Abstract
Background
Interleukin-10 (IL-10) is a cytokine whose main biological function is to suppress the immune response by induction of a signal(s) leading to inhibition of synthesis of a number of cytokines and their cellular receptors. Signal transduction is initiated upon formation of a ternary complex of IL-10 with two of its receptor chains, IL-10R1 and IL-10R2, expressed on the cell membrane. The affinity of IL-10R1 toward IL-10 is very high, which allowed determination of the crystal structure of IL-10 complexed with the extracellular/soluble domain of IL-10R1, while the affinity of IL-10R2 toward either IL-10 or IL-10/sIL-10R1 complex is quite low. This so far has prevented any attempts to obtain structural information about the ternary complex of IL-10 with its receptor chains.
Results
Structures of the second soluble receptor chain of interleukin-10 (sIL-10R2) and the ternary complex of IL-10/sIL-10R1/sIL-10R2 have been generated by homology modeling, which allowed us to identify residues involved in ligand-receptor and receptor-receptor interactions.
Conclusion
The previously experimentally determined structure of the intermediate/binary complex IL-10/sIL-10R1 is the same in the ternary complex. There are two binding sites for the second receptor chain on the surface of the IL-10/sIL-10R1 complex, involving both IL-10 and sIL-10R1. Most of the interactions are hydrophilic in nature, although each interface includes two internal hydrophobic clusters. The distance between C-termini of the receptor chains is 25 Å, which is common for known structures of ternary complexes of other cytokines. The structure is likely to represent the biologically active signaling complex of IL-10 with its receptor on the surface of the cell membrane.
Background
IL-10 is a pleiotropic cytokine (reviewed in [1,2]) that suppresses immune system response by inhibiting synthesis of proinflammatory cytokines and their cellular receptors [3,4]. In addition to immunosuppressive functions, IL-10 also possesses immunostimulatory activities, including stimulation of growth and proliferation of thymocytes, cytokine-activated T-cells, mast cells, and B-cells [5-9].
Signal transduction is initiated when IL-10 binds to its cellular receptor, which consists of two chains, IL-10R1 and IL-10R2. IL-10 is an intercalated dimer made of two identical polypeptides, each 160 amino acids long [10-12], packed in two compact, six-helix bundle domains (reviewed in [2,13-15]). The domains are related by a twofold symmetry axis passing through the interdomain interface. Both receptor chains have about a 200 amino acids-long extracellular domain, a 20 amino acids-long transmembrane helix, and an intracellular/cytoplasmic domain, which contains 322 or 62 amino acids for IL-10R1 and IL-10R2, respectively. When the ternary complex IL-10/IL-10R1/IL-10R2 is formed, tyrosine kinases Tyk2 and Jak1 become activated and phosphorylate specific tyrosine residues on signal transducers and activators of transcription, which eventually leads to activation of the transcription of corresponding genes [16,17]. Kinetic binding data on soluble IL-10 receptor chains (sIL-10R) showed that sIL-10R1 exhibits nanomolar binding constants for IL-10, while sIL-10R2 does not display any significant affinity for IL-10, although binding of sIL-10R2 to a preformed binary IL-10/sIL-10R1 complex is noticeably better [18]. It has been commonly accepted that at first IL-10 binds to its high affinity receptor IL-10R1, forming a binding site for IL-10R2, and then the binding of the latter receptor finalizes the ternary signaling complex. It has been also shown for the IFN-(/receptor complex, which is likely to be similar to the IL-10 complex, that under physiological conditions the receptor chains are already preassembled together, even before the ligand binds. Thus, binding of the ligand is necessary only for opening the intracellular domain regions to accommodate intracellular components of the downstream signal transduction pathway [19]. It was shown for IL-19 and IL-20, which also belong to the IL-10 family, that their final ternary complexes with the soluble receptors are very much alike, no matter what was the order of binding components in solution [20]. Therefore, the structure of the ternary IL-10/sIL-10R1/sIL-10R2 complex can be predicted on the basis of the structures of the intermediate binary complex IL-10/sIL-10R1 and the structure of sIL-10R2. The former, stable complex was crystallized [21] and its structure was determined [22]; however, all attempts to obtain stable ternary complexes suitable for crystallization have so far failed.
The known structures of binary and ternary cytokine-receptor complexes show that although they differ from each other in details, their overall organization is somewhat similar [22-29]. What is even more important, both ligand/receptor and receptor/receptor binding sites are located in topologically similar areas on the surface of the interacting proteins and involve similar structural elements of the molecules (Table 1). For example, in the case of the ligands, only two major receptor binding sites were found, and both included topologically conserved N- and C-terminal helices that form a left-handed four-helix bundle [30], which is a signature element of all helical cytokines, as well as loop AB. The difference was that in some cases site I was used as a high-affinity receptor binding site, while in other cases site II was the one where the high-affinity receptor was found to bind (Table 1). The crystal structure of a binary complex of IL-10 with sIL-10R1 showed that the high-affinity receptor binding site is site I, and it is reasonable to assume that sIL-10R2 will bind IL-10 in site II, making receptor/receptor contacts similar to the already published structures of ternary complexes.
Table 1.
Positions of the cytokine-receptor binding sites in known structures of ternary and binary complexes.
| Cytokine-receptor complexes (PDB code/reference) | Site I (Ligand/Receptor) | Site II (Ligand/Receptor) | Site III (Recept/Recept) | ||
| SSE* Ligand | SSE* Receptor | SSE* Ligand | SSE* Receptor | Contacting Domains | |
| Ternary complexes | |||||
| hGH/GHbp/GHbp (3HHR [23]) | A, loop AB, D | L1, L3-L6 | A, C | L1, L3-L6 | D2-D2 |
| hEPO/sEPOR/sEPOR (1EER [24]) | A, loop AB, D | L1-L6 | A, C | L1-L3, L5, L6 | no interaction |
| oPL/srPRLR/srPRLR (1F6F [25]) | A, loop AB, D | L1-L6 | A, C | L1-L4 | D2-D2 |
| hIL6/sIL6Rα /gp130 (D1-D3) (1P9M [29]) | A, D | L2-L6 | A, C | L1-L3, L5 | D2-D3 |
| Binary complexes | |||||
| hINF-γ /sINF-γ Rα ** (1FG9 [53]) | A, loop AB, F' | L2, L3, L5, L6 | |||
| hLIF/gp130 (D2-D3) (1PVH [54]) | A, C | L2, L3, L5 | |||
| G-CSF/G-CSFR (BN-BC) (1CD9 [27]) | A, C | L2-L6 | |||
| hIL4/IL4-R1 (1IAR [26]) | A, C | L1-L6 | |||
| hIL-10/sIL-10R1** (1J7V [22]) | A, loop AB, F' | L2-L6 | |||
| cmvIL-10/sIL-10R1** (1LQS [55]) | A, loop AB, F' | L2-L6 | |||
| vIL-6/gp130 (1I1R [56]) | A, C | L2, L3, L5 | |||
| Ternary hIL-10 complex | |||||
| hIL10/sIL10R1/sIL10R2**,*** | A, loop AB, F' | L2-L6 | A, D | L2, L3, L5, L6 | D2-D2 |
(*)- SSE: Secondary structure elements.
(**)- In INF-γ and IL-10 structures helix F' is a topological equivalent of helix D of other helical cytokines.
(***)- Interface areas were calculated with program Surface (CCP4) using spherical probe of the radius 1.40 Å2.
We generated a structural model of the ternary complex of IL-10 with its two soluble receptors, sIL-10R1 and sIL-10R2. The model has been built on the assumption that the structure of the binary IL-10/sIL-10R1 complex must be preserved upon the binding of the second receptor chain, the complex must possess twofold symmetry, and that the structure of the complex of one domain of IL-10 with sIL-10R1 and sIL-10R2 should be somewhat similar to the already known structures of ternary complexes having two different receptor chains. To fulfill the latter requirement, we took the structure of IL-6/IL-6Rα /gp130 [29] as an example.
Results
Structure of sIL-10R2
The structures of sIL-10R2 and sIL-10R1 appear to be very similar (Fig. 1). They are both L-shaped molecules consisting of two elongated N- and C-terminal domains (D1 and D2), oriented at about 90° with respect to each other, each having FBN-III-like topology [31-33]. Each domain consists of seven β-strands organized in two antiparallel β-sheets – A, B, E and C, C', F, and G – that pack one against another in the shape of a sandwich. Loops L2-L4 of D1 and L5-L6 of D2 (Fig. 2) are involved in ligand binding. Domains D1 and D2 of sIL-10R2 comprise 96 and 94 amino acid residues, respectively (residues 1–103 and 109–206), and are connected to each other by a 5 amino acid-long linker (residues 104–108).
Figure 1.
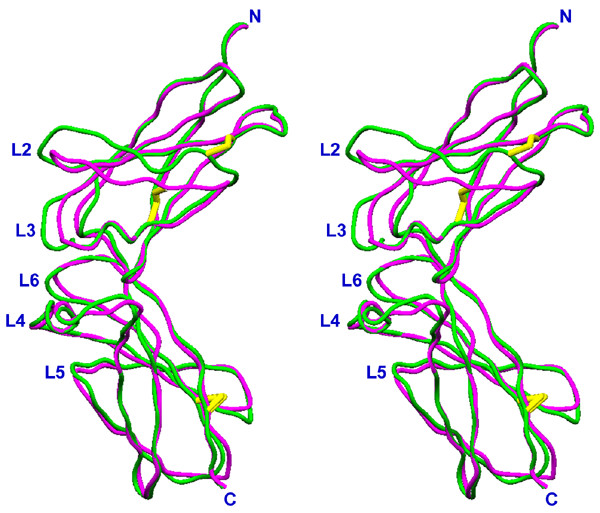
Stereo diagram of the superposition of sIL-10R1 and sIL-10R2. sIL-10R1 is green, sIL-10R2 is magenta, the disulfide bonds are shown in yellow.
Figure 2.
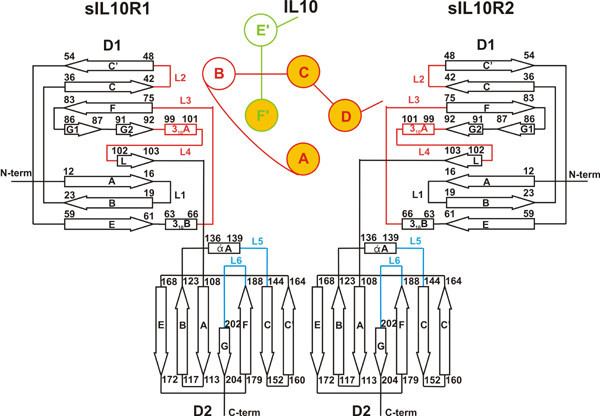
Topology diagram of a single ligand domain associated with high- and low-affinity receptors. β-strands of the receptor molecules are shown as arrows and the helices are shown as rectangles. Binding loops of the receptor D1 domain are red and the loops of the D2 domain are blue. α-helices of single ligand domain are shown as circles. Helices A, B, C, and D (red) belong to one polypeptide chain of the ligand and helices E' and F' (green) belong to another polypeptide chain of the ligand. Highlighted helices (A, C, D, F') constitute the four-helix bundle that is involved in receptor binding.
Deletions in the amino acid sequences of loops L2, L3 and β-strand C' in domain D1 of sIL-10R2 (Fig. 3) make its structure more compact than sIL-10R1. As a result, Tyr43, which is a conserved residue in class II receptors [34], is located a little higher than its counterpart in sIL-10R1. Although in sIL-10R1 Tyr43 plays a very important role in the interaction with the ligand, in sIL-10R2 it is located at the very edge of the receptor/ligand interface. Cysteines 54 and 62 of sIL-10R2 form a disulfide bond that is conserved in class II receptors [32], linking β-strand C' with the loop between the strand E and helix B. An equivalent disulfide bridge in sIL-10R1 is formed between cysteines 54 and 35 [12,22], linking together β-strands C' and C (Fig. 1).
Figure 3.
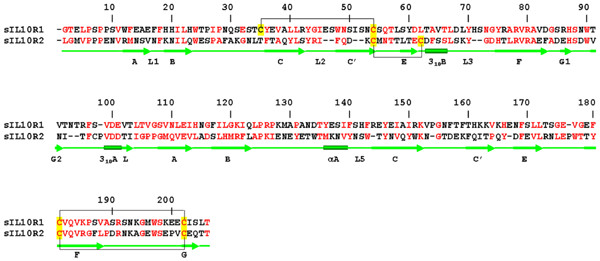
Structure-based sequence alignment of soluble receptors sIL-10R1 and sIL-10R2. Aligned molecules have 20.4% of identical residues and 53.1% of homologous residues (red). Secondary structure elements are shown in green. Cysteines involved in disulfide bonds are highlighted in yellow, disulfide bonds are shown by black lines.
Loop L3 of sIL-10R2 is two amino acids shorter in sIL-10R1 and does not protrude out of the D1 domain as much as in sIL-10R1. This alters the shape of the N-terminal part of the interdomain region and makes it complementary to the surface of the ligand-binding site II. Tyr70 is shifted ~2.5 Å inside of the D1 domain to compensate for the shortage of loop L3. In both receptors, the side chain of Tyr70 participates in the formation of a hydrophobic core of the N-terminal domain. Deletions in strand C' and loop L3 make the N-terminal domain of sIL-10R2 about 3 Å shorter than in sIL-10R1.
β-strands F (residues 77–81) of both receptors contain a conserved sequence (A/L)RVRA. Together with the sequences HSDWV in sIL-10R2 and HSNWT in sIL-10R1 (residues 87–91), this motif makes an extended π-cation system [35,36], usually found in the C-terminal domains of the type I family of cytokine receptors [37]. In sIL-10R1, a stacking structure is formed by His87, Arg80, Trp90, Arg78, Trp48, and Leu41; and is about 19 Å in length, which corresponds to roughly half of the domain length (~ 36 Å). It connects β-strands C, C', F, and G, and stabilizes the structure of the domain. In sIL-10R2, β-strand C' is located away from the β-strand F, which makes it impossible for the side chains of Phe48 and Leu41 to participate in the formation of a π-cation system, consisting in this case of only four residues – His87, Arg80, Trp90 and Arg78 – that keep together β-strands G and F. Although the π-cation system of sIL-10R2 is only ~11 Å long, which is about one-third of the total length of the domain, it still may be important for the stability of the structure of the domain. It is also likely that the π-cation system may play a dual role during the lifetime of a cytokine receptor. Mutagenesis studies of the erythropoietin receptor revealed the importance of the π-cation system for the transport of the receptor from endoplasmic reticulum to the Golgi apparatus [38,39].
In comparison to D2 of sIL-10R1, D2 of sIL-10R2 has three single amino acid deletions: Arg144, Pro154, and Glu167, as well as a single insertion of Pro176A (Fig. 3). The first deletion makes the connection between loop L5 and β-strand C shorter and moves the residues of loop L5 about 0.5 Å closer to the main body of the domain. Similarly, deletions of Pro154 and Glu167 make the loops connecting strands C and C' and strands C' and E slightly shorter (Fig. 1). β-strands F and G are held together by a disulfide bridge between Cys181 and Cys202, which is a common feature for class II cytokine receptors. Insertion of Pro176A in sIL-10R2 extends the loop between β-strands E and F by about 3 Å, bringing this part of the D2 domain closer to the cell membrane.
Hydrophobic patches on the surface and intermolecular hydrophobic clusters of the ternary IL-10/sIL-10R1/sIL-10R2 complex
Both the ligand and receptor chains have several hydrophobic patches located on the surface of the molecules (Table 2). Although the surface of IL-10 is mostly hydrophilic, with 86% of hydrophobic residues involved in the formation of the internal hydrophobic core [13], it also has three solvent-exposed hydrophobic patches (Table 2). Patch 1 is involved in the binding of sIL-10R1; patch 2 interacts with both sIL-10R1 and sIL-10R2, while patch 3 is located on the surface of helices A and D of IL-10 and participates in interactions with sIL-10R2.
Table 2.
Hydrophobic patches of IL-10, sIL-10R1 and sIL-10R2
| IL-10 | Interaction site | |
|
Patch 1 Residue Access. (%) |
Surface area 189 Å2 L46 L53 I145 A152 50 25 34 44 |
IL-10/sIL-10R1 interface, with patch 3 of sIL-10R1 |
|
Patch 2 Residue Access. (%) |
Surface area 294 Å2 H14 P16 P20 I158 73 66 71 25 |
IL-10/sIL10R1 interface, with patch 6 of sIL-10R1 IL-10/sIL10R2 interface, with Patch 7 of sIL-10R2 |
|
Patch 3 Residue Access. (%) |
Surface area 183 Å2 F36 A89 H90 33 86 45 |
IL-10/sIL-10R2 interface with Patch 2 (sIL-10R2) |
| IL-10R1 Domain D1 | ||
|
Patch 1 Residue Access. (%) |
Surface area 475 Å2 I51 Y60 A64 V65 L67 18 39 90 34 46 M129 P131 A132 60 38 100 |
No interaction |
|
Patch 2 Residue Access. (%) |
Surface area 448 Å2 P6 P9 W12 T25 P26 P28 L58 45 45 45 39 56 65 56 |
No interaction |
|
Patch 3 Residue Access. (%) |
Surface area 207 Å2 Y43 G44 I45 44 66 83 |
IL-10/sIL10R1 interface, with patch 1 of IL-10 |
| Domain D2 | ||
|
Patch 4 Residue Access. (%) |
Surface area 546 Å2 I113 H114 G116 F117 60 56 47 58 L119 L172 G175 V177 41 84 61 69 |
sIL-10R1/sIL10R2 interface, with patch 5 of sIL-10R2 |
|
Patch 5 Residue Access. (%) |
Surface area 409 Å2 V153 P154 G155 F157 F159 H161 45 56 55 73 43 56 |
sIL-10R1/sIL10R2 interface, with patch 6 of sIL-10R2 |
|
Patch 6 Residue Access. (%) |
Surface area 253 Å2 H142 F143 A189 77 62 35 |
IL-10/sIL10R1 interface, with patch 2 of IL-10 |
| IL-10R2 Domain D1 | ||
|
Patch 1 Residue Access. (%) |
Surface area 635 Å2 L2 G3 M4 P6 P7 A28 77 78 45 56 40 91 F29 G32 F83 A84 97 93 34 85 |
No interaction |
|
Patch 2 Residue Access. (%) |
Surface area 284 Å2 L41 I47 F48 29 88 62 |
IL-10/sIL10R2 interface, with patch 3 of IL-10 |
|
Patch 3 Residue Access. (%) |
Surface area 154 Å2 C98 P98A V99 52 48 42 |
No interaction |
| Domain D2 | ||
|
Patch 4 Residue Access. (%) |
Surface area 572 Å2 F160 I162 F169 L172 73 26 12 71 L175 P176A W177 49 73 57 |
No interaction |
|
Patch 5 Residue Access. (%) |
Surface area 274 Å2 V113 L114 A115 H119 47 62 100 40 |
sIL-10R1/sIL10R2 interface, with patch 4 of sIL-10R1 |
|
Patch 6 Residue Access. (%) |
Surface area 203 Å2 P107 L123 A124 69 65 54 |
sIL-10R1/sIL10R2 interface, with patch 5 of sIL-10R1 |
|
Patch 7 Residue Access. (%) |
Surface area 178 Å2 W143 P189 57 48 |
IL-10/sIL10R2 interface, with patch 2 of IL-10 |
sIL-10R1 contains six hydrophobic patches, three in the N-terminal domain and three in the C-terminal domain (Table 2). Patches 1 and 2 are similar in size and located on the opposite sides of the domain. Patch 1 is formed by five residues coming from β-strands C' and E and helix B, as well as three residues of the C-terminal domain flexible loop connecting β-strand B and helix A. This is the largest hydrophobic region in the first domain sIL-10R1, covering the surface of 475 Å2. Patch 2 is formed by seven residues coming from β-strands A, B, and E, and covers the surface of 448 Å2. Thr25 is at the center of the patch and although it is not a hydrophobic residue, its side chain is oriented in such a way that Oγ 1 points inside the protein molecule and forms a hydrogen bond with the main chain nitrogen of Ser10, whereas Cβ and Cγ 2 atoms are exposed to solvent. Patch 3 consists of three residues of loop L2 and covers the surface of 207 Å2 in the interdomain region of sIL-10R1. Patch 4 consists of eight residues covering the surface of 546 Å2 and is located close to the C-terminus, near the area which is likely to be in contact with the cellular membrane in vivo (Table 2). Patch 5 includes six residues coming from β-turn CC' and the first half of β-strand C'. It covers the surface of 409 Å2 and has an elongated shape. Patch 6 is formed by three residues of loop L5 and covers 253 Å2 of the sIL-10R1 interdomain region.
The pattern of hydrophobic patches of sIL-10R2 is similar, although not identical, to that of sIL-10R1. sIL-10R2 has seven patches, three in the N-terminal domain and four in the C-terminal domain. Patch 1 lies at the top of the sIL-10R2 N-terminal domain and is composed of 10 residues covering the surface of 635 Å2. This is the largest hydrophobic solvent-exposed area on the surface of sIL-10R2. Patches 2 and 3 (Table 2) cover 284 Å2 and 154 Å2, respectively, and are located in the proximity of the ligand/receptor interface. Patch 4 is formed by seven residues of β-strands C' and E, filling the gap between the strands and shielding the internal hydrophobic core of the molecule from the solvent. The area of the patch is 572 Å2. Patch 5 is formed by four residues coming from the β-turn connecting strands A and B, and is 274 Å2 in size. Patches 4 and 5 are located at the very bottom of the C-terminal domain and are likely to be in contact with the cellular membrane. Patch 6 covers 203 Å2 and is located in the central part of the receptor-receptor interface. Patch 7 consists of only two residues that belong to loops L5 and L6. It is 178 Å2 in size and is a part of the IL-10/sIL-10R2 interface.
Upon formation of the ternary complex, hydrophobic patches from different molecules compensate each other, creating intermolecular hydrophobic clusters inside of ligand-receptor and receptor-receptor interfaces (Table 2, Fig. 4). Thus, one IL-10 domain complexed with two receptor chains has two such clusters (1a and 2a, Fig. 4) in the IL-10/sIL-10R1 interface, two clusters (1b and 2b) in the IL-10/sIL-10R2 interface, and two clusters (1c and 2c) in the sIL-10R1/sIL-10R2 interface. Intermolecular hydrophobic clusters found in the ternary IL-10/sIL-10R1/sIL-10R2 complex are important elements that hold molecules together after they have attracted each other by long-range ionic interactions. It is worthwhile to note that patch 1 and 2 of sIL-10R1 and patches 1, 3 and 4 of sIL-10R2 remain exposed to solvent on the surface of the ternary complex.
Figure 4.
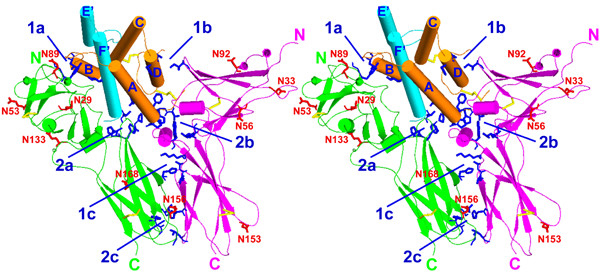
Stereo diagram of intermolecular hydrophobic clusters of the ternary IL-10/sIL-10R1/sIL-10R2 complex. Polypeptide chains of IL-10 are shown in orange and cyan, sIL-10R1 is green, and sIL-10R2 is magenta. Side chains of the residues that constitute hydrophobic clusters are shown in blue. Clusters 1a and 2a are the "top" and the "bottom" hydrophobic regions of IL-10/sIL-10R1 interface, clusters 1b and 2b are the "top" and the "bottom" hydrophobic regions of IL-10/sIL-10R2 interface and clusters 1c and 2c are the "top" and the "bottom" hydrophobic regions of sIL-10R1/sIL-10R2 interface. Disulfide bonds are shown in yellow. The potential glycosylation sites are shown in red.
The structure of the IL-10/sIL-10R1/sIL-10R2 complex
The ternary complex of IL-10/sIL-10R1/sIL-10R2 is a twofold symmetrical molecule in which each of the domains of IL-10 is bound with two receptor chains (Fig. 5). The receptor chains are bound to adjacent sides on the surface of a single IL-10 domain (Table 1). The sIL-10R1 binding site (site I) is formed by helix A, loop AB, and helix F', while the sIL-10R2 binding site (site II) is made by helices A and D (Fig. 6). sIL-10R1 and sIL-10R2, bound to the same IL-10 domain, interact with each other by their D2 domains, forming a receptor-receptor binding site (site III, Fig. 6). The distance between C-termini of sIL-10R1 and sIL-10R2 is 25.1 Å, which lies within the 25–32 Å range found in the structures of other ternary complexes [23-25,29]. The structure of the binary/intermediate complex of IL-10/sIL-10R1 [22] is the same in the ternary complex. We found only minor movements of some side chains involved in the interactions with sIL-10R2 in sites II and III. Both sIL-10R1 and sIL-10R2 receptor chains interact with ligand loops L2-L6. Site III involves interactions between residues coming from β-strands C, C', E, F and loop L6 of sIL-10R1, and residues from β-strands A, B, E and loop L5 of sIL-10R2 (Figs. 2, 6).
Figure 5.
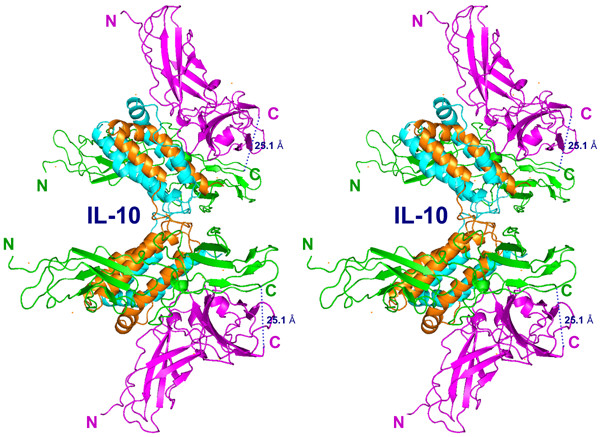
Stereo diagram of the ternary complex of human IL-10/sIL-10R1/sIL-10R2. Polypeptide chains of IL-10 are shown in orange and cyan, sIL-10R1 is green, sIL-10R2 is magenta. The hypothetical cell membrane is perpendicular to the plane of the figure on the right and the twofold symmetry axis (not shown) is horizontal.
Figure 6.
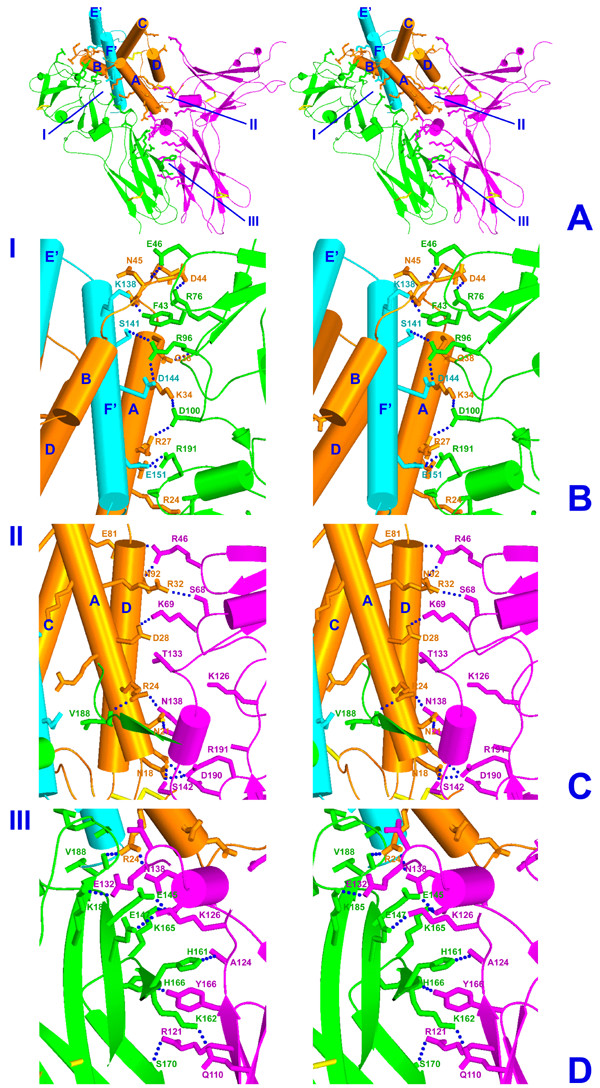
Stereo diagram of each interface within a single IL-10/sIL-10R1/sIL-10R2 signaling unit (panel A). Polypeptide chains of IL-10 are shown in orange and cyan. sIL-10R1 is green and sIL-10R2 is magenta. Panels B, C and D represent close-up view of each interface, including contact residues. (B)- an interface between IL-10 and sIL-10R1, (C)- an interface between IL-10 and sIL-10R2, (D)- an interface between sIL-10R1 and sIL-10R2. Intermolecular hydrogen bonds calculated with in 3.2 Å distance cutoff are shown as blue dotted lines.
Site I interface (IL-10/sIL-10R1)
The interaction of IL-10 with sIL-10R1 in the ternary complex is the same as that found in the crystal structure of the intermediate/binary complex [22]. Briefly, the interface is formed by the residues of the second half (middle and C-terminal) of helix A, loop AB, and helix F' of IL-10, as well as loops L2-L6 of sIL-10R1. Twenty-seven residues of the ligand interact with 23 residues of the receptor chain, creating an extensive network of hydrogen bonds (70%) (Table 3, Figs. 6A, 6B) and hydrophobic interactions (30%) (Table 2). The total change in the accessible surface for both IL-10 and sIL-10R1, calculated using a spherical probe of the radius 1.40 Å, is about 2116 Å2. Thus, the contact area between IL-10 and sIL-10R1 is about 1058 Å2. The most important residues involved in receptor-ligand binding are Pro20, Arg24, Arg27, Gln38, Glu42, Lys138, Ser141, Asp144, Glu151, and Ile158 of IL-10 and Tyr43, Arg76, Arg96, Phe143, Ser190 and Arg191 of the sIL-10R1 [22]. The IL-10/sIL-10R1 interface includes two hydrophobic clusters located at the top and at the bottom of the interface (Table 2, Fig. 4).
Table 3.
Intermolecular hydrogen bonds of the IL10/sIL10R1/sIL10R2 complex
| IL-10-IL-10R1 interface | ||||
| IL-10 | SSE | IL-10R1 | SSE | Distance Å |
| R24L NE | A | R191R O | L6 | 3.0 |
| R24L NH1 | A | V188R O | L6 | 2.9 |
| R24L NH1 | A | R191R O | L6 | 3.0 |
| R27L NE | A | S190R O | L6 | 2.9 |
| R27L NH1 | A | D100R OD2 | L4 | 3.2 |
| K34L NZ | A | D100R OD1 | L4 | 2.9 |
| Q38L OE1 | A | R96R N | L4 | 3.0 |
| D41L O | loop AB | R76R NH1 | L3 | 2.9 |
| Q42L O | loop AB | R76R NH2 | L3 | 3.0 |
| D44L OD1 | loop AB | R76R NH2 | L3 | 2.8 |
| D44L O | loop AB | G44R N | L2 | 2.9 |
| N45L N | loop AB | E46R OE2 | L2 | 3.0 |
| K138S NZ | F' | Y43R OH | L2 | 2.8 |
| S141S O | F' | R96R NH2 | L4 | 2.9 |
| D144S OD1 | F' | R96R NH2 | L4 | 2.8 |
| D144S OD2 | F' | R96R NH2 | L4 | 3.0 |
| E151S OE1 | F' | R191R NH1 | L6 | 2.9 |
| E151S OE2 | F' | R191R NH1 | L6 | 2.8 |
| E151S OE1 | F' | R191R NH2 | L6 | 2.9 |
| IL-10-IL-10R2 interface | ||||
| N18L OD1 | A | R191A NH1 | L6 | 2.7 |
| N18L ND2 | A | S142A OG1 | L6 | 2.7 |
| N21L ND2 | A | N138A O | α A | 2.8 |
| R24L NH2 | A | N138A OD1 | α A | 2.7 |
| R24L NH2 | A | T133A O | loop Bα A | 3.1 |
| D28L OD1 | A | K69A NZ | L3 | 2.8 |
| D28L OD2 | A | K69A NZ | L3 | 3.0 |
| D28L O | A | K69A NZ | L3 | 3.2 |
| R32L NH1 | A | S68A OG | L3 | 3.3 |
| E81L OE2 | loop CD | R46A NH2 | L2 | 3.3 |
| N92L OD1 | D | R46A NH1 | L2 | 2.8 |
| IL-10R1-IL-10R2 interface | ||||
| E145R OE1 | C | N138A OD1 | L5 | 3.2 |
| E145R OE2 | C | N141A ND2 | L5 | 2.7 |
| E147R OE2 | C | K126A NZ | loop Bα A | 3.2 |
| H161R NE2 | C' | A124A O | loop Bα A | 2.9 |
| K162R NZ | C' | Q110A OE1 | A | 2.6 |
| K165R NZ | loop C'E | N141A OD1 | L5 | 2.7 |
| H166R NE2 | loop C'E | Y166A OH | loop C'E | 3.0 |
| S170R OG | E | R121A NH1 | B | 3.1 |
| S170R O | E | R121A NH1 | B | 2.6 |
| K185R NZ | F | E132A OE2 | loop Bα A | 3.2 |
(*)- SSE: Secondary structure elements. (**)- In INF-γ and IL-10 structures helix F' is a topological equivalent of helix D of other helical cytokines. (***)- Interface areas were calculated with program Surface (CCP4) using spherical probe of the radius 1.40 Å2.
Site II interface (IL-10/sIL-10R2)
The interface of site II is formed by helix A, loop CD, and helix D of IL-10, as well as by loops L2, L3, L6, and α-helix A of the sIL-10R2 (Table 2, Figs. 6A, 6C). Helix A interacts with loops L3, L6, and helix A, whereas helix D interacts with loop L2 of sIL-10R2. Site II is smaller than site I; its total surface is 568 Å2, which is only 54% of the surface of site I. IL-10 and sIL-10R2 each donate 13 residues to the interface. Interacting residues are mostly polar (~80%) and link the ligand to the receptor via hydrogen bonds (Table 3) and hydrophobic interactions (Table 2). The most important interface residues are Pro16, Asn18, Asn21, Arg24, Asp28, Arg32, Glu81, Ala89, His90, and Asn92 of IL-10 and Arg46, Ile47, Ser68, Lys69, Thr133, Asn138, Ser142, Trp143, and Arg191 of sIL-10R2. Most interactions occur between IL-10 helix A and loops L3 and L6 of sIL-10R2, with only a few contacts between IL-10 helix D and loop L2 of the sIL-10R2. Arg24 of IL-10 occupies a unique position in the complex. Its side chain projects down towards the cell membrane along the receptor-receptor interface and forms hydrogen bonds with the carbonyl oxygen of Val188 of sIL-10R1 and the side-chain atom Oδ 1 of Asn138 of sIL-10R2, linking both receptor chains together. The IL-10/sIL-10R2 interface also includes a hydrophobic cluster located at the bottom part of the interface, formed by Pro16 of the ligand and Trp143 of the receptor. The upper part of the IL-10/sIL-10R2 interface also includes a hydrophobic cluster formed by Ala89 and His90 of IL-10, and I47 of sIL-10R2. Unlike in the cluster 2c (Fig. 4), the hydrophobic area 1c is weaker but still is an important element of the IL-10/sIL-10R2 interface. A smaller interface area and a reduced amount of interacting residues reflect the low affinity of the sIL-10R2 towards its binding partners.
Site III interface (sIL-10R1/sIL-10R2)
The interface between sIL-10R1 and sIL-10R2 is formed by residues that belong to β-strands C, C', E, and F of sIL-10R1 and by residues of β-strands A, B, E, and loop L5 of sIL-10R2 (Tables 2, 3; Fig. 6D). The surfaces of C-terminal domains of the receptors are complementary. Within 3.8 Å distance cutoff, receptors 1 and 2 donate 15 and 13 residues, respectively, to form a contact area that is composed of 65% hydrophilic and 35% hydrophobic residues and is about 803Å2 in size (Tables 2, 3). The most important residues are Gly116, Phe117, Glu145, Glu147, His161, Lys162, Lys165, His166, Ser170, Leu172, Gly175, and Lys185 of sIL-10R1, and Gln110, Leu114, His119, Arg121, Leu123, Ala124, Lys126, Glu132, Asn138, Asn141, and Tyr166 of sIL-10R2. The contact area between two receptors can be divided into three parts. The upper part of site III is hydrophilic and contains a wide network of hydrogen bonds. Almost all residues that are located in this area are hydrophilic. The middle part of the interface has a roughly equal number of charged and uncharged residues, while the lower part of the receptor/receptor interface is almost solely hydrophobic, with an intermolecular hydrophobic cluster at the bottom of C-terminal domains of the receptors (Figs. 4, 6).
Glycosylation sites
The receptors sIL-10R1 and sIL-10R2 contain six and four potential N-linked glycosylation sites, respectively (Fig. 4). In the complex, all sites are fully exposed to the solvent and are not part of receptor-ligand or receptor-receptor interfaces. Moreover, each particular potential carbohydrate-binding site is located at a sufficient distance from the interfaces, which eliminates the possibility of a sugar chain clashing with a neighboring protein molecule. In sIL-10R1, Asn29 flanks the top end of the receptor N-terminal domain, whereas Asn53 and Asn89 are in the central part of the domain. They are located on loop BC, β-strand C', and in the region between β-strands G1 and G2, respectively. The N-terminal domain of sIL-10R2 also has three potential glycosylation sites: Asn33, Asn56, and Asn92. Asn33 is located close to the N-terminal end of the domain, whereas Asn56 and Asn92 are found in the central part of the domain, on the opposite sides of a β-sandwich.
The C-terminal domain of sIL-10R1 contains three potential glycosylation sites at Asn133, Asn156, and Asn168, located near the α-helix A, on β-turn CC', and at the beginning of β-strand E (Figs. 2, 4), respectively. The site at Asn133 is located near the N-terminal end of the domain, close to the interdomain region of sIL-10R1. Asn156 is at the bottom of the C-terminal domain, while the oligosaccharide attached to Asn168 is in the central part of domain D2. In the ternary complex, this residue is likely to be in contact with the cellular membrane. The C-terminal domain of sIL-10R2 has only one potential carbohydrate site, Asn153, which is close to the C-terminus of the receptor. This residue is a part of β-turn CC' and is very close to position Asn156 of sIL-10R1 upon superposition of the receptor chains.
Discussion
It is clear that the quality of any theoretical model should be assessed based on its agreement with experimental data such as, for example, mutagenesis. Unfortunately, we were unable to find any such data for the IL-10/IL-10R1/IL-10R2 complex, either in literature or as personal communications. Therefore, to evaluate the correctness of the model, we can only rely on the commonly accepted criteria: the model is in the global minimum of energy; general similarity to other ternary complexes of cytokines; usage of similar, well defined receptor binding sites on the surface of the ligand (Table 1); and correlation between the available kinetic binding data and intermolecular binding surfaces and contacts. As we have already mentioned, a general similarity of the ternary IL-10 complex to known structures of other ternary complexes and involvement of the similar ligand binding sites have been postulated from the beginning; thus, these conditions have been necessarily satisfied. The model of the ternary complex does correspond to the minimum of the energy, and the pattern of the interactions of sIL-10R2 with the binary IL-10/sIL-10R1 complex agrees well with its low affinity nature. Recent study of binding of IL-10 to IL-10R2 by peptide scans [40] showed that, although IL-10R2 did not form a binary complex with IL-10, it recognized regions of helix A and loop AB of IL-10 (amino acid residues 19–43) when they were presented as 15-mer peptides. In our model, the IL-10/IL-10R2 interface includes residues 18–32 of IL-10 (Table 3). Among the residues involved in creating the IL-10R1/IL-10R2 interface, seven residues of IL-10R1 have the same residue type as residues 33–43 of IL-10 (Table 2, Table 3). This may explain the necessity of formation of IL-10/IL-10R1 binary complex before IL-10R2 can join and complete the ternary signaling complex.
IL-10 binds to the receptor chains via two sites located on the adjacent sides of its four-helix bundle [30]. Site I is formed by helices A and F', whereas site II is formed by helices A and D (Table 1). Both receptor chains interact with IL-10 through their cytokine recognition motifs, which consist of the loops and β-turns of the receptor interdomain region connecting consecutive antiparallel β-strands [22-29]. Typically, the low-affinity receptor cannot bind to a ligand by itself. Its association occurs via a cooperative binding site that is formed by both the ligand and the high-affinity receptor. With the exception of the prolactin ternary complex, C-terminal domains of the receptors contact each other in the vicinity of the cellular membrane (Table 1). Such interaction brings the intracellular parts of the receptors together, resulting in activation of signal transduction.
Receptor chains of the ternary IL-10 complex interact with each other via β-strands C, C', E, and F of sIL-10R1 and β-strands E, B, A, and loop L5 of sIL-10R2, forming an interface between the sides of their C-terminal domains Tables (2, 3; Figs. 4, 5, 6). The distance between the Cα atoms of the C-termini of IL-10R1 and IL-10R2 is 25.1 Å. In the known structures of other ternary complexes, the distances between the Cα atoms of the last residues of the extracellular parts of the receptor chains vary between 25 and 32 Å.
The surfaces of IL-10 and of both the sIL-10R1 and sIL-10R2 receptors contain several hydrophobic patches (Table 2). In the ternary complex, most of these areas face each other to form intermolecular hydrophobic clusters. Each interface contains two such areas that flank opposite sides of contact regions (Fig. 4). The bottom clusters of IL-10/sIL-10R1 and IL-10/sIL-10R2 touch each other at the point where the ligand and both receptors join, creating a larger hydrophobic area formed by all three molecules. Such a cluster is not observed in other structures of ternary complexes and is, therefore, unique for the ternary IL-10 complex.
Both receptors have several potential glycosylation sites, which are located on protein surfaces in places where they do not conflict with the three-dimensional organization of the IL-10/sIL-10R1/sIL-10R2 complex (Fig. 4). The role of carbohydrates in forming and maintaining cytokine/receptor complexes is not well understood. It was shown that the granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor α subunit requires N-glycosylation for binding and signaling [41]. Tunicamycin treatment inhibited GM-CSF binding in a dose-dependent manner, with a maximum of 85% inhibition at 3 μg/ml. Treatment of human leukemia HL-60 cells with tunicamycin completely blocked GM-CSF-induced tyrosine phosphorylation, which suggests that N-glycosylation of the receptor is necessary for intracellular signaling [41]. However, the non-glycosylated mutant of sIL-10R1 is capable of forming a stable binary complex with IL-10 [22]. GM-CSF is a glycoprotein in which glycosylation is not required for its biological activity. GM-CSF that is expressed in E. coli and thus, not glycosylated, retains full activity [42]. By contrast, the IL-10R2 binding region on IL-22 contains an N-linked carbohydrate that is likely important for binding [43]. Similarly, N-linked glycosylation of viral IL-6 is shown to increase gp130 binding and biological activity [44]. Taking into account that no carbohydrates are involved in the intermolecular interactions in the ternary IL-10/sIL-10R1/sIL-10R2 complex, as well as the fact that deglycosylated sIL-10R1 can form a quite stable binary complex with the IL-10, we may conclude that N-linked oligosaccharides are not essential for the formation of the biologically active ternary IL-10/IL-10R1/IL-10R2 complex. It is clear that more structural data are required to address the question of why some cytokines and their receptors depend upon the presence of carbohydrates, whereas others do not.
It was shown that a series of anti-human IL-10 antibodies can efficiently neutralize this cytokine [45]. Based on the recognition epitope, antibodies can be divided into three groups, A, B and C. The epitopes were identified using IL-10-derived, overlapping peptide scans prepared by spot synthesis. Antibodies of group A inhibit biological activity of IL-10 in an approximately equimolar ratio, at concentrations as low as 10 pM [45]. It has been shown that monoclonal antibody CB/RS/2 recognizes two binding regions of a discontinuous epitope on the surface of the molecule that comprises the N-terminal half of helix A and helix D [46]. Thus, this antibody binds to site II of the ligand and prevents IL-10 from association with its low-affinity receptor. An overlapping peptide scan technique has also been used for mapping IL-10 residues that bind to sIL-10R1, as well as mapping the residues of sIL-10R1 that interact with the ligand [47]. It was shown that residues of helices A and C of the ligand interact with the residues of loops L3-L6 of the high-affinity receptor. The structure of the IL-10/sIL-10R1 complex confirmed those results [22]. However, no mapping data exist for the residues that form the IL-10/sIL-10R2 interface. Such data would be very valuable to guide the design of biochemical experiments.
In the absence of sIL-10R1, IL-10 cannot bind to sIL-10R2, and the two receptor chains cannot interact with each other without a ligand. Therefore, assembly of the ternary complex consists of two steps. In the first step, IL-10 binds to sIL-10R1, forming a binary complex; subsequently, sIL-10R2 binds to the preformed IL-10/sIL-10R1 complex, completing the ternary complex. Such differences in binding abilities could be linked to the physical characteristics of the corresponding interfaces. The area of site I (1058 Å2) is about 25–45% larger than the areas of site II (568 Å2) and site III (803 Å2). The number of residues involved in interface formation is also larger for the IL-10/sIL-10R1 contact region. Site I is formed by a total of 50 residues, whereas sites II and III are composed of 26 and 28 residues, respectively. However, when combined together, sites II and III have an area and number of charged and hydrophobic interactions comparable to those of site I, which may explain the ability of the low-affinity receptor to bind to a preformed ligand-high-affinity receptor complex. This is also true if we compare the areas of binding sites I, II and III in other ternary complexes of cytokines such as GH, PL, EPO, and IL-6; there, the area of site I is usually larger than each of the areas of site II or III, whereas combined area of sites II and III are roughly equal to or larger than the area of site I [23,25,24,29].
Methods
The model of sIL-10R2, the low-affinity receptor, was generated using the published crystal structure of sIL-10R1 as a template (pdb code 1J7V ([22]). The original amino acid sequence of sIL-10R1 was mutated to that of sIL-10R2 in accordance with the alignment of their sequences shown in Figure 3 (20.4% identity and 53.1% similarity) [34,48]. Model building was followed by energy minimization, which included 500 cycles of positional refinement to optimize stereochemistry of newly built parts of the structure, followed by simulated annealing slow cool protocol and another 500 cycles of positional refinement. To preserve the overall fold, a 2.8 kcal/mole restraint has been imposed on all Cα atoms of the model. All other main and side-chain atoms were allowed to move freely within the appropriate stereochemical parameters.
A model of the ternary complex of one domain of IL-10 with sIL-10R1 and sIL-10R2 was generated on the assumption that the structure of the intermediate binary complex IL-10/sIL-10R1 does not change much upon binding of the second receptor chain, sIL-10R2, and that mutual arrangement of ligand and receptor chains is similar to what was found in the crystal structure of IL-6/IL-6-Rα /gp130 [29]. At the first step, the Cα atoms of the structure of one domain of IL-10 complexed with sIL-10R1 were superimposed onto Cα atoms of IL-6/sIL-6Rα, and then the structure of sIL-10R2 was superimposed onto gp130. Subsequently, the best fit of charge complementarity of sIL-10R2 to the intermediate binary complex was achieved upon superposition of the C-terminal domain of sIL-10R2 with the C-terminal domain of gp130, followed by rotation of sIL-10R2 as a whole around the axis perpendicular to the cell surface and passing through the center of mass of the IL-10 domain, followed again by translation along the surface of helices A and D. The second half of the hexameric IL-10/sIL-10R1/sIL-10R2 complex was generated by applying twofold symmetry (Fig. 5). Model building was followed by the energy minimization procedure under conditions described above until the drop in total energy between consecutive refinement steps was less than 1%. Root mean square deviations in positions of Cα atoms for IL-10, sIL-10R1 and sIL-10R2 in the ternary complex before and after energy minimization were 0.38 Å, 0.40 Å and 0.45 Å, respectively.
Sequence alignment was performed with program CLUSTALW 1.74 [48], model building with program CHAIN [49], and energy minimization with XPLOR 3.1 [50]; all superpositions were performed with program LSQKAB from CCP4 [51]. Figures 1, 4, and 6 were generated with program SETOR [52], while Figure 5 was generated with the program pyMOL (DeLano W.L. The PyMOL Molecular Graphics System 2002, DeLano Scientific, San Carlos, CA, USA).
Authors' contributions
SP and EM performed amino acid sequence alignment, generated the model and participated in writing the manuscript. AW participated in discussions and writing the manuscript. AZ conceived the study and participated in all stages of the work. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This publication has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade name, commercial products or organizations imply endorsement by the U.S. Government.
Contributor Information
Sergei Pletnev, Email: svp@ncifcrf.gov.
Eugenia Magracheva, Email: eugenia@ncifcrf.gov.
Alexander Wlodawer, Email: wlodawer@ncifcrf.gov.
Alexander Zdanov, Email: zdanov@ncifcrf.gov.
References
- Moore KW, de Waal MR, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and Related Cytokines and Receptors. Annu Rev Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries J. Interleukin-10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J Exp Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defrance T, Vanbervliet B, Briere F, Durand I, Rousset F, Banchereau J. Interleukin 10 and transforming growth factor beta cooperate to induce anti-CD40-activated naive human B cells to secrete immunoglobulin A. J Exp Med. 1992;175:671–682. doi: 10.1084/jem.175.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Garcia E, Defrance T, Peronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci USA. 1992;89:1890–1893. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Snipes L, Dhar V, Bond MW, Mosmann TR, Moore KW, Rennick DM. Interleukin 10: a novel stimulatory factor for mast cells and their progenitors. J Exp Med. 1991;173:507–510. doi: 10.1084/jem.173.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil IA, Suda T, Moore KW, Mosmann TR, Zlotnik A. IL-10, a novel growth cofactor for mature and immature T cells. J Immunol. 1990;145:4167–4173. [PubMed] [Google Scholar]
- Suda T, O'Garra A, MacNeil I, Fischer M, Bond MW, Zlotnik A. Identification of a novel thymocyte growth-promoting factor derived from B cell lymphomas. Cell Immunol. 1990;129:228–240. doi: 10.1016/0008-8749(90)90200-B. [DOI] [PubMed] [Google Scholar]
- Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon g. Structure. 1995;3:591–601. doi: 10.1016/S0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- Walter MR, Nagabhushan TL. Crystal structure of interleukin 10 reveals an interferon gamma- like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- Zdanov A, Schalk-Hihi C, Wlodawer A. Crystal structure of human interleukin-10 at 1.6 Å resolution and a model of a complex with its soluble receptor. Protein Sci. 1996;5:1955–1962. doi: 10.1002/pro.5560051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A. Structural features of the interleukin-10 family of cytokines. Curr Pharm Des. 2004;10:3873–3884. doi: 10.2174/1381612043382602. [DOI] [PubMed] [Google Scholar]
- Walter MR. Strucure of interleukin-10/interleukin-10R1 complex: a paradigm for class 2 cytokine activation. Immunol Res. 2002;26:303–308. doi: 10.1385/IR:26:1-3:303. [DOI] [PubMed] [Google Scholar]
- Walter MR. Crystal structures of alpha-helical cytokine-receptor complexes: we've only scratched the surface. BioTechniques. 2002;Suppl:46–57. [PubMed] [Google Scholar]
- Liu Y, Wei SHY, Ho ASY, Malefyt RW, Moore KW. Expression cloning and characterization of a human IL-10 receptor. J Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res. 2002;22:1099–1112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- Krause CD, Mei E, Xie J, Jia Y, Bopp MA, Hochstrasser RM, Pestka S. Seeing the light: preassembly and ligand-induced changes of the interferon gamma receptor complex in cells. Mol Cell Proteomics. 2002;1:805–815. doi: 10.1074/mcp.M200065-MCP200. [DOI] [PubMed] [Google Scholar]
- Pletnev S, Magracheva E, Kozlov S, Tobin G, Kotenko SV, Wlodawer A, Zdanov A. Characterization of the recombinant extracellular domains of human interleukin-20 receptors and their complexes with interleukin-19 and interleukin-20. Biochemistry. 2003;42:12617–12624. doi: 10.1021/bi0354583. http://PM:14580208 [DOI] [PubMed] [Google Scholar]
- Josephson K, McPherson DT, Walter MR. Purification, crystallization and preliminary X-ray diffraction of a complex between IL-10 and soluble IL-10R1. Acta Crystallogr D Biol Crystallogr. 2001;57:1908–1911. doi: 10.1107/S0907444901016249. [DOI] [PubMed] [Google Scholar]
- Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/S1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, Finer-Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- Elkins PA, Christinger HW, Sandowski Y, Sakal E, Gertler A, de Vos AM, Kossiakoff AA. Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat Struct Biol. 2000;7:808–815. doi: 10.1038/79047. [DOI] [PubMed] [Google Scholar]
- Hage T, Sebald W, Reinemer P. Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface. Cell. 1999;97:271–281. doi: 10.1016/S0092-8674(00)80736-9. [DOI] [PubMed] [Google Scholar]
- Aritomi M, Kunishima N, Okamoto T, Kuroki R, Ota Y, Morikawa K. Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme. Nature. 1999;401:713–717. doi: 10.1038/44394. [DOI] [PubMed] [Google Scholar]
- Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, Zauodny PJ, Narula SK. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Presnell SR, Cohen FE. Topological distribution of four-a-helix bundles. Proc Natl Acad Sci USA. 1989;86:6592–6596. doi: 10.1073/pnas.86.17.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Holm L, Sander C. The Immunoglobulin Fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF. Shared architecture of hormone binding domains in type I and II interferon receptors. Cell. 1990;61:753–754. doi: 10.1016/0092-8674(90)90182-E. [DOI] [PubMed] [Google Scholar]
- Kotenko SV. The family of IL10 related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/S1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- Ma JC, Dougherty DA. The Cationminus signpi Interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- Scrutton NS, Raine AR. Cation-pi bonding and amino-aromatic interactions in the biomolecular recognition of substituted ammonium ligands. Biochem J. 1996;319 ( Pt 1):1–8. doi: 10.1042/bj3190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton DJ, Watowich SS, Katz L, Lodish HF. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J Biol Chem. 1996;271:4699–4708. doi: 10.1074/jbc.271.23.13754. [DOI] [PubMed] [Google Scholar]
- Hilton DJ, Watowich SS, Murray PJ, Lodish HF. Increased cell surface expression and enhanced folding in the endoplasmic reticulum of a mutant erythropoietin receptor. Proc Natl Acad Sci U S A. 1995;92:190–194. doi: 10.1073/pnas.92.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K, Witte E, Reineke U, Witte K, Friedrich M, Sterry W, Asadullah K, Volk HD, Sabat R. Is there an interaction between interleukin-10 and interleukin-22? Genes Immun. 2005;6:8–18. doi: 10.1038/sj.gene.6364224. [DOI] [PubMed] [Google Scholar]
- Ding DX, Vera JC, Heaney ML, Golde DW. N-glycosylation of the human granulocyte-macrophage colony-stimulating factor receptor alpha subunit is essential for ligand binding and signal transduction. J Biol Chem. 1995;270:24580–24584. doi: 10.1074/jbc.270.41.24580. [DOI] [PubMed] [Google Scholar]
- Kaushansky K, O'Hara PJ, Hart CE, Forstrom JW, Hagen FS. Role of carbohydrate in the function of human granulocyte-macrophage colony-stimulating factor. Biochemistry. 1987;26:4861–4867. doi: 10.1021/bi00389a038. [DOI] [PubMed] [Google Scholar]
- Logsdon NJ, Jones BC, Allman JC, Izotova L, Schwartz B, Pestka S, Walter MR. The IL-10R2 binding hot spot on IL-22 is located on the N-terminal helix and is dependent on N-linked glycosylation. J Mol Biol. 2004;342:503–514. doi: 10.1016/j.jmb.2004.07.069. [DOI] [PubMed] [Google Scholar]
- Dela Cruz CS, Lee Y, Viswanathan SR, El Guindy AS, Gerlach J, Nikiforow S, Shedd D, Gradoville L, Miller G. N-linked glycosylation is required for optimal function of Kaposi's sarcoma herpesvirus-encoded, but not cellular, interleukin 6. J Exp Med. 2004;199:503–514. doi: 10.1084/jem.20031205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat R, Seifert M, Volk HD, Glaser RW. Neutralizing murine monoclonal anti-interleukin-10 antibodies enhance binding of antibodies against a different epitope. Mol Immunol. 1996;33:1103–1111. doi: 10.1016/S0161-5890(96)00072-7. [DOI] [PubMed] [Google Scholar]
- Reineke U, Schneider-Mergener J, Glaser RW, Stigler RD, Seifert M, Volk HD, Sabat R. Evidence for conformationally different states of interleukin-10: binding of a neutralizing antibody enhances accessibility of a hidden epitope. J Mol Recognit. 1999;12:242–248. doi: 10.1002/(SICI)1099-1352(199907/08)12:4<242::AID-JMR461>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Reineke U, Sabat R, Volk HD, Schneider-Mergener J. Mapping of the interleukin-10/interleukin-10 receptor combining site. Protein Sci. 1998;7:951–960. doi: 10.1002/pro.5560070412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Sack JS. CHAIN-a crystallographic modeling program. J Mol Graph. 1988;6:244–245. doi: 10.1016/s0076-6879(97)77011-3. [DOI] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR Version 31 A System for X-ray Crystallography and NMR. New Haven, Yale University Press; 1992. [Google Scholar]
- CCP4 Collaborative Computational Project, Number 4, 1994. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Evans SV. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-T. [DOI] [PubMed] [Google Scholar]
- Thiel DJ, le Du MH, Walter RL, D'Arcy A, Chene C, Fountoulakis M, Garotta G, Winkler FK, Ealick SE. Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex. Structure Fold Des. 2000;8:927–936. doi: 10.1016/S0969-2126(00)00184-2. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. 2003;12:577–589. doi: 10.1016/S1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci USA. 2002;99:9404–9409. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–2155. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]


