter. Asci octosporous, clavate to sub-ellipsoidal shaped, apical round, middle broad, base narrowed, 42.8–59 × 16.5–19.3 µm. Ascospores 4 celled, 3-septate, initially hyaline, turning brown when mature, cylindrical to oval shaped, broadly rounded at both ends, separate at mature, 8.0–9.2 × 4.1–5.1 µm. Anamorph not observed.
Notes
In 1967, Cain reported Preussia isomera, which is characterized by segmented ascospores that remain attached together until nearly mature, measuring 7–9 × 4.0–4.5 μm [14]. In this study, strain NIBRFGC000512613 also observed segmented asci that remained attached together until almost mature, and the size was similar to those in previous records (8.0–9.2 × 4.1–5.1 µm). Phylogenetic analysis using the combined ITS and LSU rDNA sequences showed that it formed the same clade as P. isomera. Based on the results of morphological and molecular analysis, we identified strain NIBRFGC000512613 as P. isomera, a species not previously recorded in Korea.
Preussia jejuensis H. Noh, H.U. Cho and S.H. Kim, sp. nov. (Figure 6)
Figure 6.
Morphological characteristics of Preussia jejuensis sp. nov. after 14 days of incubation at 25 °C on PDA (a), MEA (B), CYA (C), and OA (D). E-F: conidiomata. G-H: conidiogenous cells. I-L: conidia. Scale bar = 10 μm.
Typification
Jeju Island, the Republic of Korea, dung of horses living in Seopjikoji (33°25′48.5″N 126°56′03.5″E), collected on October 18, 2023. Holotype: NIBRFGC000512615.
Etymology
Latin, jejuensis (je.ju.en’sis. N.L. fem. adj. jejuensis) referring to Jeju, the Republic of Korea, geographical origin of the type strain of the species.
Description
On PDA, colony radial, slightly fluffy texture, clay gray color with gray-white border, 31.5–32mm in diameter. On MEA, colony radial, very fluffy texture, beige color colored, 37–42.5 mm in diameter. On CYA, colony radial, very fluffy texture, light grayish green colored with slight gray-white border, 38.5–39.7 mm in diameter. On OA, colony slightly fluffy texture, dark olive green colored, 29–30mm diameter. Conidiomata hyaline to brown, circular or oval, smooth, 67–158 × 64–152 µm. Peridium thin, hyalinem angled corner cells densely packed, 4–8 µm diam. Conidia oval, with oil droplets present at both ends or at one end, hyaline, 2.5–3.7 × 1.7–2.4 µm. Teleomorph not observed.
Notes
Morphologically, P. isomera and S. minimoides displayed a sexual stage, with ascospores formed within ascocarps. However, NIBRFGC000512615 lacked a teleomorph and instead produced conidia within conidiomata. Phylogenetic analysis using the ITS and LSU sequences revealed that it formed a clade closely related to P. isomera and S. minimoides but established an independent branch. Based on morphological and phylogenetic analyses, this study reports Preussia jejuensis NIBRFGC000512615 as a new species within the genus Preussia.
Preussia koreensis H. Noh, H.U. Cho and S.H. Kim, sp. nov. (Figure 7)
Figure 7.
Morphological characteristics of Preussia koreensis sp. nov. after 14 days of incubation at 25 °C on PDA (a), MEA (B), CYA (C), and OA (D). E-H: ascocarps without ostiole. I-M: asci. N-P: 4 celled ascospores. Scale bar: E-F = 200 μm; G-H = 100 μm; I-p = 10 μm.
Typification
Jeju Island, the Republic of Korea, dung of horses living in Seopjikoji (33°25′48.5″N 126°56′03.5″E), collected on October 18, 2023. Holotype: NIBRFGC000512614.
Etymology
Latin, koreensis, referring to the Republic of Korea, origin of the type strain of the species.
Description
Colony radial, irregular, flat, undulated margin, slightly fluffy texture, yellowish green color with cream border, 22–23mm in diameter on PDA. Colony fluffy texture, irregular, flat, undulated margin, cream color, 37–42mm in diameter on MEA. On CYA, colony irregular, flat, curled, very fluffy texture, glade green colored, fast growth, 61–63mm in diameter. On OA, colony irregular, flat, curled, brown to black colored, 25–29mm in diameter. Ascocarps black, shiny, smooth, not ostiolated, 65–140 µm in diameter. Ascomata is frequently aggregated. Peridium textura angularis, hyaline, 6.1–8.5 µm in diameter. Asci octosporous, cylindrical to clavate shaped, elongate and often curved, (50–)60–82(–100) × (10.9–)11.5–15.3 µm. Ascospores 4-celled, cylindrical to oval, straight or slightly curved, hyaline first soon turn in pale green, dark brown when mature, 26.3–36.2 × 4.2–5.8 µm. Anamorph not observed.
Notes
Morphologically, the ascocarps of S. minima and P. persica had necks, whereas strain NIBRFGC000512614 lacked an ostiole and had a different diameter range (P. koreensis = 65–140 µm; S. minima = 150–190 µm; P. persica = 120–160 µm). The size of the asci was (50–)60–82(–100) × (10.9–)11.5–15.3 µm, which was smaller than that of the other three species (P. arizonica = (98–)100–180(–185) × (16–)18–22(–24) µm; S. minima = 80–140 × 15–18 µm; P. persica = 75–90 × 13.5–15.5 µm). The ascospores, like those of the other species, were composed of four cells and measured 26.3–36.2 × 4.2–5.8 µm. This size was larger than that of P. arizonica and slightly larger than those of the other two species (P. arizonica = 18–24(–25) × 4–8(–9) µm; S. minima = 26–34 × 4–6 µm; P. persica = 28–31 × 5–6 µm). In the phylogenetic analysis, strain NIBRFGC000512614 formed a distinct branch; the node value was 100, indicating strong support. Nevertheless, morphologically, strain NIBRFGC000512614 can be distinguished from other species by its lack of a neck, smaller ascocarp diameter range, smaller asci size, and larger ascospores. Therefore, this study reports P. koreensis NIBRFGC000512614 as a new species.
4. Discussion
Coprophilous fungi, or dung-inhabiting fungi, play a critical role in natural ecosystems by breaking down animal dung and recycling nutrients into the environment [43]. These fungi are specially adapted to the transient and nutrient-rich environment of dung, which provides a favorable substrate for their growth. They also demonstrate unique reproductive strategies, including forcible spore discharge, which enables efficient dispersal to fresh dung. Genera such as Preussia, Sporormiella and Podospora are commonly associated with animal dung and are known to produce secondary metabolites with antibacterial properties, contributing not only to nutrient cycling but also to biotechnological research [24–26, 44, 45]. These secondary metabolites have been reported to exhibit high potential in various fields, including antifungal activity, antibiotic properties, plant growth promotion, and cytotoxic effects [13, 46–48]. Thus, we expect that the two new species, P. koreensis sp. nov. and P. jejuensis sp. nov., could lead to explore new secondary metabolites which have antimicrobial properties and/or plant growth enhancing ability, and enzymes which have role(s) in various physiological, cellular and biological processes in dung-inhabiting fungi. Additionally, together with the discovery of two new species along with a newly recorded species P. isomera at Seopjikoji, it is worthwhile to be noticed that Jeju Island where horses have been bred for a long period could be a new source of finding novel dung fungi in the Republic of Korea [30]. Our findings, alongside existing research on the fungal diversity of Jeju Island, a UNESCO World Natural Heritage site, contribute to enhancing the fungal diversity in this region [49, 50]. Furthermore, they provide new insights into the diversity and ecological roles of coprophilous fungi in unexplored areas, highlighting the importance of dung as an essential habitat for fungal biodiversity.
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Biological Resources, the Ministry of Environment, the Republic of Korea. The Department of Microbiology was supported through the Research Focused-Department Promotion & Interdisciplinary Convergence Research Project as a part of the Support Program for University Development for Dankook University in 2024.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Krug JC, Benny GL, Keller HW.. Coprophilous fungi. In Biodiversity of fungi. Academic Press, Burlington; 2004.467–499. [Google Scholar]
- 2.Parker AD. Associations between coprophilous ascomycetes and fecal substrates in Illinois. Mycologia. 1979;71(6):1206–1214. doi: 10.1080/00275514.1979.12021132. [DOI] [Google Scholar]
- 3.Hudson HJ. The ecology of fungi on plant remains above the soil. New Phytol. 1968;67(4):837–874. doi: 10.1111/j.1469-8137.1968.tb06399.x. [DOI] [Google Scholar]
- 4.Harley JL. Fungi in ecosystems. J. Ecol. 1971;59(3):653–668. doi: 10.2307/2258131. [DOI] [Google Scholar]
- 5.Angel SK, Wicklow DT.. Relationships between coprophilous fungi and fecal substrates in a Colorado grassland. Mycologia. 1975;67(1):63–74. doi: 10.1080/00275514.1975.12019722. [DOI] [PubMed] [Google Scholar]
- 6.Lussenhop J, Kumar R, Wicklow DT, et al. Insect effects on bacteria and fungi in cattle dung. Oikos. 1980;34(1):54–58. doi: 10.2307/3544549. [DOI] [Google Scholar]
- 7.Harper JE, Webster J.. An experimental analysis of the coprophilous fungus succession. TBMS. 1964;47(4):511–530. doi: 10.1016/S0007-1536(64)80029-2. [DOI] [Google Scholar]
- 8.Kruys A, Wedin M.. Phylogenetic relationships and an assessment of traditionally used taxonomic characters in the Sporormiaceae (Pleosporales, Dothideomycetes, Ascomycota), utilising multi-gene phylogenies. Syst Biodivers. 2009;7(4):465–478. doi: 10.1017/S1477200009990119. [DOI] [Google Scholar]
- 9.Sandberg DC, del Olmo-Ruiz M, Sykes BE, et al. Three distinctive Preussia (Sporormiaceae) from photosynthetic stems of Ephedra trifurca (Ephedraceae, Gnetophyta) in southeastern Arizona. Plant Nad Fungal Syst. 2022;67(2):63–74. doi: 10.35535/pfsyst-2022-0008. [DOI] [Google Scholar]
- 10.Nisa H, Kamili AN, Nawchoo IA, et al. Fungal endophytes as a prolific source of phytochemicals and other bioactive natural products: a review. Microb Pathog. 2015;82:50–59. doi: 10.1016/j.micpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Premalatha K, Kalra AJFE.. Molecular phylogenetic identification of endophytic fungi isolated from resinous and healthy wood of Aquilaria malaccensis, a red-listed and highly exploited medicinal tree. Fungal Ecol. 2013;6(3):205–211. doi: 10.1016/j.funeco.2013.01.005. [DOI] [Google Scholar]
- 12.Fuckel L. Fungi Rhenani Cent. 18(1), no 1702-1764. Hedwigia. 1867;6:174–175. [Google Scholar]
- 13.Mapperson RR, Kotiw M, Davis RA, et al. The diversity and antimicrobial activity of Preussia sp. endophytes isolated from Australian dry rainforests. Curr Microbiol. 2014;68(1):30–37. doi: 10.1007/s00284-013-0415-5. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Menendez V, Martin J, Siles JA, et al. Biodiversity and chemotaxonomy of Preussia isolates from the Iberian Peninsula. Mycol Progress. 2017;16(7):713–728. doi: 10.1007/s11557-017-1305-1. [DOI] [Google Scholar]
- 15.Asgari B, Zare R.. Two new species of Preussia from Iran. nova_hedwigia. 2010;90(3-4):533–548. doi: 10.1127/0029-5035/2010/0090-0533. [DOI] [Google Scholar]
- 16.Chang JH, Wang YZ.. The genera Sporormia and Preussia (Sporormiaceae, Pleosporales) in Taiwan. nova_hedwigia. 2009;88(1-2):245–254. doi: 10.1127/0029-5035/2009/0088-0245. [DOI] [Google Scholar]
- 17.Guarro J, Abdullah SK, GenÉ J, et al. A new species of Preussia from submerged plant debris. Mycol Res. 1997;101(3):305–308. doi: 10.1017/S0953756296002638. [DOI] [Google Scholar]
- 18.Cain RF. Studies of coprophilous ascomycetes VII. Preussia. Can J Bot. 1961;39(7):1633–1666. doi: 10.1139/b61-144. [DOI] [Google Scholar]
- 19.Ellis JB, Everhart BM.. The North American Pyrenomycetes. Johnson Reprint Corp., New York; 1892. [Google Scholar]
- 20.Von Arx JA. Ostiolate and nonostiolate Pyrenomycetes. K Ned Akad Wet. Proc, Ser C. 1973;76:289–296. [Google Scholar]
- 21.Von Arx JA, Van der Aa HA.. Spororminula tenerifae gen. et sp. nov. Trans Br Mycol Soc. 1987;89(1):117–120. doi: 10.1016/S0007-1536(87)80068-2. [DOI] [Google Scholar]
- 22.Guarro J, Al-Saadoon AH, Abdullah SK.. Two new coprophilous species of Preussia (Ascomycota) from Iraq. nova_hedwigia. 1997;64(1-2):177–183. doi: 10.1127/nova.hedwigia/64/1997/177. [DOI] [Google Scholar]
- 23.Nyberg Kruys Å. Phylogenetic relationships and species richness of coprophilous ascomycetes; 2005. [Doctoral dissertation]. Ekologi, miljö och geovetenskap. [Google Scholar]
- 24.Weber HA, Gloer JB.. The preussomerins: novel antifungal metabolites from the coprophilous fungus Preussia isomera Cain. J Org Chem. 1991;56(14):4355–4360. doi: 10.1021/jo00014a007. [DOI] [Google Scholar]
- 25.Paudel B, Bhattarai K, Bhattarai HD.. Antimicrobial and antioxidant activities of two polyketides from lichen-endophytic fungus Preussia sp. Z Naturforsch C J Biosci. 2018;73(3-4):161–163. doi: 10.1515/znc-2017-0105. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A. Investigation of Queensland dry rainforest endophyte and their production of antimicrobial agents; 2021. [Doctoral dissertation]. Univ Southern Queensland;. [Google Scholar]
- 27.Chen X, Shi Q, Lin G, et al. Spirobisnaphthalene analogs from the endophytic fungus Preussia sp. J Nat Prod. 2009;7:1712–1715. [DOI] [PubMed] [Google Scholar]
- 28.Talontsi FM, Lamshöft M, Douanla-Meli C, et al. Antiplasmodial and cytotoxic dibenzofurans from Preussia sp. harboured in Enantia chlorantha. Oliv Fitoterapia. 2014;93:233–238. doi: 10.1016/j.fitote.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Du L, Robles AJ, King JB, et al. Cytotoxic dimeric epipolythiodiketopiperazines from the ascomycetous fungus Preussia typharum. J Nat Prod. 2014;77(6):1459–1466. doi: 10.1021/np5002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Youn UJ, Seo SS, Yim JH, et al. Chemical constituents from the culture filtrate of a Himalayan soil fungus, Preussia sp. and their anti-inflammatory activity. Kor J Microbiol. 2018;54(1):18–23. [Google Scholar]
- 31.Mun HY, Oh Y, Goh J.. Evaluation of extracellular enzyme activity of fungi from freshwater environment in South Korea. Kor J Mycol. 2023;51(4):265–276. 55. [Google Scholar]
- 32.UNESCO World Heritage Centre . Jeju Volcanic Island and Lava Tubes; 2024. UNESCO. https://whc.unesco.org/en/list/1264.2007.
- 33.Seo JH, Park KD, Lee HK, et al. Genetic diversity of Halla horses using microsatellite markers. J Anim Sci Technol. 2016;58(1):40. doi: 10.1186/s40781-016-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Han A, Kronstad J, et al. Differentiation of sapstain fungi by restriction fragment length polymorphism patterns in nuclear small subunit ribosomal DNA. FEMS Microbiol Lett. 1999;177(1):151–157. doi: 10.1111/j.1574-6968.1999.tb13726.x. [DOI] [Google Scholar]
- 35.Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho A. Constructing phylogenetic trees using maximum likelihood; 2012. Scripps Senior Theses. Paper 46. [Google Scholar]
- 37.Kimura MA. Simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinform. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Ronquist F, Teslenko M, Van Der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White T, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322. [Google Scholar]
- 41.Cubeta MA, Echandi E, Abernethy T, et al. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathol. 1991;81(11):1395–1400. doi: 10.1094/Phyto-81-1395. [DOI] [Google Scholar]
- 42.Vilgalys R, Hester M.. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dix NJ, Webster J.. Coprophilous fungi. In Fungal ecology. Cambridge, MA: Chapman & Hall; 1995; 203–224. [Google Scholar]
- 44.Ahmed SI, Cain RF.. Revision of the genera Sporormia and Sporormiella. Can J Bot. 1972;50(3):419–477. doi: 10.1139/b72-061. [DOI] [Google Scholar]
- 45.Weber RW, Meffert A, Heidrun ANKE, et al. Production of sordarin and related metabolites by the coprophilous fungus Podospora pleiospora in submerged culture and in its natural substrate. Mycol Res. 2005;109(Pt 5):619–626. doi: 10.1017/s0953756205002765. [DOI] [PubMed] [Google Scholar]
- 46.Weber HA, Gloer JB.. Interference competition among natural fungal competitors: An antifungal metabolite from the coprophilous fungus Preussia fleischhakii. J Nat Prod. 1988;51(5):879–883. doi: 10.1021/np50059a011. [DOI] [PubMed] [Google Scholar]
- 47.Al-Hosni K, Shahzad R, Khan L, et al. Preussia sp. BSL-10 producing nitric oxide, gibberellins, and indole acetic acid and improving rice plant growth. J. Plant Interact. 2018;13(1):112–118. doi: 10.1080/17429145.2018.1432773. [DOI] [Google Scholar]
- 48.Chen X, Shi Q, Lin G, et al. Spirobisnaphthalene analogues from the endophytic fungus Preussia sp. J Nat Prod. 2009;72(9):1712–1715. doi: 10.1021/np900302w. [DOI] [PubMed] [Google Scholar]
- 49.Halda JP, Woo JJ, Liu D, et al. Jejulea byssolomoides gen. et sp. nov., a remarkable Pilocarpaceae (Lichen-Forming Ascomycetes) from Jeju Island. South Korea. Mycobiology. 2022;50(3):172–180. doi: 10.1080/12298093.2022.2081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee W, Kim DG, Perera RH, et al. Diversity of Nigrospora (Xylariales, Apiosporaceae) species identified in Korean macroalgae including five unrecorded species. Mycobiology. 2023;51(6):401–409. doi: 10.1080/12298093.2023.2283272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.