Abstract
‘Ki-energy’ (life-energy) is believed to increase the immune activity of its practitioners. It has also been shown to cause neuropsychological effects. We undertook this study to obtain objective and scientific evidence as to whether or not a ‘Ki-effect’ could inhibit the growth of cultured cancer cells. Cultured human liver carcinoma cells, HepG2, were used. A Japanese Ki-expert held his fingers toward the cells in culture dishes for 5 or 10 min. After culturing for 24 h, we measured cell numbers, protein concentration per cell, certain mRNA expressions and the synthesis of regucalcin. The results were compared with those for control cells (non-treated cells). We found that the number of cells in the Ki-exposed groups were less than those in the controls by 30.3 and 40.6% with 5 and 10 min Ki-exposure, respectively. The protein content per cell in the Ki-exposed groups (5 and 10 min) was higher than that in the control groups by 38.8 and 62.9%, respectively. These results were statistically significant. Using RT–PCR, we found that the mRNA expression for c-myc, a tumor stimulator gene, was decreased, while that for regucalcin, which suppresses DNA synthesis, was increased. Our molecular biological studies and mathematical model analysis demonstrated that Ki-energy inhibited cancer cell division. The data also indicate that the Ki-effects involve some form of infrared radiation from the human body. This study suggests the possibility that Ki-energy may be beneficial for cancer patients because it suppresses cancer cell growth, and at the same time, it stimulates immune functions of the patients.
Keywords: breathing exercise, c-myc, cancer cells, HepG2, infrared radiation, regucalcin
Introduction
Many people believe that ‘Ki-energy’ (life-energy) is a ‘superpsychological’ or ‘supernatural’ phenomenon. The aim of this paper is to provide evidence that ‘Ki’ is a ‘natural’ phenomenon. If it is a ‘natural’ phenomenon, it can be studied scientifically. In order to accomplish this goal, we formed a research team consisting of two biophysicists, two molecular biologists and a Japanese Ki-expert, Kozo Nishino (who was once a medical student, then a ballet choreographer and finally became a master of Aikido, a Japanese martial art). Nishino has developed a breathing method of enhancing the level of life-energy or vitality called ‘Ki’ (1–6). Many of his students experienced improvement in their health in areas such as high blood pressure, osteoporosis, arthritis and heart ailments. A dozen students overcame cancer without surgery (5–9). A study demonstrated that this breathing method increased the immune activity of the practitioners as judged by the blood level of NK cell activity (10). Nishino developed his Breathing Method independently of the Chinese Qigong practice.
Qigong practice has been known to raise energy levels (called ‘Qi’ in Chinese) and improve health (11–18). A group of Japanese scientists made a pioneering observation that the Chinese Qigong practice involved a neuropsychological effect in which the brain wave activities of both a Qigong healer and a volunteer who received his Qi-energy were synchronized (19).
Using human liver carcinoma cells, HepG2, we performed a series of simple, straightforward experiments to test whether the Ki-effect could be studied scientifically. Namely, cells in culture dishes were exposed to 5 or 10 min of Ki-emission from Nishino's fingers. They were subsequently incubated for 24 h to examine whether cell growth was reduced as compared to those in the control dishes to which no Ki was applied.
We searched to see if a specific protein was expressed in Ki-exposed cells, since that might relate to a mechanism of the Ki-effect. We also tested the possibility of whether or not Nishino's Ki-energy might penetrate skin and muscle to reach cancer-afflicted organs and attack cancer cells. From the study of the materials that pass or block the Ki-effect, we also tried to search for the nature of the Ki-energy.
Materials and Methods
Chemicals
α-Minimum essential medium (α-MEM) and penicillin (5000 U ml−1)–streptomycin (5000 μg ml−1) solution were obtained from Gibco Laboratories (Grand Island, NY, USA). Fetal bovine serum (FBS) was purchased from Sigma-Aldrich (St Louis, MO, USA).
Cell Culture
All experiments were performed at the Graduate School of the University of Shizuoka. The cloned human hepatoma cells, HepG2, were maintained for 24–72 h in α-MEM supplemented with 10% FBS, 50 U ml−1 penicillin and 50 μg ml−1 streptomycin in humidified 5% CO2 or 95% air at 37°C. We normally seeded 1 × 105 cells per ml per dish and cultured until the start of the experiment. (When we needed a larger number of dishes for an experiment, the seeded cell number per dish was decreased accordingly.) The cells were settled and attached onto the dishes in ∼3 h and proliferated. We performed the Ki-exposure experiments between 24 and 48 h after seeding, in other words, long after the cells were settled. Before the experiment, we took several dishes and counted the cell numbers of each dish to confirm that the numbers were equal for all dishes. The number was normally ∼2–4 × 105 cells per ml per dish. Under these conditions, the dishes were <20% confluency (in 35 mm diameter culture dishes) (20). Since this was much less than the full confluent condition, the cell doubling time, which was 24 h under our conditions, did not change with the cell number.
Application of Ki-energy
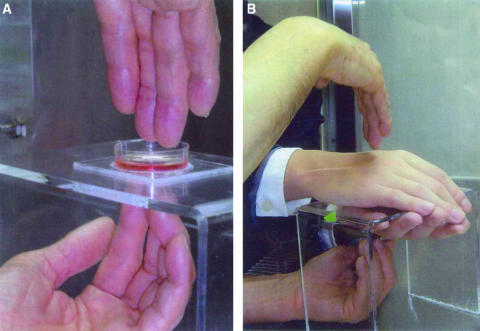
Cultured dishes were randomly distributed into the control and Ki-exposure groups. Then, each culture dish was taken out to a sterilized hood and the lid was removed. The dish was exposed to the Ki-energy emitted from Nishino's fingers (Fig. 1A) for 5 or 10 min. His fingers were sterilized with 70% ethanol before each exposure in order to prevent possible bacterial contamination. The control dish was handled similarly without the Ki-exposure. The culture dishes were returned to the incubator and incubated for 24 h. We chose 24 h because the cell doubling time was 24 h. Then, cell counting, protein studies and/or RT–PCR were performed. Since both control and Ki-exposed groups had the same number of cells at the start of the experiment, the difference between these two groups were caused by the Ki-effect.
Figure 1.
(A) Ki-energy was applied to a culture dish through Nishino's fingers. (B) The culture dish was covered by both hands of another individual through which the Ki-energy was applied. Cells were subsequently cultured for 24 h.
We also tested the effect of the hands of another person (who had no Ki training and who covered the dish as shown in Fig. 1B) on the Ki inhibition of cell growth. This is a test for evaluating the effect of an intervening human tissue when Ki is emitted aiming at an internal organ afflicted by cancer. Our experiments per day consisted of three sets of 5 min control and 5 min Ki-exposure experiments (n = 3), and one or two sets of 10 min experiments (n = 1 or 2). We limited the amount of Ki-emission for the purpose of maintaining the level of the emitter's Ki-energy. In order to accumulate more data points, we performed 5 min experiments on three different days (total n = 9). Although the exact cell numbers per dish were slightly different in each experiment, by taking the ratio between the control and the Ki-exposed group, and by analyzing it with appropriate statistics we were able to obtain statistically meaningful data.
Cell Counting
After trypsinization of the cells in each culture dish using a Ca2+/Mg2+-free phosphate-buffered saline (PBS) containing 0.2% trypsin and 0.02% EDTA for 2 min at 37°C, cells were collected and wash-centrifuged in a PBS solution at 100 g for 5 min. The cells were resuspended in a 0.5 ml PBS solution, and an aliquot was stained with eosin. The cells were counted under a microscope using a hemocytometer plate. For each dish, we took the average of two counts.
Protein Studies
After culture, cells were washed three times with PBS, scraped with 0.5 ml of ice-cold 0.25 M sucrose solution containing 1 mM phenylmethylsulfonyl fluoride and 10 μg ml−1 leupeptin, and sonicated for 60 s. Then, the samples were used for protein quantitation by the method of Lowry (21) or for immunoassay of regucalcin by western blot analysis.
Quantification of Specific mRNA by RT–PCR
In order to investigate the molecular mechanism behind the Ki-effect, we examined the change in tumor-related gene expression in the cloned human hepatoma HepG2 cells exposed to Ki-energy. The cells were exposed to Ki-energy for 5 min, and were cultured for another 24 h. The changes in mRNAs for c-myc [a tumor stimulator gene, (22)], p53 [a tumor suppressor gene, (23)], regucalcin [a protein which suppresses DNA synthesis, (24)] and β-actin (a protein not related to cancer) were analyzed by using RT–PCR. Total RNAs were prepared using the method of Chomczynski and Sacchi (25) from the cloned human hepatoma HepG2 cells. RT–PCR was performed with a Titan™ One Tube RT–PCR Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) as recommended by the supplier to determine the gene expression of c-myc, p53, regucalcin or β-actin.
The primers were generated based upon the published human sequences. The primers for the amplification of c-myc cDNA were 5′-(692)-CCAGCGAGGATATCTGGAAG-(712)-3′ and 5′-(1239)-CGTCGAGGAGAGCAGAGAAT-(1259)-3′ (22). The primers for p53 cDNA were 5′-(369)-CCAGCTTCGGAACAAGAGAC-(389)-3′ and 5′-(910)-CACAGAGCCAGGCTTTCATC-(930)-3′ (23). The primers for regucalcin were 5′-(316)-GGAGGCTATGTTGCCACCATTGGA-(317)-3′ and 5′-(850)-CCCTCCAAAGCAGCATGAAGTTG-(872)-3′ (26). The primers for β-actin cDNA were 5′-(410)-CCAAGGCCAACCGCGAGAAGATGAC-(434)-3′and 5′-(996)-AGGGTACATGGTGGTGCCGCCAGAC-(996)-3′ (27).
RT–PCR was performed using a reaction mixture (20 μl) containing 1 μg of total RNAs, the RT–PCR buffer supplied, the Titan™ enzyme mix (AMV and Expand™ High Fidelity), 0.2 mM deoxynucleotide triphosphate, 5 mM dithiothreitol, 5 U RNase inhibitor, 2.5 U Taq DNA polymerase and 0.3 μM primers. Samples were incubated at 50°C for 30 min and at 94°C for 2 min, and then amplified for 25 sets under the following conditions; denaturation for 30 s at 94°C, annealing for 30 s at 60°C and extension for 60 s at 68°C. The amplified PCR products were separated using electrophoresis on a 1.5% agarose gel, and visualized by ethidium bromide staining and quantitated using a densitometer.
Western Blot Analysis
The homogenate of cultured cell was centrifuged for 10 min at 5500 g at 4°C, and the supernatant was used for western blot analysis (28). An aliquot of protein (10 μg) was subjected to SDS–PAGE (12% polyacrylamide gel). After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane at 100 mA for 4 h. The membranes were incubated for 1 h with a polyclonal rabbit anti-regucalcin antibody (29), which was diluted 1:2000 with a washing buffer [10 mM Tris–HCl, pH 8, containing 150 mM NaCl, 0.1% (w/v) Tween-20] containing 5% (w/v) skim milk. The membrane was then washed four times with the washing buffer and, subsequently, the membrane was incubated for 1 h with horseradish peroxidase-linked anti-rabbit IgG, which was diluted 1:5000 with the washing buffer containing 5% (w/v) skim milk. After washing the membrane, protein bands were detected and quantitated using an enhanced chemiluminescent kit (Biosciences, Piscataway, NJ, USA) following the manufacture's instruction. The molecular size of the detecting protein was determined by running the standard proteins with known sizes in parallel.
Statistical Analysis
This was done using StatView™ software. Data were expressed as the mean ± SEM. For the data shown in Fig. 2, the significance of the differences between the control and the Ki-exposed groups was determined by the Student's t-test for comparison of two groups, and by analysis of variance with the Fisher's PLSD test for comparison of multiple groups. For the data shown in Table 1, data were collected from tests performed on different days. Therefore, we first expressed the Ki-effect by the percent change in each set, which consisted of a control dish and a Ki-exposed dish. Then, we analyzed them using the Mann–Whitney test (because of the non-parametric nature of the data) to determine the significance of the differences. These data expressed in terms of ratios were used for the mathematical model analysis. In all experiments, the difference was considered statistically significant when P < 0.05.
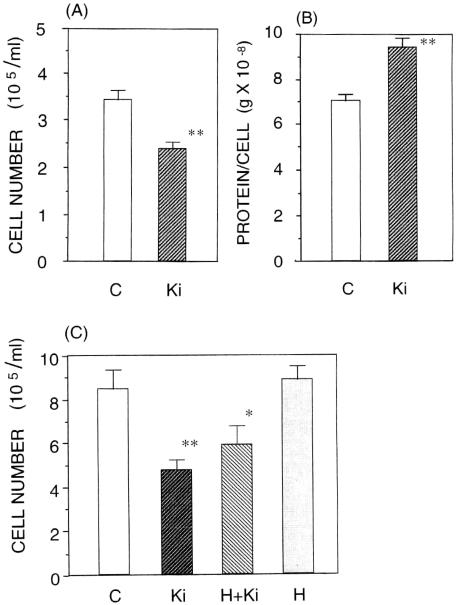
Figure 2.
Effects of 5 min of Ki-emission on cell growth in the cloned human hepatoma HepG2 cells measured after a 24 h incubation. C indicates control group while Ki indicates Ki-exposed group. Each column represents the mean±SEM of three experiments with separate dishes of cultured cells. *P < 0.05, compared with the control value. **P < 0.01, compared with the control value. (A) Inhibition of cell growth by Nishino's Ki-energy. (B) Increase of protein content by the Ki-energy (data taken on November 5, 2002). (C) Effect of human tissue (both hands of another individual without Ki training) on the Ki-induced inhibition of cell growth. Nishino applied his Ki-energy to cultured cells with and without the shielding by another individual's hands as shown in Fig. 1B (data taken on February 5, 2004). H + Ki indicates Ki being applied through hands while H indicates the effect of shielding hands alone.
Table 1.
Summary of the growth inhibition and the protein content increase by 5 and 10 min Ki-exposure
| Date of experiments | Growth inhibition (%) | Protein content increase (%) |
|---|---|---|
| (A) 5 min Ki-exposure (n = 9) | ||
| From percent change data measured on October 30, 2002, November 5, 2002 and February 5, 2004 | 30.3 ± 5.1* | 38.8 ± 3.4* |
| (B) 10 min Ki-exposure (n = 5) | ||
| From percent change data measured on September 18, 2002, October 30, 2002, December 5, 2003 and February 5, 2004 | 40.6 ± 3.4* | 62.9 ± 7.0** |
All percent change data were analyzed using the non-parametric Mann–Whitney test.
*P < 0.01 and
**P < 0.05.
Results
Ki-effects on Cell Number and Protein Content
First, we performed an experiment with 5 min of Ki-exposure. Figure 2A and B show typical data. In this experiment, the decrease in cell numbers as counted 24 h later was 30.5% (P < 0.01), and the increase in the protein content per cell was 33.6% (P < 0.01). When a person who had not practiced Nishino's method imitated the action of Nishino, no effect was observed (data not shown).
All experiments gave statistically significant results. The results for one group are shown in Fig. 2A and B and for the second group in Fig. 2C. The results for the third experiment were similar (data not shown). For the 5 min experiments, we had nine sets. For each set, we expressed the effect of Ki-exposure by a percent change. We then analyzed the effect of Ki statistically using a non-parametric Mann–Whitney test. As shown in Table 1 (A), the differences in both cell numbers and protein contents between the control and the Ki-exposed groups were statistically significant (n = 9). For 10 min Ki-experiments, we performed altogether five sets of the control and Ki-exposure experiments. We analyzed them in the same fashion. As shown in Table 1 (B), the results were also statistically significant (n = 5).
Figure 2C shows that the emission of Ki through the hand of another person still inhibited the growth (P < 0.05), although the degree of inhibition was somewhat less. The figure also shows that the shielding hands alone did not reduce the number of cells.
Mathematical Model Analysis
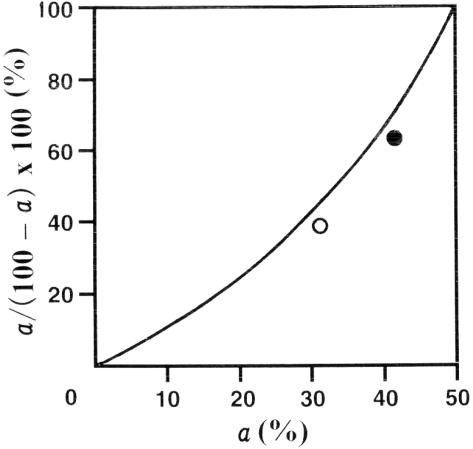
We examined the validity of our data using a simple cell division model. Suppose we start with 100 cells. When cell division is finished in 24 h, the number will be 200. If the number is reduced by Ki-energy by a%, then the number is 200 (1 − a/100). If we assume that the protein content per undivided cell and that per divided cell are 2 and 1, respectively, then the total protein amount is (100 − a) × 2 + 2a × 1 = 200 (this simple model assumes that all undivided cells reach the protein content of 2 in 24 h. This may overestimate the protein content per cell because the value for some cells would be <2). Then, the protein content per cell is 1/(1 − a/100). The percent increase in protein content per cell is expressed by a/(100 − a) × 100 (%). Figure 3 shows the relationship between the growth inhibition (a%) and the protein content increase (%). As shown in the figure, the actual values for growth inhibition and protein content increase for 5 and 10 min of Ki-exposure (taken from Table 1) are situated relatively close to this theoretical curve.
Figure 3.
Theoretical relationship between the decrease (%) in cell counts (abscissa) and the increase (%) in the protein content per cell (ordinate). The mean values for 5 and 10 min of Ki-exposure experiments are shown with open and filled circles, respectively. See details in the text.
With this model, we can also estimate the percentage of cells that have finished cell division. Let that percentage be b, then b = 100 − 2a. Therefore, with 5 and 10 min of Ki-exposure, 39.4 and 18.8% of the cells underwent cell division in 24 h, respectively. With no Ki-exposure, a = 0 and thus, b = 100%. This result supports our notion that the mechanism of Ki-inhibition is to slow down cell division in cancer cells.
Analyzing the Molecular Mechanism of the Ki-effect
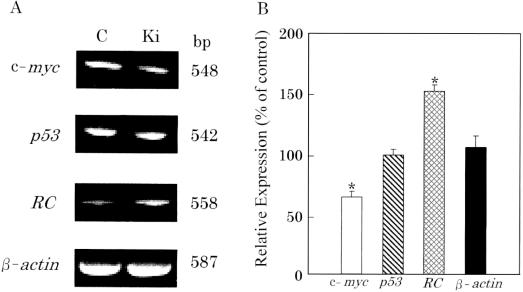
As shown in Fig. 4, the expression of c-myc mRNA was significantly suppressed (P < 0.01), while the expression of regucalcin mRNA was significantly increased (P < 0.01). The expressions of p53 and β-actin mRNAs were not altered with the Ki-exposure.
Figure 4.
Analysis of the Ki-effects on tumor-related gene expressions (c-myc, p53 and regucalcin) in the cloned human HepG2 cells by RT–PCR. Ki-energy was applied for 5 min, and cells were cultured for 24 h. Total RNAs (1 μg) extracted from the cells were analyzed by RT–PCR using specific primers (see the text). (A) A typical example of gel electrophoresis results for RT–PCR. Similar figures were obtained in all the four experiments with separate samples. (B) The densitometric data for these gene expressions are shown as a percentage of the control (mean ± SEM of four experiments). *P < 0.01, compared with the control value. RC, regucalcin; C, control experiments; Ki, Ki-exposed experiments.
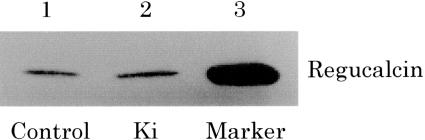
Besides its mRNA level, we also determined the protein level of regucalcin in the hepatoma cells by the western blot analysis. The level in the Ki-exposed cells was 125.1 ± 2.5% of the control (mean ± SEM; n = 4) (Fig. 5). It was statistically significant (P < 0.05). The increase in the regucalcin protein level must have resulted from the enhancement of regucalcin mRNA induced by the Ki-energy.
Figure 5.
Western blot indicating an increase of regucalcin content of the cell by Ki-energy. Lane 2 had an average of 25.1% more material than lane 1 (n = 4).
The Nature of Ki-energy
We also studied the effects of materials that could interfere with the Ki-effects. As shown in Table 2, when the culture dish was covered with aluminum foil (0.05 mm thick), the Ki-effect was completely blocked. Table 2 also shows the experiments to test the effects of plastics. Both culture dish and lid are made of transparent polystyrene (thickness 0.6 mm). Normally, we exposed culture dishes to Ki-energy without the lid. For this test, we left the lid closed and the Ki-energy was applied. As shown in this table, leaving the lid closed did not interfere with the Ki-effect. However, when we placed two plates made of black acrylic (1.6 mm thick; one on top and the other under the culture dishes) while the Ki-energy was being applied, the Ki-effect was completely blocked.
Table 2.
Effects of materials, which would intervene the Ki-effects while Ki was applied for 5 min, and cell growth was measured 24 h later
| Conditions | Cell numbers (105 per ml) |
|---|---|
| Experiment 1 (n = 3) | |
| No Ki was applied | 7.34 ± 0.16 |
| Ki was applied without the polystyrene lid | 5.95 ± 0.37* |
| Ki was applied when the dish was wrapped up with aluminum foil | 7.26 ± 0.40# |
| Experiment 2 (n = 4) | |
| No Ki was applied | 5.89 ± 0.43 |
| Ki was applied without the polystyrene lid | 3.89 ± 0.34* |
| Ki was applied with the polystyrene lid closed | 3.93 ± 0.37* |
| Ki was applied when two black acrylic plates were placed on top and under the dish | 6.06 ± 0.66# |
Thickness of the materials: aluminum foil, 0.05 mm; polystyrene lid, 0.8 mm; black acrylic, 1.6 mm.
*P < 0.05 from the control.
#P > 0.8 from the control, and P < 0.05 from the experiment where Ki was applied without the lid.
Discussion
We have conducted simple and straightforward experiments to investigate the effects of Ki-energy using cultured human carcinoma cells. The results for the measurement of both cell numbers and protein contents were statistically significant with n = 9 for 5 min Ki-exposure and n = 5 for 10 min Ki-exposure (Table 1). In separate experiments where only the cell numbers were counted, the results were also statistically significant with n = 7 (Table 2). The total number of our experiments is still small. We have to increase the number and should adopt a blind test procedure. These are goals of our future investigation.
It is interesting to note that the protein content of Ki-exposed cells was remarkably higher than that of the controls. It is well known that the content of a cell increases when the cell is preparing for cell division, and that the amount is almost doubled right before cell division. Therefore, the observed increase in the protein content of Ki-exposed cells suggests that cell division was delayed by Ki-energy. The mathematical model analysis supported this conclusion (Fig. 3).
Regucalcin, a regulatory protein in the intracellular signaling system (30), exerts a suppressive effect on proliferation of cloned rat hepatoma cells (26). Regucalcin is present in the nucleus of rat liver (31). Moreover, regucalcin suppresses the expression of c-myc and Ha-ras mRNAs in cloned rat hepatoma cells (32). Presumably, the Ki-energy suppressed proliferation of human hepatoma cells by influencing gene expression in the nucleus.
Our data indicated that the level of p53 did not change with the Ki-exposure, but the levels of c-myc and regucalcin were changed. Both hyperthermia therapy (42°C) and X-ray radiation are known to increase the p53 level in nude mice bearing human tumor cells, and consequently, these therapies inhibit the growth of the tumor (33,34). Our result implies that the observed Ki-effect is different from that caused by high temperature treatment or high energy emission. This raises the interesting possibility of using Ki-energy in cancer therapy. If the Ki-therapy is combined with hyperthermia and/or X-ray radiation, it may produce synergistic effects to inhibit tumor growth more effectively. Indeed, a Chinese medical study reported that the combination of the Qigong therapy and modern medical treatment produced beneficial effects (12). Several other scientific studies also demonstrated that Qi-energy has detectable biological and physiological effects (13,35–37).
What is the nature of observed Ki-energy? It passes through a clear plastic plate, but was blocked by aluminum foil. This suggests that it may be a form of electromagnetic wave. In this regards, it is interesting to note that Kawano et al. (19) found that a form of energy was transmitted through the air from a Chinese Qigong practitioner to another person who was sitting in front of him.
Then, a question to be asked is ‘What is the frequency?’ Since the Ki-energy goes through a clear plastic plate it cannot be a UV radiation, because plastics generally absorb ultraviolet. The fact that black acrylic plates effectively block the Ki-energy indicates that it may be a form of infrared radiation, because if this is in a radio or TV frequency range, it can go through a black plastic plate. A human body radiates infrared by virtue of its temperature. In 1978, Chinese scientists published the finding of infrared radiation from the hand of a Qigong practitioner (38). Since then, several investigators reported that the Ki-energy may involve infrared radiation (39–44).
In our experiments, if the infrared radiation was blocked, then the Ki-effect of inhibiting cell growth was lost. In other words, there was a direct linkage between the two. If the Ki-energy is a form of infrared radiation it could penetrate human tissue, therefore explaining Ki-energy's ability to inhibit cell growth through human hands (Fig. 2C).
The level of p53 was known to increase by a high temperature treatment (42°C) or by X-ray radiation (33,34). However, since the level did not change in our case (Fig. 5), the intensity of infrared radiation must be very low. We still do not know the mechanism of how such a small amount of energy from the human body could cause a tangible change in the cells.
In conclusion, we have shown evidence that the effects of Ki-energy can be analyzed scientifically using an established cell culture model. Our experiments using cultured cancer cells demonstrated that cell growth was delayed by exposure to Ki-energy for 5–10 min. The mechanism seems to be related to a decrease in mRNA expression for c-myc, a tumor stimulator gene, and to an increase of that for regucalcin, which suppresses DNA synthesis. Our data suggest that infrared radiation may be involved in the observed Ki-effects.
References
- 1.Nishino K. Ki-energy in health. Proceedings of the 4th International Congress on Traditional Asian Medicine; 1994. pp. 148–70. [Google Scholar]
- 2.Nishino K. The Breath of Life: Using the Power of Ki for Maximum Vitality. Tokyo, NY, London: Kodansha International; 1997. [Google Scholar]
- 3.Nishino K. Nishino Breathing Method (in Japanese) Tokyo: Kodansha; 1987. [Google Scholar]
- 4.Nishino K. Vital Power: Nishino Breathing Method (in Japanese) Tokyo: Mikasa-Shobo; 2003. [Google Scholar]
- 5.Nishino K. Enhancing Life Energy: Nishino Breathing Method (in Japanese) Tokyo: Chikuma-Shobo; 2003. [Google Scholar]
- 6.Nishino K. Nishino Breathing Method: Activation of Life Energy (in Japanese) Tokyo: Kodansha; 2004. [Google Scholar]
- 7.Nishino K. Vitality is Determined by Breathing (in Japanese) Tokyo: Jitsugyo no Nipponsha; 2001. [Google Scholar]
- 8.Yumi K. The Beauty Spark of the Youth Produced by the Nishino Breathing Method (in Japanese) Tokyo: Take Shobo; 2003. [Google Scholar]
- 9.Namae Y. The Power of Ki (in Japanese) Tokyo: Shogakkan; 1990. [Google Scholar]
- 10.Kimura H, Nagao F, Tanaka Y, Sakai S, Ohnishi ST, Okumura K. Beneficial effects of the Nishino breathing method on the immune activity and stress level. J Altern Complement Med. 2005;11:285–91. doi: 10.1089/acm.2005.11.285. [DOI] [PubMed] [Google Scholar]
- 11.Li XF, Bi ZX, Cheng ZY. The antitumor effects of qigong-emitted external Qi and its influence on the immunologic functions of tumor-bearing mice. J Tongli Med Univ. 1991;11:253–6. doi: 10.1007/BF02888162. [DOI] [PubMed] [Google Scholar]
- 12.Sancier KM. Therapeutic benefits of qigong exercises in combination with drugs. J Altern Complement Med. 1999;5:383–9. doi: 10.1089/acm.1999.5.383. [DOI] [PubMed] [Google Scholar]
- 13.Lee M. Effect of emitted qi on in vitro natural killer cell cytotoxic activity. Am J Chin Med. 2001;29:17–22. doi: 10.1142/S0192415X01000034. [DOI] [PubMed] [Google Scholar]
- 14.Xin Y, Fong YT, Wolf G, Wolf D, Cao W. Protective effect of XY99-5038 on hydrogen peroxide induced cell death in cultured retinal neurons. Life Sci. 2001;69:289–99. doi: 10.1016/s0024-3205(01)01122-5. [DOI] [PubMed] [Google Scholar]
- 15.Chen KW, Shiflett SC, Ponzio NM, He B, Elliott DK, Keller SE. A preliminary study of the effect of external qigong on lymphoma growth in mice. J Altern Complement Med. 2002;8:615–21. doi: 10.1089/107555302320825138. [DOI] [PubMed] [Google Scholar]
- 16.Xin Y, Fong YT, Wolf D, Wolf G, Shen H, Zahara M, et al. XY99-5038 promotes long-term survival of cultured retinal neurons. Int J Neurosci. 2002;112:1209–27. doi: 10.1080/00207450290026166. [DOI] [PubMed] [Google Scholar]
- 17.Lee M. Effects on in vitro and in vivo qi-therapy on neutrophil superoxide generation in healthy male subjects. Am J Chin Med. 2003;31:623–8. doi: 10.1142/S0192415X03001119. [DOI] [PubMed] [Google Scholar]
- 18.Chen K. An analytic review of studies on measuring effects of external QI in China. Altern Ther Health Med. 2004;10:38–50. [PubMed] [Google Scholar]
- 19.Kawano K, Koito H, Fujiki T, Shinagawa Y. EEG and topography during Chinese ‘Qigong’ training. Neurosciences. 1990;16:503–8. [Google Scholar]
- 20.Murata T, Shinya N, Yamaguchi M. Expression of calcium-binding protein regucalcin mRNA in the cloned human hepatoma cells (HepG2): stimulation by insulin. Mol Cell Biochem. 1997;175:163–8. doi: 10.1023/a:1006844815743. [DOI] [PubMed] [Google Scholar]
- 21.Lowry OH, Rosebrough NH, Farr AL, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:263–73. [PubMed] [Google Scholar]
- 22.Watt R, Stanton LW, Marcu KB, Gallo RC, Croce CM, Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983;303:725–8. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- 23.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplication of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–3. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 24.Misawa H, Yamaguchi M. Transcript heterogeneity of the human gene for Ca2+-binding protein regucalcin. Int J Mol Med. 2000;5:283–7. [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinum thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Misawa H, Inagaki S, Yamaguchi M. Suppression of cell proliferation and deoxyribonucleic acid synthesis in cloned rat hepatoma H4-II-E cells overexpressing regucalcin. J Cell Biochem. 2002;84:143–9. doi: 10.1002/jcb.1274. [DOI] [PubMed] [Google Scholar]
- 27.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessendolf JHM, Ganfinkel S, Zhan X, Brown S, Maciag T. Identification of a nuclear localization sequence within the structure of the human interleukin-1 alpha precursor. J Biol Chem. 1993;268:22100–4. [PubMed] [Google Scholar]
- 29.Yamaguchi M, Isogai M. Tissue concentration of calcium-binding protein regucalcin in rats by enzyme-linked immunoadsorbent assay. Mol Cell Biochem. 1993;122:65–8. doi: 10.1007/BF00925738. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi M. The role of regucalcin in nuclear regulation of regenerating liver. Biochem Biophys Res Commun. 2000;276:1–6. doi: 10.1006/bbrc.2000.3359. [DOI] [PubMed] [Google Scholar]
- 31.Tsurusaki Y, Misawa H, Yamaguchi M. Translocation of regucalcin to rat liver nucleus: involvement of nuclear protein kinase and protein phosphatase regulation. Int J Mol Med. 2000;6:655–60. doi: 10.3892/ijmm.6.6.655. [DOI] [PubMed] [Google Scholar]
- 32.Tsurusaki Y, Yamaguchi M. Overexpression of regucalcin modulates tumor-related gene expression in cloned rat hepatoma H4-II-E cells. J Cell Biochem. 2003;90:619–26. doi: 10.1002/jcb.10652. [DOI] [PubMed] [Google Scholar]
- 33.Asakawa I, Yoshimura H, Takahashi A, Ohnishi K, Nakagawa H, Ota I, et al. Radiation-induced growth inhibition in transplanted human tongue carcinomas with different p53 gene status. Anticancer Res. 2002;22:2037–44. [PubMed] [Google Scholar]
- 34.Takahashi A, Ota I, Tamamoto T, Asaka I, Nagata Y, Nakagawa H, et al. p53-dependent hyperthermic enhancement of tumour growth inhibition by X-ray or carbon-ion beam irradiation. Int J Hyperthermia. 2003;19:145–53. doi: 10.1080/02656730210166131. [DOI] [PubMed] [Google Scholar]
- 35.Sancier KM. Medical applications of Qigong and emitted Qi on human, animals, cell cutures and plants: review of selected scientific research. Am J Acupunct. 1991;19:367–77. [Google Scholar]
- 36.Yount G. In vitro test of external qigong. BMC Complement Altern Med. 2004;4:5. doi: 10.1186/1472-6882-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount G. Biofield perception: a series of pilot studies with cultured human cells. J Altern Complement Med. 2004;10:463–7. doi: 10.1089/1075553041323885. [DOI] [PubMed] [Google Scholar]
- 38.Kiang T. Chinese ‘Nature Magazine’: Chinese style. Nature. 1978;275:697. [Google Scholar]
- 39.Yuasa Y. Qi and Human Science (in Japanese); Proceeding of Sino-Japan Qigong Conference (in Japanese); Tokyo, Japan: Hirakawa pub. Co.; 1990. [Google Scholar]
- 40.Chien CH, Tsuei JJ, Lee SC, Huang YC, Wei YH. Effect of emitted bioenergy on biochemical functions of cells. Am J Chin Med. 1991;19:285–92. doi: 10.1142/S0192415X91000375. [DOI] [PubMed] [Google Scholar]
- 41.Yuasa Y. In: The Body, Self-Cultivation and Ki-energy. Nagatomo S., Hull M. S., translators. Albany, NY: State University of New York Press; 1993. [Google Scholar]
- 42.Machi Y. The Science of Ki (in Japanese) Tokyo: Tokyo Denki University Press; 1993. [Google Scholar]
- 43.Eisenberg D, Wright TL. Encounter with Qi: Exploring Chinese Medicine. NY and London: WW. Norton & Company, Inc.; 1995. [Google Scholar]
- 44.Fukushima M, Kataoka T, Hamada C, Matsumoto M. Evidence of Qi-gong energy and its biochemical effect on the enhancement of the phagocytic activity of human PMN leukocytes. Am J Chin Med. 2001;29:1–16. doi: 10.1142/S0192415X01000022. [DOI] [PubMed] [Google Scholar]