Abstract
Dipyridamole is an effective inhibitor of cardiovirus growth in cell culture. The effects of dipyridamole on mengovirus replication in vivo and in vitro were examined in the hope the drug could be used as an experimental analog of the poliovirus inhibitor guanidine. Guanidine selectively inhibits poliovirus RNA synthesis but not RNA translation, and as such, has been a valuable research tool. Although guanidine does not inhibit cardiovirus infection, a compound with similar discriminatory characteristics would be experimentally useful for parallel work with these viruses. We found that mengovirus plaque formation in HeLa or L cells was inhibited nearly 100% by the presence of 80 μM dipyridamole. The inhibitory effect was reversible and targeted an early step in the replication cycle. Studies with luciferase-expressing mengovirus replicons showed that viral protein synthesis was unaffected by dipyridamole, and rather, RNA synthesis was the step targeted by the drug. This assessment was confirmed by direct analyses of viral translation and RNA synthesis activities in a Krebs-2-derived in vitro system that supported complete, infectious cardiovirus replication. In Krebs extracts, dipyridamole specifically inhibited viral RNA synthesis to more than 95%, with no concomitant effect on viral protein translation or polyprotein processing. The observed inhibition reversibly affected an early step in both minus-strand and plus-strand RNA synthesis, although inhibition of plus-strand synthesis was more profound than that of minus-strand synthesis. We conclude that dipyridamole is a potent experimental tool that readily distinguishes between cardiovirus translation and RNA replication functions.
The positive-sense RNA genomes of all members of the family Picornaviridae have 5′ untranslated regions, followed by single, long open reading frames, 3′ untranslated regions, and poly(A) tails. Translation of the reading frame is directed by an internal ribosome entry site, located as part of the 5′ untranslated region (14). The consequent polyproteins undergo a progression of cis and trans proteolytic cleavages, generating a cascade of polyprotein intermediates and individual, mature proteins. The P1 region proteins (1A, 1B, 1C, and 1D) become the structural units for new virions. The P2 (2A, 2B, and 2C) and P3 (3A, 3BVPg, 3Cpro, and 3Dpol) proteins are nonstructural. During infection, P2 and P3 region precursors and mature proteins interact with each other, with cellular proteins, and with particular cis-acting genome elements to direct the synthesis of new viral RNA.
Much of our current understanding of the processes involved in picornavirus RNA replication is based on studies with poliovirus (reviewed in reference 23). Among the described steps are a discrete series of protein-RNA interactions that help regulate genome conversion from translation to replication templates. For example, cellular poly(C)-binding protein and viral polymerase precursor 3CD are known to bind an RNA cloverleaf motif near the 5′ end of the poliovirus genome (9, 10, 13, 21). In combination with cellular poly(A) binding protein PABP1, which links to the 3′ untranslated region, the intact complex purportedly helps circularize the genome and orient the viral polymerase (3Dpol) for minus-strand initiation (13). Among other obligate steps, viral protein VPg (3B) is uridylylated by 3Dpol to form VPg-pUpU, in reactions templated by an internal cis-acting replication element (cre), which, again, is part of the RNA genome (16, 28, 37).
Recent studies suggest further that VPg uridylylation may not be strictly required for the initiation of minus-strand synthesis, but rather, unmodified VPg may act directly with the 3′ untranslated region (19, 20). Regardless, once full-length minus strands have been synthesized, it is generally accepted that for poliovirus, new plus-strand RNA initiation requires the uridylylation of VPg, or VPg-containing precursors, using the internal cre as template. RNA elongation after the VPg-primed reaction results in new, full-length, plus-strand molecules. Multiple or reiterative polymerase initiations on the same minus-strand templates produce distinctive, branched replicative intermediate structures. The new plus strands are released from the complex and they serve as mRNAs for the translation of more viral proteins, as templates for additional rounds of genome replication, or as virion RNAs encapsidated into mature progeny particles.
As elegant as this model is, several of the proposed features in the poliovirus (enterovirus) replication cycle, do not have obvious homologs or analogs among other genera of picornaviruses. Aphthoviruses, cardioviruses, kobuviruses, teschoviruses, and hepatoviruses, for example, lack 5′ cloverleaf motifs. They have different 3′ untranslated region sequences, and their presumptive cre structures are distributed at various locales in each different genome. The polyprotein processing patterns are also unique for each type of virus, including VPg and its observed precursors. The ability to tease apart the essential characteristics of these parallel replication schemes has been hindered somewhat by the lack of common molecular tools that have proven essential to the poliovirus systems. In particular, the antiviral agent guanidine-HCl is widely recognized as a potent inhibitor of poliovirus replication and is creatively employed in many poliovirus-based experiments.
Guanidine specifically prevents the initiation of minus-strand RNA synthesis (3), thereby allowing an easy experimental separation of poliovirus RNA translation functions from RNA synthesis functions. Moreover, since the action of guanidine is reversible, drug withdrawal can initiate synchronous commencement of RNA replication for in vitro experiments. Guanidine resistance in poliovirus is associated with nucleotide mutations in the 2C gene of the P2 region, and the drug inhibits the ATPase activity of recombinant poliovirus 2C (2, 25, 26). Unfortunately, guanidine does not have equivalent activities against most other picornaviruses, including cardioviruses (15, 27). This specificity is puzzling because the 2C and 3Dpol proteins share definitive sequence and mechanistic similarities among all members of the family.
As an initial probe into the fundamentals of the mengovirus-specific replication scheme, we employed a cell-free replication system based on Krebs-2 cell extracts, similar to that recently described by Yuri Svitkin (32). Akin to the HeLa cell-based systems for de novo poliovirus replication (17), Krebs-2 extracts support all functions necessary for infectious cardiovirus synthesis, including internal ribosome entry site-directed protein translation, polyprotein processing, viral RNA replication, and virion formation (32). The cell-free nature of the reactions permits direct experimental access to every biochemical pathway. In the absence of cellular membranes, drug or antibody additions are also freely manipulated.
We now report use of the Krebs-2 system to characterize the antiviral activity of dipyridamole, one of the few drugs known to affect cardiovirus growth in tissue culture. Dipyridamole is a modified purine with commercially therapeutic applications as an antiplatelet agent (8). It is also a potent inhibitor of mengovirus infectivity to FL and L cells (35). We have traced the source of this inhibition to a reversible, molecular step early in the mengovirus RNA replication pathway. Dipyridamole did not affect internal ribosome entry site-dependent translation or polyprotein processing in vitro or in vivo, and in that regard, it behaved like guanidine in poliovirus replication systems. Dipyridamole is clearly a powerful, practical new reagent that should be extremely useful in subsequent experiments to compare and contrast the specific events in cardiovirus RNA synthesis with those from other picornaviruses.
MATERIALS AND METHODS
Virus and cells.
H1-HeLa cells (ATCC CRL1958) were grown in suspension cultures with modified Eagle's medium and 10% calf serum. Mouse L929 cells were grown in RPMI medium supplemented with 10% fetal calf serum. Recombinant mengovirus vMwt (6) propagation in HeLa cells and virus titration by plaque assay were as described (29). In dipyridamole inhibition assays, the cell monolayers (3 × 106 cells) were exposed to virus (200 PFU), for 30 min at 20°C. The inocula were removed and the cells were covered with agar (1%), followed by a liquid medium overlay containing dipyridamole (0 to 80 μM, Sigma) solubilized in ethanol. After 30 h of incubation (37°C under 5% CO2) the cells were stained with crystal violet to visualize plaques.
Drug efficiency (percent inhibition) was defined as the ratio of plaques formed on treated versus untreated cells. Drug reversibility and timing assays were similar, except the infection was with 50 PFU/cell (3 × 106 cells), the liquid medium overlay was replaced as indicated with fresh medium (with or without 80 μM dipyridamole), and the samples were harvested after 8 h of incubation (37°C, under 5% CO2). The cells were subjected to three freeze-thaw cycles and the clarified supernatants were then titrated for infectivity by plaque assay.
Ribozyme cDNAs.
The mengovirus replicon pMluz has been described (7). It encodes a firefly luciferase gene (luc) that substitutes for a portion of the viral capsid-coding region in the context of the mengovirus plasmid pMwt. Polymerase T7 transcripts templated by pMluz begin with two 5′ nonviral G residues, before the viral genome/reporter, and end on the 3′ side, with a poly(A)23-CG sequence (7). To create plasmids that expressed viral transcripts without the exogenous 5′ bases, a self-cleaving ribozyme cassette (4, 12) was constructed from overlapping cDNA primers and then engineered into pMluz. Primers P1 to P8 (Table 1) were reacted with T7 polynucleotide kinase (Promega). Complementary pairs, P1+P2, P3+P4, and P7+P8, were combined, denatured (95°C), and allowed to hybridize. The product fragments (three pairs) were mixed, treated with T4 DNA ligase (Promega), and then digested with RsrII and NdeI (New England Biolabs), creating a ribozyme-encoding fragment that could be substituted for the analogous fragment in pMluz. After transformation into Escherichia coli MVII90, plasmid Rz-pMluz was amplified and then screened by sequencing throughout the regions of interest (T7 promoter, ribozyme, 5′ end of mengovirus genome). Plasmid Rz*-pMluz, with a point mutation inactivating the ribozyme sequence (highlighted bases in Table 1), was of identical design except primer pair P5′P6 replaced P3′P4 during the construction. Plasmids Rz-pMwt and Rz*-pMwt were also similar except they linked, respectively, the wild-type (Rz) and mutant (Rz*) ribozyme sequences to an intact, infectious pMwt genome sequence.
TABLE 1.
Synthetic primers
| Primer | Sequence |
|---|---|
| P1 | CATATGCGGTTGTAATACGACTCACTATAGGGCTTTCAAA |
| P2 | ATCAGTTTGAAAGCCCTATAGTGAGTCGTATTACAACCGCATATG |
| P3 | CTGATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTCTTTGAAAGC |
| P4 | AAAGAACCCGGGATACCGGGTTTCGGCCTTTCGGCCTC |
| P5 | CTGATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTGTTTGAAAGC |
| P6 | AAACAACCCGGGATACCGGGTTTCGGCCTTTCGGCCTC |
| P7 | CGGGGGTGGGAGATCCGGATTGCCGGTCCGCTCG |
| P8 | CGAGCGGACCGGCAATCCGGATCTCCCACCCCGGCTTC |
Replicon assays.
Plasmids pMluz, Rz-pMluz, and Rz*-pMluz were digested with BamHI before transcription reactions with T7 RNA polymerase. The product RNAs were extracted with phenol-chloroform and then precipitated with ethanol. After resuspension in water, the RNA concentration was determined by absorbance at 260 nm. Confluent HeLa cell monolayers (1.7 × 106 cells per 35-mm plate) were incubated (30 min, 20°C) with RNA (1 μg) and liposomes. The plates were washed and then overlaid with maintenance medium containing dipyridamole (0 to 80 μM). Following incubation (37°C under 5% CO2), the plates were rinsed with phosphate-buffered saline and the cells were lysed with luciferase lysis reagent (Promega). Luciferase activity was determined in standard assays (Luciferase Assay System, Promega) using a Monolight 2010 luminometer.
Isolation of Krebs-2 S10 lysates.
Krebs-2 ascites cell propagation in mice and the isolation of S10 lysates were as described (32). Briefly, Krebs-2 cell inoculants (0.4 ml), kindly provided by Yuri Svitkin of McGill University, were injected in the peritoneal cavities of mice (6 weeks old, female, BALB/c). After 7 days, the mice were euthanized, the ascites fluids were harvested and then transferred into Earl's balanced salt solution on ice. After two washes with Earl's balanced salt solution, the pelleted cells were suspended in Dulbecco's modified Eagle's medium without methionine and incubated with gentle agitation (2 h, 37°C). The suspension was filtered through cheesecloth to remove particulates, and then the cells were collected by centrifugation and washed twice with HNG buffer (35 mM HEPES-KOH, pH 7.3, 146 mM NaCl, 11 mM d-glucose). Cell pellets were resuspended in hypotonic buffer (25 mM HEPES-KOH, pH 7.3, 50 mM KCl, 1.5 mM MgCl2) and placed on ice (20 min). The cells were broken by Dounce homogenization (15 strokes) and then supplemented with 1/10th volume of concentrated buffer (25 mM HEPES-KOH, pH 7.3, 1 M KCH3COO, 30 mM MgCl2, 30 mM dithiothreitol). After centrifugation (10,000 × g), aliquots of the supernatants (S10 fraction) were flash frozen on dry ice before storage (−80°C).
Protein synthesis, RNA synthesis, and VPg uridylylation in Krebs-2 S10 lysates.
Viral RNA (vMwt) was isolated from sucrose-purified virions (29). The particles were disrupted with sodium dodecyl sulfate (SDS, 1%) and proteinase K (20 μg/ml), followed by extraction with phenol-chloroform and precipitation with ethanol. Recombinant viral transcripts were prepared as described for replicon assays. Cardioviral translation and replication in Krebs-2 lysates programmed with these RNAs were essentially as described (32). The lysates (200 μl) were treated with micrococcal nuclease (150 units/ml) in the presence of CaCl2 (75 mM, 20°C, 20 min) before EGTA was added (to 2 mM), to quench the nuclease.
Protein/RNA synthesis reactions contained nuclease-treated lysate (20 μl, or 50% by volume), nucleotides (1 mM ATP, 0.2 mM GTP, 0.2 mM CTP, 0.2 mM UTP), creatine phosphate (10 mM), creatine kinase (0.2 mg/ml), l-amino acids (0.2 mM each, without methionine), salt buffer (75 mM KCH3CO2, 1 mM MgCl2, 0.25 mM spermidine), and either virion RNA (0.5 μg) or transcript RNA (1 μg). When protein label was required, the mix was supplemented with [35S]methionine (2 μl, 10 μCi/μl; Amersham), and incubation was for 3 h (32°C), after which Laemmli loading buffer (8 μl, with 1% SDS) was added. Protein bands were visualized after PAGE fractionation and autoradiography. When RNA label was required, α[32P]CTP (1.5 μl, 10 μCi/μl, >3,000 Ci/mmol; Perkin Elmer) was added to the reaction mix after 4 h of incubation. SDS (to 1%) and proteinase K (to 20 μg/ml) were added 1 h later. The samples were extracted twice with phenol-chloroform, before precipitation with ethanol. RNA products were fractionated by agarose electrophoresis (1% gels in 90 mM Tris-borate, 2 mM EDTA). The labeled bands were visualized by autoradiography and quantitated by phosphorimaging. VPg uridylylation was assayed as described (20) with minor modifications. Reaction mixtures programmed with viral RNA were supplemented with α[32P]UTP (5 μl, 10 μCi/μl, >3,000 Ci/mmol; Perkin Elmer) after 3.5 h of incubation at 32°C. One hour later, replication complexes were collected by centrifugation (16,000 × g for 15 min). The pellets were resuspended in 1x Tricine sample buffer and analyzed by fractionation on Tris-Tricine-polyacrylamide (12%) gels. The labeled bands were visualized by autoradiography and quantitated by phosphorimaging.
Recombinant 3Dpol.
Recombinant mengovirus 3Dpol was expressed and isolated using procedures similar to those for poliovirus (11). Craig Cameron, Penn State University, generously provided plasmid pM3D encoding a ubiquitin-linked mengovirus 3Dpol fusion protein. Protein induction after pM3D transformation of E. coli BL21(pCG1) (also provided by Cameron) results in cleavage of the ubiquitin moiety and accumulation of 3Dpol. The transformed cells were grown in 2 × YT broth supplemented with chloramphenicol and kanamycin at 37°C (A600 of 0.1). Isopropylthiogalactopyranoside (IPTG) was added (to 500 μM) and incubation was continued (3.5 h). The cells were collected by centrifugation, washed twice with TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), before resuspension (to 4 mg/ml) in lysis buffer (100 mM KPO4, 60 μM ZnCl2, 4 μg/ml leupeptin, 10 mM dithiothreitol, and 2 mM phenylmethylsulfonyl fluoride). Lysis was by sonication (four times for 30 seconds each).
Polyethyleneime (5%, 0.0534 μl/ml cell lysate) was added and the cell debris was removed by centrifugation (100,000 × g, 30 min at 4°C). Ammonium sulfate was added (314 g/liter) and the insoluble protein was collected by centrifugation (12,000 × g, 30 min, 4°C). The pellet was resuspended (50 mM Tris-HCl, pH 8.0), dialyzed against buffer C (12 to 14 h,100 mM Tris-HCl, pH 8.0, 20% glycerol, 60 μM ZnCl2, 0.1% NP-40, 50 mM NaCl) and then applied to a Cibacron blue column (5 ml, Bio-Rad) which had been equilibrated in buffer C. Elution was with a NaCl gradient (50 to 1000 mM). Fractions containing 3Dpol, as observed by gel electrophoresis, were pooled and loaded onto a Q-Sepharose column (5 ml, Bio-Rad). A linear gradient of NaCl (50 to 200 mM) was applied. Fractions containing 3Dpol were pooled, reapplied to a fresh Q-Sepharose column (0.5 ml, Bio-Rad), and then eluted with buffer (50 mM HEPES-KOH, pH 8.0, 20% glycerol, 60 μM ZnCl2, 10 mM β-mercaptoethanol, 0.1% NP-40, 500 mM NaCl).
The recombinant enzyme was tested in reactions (50 μl) containing 3Dpol protein (1 μg), oligo(G) primer (25 μg), poly(C) template (5 μg), α[32P]GTP (1 μl, 10 μCi/μl, >3,000 Ci/mmol, Amersham), and buffer (50 mM HEPES-KOH, pH 7.5, 10 mM β-mercaptoethanol, 5 mM MnCl2, 60 μM ZnCl2, 0.5 mM GTP). Samples were incubated for 30 min (30°C) before the addition of EDTA (to 4 mM). The acid-insoluble incorporation of label was determined (duplicate 5-μl aliquots) by filter assay.
RESULTS
Dipyridamole affects mengovirus plaques.
Mengovirus infection of confluent HeLa or L cell monolayers produces plaques of about 2 mm in diameter after 30 h of incubation. For both types of cells, the presence of dipyridamole significantly reduced the number of plaques and the plaque size in a dose-dependent manner (Table 2). Although drug solubility limited the maximum dose tested to 80 μM, at this level no plaques were evident in HeLa cells even after 72 h (not shown), and the few that formed in L cells were minute in size and difficult to detect. Cells treated with dipyridamole alone were indistinguishable from untreated cells as determined by standard light microscopy (data not shown).
TABLE 2.
Mengovirus plaque phenotypes in the presence of dipyridamole
| Dipyridamole (μM) | HeLa cells
|
L cells
|
||
|---|---|---|---|---|
| Plaque reduction (%)a | Relative plaque sizeb | Plaque reduction (%) | Relative plaque sizeb | |
| 80 | 100 | N/A | 91 | Minute |
| 60 | 98 | Minute | 87 | Minute |
| 40 | 93 | + | 64 | + |
| 20 | 68 | ++ | 48 | ++ |
| 10 | 25 | ++ | 24 | +++ |
| 0 | 0 | ++++ | 0 | ++++ |
Values represent the average of two experiments, each done in triplicate.
Plaques in the absence of drug averaged about 2 mm in diameter.
Plaque inhibition is reversible.
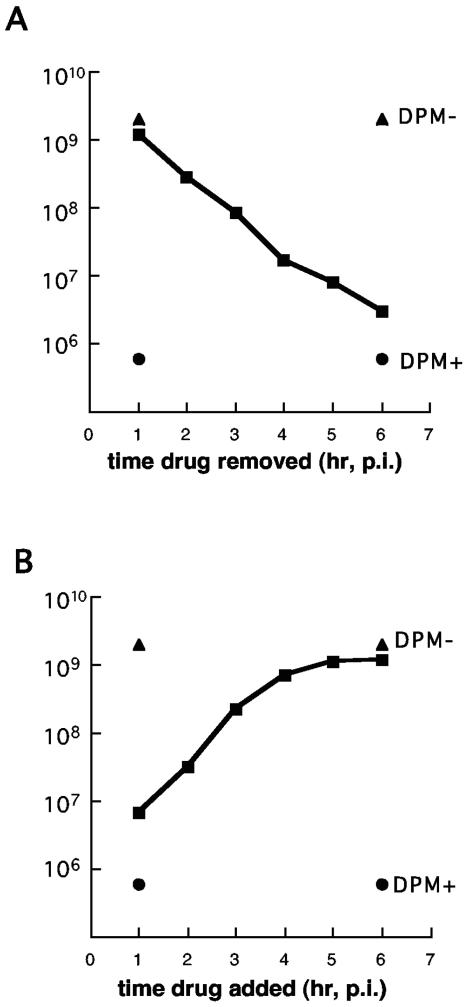
To determine if dipyridamole inhibition was reversible, the drug was removed from infected cultures at various times postinfection by exchange with drug-free medium (Fig. 1A). When HeLa cells were freed from drug at 1 h postinfection, the virus recovered 7 h later titered almost as high as drug-free controls. Exposure to the drug for longer times progressively dropped the apparent titers by up to 104-fold if the samples were analyzed at 8 h postinfection. However, if the drug was removed between 2 and 6 h postinfection, and the samples were allowed to recover for a full 7 to 8 h, virus production typically recuperated to nearly drug-free levels (not shown). For example, if the drug was removed at 2 h postinfection, the virus recovered at 9 h postinfection titered at 9.2 × 108 PFU/ml. When the drug was removed at 6 h postinfection, the titer reached 5.1 × 108 PFU/ml by 13 h postinfection. Considering these experiments were initiated at 50 PFU per cell, the results strongly corroborate previous reports that dipyridamole is a potent inhibitor of mengovirus growth (35). Moreover, they indicate the drug effect was reversible if the infection was allowed sufficient time to recover.
FIG. 1.
Dipyridamole and virus yield. HeLa cell monolayers were infected at a multiplicity of 50 PFU/cell. Dipyridamole-treated samples (DPM+) had 80 μM dipyridamole (in ethanol) added to the medium at the time of infection. Dipyridamole-untreated samples (DPM−) were dosed with an equivalent volume of ethanol. (A) Medium from dipyridamole-treated cultures was exchanged with drug-free medium at the indicated times. Virus was harvested at 8 h postinfection and the titer was determined by plaque assay. (B) Medium from dipyridamole-untreated cultures was exchanged with medium containing dipyridamole (80 μM) at the indicated times. Virus was harvested at 8 h postinfection and then titered.
The converse experiment of adding the drug at progressively later times after infection initiated revealed that the drug activity targeted an early step in the viral replication cycle (Fig. 1B). The longer dipyridamole was absent from the medium, the higher the titer of recovered virus. Once the infection proceeded past 3 to 4 h, dipyridamole became less effective at inhibiting viral processes. Drug added after 6 h postinfection was nearly ineffective on samples titered at 8 h postinfection. Similar results were observed for mengovirus-infected L cells (not shown).
Mengovirus replicons.
Drug-induced defects on input RNA translation or on the initiation of RNA synthesis would manifest equivalently in the previous assays as early blocked steps in the viral replication cycle. We previously reported the successful use of a mengovirus replicon in which a luciferase reporter gene (luc) replaced part of the capsid-coding region. This system was used to tease apart the contributions of genome translation and RNA synthesis on the cardioviral growth cycle (7).
Transfection of pMluz RNA into HeLa cells gives a biphasic luciferase response (e.g., Fig. 2A) as protein is first translated from the input transcripts (1 to 3 h), and then from replicated viral RNA (3 to 8 h). Recent work with similar replicons from poliovirus, however, has called this interpretation into question. For poliovirus, the presence of two 5′ nonviral guanosine residues, the engineered artifacts of T7 polymerase reactions, was shown to influence the biphasic luciferase response of poliovirus replicons. Alternate replicon transcripts tailored with precise viral 5′ ends, through the use plasmid-encoded ribozyme sequences, were found to initiate replication at earlier times, and the lag between input RNA translation and new RNA translation was relatively diminished (12). Before testing dipyridamole with mengovirus replicons, it seemed prudent to first investigate whether the transcript 5′ ends would likewise influence analogous assays with cardiovirus sequences.
FIG. 2.
Replicon assays. (A) HeLa cell monolayers were transfected with RNA transcribed from mengovirus replicon plasmids or mock transfected (mock). At the indicated times posttransfection, cells were harvested and lysed and the luciferase activity in the cytoplasm was determined. Relative light units (RLU) are averaged values for duplicate (20 μl) samples. (B) Samples were treated as in A, except that dipyridamole (80 μM in ethanol) was added to the medium of an Rz-pMluz sample series at the time of transfection. Control samples had equivalent volumes of ethanol added to the medium.
Accordingly, active (Rz-pMluz) and inactive (Rz*-pMluz) ribozyme cassettes were incorporated into pMluz plasmids. The resultant RNAs had 0 or 50 nonviral 5′ bases, compared to the two nonviral bases (GG) at the 5′ end of pMluz. When tested in cells, and in contrast to poliovirus reports (12), all replication-competent mengovirus transcripts still showed biphasic luciferase production (Fig. 2A) that discriminated input from replicated RNA translation. The initial lag phase for Rz-pMluz ended at about 3 h postinfection, roughly the same time as the lag displayed by the other replicating transcripts containing nonviral nucleotides. Therefore, the presence of an authentic viral 5′ end did not alter the lag phase, as has been reported for poliovirus (12). Like poliovirus, however, mengovirus RNAs with precise viral ends (Rz-pMluz) then became 5- to 10-fold more effective replication templates relative to those with two (pMluz) or 50 (Rz*-pMluz) heterologous bases. Control samples with defective polymerases (pMluz-Age) failed to convert translating RNAs into replication templates, regardless of the incubation time. As expected, low levels of luciferase continued to be synthesized from such transcripts, until, presumably, they degraded (+6 h).
The highly active mengovirus ribozyme replicon was subsequently tested for dipyridamole sensitivity (Fig. 2B). Initially, luciferase was produced from the input RNA at levels similar to non-drug-treated samples, suggesting that initial genome translation was not the step affected by the drug. Clearly, though, dipyridamole prevented the next, replication-dependent phase of protein synthesis, and the drug curve essentially paralleled that of control samples programmed with inactive polymerase (pMwt-Age).
Krebs-2 assays with virion RNA.
In 2003, Svitkin and Sonnenberg (32) described a highly effective Krebs-2 lysate system that allowed in vitro examination of all steps in the encephalomyocarditis virus replication cycle. With advice and reagents generously provided by Y. Svitkin, we reproduced this system, and established that when programmed with mengovirion RNA or with Rz-pMwt transcripts, Krebs-2 lysates synthesized at least 108 PFU/ml of infectious mengovirus progeny within 20 h (not shown).
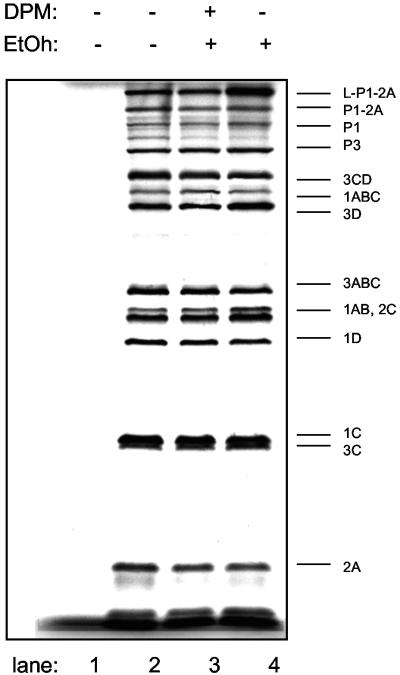
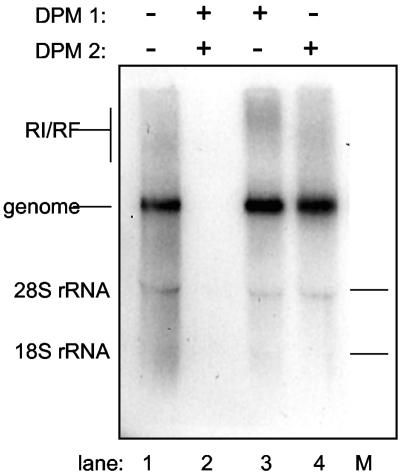
A key feature of any cell-free assay is that viral translation and replication products can be readily labeled and monitored. The addition of [35S]methionine to extracts programmed with mengovirus virion RNA gave the full panel of expected viral precursors and cleavage products (Fig. 3, lane 2). The addition of 80 μM dipyridamole or its solvent, ethanol, did not alter this pattern significantly (Fig. 3, lanes 3 and 4). But, when RNA synthesis was monitored by the addition of [32P]CTP, the drug was clearly inhibitory (Fig. 4). Standard reactions (Fig. 4, lane 1), or standard reactions supplemented with ethanol (Fig. 4, lane 2), gave strong bands of genome RNA as well as replicative intermediate and replicative form complexes. Samples to which 20, 40, or 80 μM dipyridamole had been added (Fig. 4, lanes 4, 5, and 6, respectively), gave sequentially weaker bands, indicating a drug-dependent inhibition of viral RNA synthesis. At 80 μM dipyridamole, single-stranded mengovirus RNA synthesis was inhibited >96%.
FIG. 3.
Protein synthesis in cell-free assays. Krebs-2 lysates programmed with mengovirus RNA were labeled with [35S]methionine as described in Materials and Methods. The proteins were fractionated by polyacrylamide gel electrophoresis and detected by autoradiography. Reactions contained no RNA (lane 1); no additions (lane 2); 80 μM dipyridamole (DPM, lane 3); or 1.25% ethanol (EtOh, lanes 3 and 4). The migration positions of known mengovirus proteins are indicated.
FIG. 4.
RNA synthesis in cell-free assays. Krebs-2 lysates programmed with mengovirus RNA were pulse-labeled with [32P]CTP as described in Materials and Methods. Reactions contained no RNA (lane 6); no additions (STD, lane 1); 1.25% ethanol (lanes 2 to 5); or 20, 40, or 80 μM dipyridamole (DPM) (lanes 3 to 5). Samples were fractionated on 1% native agarose gels, and the labeled bands were visualized by autoradiography. % of STD is the relative intensity of single-stranded genome bands, as determined by phosphorimaging and ImageQuant software. RI/RF, replicative intermediate/replicative form.
The fact that RNA replication but not translation was affected by dipyridamole in vitro was consistent with the replicon results and plaque reduction assays. But, if dipyridamole specifically affected RNA synthesis, the targeted step should also account for drug reversibility. Accordingly, Krebs-2 lysates programmed with virion RNA were incubated in the presence or absence of dipyridamole. After 4 h, the replication complexes were collected by centrifugation, resuspended in fresh lysates (with or without drug), and then allowed to synthesize labeled RNA for an additional hour (Fig. 5). Unless dipyridamole was present during both incubation periods (Fig. 5, lane 2), the samples accumulated new viral RNA to (roughly) equivalent amounts. When dipyridamole was removed after the first incubation, RNA synthesis recovered to normal levels (Fig. 5, compare lanes 1 and 3), confirming the in vitro effect on viral RNA synthesis was indeed reversible. Moreover, even when dipyridamole was present during the second incubation period, if the previous incubation had been drug free (Fig. 5, lane 4) RNA synthesis was able to continue. This pattern strongly suggests dipyridamole targeted the formation or activation of newly assembled replication complexes, but not the progression of preformed complexes.
FIG. 5.
Dipyridamole reversibility in Krebs-2 assays. Krebs-2 lysates programmed with mengovirus virion RNA were incubated for 4 h at 32°C in the presence (lanes 2 and 3) or absence (lanes 1 and 4) of 80 μM dipyridamole (DPM). The replication complexes were collected by centrifugation at 16,000 × g and then resuspended in fresh lysates in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 80 μM dipyridamole. [32P]CTP was added and incubation was continued for 1 h. The samples were fractionated on 1% native agarose gels, and the labeled bands were visualized by autoradiography.
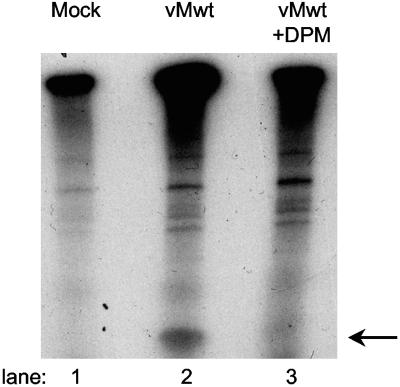
Uridylylation of VPg and/or its precursors is one of the key events required early in picornaviral RNA replication (19, 20). To determine if dipyridamole interfered with this step, we examined the synthesis of VPg-pUpU in Krebs-2 lysates programmed with virion RNA in the presence and absence of drug (Fig. 6). After 3.5 h of incubation, standard reactions were pulse-labeled with [32P]UTP for an additional hour. Replication complexes, including VPg-pUpU, were collected by centrifugation, fractionated on gels, and analyzed for label. While uridylylated (labeled) VPg was detected in the standard reaction (Fig. 6, lane 2), little or no VPg label was evident in the presence of dipyridamole (Fig. 6, lane 3), or the mock-infected sample (Fig. 6, lane 1). This experiment is consistent with the idea that dipyridamole targeted an early step in the replication cycle, but does not clarify whether inhibition of uridylylation is the cause or consequence of more global drug-dependent defects, such as improper assembly or processing of replication complexes.
FIG. 6.
Dipyridamole and VPg uridylylation. Krebs-2 lysates programmed with: no RNA (lane 1), vMwt RNA (lane 2), or vMwt RNA plus dipyridamole (DPM) (80 μM, lane 3) were pulse-labeled with [α-32P]UTP for 1 h as described in Materials and Methods. Replication complexes were collected by centrifugation, fractionated on Tris-Tricine gels, and visualized by autoradiography. The arrow indicates the known migration of VPg-pUpU.
Krebs-2 assays with transcript RNA.
As discussed above, the 5′ ends of poliovirus replicon transcripts are known to affect the overall efficiency and timing of RNA synthesis, after transfection into cells. The extra nonviral 5′ bases have even stronger effects on poliovirus plus or minus-strand template selection in cell-free reactions, where fortuitous repair of defective ends is less likely (12). Specifically, poliovirus templates with 5′ extensions are reported to support minus-strand synthesis to give replicative intermediate and replicative form products in HeLa extracts, but only those templates with properly truncated 5′ ends (i.e., with ribozymes) can direct subsequent synthesis of new plus-strands (12).
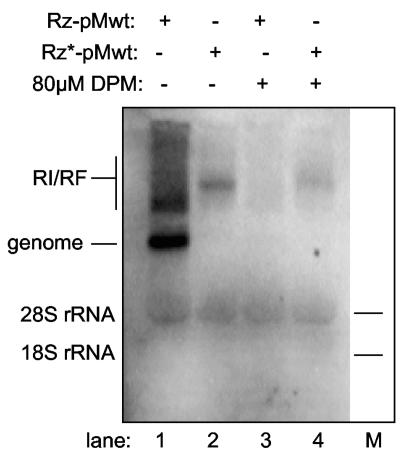
Since mengovirus replicons also seemed sensitive to exogenous 5′ sequences, active (Rz-pMwt) and inactive (Rz*-pMwt) ribozyme segments were linked 5′ to recombinant wild-type mengovirus cDNAs (pMwt), and the resulting genome transcripts were tested in Krebs-2 assays for their ability to produce plus- and minus-strand products (Fig. 7). Lysates programmed with Rz-pMwt RNA synthesized large amounts of single-strand product as well as replicative intermediate/replicative form material. When the ribozyme was inactivated, however, a much weaker band of replicative form material was the only product (Fig. 7, lane 2). If indeed the mengovirus polymerase behaves like poliovirus in this regard, it is likely the majority of label in this band is of minus-strand origin (characterization is under way).
FIG. 7.
Effect of dipyridamole on Rz-pMwt and Rz*-pMwt RNA synthesis. Krebs-2 lysates were programmed with Rz-pMwt or Rz*-pMwt transcript RNAs and then pulse-labeled with [32P]CTP in the presence or absence of dipyridamole (DPM) (80 uM). Samples were fractionated on native agarose gels and visualized by phosphorimaging. Lanes: 1, Rz-pMwt RNA; 2, Rz*-pMwt RNA; 3, Rz-pMwt RNA with dipyridamole; 4, Rz*-pMwt RNA with dipyridamole.
The absence of a strong band of single-stranded, genome-length product suggests that mengovirus plus-strand synthesis was especially sensitive to the 50 extra nonviral 5′ bases. When dipyridamole was added to similar reactions, Rz-pMwt replication (Fig. 7, lane 3), including replicative form/replicative intermediate and single-stranded RNA synthesis, was completely abrogated by the drug. The Rz*-pMwt reactions were likewise impaired, producing only about 40% of the amount of replicative form RNA as in the presence of dipyridamole (Fig. 7, compare lanes 2 and 4). While significant, this reduction in minus-strand synthesis cannot account for the nearly complete inhibition (>95%) observed in reactions programmed with Rz-pMwt or virion RNA. Further experiments are now under way to better characterize the precise drug effects on minus-strand versus plus-strand RNA synthesis inhibition. Nonetheless, all data clearly indicate dipyridamole is a potent inhibitor of mengovirus RNA replication and is readily useful to distinguish between RNA synthesis and translation events during the replication cycle.
Dipyridamole does not inhibit 3Dpol elongation.
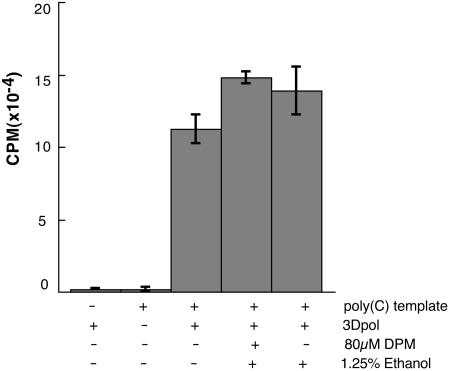
Picornavirus 3Dpol polymerases can bypass normal RNA synthesis initiation events, as well as the requirements for additional host and viral factors, if cell-free reactions are programmed with appropriate template-primer complexes (24). Elongation on oligo(G)-primed, poly(C) templates is an especially efficient activity of recombinant mengovirus 3Dpol (C. Cameron, personnel communication). Full-length mengovirus 3Dpol was expressed from plasmid pM3D in E. coli (11) (C. Cameron, personal communication), purified to >90% homogeneity, and then tested for poly(G) synthesis activity in the presence and absence of dipyridamole. Robust incorporation of [32P]GTP was dependent on the presence of both enzyme and template (Fig. 8). Dipyridamole and its solvent, ethanol, were not inhibitory to these reactions, and if anything, slightly stimulated label incorporation. Consistent with other dipyridamole results, these data point to a genome-dependent RNA synthesis initiation event, as the mechanistic target of the drug.
FIG. 8.
3Dpol reactions. Recombinant mengovirus 3Dpol was tested for poly(G) synthesis activity as described in Materials and Methods. The reactions contained template, polymerase, dipyridamole (DPM) (80 μM), and ethanol (1.25%) as indicated. Bars show the averaged values of acid-insoluble [32P]GTP incorporation from duplicate aliquots (5 μl).
DISCUSSION
Guanidine-HCl is a potent, reversible inhibitor of poliovirus minus-strand RNA synthesis, and the use of this drug has contributed greatly to experiments dissecting the molecular mechanisms of picornaviral synthesis. However, not all poliovirus-discovered mechanisms or the action of guanidine is germane to other viruses. For unknown reasons, only certain picornaviruses are susceptible to guanidine. Our interest in the purine analog dipyridamole was instigated by reports describing it as a broad-spectrum inhibitor of multiple viruses, including mengovirus, poliovirus (another picornavirus), Semliki Forest virus (Togavirus), and influenza A virus (Orthomyxovirus), among others (34, 36). Our mengovirus assays confirmed a strong inhibitory activity on plaque formation in both HeLa and L cells and further showed the effect was reversible. Preliminary experiments with rhinovirus 16 (in HeLa cells) also suggest a similar pattern of inhibition (data not shown). The antimengovirus dipyridamole effect was strongest if the drug was present during the early stages of infection, consistent with the idea that it impeded the formation or function of newly formed replication complexes.
Mengovirus replicon experiments and in vitro assays with Krebs-2 lysates confirmed that viral translation and polyprotein processing were indeed unaffected by the drug. Rather, dipyridamole seemed specifically to inhibit viral RNA synthesis in an unknown, albeit reversible manner. Both minus-strand and plus-strand synthesis was significantly reduced in the Krebs-2 system when the drug was added early in the replication cycle. The presence of the drug also prevented the accumulation of uridylylated VPg, one of the early events in RNA replication. The observed inhibition could not be attributed to defects in RNA elongation, since dipyridamole did not hinder poly(G) polymerase activity by recombinant 3Dpol. We conclude that dipyridamole must interfere with one or more steps in the initiation of RNA synthesis, and moreover, that this interference was not permanent and could be relieved if the drug was removed from replication complexes.
Several molecular functions ascribed to dipyridamole warrant discussion relative to potential antiviral mechanisms. Historically, the drug has seen extensive medical use as a coronary vasodilator and an antithrombic agent, and at one point, under the trade name of Persantin, it was one of the 50 most widely prescribed drugs in the United States (8). Dipyridamole is very effective at inhibiting nucleoside transport into cells, and this is the presumed mechanism for its medical efficacy. Its activity was also evaluated for other applications, including use as an inhibitor of herpes simplex virus reactivation (33) and as a potentiator of the antiviral effects of 3′azido-3′deoxythymidine against human immunodeficiency virus (22). Neither of these effects proved of significantly therapeutic value, but similar experiments did suggest dipyridamole might have additional, modest antiviral effects against human immunodeficiency virus even when administered in the absence of 3′azido-3′deoxythymidine (22). Our mengovirus studies found dipyridamole to be equally effective in infected cells and in vitro replication reactions where nucleoside uptake was not an issue. Therefore, nucleoside transport defects are unlikely to be responsible for the drug's activity against mengovirus. Rather, the mengovirus effects and the 3′azido-3′deoxythymidine-independent activity against human immunodeficiency virus probably originate from some alternate molecular mechanism.
Another dipyridamole activity is the stimulation of prostacyclin synthesis, with a correlate, concentration-dependent enhancement of the action of prostaglandins on cells (18). These fatty acid derivatives are synthesized from arachidonic acid by clooxygenases and help mediate cellular inflammatory responses, including some reactions shown to have minor antiviral effects on adenovirus, parainfluenza virus, and measles virus growth (reviewed in reference 31). While certain prostaglandins can partially inhibit poliovirus replication in cells without affecting viral protein synthesis (5), again, it is unlikely that upregulation of the clooxygenase enzyme is a key factor in dipyridamole inhibition of mengovirus. Our in vitro systems use enucleated cell lysates treated with micrococcal nuclease, where no host-encoded proteins are synthesized. Although it remains a formal possibility that dipyridamole could stimulate an endogenous clooxygenase activity, the reversible effects in our lysate experiments still beg other explanations.
The hypothesis we currently favor derives from a third activity ascribed to dipyridamole, its ability to inhibit phosphodiesterases. Phosphodiesterase 4, for example, a known dipyridamole target, is the major cyclic adenosine-3′,5′-monophosphate-metabolizing enzyme in eukaryotic cells. Phosphodiesterase 4 mediates the rate at which cyclic 3′,5′-AMP is converted into 5′-AMP (8). It would be surprising if fluctuations in cellular AMP concentrations per se were responsible for the drug-dependent changes in picornaviral RNA synthesis rates. But other phosphodiester events have special roles in the initiation of plus- and minus-strand synthesis for every picornavirus. In particular, 3Dpol-mediated, VPg uridylylation to VPg-pUpU precedes every initiation and is the key step in the linkage of this protein to new RNAs (37).
If dipyridamole were to interfere or even influence the dynamics of this pathway through reaction with 3Dpol, VPg, UTP, or the cre element, one might expect inhibition of initiation events but not elongation events, as in our experiments. Indeed, we have demonstrated that VPg uridylylation is one of the steps inhibited by the presence of dipyridamole. Further experiments are under way to determine the precise molecular components and their mode of action. Regardless of the ultimate mechanism, however, or drug-targeted step, our experiments confirm that dipyridamole is an effective experimental tool which can be wielded like guanidine, to regulate the mengoviral replication cycle, both in cells and in cell-free lysates that reproduce the infectious cycle.
Another conclusion from this work addresses the functional characteristics of the 5′ ends of mengovirus template RNAs. In luciferase replicon assays, the translation activities of input transcripts were readily distinguished from those of newly synthesized RNAs. While the slope and height of the exponential phase of luciferase activity were responsive to the 5′ context, the timing of the initial lag period was not. In contrast to reports with ribozyme-cleaved poliovirus replicons (12), the initial 3-h lag phase was not eliminated when ribozyme-cleaved mengovirus replicons were compared to those with 2 or 50 heterologous nucleotides at the 5′ end. Herold and Andino reported that poliovirus transcripts with authentic 5′ ends showed “no initial translation phase of the input viral genome for 2 to 3 h before a switch to a replicative state” (12). Clearly, this is not the case for mengovirus and we suspect the two-phase pattern of luciferase production indicates a time-sensitive trigger that naturally coordinates the onset of RNA synthesis. Encephalomyocarditis virus replicons with defects in the 2A-dependent shutoff of host transcription and translation (e.g., pE-luc-2AΔ58) have longer lag periods than wild-type replicons, perhaps implicating a feedback signal generated by these processes (1).
The Krebs-2 assays were also sensitive to 5′ context, in that virion RNA and Rz-pMwt transcripts replicated and produced high-titer infectious progeny, while other transcripts did not. Virion RNAs are linked by 5′ phosphodiester bonds to a tyrosine within VPg. Rz-pMwt transcripts and related replicons terminate with 5′ hydroxyl groups as the result of their ribozymes (30). On the other hand, pMwt and Rz*-pMwt transcripts have 5′ triphosphates and carry 2 to 50 additional nonviral bases, templated by their plasmids. Since all nonribozyme replicons we have tested seem to come out of the lag phase with similar kinetics and activities, we suspect it might be the triphosphates, and not the extra bases, that contribute to their less effective replication. We now intend to use dipyridamole to tease apart these essential initiation step questions and perhaps explain the crucial 5′ recognition features required by mengovirus 3Dpol.
Acknowledgments
This work was supported by NIH grant AI-17331 to A.C.P. and NIH postdoctoral training grant GM-069311 to C.F.-H.
We thank Marchel Hill for her excellent technical assistance with cell cultures and Yuri Svitkin for his generous, repeated gifts of Krebs-2 cells and his invaluable suggestions regarding the activities of the related cell-free system. We also thank Craig Cameron for plasmid pM3D and his expertise related to the preparation of purified, active enzyme.
REFERENCES
- 1.Aminev, A. G., S. P. Amineva, and A. C. Palmenberg. 2003. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 95:45-57. [DOI] [PubMed] [Google Scholar]
- 2.Anderson-Sillman, K., S. Bartal, and D. R. Tershak. 1984. Guanidine-resistant poliovirus mutants produce modified 37-kilodalton proteins. J. Virol. 50:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowrira, B. M., P. A. Pavco, and J. A. McSwiggen. 1994. In vitro and in vivo comparison of hammerhead, hairpin, and hepatitis delta virus self-processing ribozyme cassettes. J. Biol. Chem. 269:25856-25864. [PubMed] [Google Scholar]
- 5.Conti, C., P. Mastromarino, P. Tomao, A. De Marco, F. Pica, and M. G. Santoro. 1996. Inhibition of poliovirus replication by prostaglandins A and J. in human cells. Antimicrob. Agents Chemother. 40:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 63:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duque, H., and A. C. Palmenberg. 2001. Phenotypic characterization of three phylogenetically conserved stemloop motifs in the mengovirus 3′ untranslated region. J. Virol. 73:3111-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.FitzGerald, G. A. 1987. Dipyridamole. N. Engl. J. Med. 316:1247-1257. [DOI] [PubMed] [Google Scholar]
- 9.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gohara, D. W., C. S. Ha, S. Kumar, B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D[pol]) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 12.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang, S. K., M. V. Davies, R. J. Kaufman, and E. Wimmer. 1989. Initiation of protein synthesis by the internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J. Virol. 63:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick, W., and S. Penman. 1968. Replication of mengovirus in HeLa cells preinfected with nonreplicating poliovirus. J. Virol. 2:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKnight, K. L., and S. M. Lemon. 1999. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 18.Moncada, S., and R. Korbut. 1978. Dipyridamole and other phosphodiesterase inhibitors act as antithrombotic agents by potentiating endogenous prostacyclin. Lancet i:1286-1289. [DOI] [PubMed] [Google Scholar]
- 19.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, S. S., J. Szebeni, L. M. Wahl, and J. N. Weinstein. 1991. Differential inhibition of 2′-deoxycytidine salvage as a possible mechanism for potentiation of the anti-human immunodeficiency virus activity of 2′,3′-dideoxycytidine by dipyridamole. Antimicrob. Agents Chemother. 35:1250-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, A. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 24.Paul, A. K., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3D. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 25.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 26.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prather, S. O., and M. W. Taylor. 1975. Host-dependent restriction of mengovirus replication. IV. Effect of some quaternary ammonium ions on the restricted replication of mengovirus in Madin-Darby bovine kidney cells. J. Virol. 15:1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reider, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 30.Sigurdsson, S. T., and F. Eckstein. 1995. Structure-function relationships of hammerhead ribozymes: from understanding to applications. Trends Biotechnol. 13:286-289. [DOI] [PubMed] [Google Scholar]
- 31.Steer, S. A., and J. A. Corbett. 2003. The role and regulation of COX-2 during viral infection. Viral Immunol. 16:447-460. [DOI] [PubMed] [Google Scholar]
- 32.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenser, R. B., A. Gaydos, and K. A. Hay. 2001. Inhibition of herpes simplex virus reactivation by dipyridamole. Antimicrob. Agents Chemother. 45:3657-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonew, E., M. K. Indulen, and D. R. Dzeguze. 1982. Antiviral action of dipyridamole and its derivatives against influenza virus A. Acta Virol. 26:125-129. [PubMed] [Google Scholar]
- 35.Tonew, M., and D. Dzeguze. 1977. Dipyridamole, an inhibitor of mengovirus replication in FL and L cells. Chemotherapy 23:149-158. [DOI] [PubMed] [Google Scholar]
- 36.Tonew, M., E. Tonew, and R. Mentel. 1977. The antiviral activity of dipyridamole. Acta Virol. 21:146-150. [PubMed] [Google Scholar]
- 37.Wimmer, E. 1982. Genome-linked proteins of viruses. Cell 28:199-201. [DOI] [PubMed] [Google Scholar]