Abstract
Protein design studies using coiled coils have illustrated the potential of engineering simple peptides to self-associate into polymers and networks. Although basic aspects of self-assembly in protein systems have been demonstrated, it remains a major challenge to create materials whose large-scale structures are well determined from design of local protein–protein interactions. Here, we show the design and characterization of a helical peptide, which uses phased hydrophobic interactions to drive assembly into nanofilaments and fibrils (“nanoropes”). Using the hydrophobic effect to drive self-assembly circumvents problems of uncontrolled self-assembly seen in previous approaches that used electrostatics as a mode for self-assembly. The nanostructures designed here are characterized by biophysical methods including analytical ultracentrifugation, dynamic light scattering, and circular dichroism to measure their solution properties, and atomic force microscopy to study their behavior on surfaces. Additionally, the assembly of such structures can be predictably regulated by using various environmental factors, such as pH, salt, other molecular crowding reagents, and specifically designed “capping” peptides. This ability to regulate self-assembly is a critical feature in creating smart peptide biomaterials.
Keywords: biomaterial, coiled coil, protein design, circular dichroism, atomic force microscopy
Designed proteins are valuable paradigms for engineering at the nanoscale, offering favorable properties such as atomic-level precision and tight regulation of self-assembly by using a variety of environmental cues (i.e., pH, ionic strength, temperature) (1–3). As one of the most well studied and naturally abundant structural motifs in proteins, coiled coils are particularly suited to protein design. Their basic sequence feature, the heptad repeat, is a seven-residue pattern (abcdefg)n of nonpolar and polar residues that gives rise to amphipathic α-helices. The hydrophobic effect drives the burial of nonpolar residues at the helix-pairing interface and influences geometric details of resulting structures (4). Electrostatic and polar residues at buried or solvent-exposed locations provide additional means to manipulate structural features by stabilization or destabilization (negative design) of key interactions (5–8).
Recent attempts to design self-assembling protein networks by using coiled coils have focused on simple systems, such as linear and branched fibrils (9–14), planar assemblies (15), and hydrogels (16–18). Specifically, filament formation has been achieved by stabilization of intermediate structures that foster intermolecular coiled-coil interactions (11–14). Previous studies have reported the design of short helical peptides, which interact via a dimeric coiled-coil motif and are stabilized by a combination of overlapping hydrophobic and electrostatic intermolecular interactions (9, 13, 14). These peptides do indeed self-assemble into filaments; however, these filaments also show evidence of extensive lateral association, to form fibrils; problems with such aggregation continue to pose a major obstacle toward the formation of well controlled nanoscale structures (9, 14). Here, we present a dual-component design strategy for the engineering of self-assembling peptides, using hydrophobic interactions to favor axial assembly and electrostatic forces to regulate lateral assembly in the formation of nanoropes. Evidence for self-assembled polymers in solution is provided by analytical ultracentrifugation and dynamic light scattering (DLS) experiments. The solution studies are complemented by atomic force microscopy (AFM) imaging of polymers deposited on mica surfaces. Additionally, CD experiments demonstrate that our system also fulfills another tenet of nanoengineering: reversible regulation of self-assembly using a variety of mechanisms, such as solvent or temperature factors.
Materials and Methods
Molecular Modeling. The starting molecular model for energy minimizations was the CpA helix interface as generated from the C2 symmetric crystal structure of GCN4 (19) by using allbuild and generate algorithms from the DeGrado laboratory (20). Energy minimizations were run by using insightii 2000 (Biosym Technologies, San Diego). Side chains were manually adjusted to the most energetically favorable rotamer, and molecular mechanics were run in discover by using a combination of steepest descents and conjugate gradients to optimize the modified structure. Some additional modeling was carried out to test the effect of introducing the S-acetamidomethyl (Acm) group on the cysteines on the structure. The N-terminal cysteine placed the polar Acm group in a highly solvated position at the expense of minor unraveling of the first few residues of the helix. However, this unraveling was not caused by significant steric overlap because the more centrally located cysteine maintained the burial of the Acm group; furthermore the Acm group was rotated to a position suggestive of an H bond with a free amide group on the neighboring peptide, thus possibly acting to stabilize the staggered dimer intermediate.
Peptide Synthesis and Purification. The synthesis of CpA was carried out on an Applied Biosystems 433A peptide synthesizer (PerSeptive Biosystems, Framingham, MA) by using standard N-α-(9-fluorenylmethyloxycarbonyl) chemistry and 5-(4′-aminomethyl-3′,5′-dimethoxyphenoxy)-valeric acid resin (Advanced ChemTech), which provides an amide at the carboxyl terminus. CpA was synthesized in two forms, using either Cys(Acm) or Cys(trityl). Cys(Acm) uses a side-chain blocking group that is not removed during trifluoroacetic acid (TFA) cleavage and protects the side chain from unwanted oxidation. Cys(trityl) can be removed with TFA and was used to test the effects of disulfide crosslinking on polymerization. The peptides were acetylated at the amino terminus before TFA cleavage. The peptides were purified by RP-HPLC, and the identities of the products were verified by MALDI-TOF MS. Stock solutions of peptides were prepared by lyophilization and dissolution in MilliQ-purified water to ≈1–2 mM concentration, followed by filtration (0.2 μm) to remove large contaminants. Concentration of the peptide solutions was determined by a modified ninhydrin procedure (21).
Analytical Ultracentrifugation. All ultracentrifuge experiments were performed in a Beckman model Optima XL-A analytical ultracentrifuge with an An-60 Ti rotor. Velocity experiments used two-channel Epon, charcoal-filled centerpieces with 12-mm pathlengths containing 435-μl samples and 450-μl buffer references. Sedimentation velocity (SV) of the solute boundary was assayed at a speed of 50,000 rpm and temperatures of 4°C or 25°C. Absorbance data were collected with a radial step size of 0.003 cm. Using the dcdt+ program v. 1.16 (John Philo, Thousand Oaks, CA), multiple species models were fit to the processed data set to acquire sedimentation and diffusion constants. Molecular weights for analyzed species were calculated from these constants by using the Svedberg equation. Parameter values for peptide partial specific volume, solvent densities, and viscosities were calculated with the sedntrp program (22). The lengths and diameters of the filaments were estimated by using a cylindrical model for the frictional coefficient in the Svedberg equation. The Svedberg equation, in combination with a volume equation based on the molecular weight of the filament, was used to solve for values of diameter (d) and length (L); this calculation was performed by using sedntrp (22).
DLS. DLS data were collected on a DynaPro-MS/X molecular sizing instrument (Protein Solutions, Lakewood, NJ), which uses an 825-nm laser (used at 10% power) and a fixed scattering vector of 90°. Instrument operation, data collection, and analysis were managed through the dynamics (V6) software interface. Solution volumes of 15 μl were measured in a freshly cleaned cuvette to reduce dust contamination. Samples were studied at 4°C, 12°C, or 25°C. An autocorrelation function, based on the raw scattering data, was fit to a distribution of species decay times, which were used to calculate the diffusion coefficient distributions by using a simple theoretical model. For each analyzed sample, ≈25 s of scattering data were collected per measurement to yield a value for the diffusion coefficient. Several measurements were taken, and the set of diffusion coefficients was statistically analyzed.
AFM. Data were collected in tapping mode by using a Bioscope atomic force microscope (Digital Instruments, Santa Barbara, CA) to obtain size and shape information for polymers deposited under various conditions. Cantilevers were from Nanodevices Metrology Probes, (Redding, CA). Samples were prepared in 10 mM Tris·HCl, pH 8.0 and the appropriate salt and incubated on a freshly cleaved mica surface for 10 min at 4°C or 25°C to foster polymer deposition. Imaging was performed after sample blow drying and desiccation. Data analysis used standard Digital Instruments software and the wsxm scanning probe microscopy program.
CD Spectropolarimetry. CD data were collected on an Aviv Associates (Lakewood, NJ) 62A CD spectropolarimeter. Thermal denaturations were monitored from 2° to 98°C at 222 nm, with a step size of 2°C and equilibration time delay of 2 min. Wavelength scans were performed at 25°C and monitored from 198 to 250 nm, using a step size of 0.5 nm and a signal averaging time of 3 s. Standard conditions for most experiments used 10 mM Tris·HCl, pH 8.0. All measurements were taken by using a bandwidth of 1.5 nm.
Results and Discussion
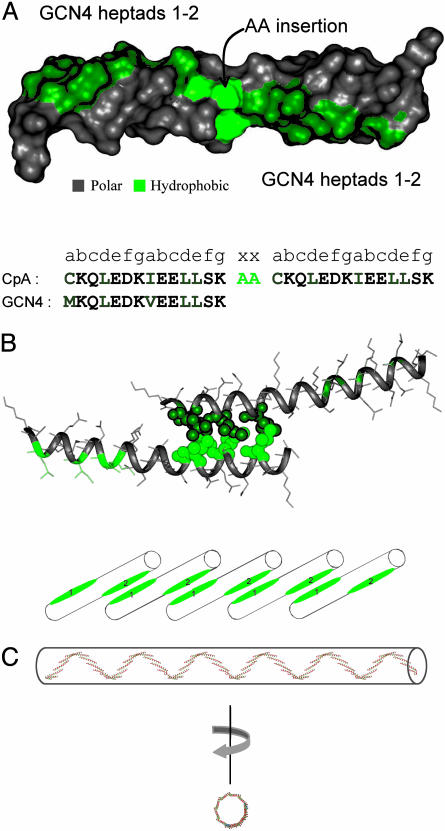
Borrowing from the leucine zipper motif of the yeast transcription factor, GCN4, our design strategy implements a well studied parallel coiled coil that facilitates protein dimerization via helix pair interactions. As with other coiled-coil systems, assembly is a highly cooperative process involving concomitant helix formation and binding between partner chains. We designed a peptide, CpA, consisting of two identical GCN4 sequence elements separated by a two-residue alanine insertion. The insertion introduces a phase shift in the heptad repeat, generating two hydrophobic ridges oriented at ≈200° with respect to each other on the α-helix (Fig. 1A). Blunt-end dimers, as seen in native GCN4, are no longer possible in this configuration (Fig. 1B); rather, dimers with exposed hydrophobic “sticky ends” form instead, initiating the template for an open-assembly polymerization reaction (Fig. 1C). CpA self-assembly is engineered to require an enhancement of the hydrophobic effect. Its two-heptad coiled-coil interface provides insufficient surface area to induce structure under typical aqueous conditions (23). Additives such as salt or glycerol, which are known to enhance the hydrophobic effect, thus provide means to activate a solution of free monomers to form polymers. Additionally, native residues have been replaced with: (i) cysteines at the first a position of each GCN4 module to provide the potential for covalent crosslinking of polymers; and (ii) isoleucines at the second a position in each GCN4 module to increase the specificity for dimerization (4).
Fig. 1.
Protein design of a phased-hydrophobic polymer. (A) Surface representation of CpA, color-coded in green to highlight the hydrophobic residues. The amino acid sequence represents a tandem repeat of the first two heptads from the coiled-coil domain from the yeast transcription factor, GCN4, with a two-alanine insertion between the two pairs of heptads. The sequence of the designed peptide is shown in juxtaposition to the native sequence from GCN4 to highlight the differences. The methionine at the first position has been mutated to cysteine to allow crosslinking of the polymers through the formation of disulfide bonds. (B) Molecular model of the CpA dimer interface, showing the packing interactions between residues highlighted as space-filling representations. A staggered arrangement of individual helix interactions fosters polymerization as each unsatisfied hydrophobic surface on the growing chain recruits additional peptide units, as shown by the cylinders below the model. (C) Molecular model of a self-assembled polymer, illustrating several higher-order structural features. The two-alanine insertion results in a phase shift of ≈200° of the C-terminal hydrophobic surface, relative to the N-terminal hydrophobic surface, leading to a third level of supercoiling. Rotation about an axis perpendicular to the direction of coiling allows for a view down the supercoil, allowing for a visual representation of the width (≈6.7 nm).
To model the structure of the CpA polymer, we first assume that the geometry of the helix-pairing interaction in our peptide is identical to the WT interaction (19, 24). Idealized geometric parameters describing the GCN4 coiled coil were extracted and then used to model a two-heptad helical dimer interface (20). This structure was extended and energy was minimized to model a short polymer in which higher-order features that emerge from the geometric constraints of monomer–monomer interactions could be more easily visualized (Fig. 1C). Features of the model, such as the axial and lateral dimensions of resultant polymers, as well as the degree to which control can be exerted over higher-order self-assembly, are tested here by a variety of experimental approaches.
Much of our initial effort focused on the study of a peptide containing an Acm-protected cysteine to avoid unwanted oxidation; this group can also be readily removed to generate the free thiol. We modeled the effect of placing the bulky Acm groups in a core position and found that it could be well accommodated (see details in Materials and Methods).
We first probed for the presence of self-assembled polymers in solution by using CpA with the Acm-modified cysteine. SV data collected in 1.5 M NaCl revealed polydisperse, large polymers with s20,w values in the range of 6.1–6.9, depending on conditions (Table 1). Size distribution within the folded ensemble, as determined from g(s*) vs. s* plots from SV experiments, shows an asymmetric distribution function leaning toward smaller s values or smaller sizes. An upper limit to polymer size is clearly marked by the presence of a dominant peak in the distribution, suggesting that there is an intrinsic polymer size limit. This limit may be related to the mechanical stability of the polymer nanofilaments or kinetic assembly effects. Although the limit in assembly may be kinetically controlled, the actual assembly of individual peptides into polymers is driven thermodynamically by the burial of hydrophobic regions (see CD temperature experiments below). Each species in solution, polymer or monomer, has two hydrophobic ends capable of joining it to two other species in solution. For this reason, we believe the predominant assembly mechanism is akin to step-growth polymerization rather than the more familiar chain-growth polymerization mechanism wherein monomer units are added only to actively growing polymer chain sites (25).
Table 1. Polymerization of CpA depends on salt, temperature, and peptide concentration.
| 1.5 M NaCl
|
|||||
|---|---|---|---|---|---|
| 25°C
|
0.75 M (NH4)2SO4
|
||||
| Parameters | 100 μM | 150 μM | 4°C | 25°C | 4°C |
| s20,W | 6.1 | 6.3 | 6.9 | 5.0 | 5.2 |
| D20,W | 1.38 | 0.97 | 0.38 | 0.62 | 0.11 |
| Molecular mass, kDa | 420 | 630 | 1,800 | 780 | 4,600 |
| No. of monomers | 120 | 180 | 500 | 220 | 1,300 |
| f/f0 | 3.3 | 4.2 | 7.7 | 6.1 | 19 |
| L × d | 180 × 1.9 | 288 × 1.8 | 435 × 2.5 | 513 × 1.5 | 1,955 × 1.9 |
The samples contained 100 μM peptide (unless stated otherwise) and 10 mM Tris·HCl, pH 8.0. s20,W and D20,W are standardized values for pure water at 20°C for both the Svedberg values (in units of 10-13 s) and diffusion constants [in units of cm2·s-1 (× 10-7)], respectively, as measured by sedimentation velocity experiments (Optima XLA analytical ultracentrifuge, Beckman Instruments). The approximate molecular mass (103 Da) was calculated from the Svedberg values and diffusion constants by using dcdt+. The number of monomers is calculated by dividing the measured molecular mass with the monomer theoretical molecular mass. f/f0 is the frictional ratio relative to that expected for an anhydrous sphere. L × d are the axial and lateral dimensions of the polymers, in nm, when the data are modeled as a cylinder.
Plots of g(s*) vs. s* also provide diffusion constants for the distributions. It is well known that polydisperse dynamic equilibrium systems are refractory to calculation of accurate diffusion constants; so the values reported in Table 1 must be treated as approximations only. Nevertheless, DLS measurements of the diffusion constants under the same conditions provided values in good agreement. As a result, the molecular weight and shape of the assemblies in solution could be calculated by using the s20,w and D20,w parameters. The molecular masses of the assembled species ranged from 420 to 1,800 kDa, depending on conditions (Table 1), representing the self-assembly of hundreds of peptides per polymer. The frictional coefficients for the polymers were much greater than expected for an anhydrous sphere, suggesting elongated structures. Assuming that the polymer shape in solution can be approximated by a cylindrical model, it is straightforward to calculate the average lengths and diameters for the polymers (L × d; Table 1). Given the complexity of the mixtures, the absolute values of the lateral dimensions should be interpreted with caution. Although these values are significantly smaller than predicted by our computer model (the diameter of the superhelix is predicted to be 6.9 nm), we can say with some confidence that there is little, if any, lateral association of individual filaments. Peptides containing the cysteine without the Acm-protecting group are also able to polymerize in the presence of salt, indicating that the Acm-protecting group is not solely responsible for polymerization (Table 3, which is published as supporting information on the PNAS web site).
To test the hypothesis that polymerization is being driven by salting-out effects (enhancement of the hydrophobic contribution to stability), we tested self-assembly in ammonium sulfate, a much better salting-out agent than NaCl. High concentrations of ammonium sulfate led to the formation of much larger species than did NaCl (Table 1). This effect is studied more carefully in AFM and CD experiments described below.
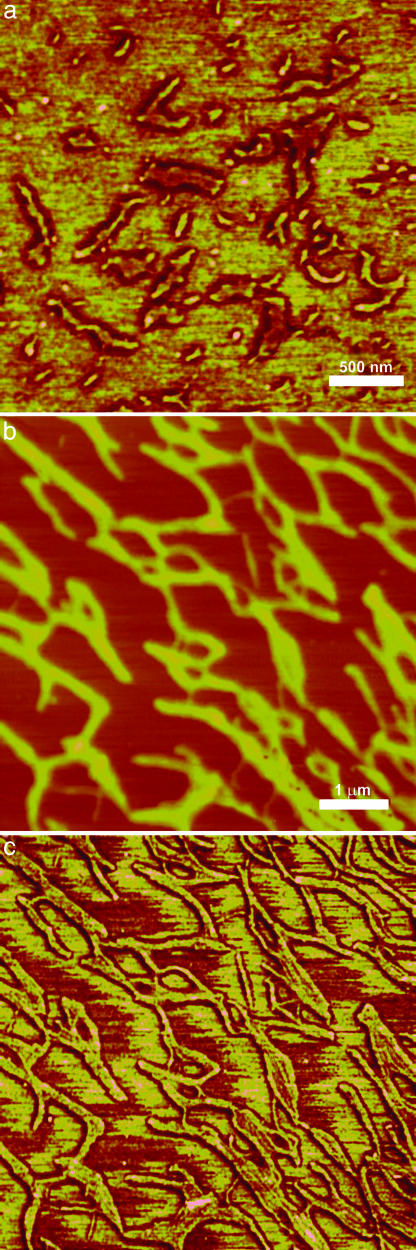
To more explicitly test the predictions from SV measurements for the axial and lateral dimensions, we took tapping-mode AFM images of CpA polymers. Polymers deposited on a mica surface in the presence of NaCl reveal nanofilaments 86–376 nm in length (Fig. 2a), in surprisingly good agreement with the lengths predicted from SV. There is significant heterogeneity in nanofilament length, certainly owing to both solution properties (as seen by SV) and the details of sample preparation on a surface. During the drying process, solvent evaporation leads to increased salt and peptide concentration, which should significantly affect polymer size. Indeed, in other images, we see local dense pockets of longer nanofilaments on the surface that we attribute to such effects. Nevertheless, despite the heterogeneity in length, we observe a uniform width with little evidence for lateral association, again in strong agreement with the SV measurements. We see fairly straight rods in this length regime, with minor defects caused by kinks or curvature. Both of these features are consistently observed with other designed coiled coils that form nanofilaments in this size range and probably reflects the natural persistence length of such structures (9, 13, 14).
Fig. 2.
Tapping-mode AFM images reveal filamentous polymers and fibrils (nanoropes). Samples were prepared by using 144 μM peptide and 10 mM Tris·HCl, pH 8.0. (a) Phase image of nanofilaments formed in 1.5 M NaCl. (b) Topography image of nanofilaments formed in 0.75 M (NH4)2SO4; z-scale, 25 nm. The nanofilaments are much longer than those in a and show greater tendency to associate. (c) Phase image showing the same area and magnification as in b. The internal structure is better resolved in the phase images.
Interestingly, when ammonium sulfate is used to induce polymerization in place of NaCl, the nanofilaments imaged with AFM were up to several micrometers in length and showed more proclivity for lateral association, resulting in the formation of fibrils or nanoropes (Fig. 2 b and c). Indeed, SV data show polymers formed from ammonium sulfate are larger than those formed in NaCl (Table 1). We suggest that ammonium sulfate fosters lateral association of filaments, as the sulfate ion can bridge solvent-exposed lysine residues between individual nanofilaments.
Polymer height measurements (2.9 ± 0.6 nm in NaCl and 3.2 ± 0.8 nm in ammonium sulfate) are in reasonable agreement with the lateral dimensions calculated from the SV experiments. However, these values are significantly smaller than the lateral dimensions predicted by our model. Although our model may require further refinement for predicting the lateral dimension, it is not clear how accurate the lateral dimension predicted from SV analysis is because a cylinder may not correctly approximate the sedimentation or diffusion behavior of a supercoiling rod. For AFM, the heights of protein structures are consistently smaller than the expected diameters of such structures in solution (26, 27). Here, we suggest that interactions with the mica surface might cause a partial collapse of the suprahelical structure, which, when combined with forces exerted by the AFM tip to compress the material (Fig. 1C), result in undersized height measurements.
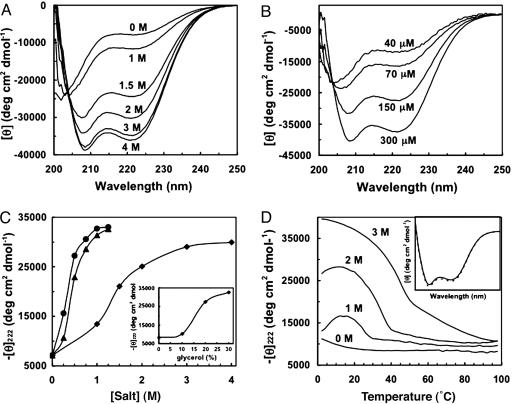
Having established the ability of the CpA peptide to form polydisperse filaments with well defined lateral dimensions, we wanted to verify that the polymers are indeed helical in nature, and then explore the underlying physicochemical principles governing self-assembly. CD measurements confirm the helical nature of the polymers in solution. Furthermore, acquisition of helical structure is rapid, occurring within seconds (samples obtain a constant CD signal within the dead time of manual mixing). Formation of a helical structure highly depends on both NaCl concentration (Fig. 3A) and peptide concentration (Fig. 3B), mirroring the effects of salt and peptide concentration on polymerization as seen in our SV experiments (Table 1).
Fig. 3.
The stability of the polymers can be modulated by environmental conditions. (A) Spectra were measured at 25°C by using 102 μM peptide and 10 mM Tris·HCl, pH 8.0. (B) Polymerizing systems under thermodynamic control depend on monomer (or peptide) concentration. Samples were measured at 25°C and contained 2 M NaCl and 10 mM Tris·HCl, pH 8.0. (C) Comparison of salt effects on helix stability suggests sulfate preferentially stabilizes structure. Samples were measured at 25°C by using 144 μM peptide and 10 mM Tris·HCl, pH 8.0. NaCl (▪), (NH4)2SO4 (▴), and Na2SO4 (•). (Inset) The effect of glycerol on helix content is shown. (D) Temperature unfolding experiments reveal the stability of the CpA polymers as a function of NaCl concentration. Samples were measured by using 144 μM peptide and 10 mM Tris·HCl, pH 8.0. At the highest NaCl concentration, a single transition is seen whose midpoint is ≈55°C. (Inset) The helical spectrum before (solid line) and after (dashed line) thermal denaturation, demonstrating quantitative reversibility, is shown.
At low salt concentrations, no helix content is observed, consistent with the well known observation that helical structure is tightly linked to coiled-coil formation (and in our case, concomitant self-assembly). The folding appears to occur in a concerted fashion in response to salt, suggesting that the self-assembly process, as monitored by the acquisition of helix content, is cooperative, similar to general protein folding principles (Fig. 3C). We hypothesize that the salt induces structure through an enhancement of hydrophobic interactions because of the salting-out effect. This hypothesis is supported by the fact that ammonium sulfate, a stronger salting-out agent, can induce structure at lower concentrations (Fig. 3C). Surprisingly, the effect of sodium sulfate is comparable to the effect seen with ammonium sulfate, suggesting that the enhanced salting-out effect is largely owing to the sulfate dianion. Additionally, the sulfate may play a role in inducing structure through filament crosslinking, as seen in the AFM images; there is no reason to believe that these dual roles should be mutually exclusive. The helical structure was found to be strongly temperature-dependent, reflecting the temperature dependence of self-assembly (SV experiments, Table 1). Thermal unfolding profiles were measured as a function of increasing NaCl concentration (Fig. 3D). At low salt concentrations, two transitions are evident, because of both heat- and cold-denaturation processes, a commonly observed phenomenon in coiled-coil assembly (28). At the highest NaCl concentration, where the peptide is almost exclusively found in polymer form, only a single thermal unfolding transition is present because the cold-denaturation midpoint falls below measurable limits. These thermal unfolding experiments also reveal that polymer assembly is highly reversible upon cooling, as measured by recovery of helical signal after thermal unfolding (Fig. 3D Inset). Similar CD experiments were carried out with CpA containing a free cysteine to test the influence of the Acm group: salts, specifically NaCl (data not shown) and ammonium sulfate (Fig. 4A, which is published as supporting information on the PNAS web site), increase helix content, presumably because of increased polymerization, and the stability is temperature-sensitive (Fig. 4B).
The NaCl experiments described above strongly suggest that structure-stabilizing salt effects can be ascribed to enhancement of the hydrophobic effect; this hypothesis is borne out by experiments showing that glycerol, a nonelectrolyte, also known to enhance the hydrophobic effect, can induce helix content, and consequent polymerization (Fig. 3C Inset). No significant pH dependence (pH range of 2 to 12) was observed in the helical signal as monitored by CD, suggesting that polymer stability is largely insensitive to electrostatic effects (data not shown). To further demonstrate that the hydrophobic effect is responsible for the structure and stability of the polymer, we designed additional peptides with modifications to the hydrophobic core (a heptad position). Replacement of the two cysteines at the a heptad positions with a more polar residue, serine, results in a peptide that is much more resistant to polymerization, whereas replacement with a nonpolar residue, isoleucine, greatly facilitates polymerization in response to NaCl as judged by CD experiments. The peptide with isoleucine in place of cysteine in the core has a sedimentation coefficient, S20,w, of 5.5 as measured in 1.5 M NaCl at 25°C, which, when coupled with the measured diffusion constant, gives an average molecular mass of 674 kDa. The molecular mass of CpA under identical conditions is only 420 kDa. Thus, we conclude from these experiments that CpA polymerization is a rapid, fully reversible, highly cooperative process and a dynamic equilibrium between polymer assembly and disassembly leading to a heterogeneous population of higher-order structures.
Given the similarities between CpA assembly and step-growth polymerization, we felt an engineered peptide capable of binding to only a single CpA molecule, rather than two, could be used to control the average size of the nanofilament assemblies. Such a peptide, called CpAcap, was synthesized by incorporating only the first two heptad repeats of CpA. Binding of this peptide to the hydrophobic “sticky ends” of a CpA polymer ultimately should lead to polymers of smaller size. Consistent with these predictions, it was found that polymer size decreases and diffuses more quickly as the molar excess of CpAcap is increased relative to CpA, as seen by DLS (Table 2). The scattering intensity caused by the presence of CpA polymers is also reduced and becomes extremely scarce in solutions of ×6 molar capping excess, disappearing entirely at higher concentrations. This finding confirms that the CpAcap peptide is fully capable of reducing average polymer length, consistent with the effect of mixing monofunctional and bifunctional monomers in a step-growth polymerization (25).
Table 2. Polymerization of CpA can be regulated by CpAcap, a capping peptide.
| [CpAcap]/[CpA] | D20,w (cm2·s-1) × 10-7 |
|---|---|
| 1 | 1.30 ± 0.23 |
| 2 | 1.44 ± 0.34 |
| 4 | 2.34 ± 1.12 |
| 6 | 2.56 |
Diffusion constants were measured by DLS using 100 μM CpA in 1.5 M NaCl, 10 mM Tris·HCl, pH 8.0. The scattering intensity was found to decrease with increasing [CpAcap]/[CpA] becoming too low for the 6× sample to get a meaningful error value and disappearing entirely at 8× and 12× molar excess of CpAcap over CpA.
We have demonstrated that our designed peptides fold into well defined structures that are consistent with our design work and molecular modeling studies. Using the hydrophobic interaction in place of electrostatics as the driving force for polymerization, we avoid previously noted problems with nonspecific aggregation to form nanofilaments. We propose that electrostatic interactions can be used to modulate higher-order, lateral association to form nanoropes, as suggested by our experiments with ammonium sulfate. Polymer structure can be regulated by a number of factors, including salt, temperature, and peptide concentration. Additionally, capping agents demonstrate significant concentration-dependent regulatory effects on polymer assembly. Peptide biomaterials have been well recognized as offering unique opportunities for regulation and responsiveness to environmental cues, thus making them ideally suited as smart biomaterials (2, 3). Our work provides a concerted approach to identifying simple smart responses and provides a foundation for future, more elaborate mechanisms to impart smart behavior. This work is a prelude to future protein design and nanoengineering studies, in which peptide polymer scaffolds can be used for the attachment of chemical groups to the peptides that will confer functionality to the structure of the polymer (29).
Supplementary Material
Acknowledgments
We thank Bill DeGrado and Ben North for help with implementation of computer algorithms for coiled-coil parameterization and Lawrence Lee, Kevin Monahan, and Bala Tanjore for technical assistance. This work was supported by National Science Foundation Grants DBI-9970203, MCB-9817188, and MCB-0211754; a grant from the David and Lucile Packard Foundation; and the Howard Hughes Medical Institute.
Author contributions: D.E.W., W.M.A., G.E.N., A.D.S., W.F.S., and R.F. designed research; D.E.W., C.L.P., W.M.A., G.E.N., E.D.C., A.D.S., and R.F. performed research; D.E.W., A.D.S., W.F.S., and R.F. analyzed data; D.E.W., A.D.S., W.F.S., and R.F. wrote the paper; and W.F.S. supervised AFM work.
Abbreviations: AFM, atomic force microscopy; DLS, dynamic light scattering; Acm, S-acetamidomethyl; SV, sedimentation velocity.
References
- 1.Whitesides, G. M., Mathias, J. P. & Seto, C. T. (1991) Science 254, 1312–1319. [DOI] [PubMed] [Google Scholar]
- 2.Yeates, T. O. & Padilla, J. E. (2002) Curr. Opin. Struct. Biol. 12, 464–470. [DOI] [PubMed] [Google Scholar]
- 3.MacPhee, C. E. & Woolfson, D. N. (2004) Curr. Opin. Solid State Mater. 8, 141–149. [Google Scholar]
- 4.Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. (1993) Science 262, 1401–1407. [DOI] [PubMed] [Google Scholar]
- 5.Fairman, R., Chao, H. G., Lavoie, T. B., Villafranca, J. J., Matsueda, G. R. & Novotny, J. (1996) Biochemistry 35, 2824–2829. [DOI] [PubMed] [Google Scholar]
- 6.Akey, D. L., Malashkevich, V. N. & Kim, P. S. (2001) Biochemistry 40, 6352–6360. [DOI] [PubMed] [Google Scholar]
- 7.Zhu, H., Celinski, S. A., Scholtz, J. M. & Hu, J. C. (2000) J. Mol. Biol. 300, 1377–1387. [DOI] [PubMed] [Google Scholar]
- 8.Oakley, M. G. & Kim, P. S. (1998) Biochemistry 37, 12603–12610. [DOI] [PubMed] [Google Scholar]
- 9.Ryadnov, M. G. & Woolfson, D. N. (2003) Nat. Mater. 2, 329–332. [DOI] [PubMed] [Google Scholar]
- 10.Ryadnov, M. G. & Woolfson, D. N. (2003) Angew. Chem. Int. Ed. Engl. 42, 3021–3023. [DOI] [PubMed] [Google Scholar]
- 11.Ogihara, N. L., Ghirlanda, G., Bryson, J. W., Gingery, M., DeGrado, W. F. & Eisenberg, D. (2001) Proc. Natl. Acad. Sci. USA 98, 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandya, M. J., Spooner, G. M., Sunde, M., Thorpe, J. R., Rodger, A. & Woolfson, D. N. (2000) Biochemistry 39, 8728–8734. [DOI] [PubMed] [Google Scholar]
- 13.Potekhin, S. A., Melnik, T. N., Popov, V., Lanina, N. F., Vazina, A. A., Rigler, P., Verdini, A. S., Corradin, G. & Kajava, A. V. (2001) Chem. Biol. 8, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 14.Zimenkov, Y., Conticello, V. P., Guo, L. & Thiyagarajan, P. (2004) Tetrahedron 60, 7237–7246. [Google Scholar]
- 15.Prive, G. G., Anderson, D. H., Wesson, L., Cascio, D. & Eisenberg, D. (1999) Protein Sci. 8, 1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, C., Kopecek, J. & Stewart, R. J. (2001) Biomacromolecules 2, 912–920. [DOI] [PubMed] [Google Scholar]
- 17.Wang, C., Stewart, R. J. & Kopecek, J. (1999) Nature 397, 417–420. [DOI] [PubMed] [Google Scholar]
- 18.Petka, W. A., Harden, J. L., McGrath, K. P., Wirtz, D. & Tirrell, D. A. (1998) Science 281, 389–392. [DOI] [PubMed] [Google Scholar]
- 19.O'Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. (1991) Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- 20.North, B., Summa, C. M., Ghirlanda, G. & DeGrado, W. F. (2001) J. Mol. Biol. 311, 1081–1090. [DOI] [PubMed] [Google Scholar]
- 21.Rosen, H. (1957) Arch. Biochem. Biophys. 67, 10–15. [DOI] [PubMed] [Google Scholar]
- 22.Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Biopolymer Science, eds. Harding, S. E., Rowe, A. J. & Horton, J. C. (Royal Society of Chemistry, Cambridge, U.K.), pp. 90–125.
- 23.Su, J. Y., Hodges, R. S. & Kay, C. M. (1994) Biochemistry 33, 15501–15510. [DOI] [PubMed] [Google Scholar]
- 24.O'Shea, E. K., Rutkowski, R. & Kim, P. S. (1989) Science 243, 538–542. [DOI] [PubMed] [Google Scholar]
- 25.Odian, G. G. (1991) Principles of Polymerization (Wiley, New York).
- 26.Umemura, K., Sutoh, K., Tokunaga, F., Kataoka, M., Kamikubo, H., Arakawa, H. & Ikai, A. (1996) Scanning 18, 275–280. [Google Scholar]
- 27.Ji, X., Oh, J., Dunker, A. K. & Hipps, K. W. (1998) Ultramicroscopy 72, 165–176. [DOI] [PubMed] [Google Scholar]
- 28.Boice, J. A., Dieckmann, G. R., DeGrado, W. F. & Fairman, R. (1996) Biochemistry 35, 14480–14485. [DOI] [PubMed] [Google Scholar]
- 29.Vandermeulen, G. W. & Klok, H. A. (2004) Macromol. Biosci. 4, 383–398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.