Abstract
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) late expression factor 3 (LEF-3) is an essential protein for DNA replication in transient assays. P143, a large DNA-binding protein with DNA-unwinding activity, is also essential for viral DNA replication in vivo. Both LEF-3 and P143 are found in the nucleus of AcMNPV-infected cells, but only LEF-3 localizes to the nucleus when expressed in transfected cells on its own from a plasmid expression vector. P143 requires LEF-3 as a transporter to enter the nucleus. To investigate the possibility that LEF-3 carries a nuclear localization signal domain, we constructed a series of LEF-3 deletion mutants and examined the intracellular localization of the products in plasmid-transfected cells. We discovered that the N-terminal 56 amino acid residues of LEF-3 were sufficient for nuclear localization and that this domain, when fused with either the green fluorescent protein reporter gene or P143, was able to direct these proteins to the nucleus. Transient DNA replication assays demonstrated that fusing the LEF-3 nuclear localization signal domain to P143 did not alter the function of P143 in supporting DNA replication but was not sufficient to substitute for whole LEF-3. These data show that although one role for LEF-3 during virus infection is to transport P143 to the nucleus, LEF-3 performs other essential replication functions once inside the nucleus.
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the type species of the genus Nucleopolyhedrovirus in the family Baculoviridae (1). Baculoviruses are large, enveloped, double-stranded DNA-containing viruses that are pathogenic only to invertebrates, mainly insects of the order Lepidoptera. The replication cycle of AcMNPV in Spodoptera frugiperda cells is controlled mainly at the transcriptional level and occurs in an ordered cascade of early and late phases, roughly divided by the initiation of viral DNA replication at about 6 to 8 h postinfection (6).
The expression of early proteins is largely controlled by an immediate-early protein called IE-1, which regulates viral transcription and is essential for viral DNA replication (5, 19). In addition to the ie-1 gene, transient DNA replication assays have identified at least eight other AcMNPV genes involved in DNA replication, including ie-2, p143, dnapol, lef-1, lef-2, lef-3, pe-38, and p35 (reviewed in reference 5). The p143 gene has been shown to be essential for viral DNA replication by characterization of a temperature-sensitive AcMNPV mutant called ts8 (7, 18). Because baculovirus DNA replication takes place in the nucleus, these proteins must be imported to this location following their synthesis in the cytoplasm. LEF-3, a single-stranded DNA binding protein that self-localizes to the nucleus, is also essential for nuclear localization of P143 (8, 27). Along with IE-1, P143 and LEF-3 form a complex on closely linked sites on viral DNA, suggesting that P143 and LEF-3 form a stable complex that plays a role during viral DNA replication (12). This complex likely forms in the cytoplasm soon after their synthesis through an LEF-3-P143-interacting domain. Once the complex is formed, a nuclear localization signal domain (NLS) on LEF-3 may be responsible for transporting the complex to the nucleus.
In eukaryotic cells, there is orderly traffic of proteins between the cell nucleus and the cytoplasm. Small proteins (less than approximately 40 kDa) can diffuse through the nuclear pore, but larger proteins require an energy-dependent process for active recruitment to and translocation across the nuclear membranes through nuclear pores (9). The import of proteins typically requires a NLS. Little is known of the transport pathways in insect cells, but they are probably mediated by importin-like transport receptor family molecules (20). These proteins act as shuttles, interacting with specific NLS sequences on cargo proteins that must be actively moved through nuclear pores into the nucleus.
In this study, we investigated the domains involved in nuclear localization of AcMNPV LEF-3. A number of deletion mutants were prepared that would express portions of the lef-3 open reading frame when transfected into insect cells. The expressed proteins were characterized for intracellular localization by immunofluorescence microscopy and biochemical fractionation. In addition, by using a similar approach, we investigated the domains of LEF-3 that are required to transport P143 into the nucleus. Finally, we generated a fusion protein that resulted in the self-localization of P143 to the nucleus and investigated the ability of this protein to support plasmid replication in transient expression assays. Our results show that LEF-3 is a multifunctional protein with an NLS domain that is conserved in the group I nucleopolyhedroviruses.
MATERIALS AND METHODS
Cells and viruses.
The Spodoptera frugiperda continuous cell line IPLBSF-21 (Sf21 cells) was maintained at 28°C in TC100 medium supplemented with 10% fetal calf serum. AcMNPV (strain HR3) was prepared and titrated as previously described (18).
Plasmids.
All LEF-3 deletion plasmid constructs were derived from pHSEHLEF3, a plasmid vector in which the Drosophila melanogaster heat shock 70 promoter drives the expression of six-His-tagged AcMNPV LEF-3, beginning with amino acid residue 2 (24). This vector also includes an in-frame HA.11 epitope. Specific deletions within the lef-3 open reading frame were generated by a PCR-fusion mutagenesis protocol (10). Two nested primers were designed to be complementary and to carry two segments corresponding to flanking sequences upstream and downstream of the region to be deleted. The sequence of the nested primers is summarized in Table 1. Each of these primers was used in reactions with upstream or downstream outside primers C-22910 (5′-CAA ACC CTT CGA TTA TCT CTA AC-3′) or C-22911 (5′-AAC AGT TCA CCT CCC TTT TC-3′) to amplify products upstream and downstream of the region to be deleted. The products were then mixed, denatured, and used as templates in a third PCR amplification using only the outside primers. The final PCR product was expected to contain a specific deletion of the region specified by the overlapping region of the nested primers. The final PCR products were digested with XbaI and NotI, and then cloned into XbaI- and NotI-digested vector pHSEHLEF3 so that each coding sequence was fused in-frame with both the influenza virus hemagglutinin (HA) epitope and a six-histidine tag at the N terminus.
TABLE 1.
Sequences of mutagenic primersa
| Primer sequence | Amino acids of mutant | Primer name |
|---|---|---|
| CAAACCCTTCGATTATCTCTAAC | Outside primers(up stream) | C-22910 |
| AACAGTTCACCTCCCTTTTC | Outside primers(down stream) | C-22911 |
| TCATCACAGATCCATGGACGACATCAATGAGGTG | aa190-385 | C-22912 |
| CCTCATTGATGTCGTCCATGGATCTGTGATGATG | aa190-385 | C-22913 |
| AATATAAAGGGGTCTTAATTCTATTCGACGTTTG | aa2-189 | C-22914 |
| CGTCGAATAGAATTAAGACCCCTTTATATTGTTC | aa2-189 | C-22915 |
| ACTAACAAACATTTTTAATTCTATTCGACGTTTG | aa2-125 | C-24270 |
| CGTCGAATAGAATTAAAAATGTTTGTTAGTCAAAT | aa2-125 | C-24271 |
| GAAGGCAAGTGCTATTAATTCTATTCGACGTTTG | aa2-83 | C-23424 |
| CGTCGAATAGAATTAATAGCACTTGCCTTCTTC | aa2-83 | C-23425 |
| TACCATTACACTTTTGACGACATCAATGAGGTG | aa2-56/190-385 | C-23573 |
| CTCATTGATGTCGTCAAAAGTGTAATGGTATTC | aa2-56/190-385 | C-23574 |
| TCATCACAGATCCGACATTAGTTTAAATTATG | aa84-189 | C-23422 |
| ATTTAAACTAATGTCGGATCTGTGATGATGATG | aa84-189 | C-23423 |
| AAGGCAAGTGCTATGAAAATGAAGATGGCGTC | aa2-83/126-385 | C-23964 |
| CCATCTTCATTTTCATAGCACTTGCCTTCTTC | aa2-83/126-385 | C-23965 |
To generate LEF-3 mutants, two rounds of PCR were carried out. In the first PCR, fragments were amplified by either outside primer C-22910 or C-22911 plus one nested primer. Second-round PCR was performed by using only the outside primers C-22910 and C-22911. The inside primers for each mutant are summarized in the table. aa, amino acids
The gene coding for enhanced green fluorescent protein (EGFP) was amplified from pEGFP-1 (Clontech) by PCR using BglII containing (underlined) primers C-23120 (5′-CAA CAG AGATCT ACC ATG GTG AGC AAG GGC-3′) and C-14112 (5′-TCG AGATCT CTT GTA CAG CTC GTC C-3′). The PCR product was digested with BglII and the resulting 730-bp fragment was ligated into BglII-digested pHSEHP143 (24) to generate pHSEHGFP-P143. pEGFP-1 was digested with EcoRI, blunt-ended with Klenow polymerase, and then digested with NotI to release the GFP open reading frame, which was then cloned into pHSEH digested with BglII, blunt ended with Klenow polymerase, and then digested with NotI to generate pHSEHGFP.
The region coding for amino acids 2 to 56 of LEF-3 in pHSEHLEF3 was amplified with primers C-24026 (5′-CCT CAC CTGCAG GCG ACC AAA AGA TCT TTG-3′) and C-24027 (5′-CGT CTC CCCGGG CAA AAG TGT AAT GGT ATT C-3′). The 190-bp PCR product was digested with PstI and SmaI and cloned into PstI- and SmaI-digested pHSEHGFP to generate pHSEHLEF3(2-56)-GFP. The region coding for amino acids 2 to 56 of LEF-3 fused with GFP in pHSEHLEF3(2-56)-GFP was amplified by PCR using primers C-24026 (5′-CCT CAC CTG CAG GCG ACC AAA AGATCT TTG-3′) and C-14112. The 921-bp PCR product was digested with BglII and cloned into BglII-digested pHSEHP143 to generate pHSEHLEF3(5-56)-GFP-P143. The same region of lef-3 was also PCR amplified with primers C-25030 (5-CCT CGC GGA TCC ATG GCG ACC AAA AGA TCT TTG −3′) and C-25031 (5′-CGT CGC GGA TCC AAA AGT GTA ATG GTA TTC-3′). The PCR product was digested with BamHI and cloned into BglII-digested pHSEHP143 to produce pHSEHLEF3(1-56)-P143.
PCR fusion was used to produce a clone under the control of the endogenous p143 promoter. Primers C-25393 (5′-CGG CTC GTA TGT TGT GTG G −3′) and C-25396 (5′-TCT TTT GGT CGC CAT GTT GGC TAT CGT GTT TGT −3′) were used to amplify a region in pAcP143 including 787 bp of the p143 promoter. Primers C-25394 (5′-ACT TTC CAA CAC CCA GTC GTG-3′) and C-25395 (5′-ACA CGA TAG CCA ACA TGG CGA CCA AAA GAT CTT TG-3′) were used with pHSEHLEF3(1-56)-P143 to amplify a 462-bp product carrying the lef-3 sequences (amino acids 1 to 56) fused with p143 sequences (amino acids 2 to 99). The two PCR products were diluted, mixed, and used as the template in a third PCR with primers C-25393 and C-25394 to produce a 1,249-bp product. Following digestion with PstI, the 1,105-bp fragment was ligated to a 6,399-bp PstI fragment from pAcP143 to produce pAcLEF3(1-56)-P143. All clones were confirmed by restriction enzyme and nucleotide sequence analysis.
Infections, transfections, immunoblotting, and immunofluorescence microscopy.
Sf21cells (106), seeded into 35-mm dishes (with or without coverslips) were infected or transfected as previously described (2) and incubated at 28°C for 24 to 48 h. In some cases, the cells were heat shocked at 42°C for 30 min at 20 h posttransfection, and returned to 28°C for 3.5 h. Cells transfected with 2 μg of plasmid DNA were harvested at 48 h postinfection and biochemically fractionated exactly as described (23).
Cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Hybond-C). The membranes were blocked with 5% skimmed milk powder overnight at 4°C and then incubated with primary polyclonal antibody against AcMNPV LEF-3 (2) (1:2,500), monoclonal antibody against P143 (15) (1:3,000), or monoclonal antibody against the six-His tag (Sigma) (1:10,000) for 1 h at room temperature. Following three washes with PBS-T (phosphate-buffered saline plus 0.05% Tween 20), the membranes were incubated for 30 min with the appropriate goat secondary antibodies (Molecular Probes) conjugated with horseradish peroxidase (1:50,000 dilution). The immunoreactive proteins were detected with a chemiluminescent detection system (New England Nuclear).
The cells on coverslips were washed with PBS and fixed with 2% paraformaldehyde for 10 min at room temperature, washed again with PBS, and then permeabilized in 100% methanol for 20 min at 4°C. Following three washes with PBS-T, the cells were blocked for 1 h in 1% goat serum in PBS-T, and then incubated with polyclonal antibody against AcMNPV LEF-3 (1:1,000), a monoclonal antibody against AcMNPV P143 (1:1,000), or a monoclonal anti-polyhistidine antibody (1:1,000) for 1 h at room temperature. Following four washes with PBS-T, the coverslips were incubated for 1 h with goat anti-rabbit immunoglobulin G conjugated with Alexa Fluor 488 (Molecular Probes), goat anti-rabbit immunoglobulin G conjugated with Alexa Fluor 568 (Molecular Probes) or goat anti-mouse immunoglobulin G conjugated with Alexa Fluor 568 (Molecular Probes). Following four washes with PBS-T, the cells were stained with 5% (vol/vol) DAPI (4′,6′-diamidino-2-phenylindole) (Molecular Probes) for 2 min at room temperature. Cells were viewed with a Leitz Aristoplan microscope with an I3 Leitz band pass filter for Alexa Fluor 488 detection, an N3 Leitz band pass filter for Alexa Fluor 568 detection, and an A Leitz filter for DAPI stain. Transfection efficiencies as judged by the percentage of positively fluorescing cells were routinely between 15 and 40%. Photographs were taken with a Nikon COOLPIX 995 digital camera with a 100X objective lens and complied with Thumbs Plus 4.0 software (Cerious Software Inc).
Transient replication assays.
Equal molar amounts of plasmids expressing all of the AcMNPV genes essential for viral DNA replication (AcMNPV replication library: pAcIE-1, pAcIE-2/PE38, pAcLEF-1, pAcLEF-2, pAcLEF-3, pAcP143, pAcDNAPOL, and pAcP35) were mixed and transfected into Sf21 cell as previously described (2). At 48 h postinfection, total infected cell DNA was purified, aliquots were digested with HindIII and/or DpnI for 6 h, and the DNA fragments were electrophoresed through 0.7% agarose gels. Following transfer to Nytran Plus positive charged nylon membranes (Schleicher & Schuell), the filters were hybridized with probes prepared by random-primed labeling of a PCR fragment derived from the AcMNPV ie-1 gene using primers C-24732 (5′-CTT GAG CAG CCT GTT GTG-3′) and C-24733 (5′-TGT GAC CCT TTA CAT TAT C-3′) with digoxigenin-labeled dUTP (Roche).
RESULTS
Expression and localization of mutated LEF-3.
AcMNPV lef-3 encodes a 385-amino-acid protein (LEF-3), originally described as an essential late expression factor (17) and later shown to be essential for viral DNA replication in transient assays (14). The specific role of LEF-3 in this process is unknown. We have shown that LEF-3 is translocated to the nucleus soon after expression and is required for active transport of P143, another viral protein essential for DNA replication (27). Based on these results, we predicted that LEF-3 would carry an amino acid domain required for efficient transport of LEF-3 into the nucleus.
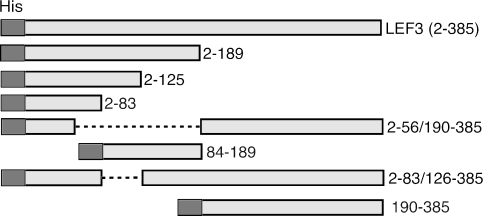
To identify a LEF-3 NLS domain, a number of LEF-3 deletion mutants were constructed (Fig. 1). First, constructs expressing only the N terminus (pHSEHLEF3-N; amino acids 2 to 189) or the C terminus (pHSEHLEF3-C; amino acids 190 to 385) were made. We confirmed by immunoblotting extracts from plasmid-transfected Sf21 cells that these constructs expressed polypeptides of the predicted sizes: 24.7 kDa for the N-terminal and 25.5 kDa for the C-terminal polypeptide (Fig. 2). The plasmid expressing full-length LEF-3 (pHSEHLEF3) (amino acids 2 to 385) produced the expected band of 47.3 kDa, slightly larger than the endogenous LEF-3 produced by pIE1-LEF-3 or AcMNPV infection (44.5 kDa) because of the EpiHA and 6XHis tags fused at the N-terminal region of the expressed protein. These results also demonstrated that a polyclonal anti-LEF-3 antibody (2) recognized epitopes in both half domains of LEF-3.
FIG. 1.
Schematic diagram of LEF-3 deletion mutants. The light shaded boxes represent regions of LEF-3 (as indicted by amino acid numbers on the right) expressed by plasmids under the control of the Drosophila heat shock 70 promoter. Each protein was fused at the N terminus with the same 22-amino-acid sequence including the 6XHis tag (His, dark box). Dashed lines represent deleted amino acids.
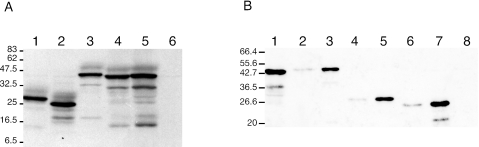
FIG. 2.
Immunoblot analysis of mutant LEF-3 expression in transfected versus infected cells. (A) Sf21 cells were transfected with pHSEHLEF3-C (lane 1), pHSHLEF3-N (lane 2), pHSEHLEF3 (lane 3), or pIE-1hrlef3 (lane 4) or infected with AcMNPV (lane 5, multiplicity of infection of 1, 24 h postinfection). The transfected cells were heat shocked at 20 h posttransfection at 42°C for 30 min and returned to 28°C for 3.5 h before harvesting. Extracts were analyzed by SDS-PAGE and immunoblotting using a polyclonal antibody against LEF-3. Mock-transfected cell extracts are also shown (lane 6). (B) Sf21 cells were transfected in duplicate with pHSEHLEF3 (lane 2 and 3), pHSEHLEF3-C (lane 4 and 5) or pHSHLEF3-N (lane 6 and 7). One set of cells was heat shocked (lanes 3, 5, and 7) while the others were not (lanes 2, 4, and 6). At 24 h posttransfection, extracts were prepared, and immunoblotted and probed as described for panel A. Cells infected with AcMNPV (lane 1, multiplicity of infection of 1, 24 h postinfection) or mock transfected (lane 8) are also shown.
We also determined the effect of heat shock on expression of these constructs since the inserts were under the control of the Drosophila heat shock 70 promoter. Heat shocking the transfected cells resulted in high levels of LEF-3 expression, but slightly lower than the amount of LEF-3 expressed in AcMNPV-infected cells (multiplicity of infection of 1). Both deletion mutants were expressed at similar levels following heat shock. Therefore, to enhance the detection levels, in the following experiments using plasmids expressing proteins under the control of the heat shock promoter, all transfected cells were heat shocked prior to harvesting and analysis.
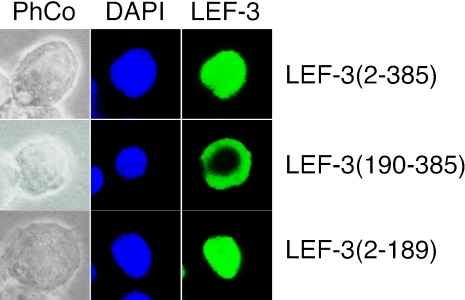
We used immunofluorescence microscopy with the same anti-LEF-3 antibody to detect the expressed proteins in transfected cells. pHSEHLEF3 or pHSEHLEF3-N expressed polypeptides that were localized in the nucleus but cells transfected with pHSEHLEF3-C expressed a polypeptide that remained cytoplasmic (Fig. 3). These results were consistent in 90 to 100% of the positively fluorescing cells and indicated that the N-terminal half of LEF-3 contains a domain sufficient for nuclear localization.
FIG. 3.
Intracellular localization of LEF-3 and LEF-3 deletion mutants in transfected Sf21 cells. Sf21 cells were transfected with pHSEHLEF3 (LEF-3 amino acids 2 to 385), pHSEHLEF3-C (LEF-3 amino acids 190 to 385), or pHSHLEF3-N (LEF-3 amino acids 2 to 189). At 24 h posttransfection and heat shock as described for Fig. 2, the cells were prepared for immunofluorescence microscopy by probing with polyclonal anti-AcMNPV LEF-3 (1:1,000) antibody and detected with goat anti-rabbit immunoglobulin G conjugated with Alexa Fluor 488. Cell nuclei were stained with DAPI. Phase contrast images of the same cells are also shown (PhCo). All fluorescent cells revealed the pattern shown in these representative micrographs.
To narrow down the region containing the nuclear localization domain, a series of deletion mutants in the N-terminal half of LEF-3 were generated. Some deletions which retained the C-terminal region to maintain enough of the LEF-3 open reading frame to react with the LEF-3 antibody were also prepared (Fig. 1). Sf21 cells were transfected with the various expression plasmid DNAs and examined by immunofluorescence microscopy. The results reproducibly revealed that proteins that retained amino acids 2 to 56 of LEF-3 were successfully transported to the nucleus, while constructs that produced proteins lacking this region remained cytoplasmic (Table 2). These results clearly indicated that the nuclear localization domain is located within the first 56 amino acids of LEF-3.
TABLE 2.
Intracellular localization of LEF-3 deletion mutants and GFP-P143a
| LEF-3 mutant | Intracellular localization
|
|
|---|---|---|
| LEF-3 alone (immunofluorescence) | LEF-3 + GFP-P143 (GFP fluorescence) | |
| LEF-3 | N | N |
| LEF-3(2-189) | N | N |
| LEF-3(2-125) | N | N |
| LEF-3(2-83) | N* | C |
| LEF-3(2-56/190-385) | N | C |
| LEF-3(2-83/126-385) | N | C |
| LEF-3(84-189) | C | C |
| LEF-3(190-385) | C | C |
Sf21 cells were transfected with the expression plasmid DNA. Following heat shock, intracellular localization of LEF-3 or GFP P143 was viewed by fluorescence microscopy. Transfection efficiencies were between 20 and 40%. Of these positively transfected cells, 90 to 100% revealed the indicated intracellular localization. N, nucleus; C, cytoplasm. *, detected with anti-His tag antibody.
LEF-3 NLS transports GFP and P143 to the nucleus.
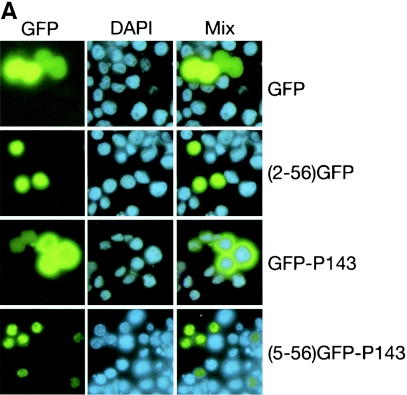
To support the hypothesis that the N-terminal region of LEF-3 was critical for nuclear localization, constructs were made in which the N-terminal 56 amino acids of LEF-3 were fused in-frame with the green fluorescence reporter protein GFP (Fig. 4). Following transfection of Sf21 cells with pHSEHLEF3(2-56)-GFP DNA, fluorescence microscopy revealed that the LEF-3 GFP fusion protein colocalized with the DAPI nuclear stain in 100% of the GFP-positive cells while GFP alone was distributed diffusely throughout the cells (Fig. 5A). A similar construct fusing LEF-3 amino acids 5 to 56 with GFP gave the same results (data not shown). We corroborated these studies by biochemical fractionation of similarly transfected cells and immunoblotting analysis. All the His-tagged proteins consisting of GFP fused with the LEF-3 amino acid 2 to 56 domain were detected in the nuclear fraction, while GFP on its own was found in both cytoplasmic and nuclear extracts (Fig. 5B). These data strongly support our hypothesis that a nuclear localization domain is located within the first 56 amino acids of LEF-3.
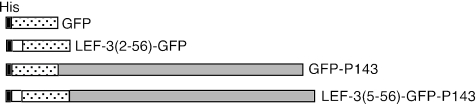
FIG. 4.
Schematic diagram of GFP fusion constructs. Plasmids were constructed that expressed GFP (represented by the stippled areas), LEF3 amino acids 2 to 56 (represented by the white areas) fused with GFP [LEF-3 (2-56)-GFP], GFP fused with P143 (represented by the gray areas) (GFP-P143), or LEF3 amino acids 5 to 56 fused with GFP and P143 [LEF-3 (5-56)-GFP-P143]. Each protein was fused at the N terminus with the same 22-amino-acid sequence including the 6XHis tag (His, black areas). The length of each area is drawn to the same scale according to the number of amino acids contained in each region of the fusion proteins.
FIG. 5.
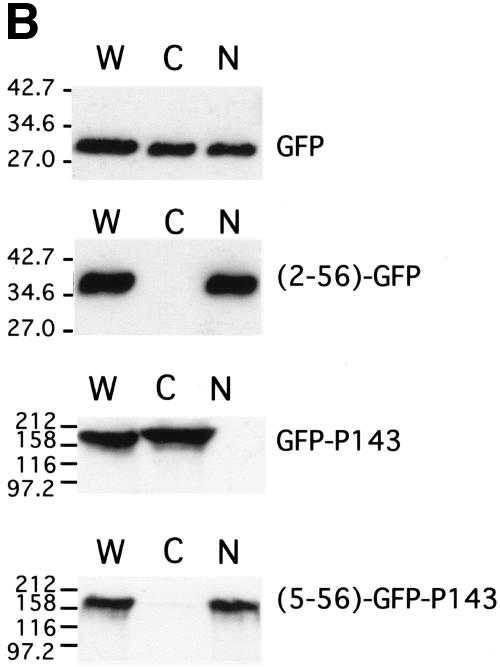
Intracellular localization of GFP fusion protein in transfected cells. Sf21 cells were transfected with plasmid pHSEHGFP (GFP), pHSEHLEF3(2-56)-GFP [(2-56)-GFP], pHSEHGFP-P143 (GFP-P143), and pHSEHLEF3(5-56)-GFP-P143 [(5-56)-GFP-P143]. (A) At 20 h posttransfection, the cells were heat shocked at 42°C for 30 min and returned to 28°C for 3.5 h. Then the cells were fixed, and cell nuclei were stained with DAPI. GFP fluorescence was directly observed. Fifteen to 40% of the total cells were positive for GFP fluorescence, and 100% of these positive cells showed the patterns represented in each panel. Representative fields of cells are shown, with the blue DAPI (nuclear stain) and green GFP fluorescence superimposed. (B) Similarly transfected cells were harvested at 48 h posttransfection and then extracted into cytoplasmic (C) and nuclear (N) fractions. Equivalent amounts of cytoplasmic and nuclear fractions were immunoblotted and probed with either antipolyhistidine (top two panels) or anti-P143 (bottom two panels) antibodies. Whole-cell extracts (W) were included as controls.
Because we have shown that LEF-3 is responsible for transporting P143 to the nucleus, we tested the hypothesis that only the N-terminal region of LEF-3 was required for this function by also fusing this region of LEF-3 with P143. To easily observe the intracellular location of P143, we also fused GFP to P143 [pHSEHLEF3(5-56)-GFP-P143] (Fig. 4). Following transfection of pHSEHLEF3(5-56)-GFP-P143 into Sf21 cells, GFP fluorescence was detected exclusively in the nucleus (Fig. 5A). A GFP-P143 fusion protein without the N-terminal region of LEF-3 (pHSEH-GFP-P143) revealed exclusive cytoplasmic fluorescence (Fig. 5A). When similarly transfected cells were biochemically fractionated and analyzed by immunoblotting, GFP-P143 was detected only in the cytoplasmic fraction, while GFP-P143 fused with the LEF-3 domain (5 to 56) was found exclusively in the nuclear fraction (Fig. 5B). Therefore, LEF-3 contains a true nuclear localization signal domain that can function independently of the rest of the LEF-3 open reading frame.
Interaction of LEF-3 mutants and P143.
We have previously shown that LEF-3 is essential for the transport of P143 from the cytoplasm to the nucleus during virus infection, suggesting that LEF-3 also contains a P143-interacting domain. We therefore investigated the ability of the LEF-3 deletion mutants to interact with and transport P143 to the nucleus. In order to monitor the expression and localization of P143 in transfected cells, an expression plasmid with GFP fused to the N terminus of P143 (pHSEH-GFP-P143) was used (Fig. 4). This construct was transfected into Sf21 cells and cell extracts were examined by Western blotting using anti-P143 antibody to confirm the expression of P143 (data not shown). The transfected cells were also examined for GFP fluorescence as described above. The GFP-P143 reporter remained in the cytoplasm, as expected (Fig. 5).
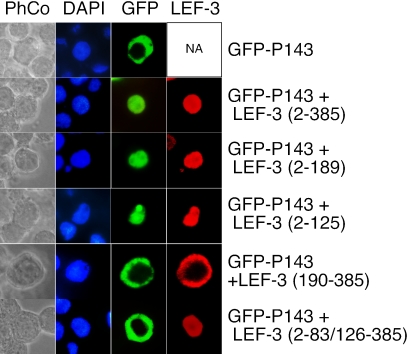
To identify the region in LEF-3 that is responsible for interacting with P143, LEF-3 deletion mutants were cotransfected with pHSEH-GFP-P143 and transfected cells were examined for GFP fluorescence. Under our transfection conditions, almost all cells that were GFP positive were also positive for LEF-3 by immunofluorescence. Cells representative of 90 to 100% of the positive cells were photographed. Mutants expressing the N-terminal half (pHSEHLEF3-N) or the N-terminal 125 amino acids (pHSEHLEF3)(2-125) resulted in nuclear GFP fluorescence, but other mutants not expressing this region failed to transport GFP-P143 into nucleus (Fig. 6). These results indicate that although only LEF-3 amino acids 2 to 56 are required for LEF-3 nuclear localization (Fig. 5, Table 2), a domain of LEF-3 including amino acids 2 to 125 is necessary for interacting with P143 and transporting it to the nucleus. A deletion mutant expressing the N-terminal 83 amino acids (pHSEHLEF3)(2-83/126-385) failed to transport P143 to the nucleus (Fig. 6), suggesting that the region of LEF-3 between amino acids 83 and 125 may interact directly with P143.
FIG. 6.
Intracellular localization of GFP-P143 in the presence of LEF-3. Sf21 cells were transfected with pHSEHGFP-P143, or cotransfected with plasmids pHSEHGFP-P143 plus pHSEHLEF3, pHSEHLEF3-N, pHSEHLEF3(2-125), pHSHLEF3-C, or pHSEHLEF3(2-83/126-385). The cells were harvested at 24 h posttransfection, fixed, and either observed directly for GFP fluorescence or probed with polyclonal anti-AcMNPV LEF-3 (1:1,000) antibody. Immunofluorescence was detected with secondary goat anti-rabbit immunoglobulin G conjugated with Alexa Fluor 568. Cell nuclei were detected by DAPI staining. Ninety to 100% of the positively transfected cells showed the patterns represented in each panel.
DNA replication assay.
Both LEF-3 and P143 are essential for viral DNA replication as suggested by transient DNA replication assays. However, with the discovery that LEF-3 was essential for nuclear localization of P143, the possibility existed that this was the only function of LEF-3 during infection. By constructing a P143 fusion protein that self localized to the nucleus, we had the opportunity to test whether this protein could support DNA replication. To avoid any potential interference that the GFP fusion might have with the replication function of P143, we prepared another construct in which only the amino acid 1 to 56 domain of LEF-3 was fused with P143. A variety of transient DNA replication assays were conducted, taking advantage of a library of plasmids that together express nine AcMNPV genes (ie-1, ie-2, p143, dnapol, lef-1, lef-2, lef-3, pe38, and p35) necessary for viral DNA replication. All these genes, expressed under their endogenous promoters, were transfected into Sf21 cells and total intracellular DNA was analyzed by DpnI digestion to distinguish between unreplicated input plasmid and newly replicated DNA (Fig. 7A).
FIG. 7.
Transient DNA replication assays and expression of P143. (A) Sf21 cells were transfected with a collection of plasmids which together expressed AcMNPV genes necessary for plasmid DNA replication (ie1, dnapol, lef1, lef2, p35, pe38, and ie2) plus p143 and lef-3 (lane 1), only p143 (lane 2; no LEF-3), only lef-3 (lane 3; no P143), lef-3 plus lef3(2-56)p143 (lane 4) or only lef-3(2-56)p143 (lane 5; no LEF-3). All of these constructs expressed proteins under their endogenous promoters. Following incubation for 48 h, total intracellular DNA was prepared and digested with HindIII to linearize the plasmids, and DpnI to digest nonreplicated plasmid DNA. Southern blots of these restriction-digested DNA preparations were probed with a digoxigenin-labeled PCR product of the AcMNPV ie-1 gene. (B) Sf21 cells were transfected with pAcP143 (P143) or pAcLEF3(1-56)-P143 (1-56-P143). At 24 h posttransfection, the cells were prepared for immunofluorescence microscopy and probed with monoclonal anti-AcMNPV P143 antibody. Immunofluorescence was detected using a secondary goat anti-mouse immunoglobulin G conjugated with Alexa Fluor 568. Nuclei were detected by DAPI staining. The red immunofluorescence P143 stain is superimposed with the blue DAPI nuclear stain. (C) Similarly transfected cells were harvested at 48 h posttransfection, and biochemically fractionated into cytoplasmic (C) and nuclear (N) extracts that were analyzed by immunoblotting and probing with anti-P143 antibody. Whole-cell extracts (W) were included as controls.
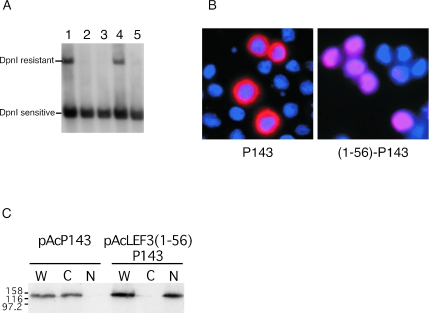
As expected, cotransfection of these plasmids resulted in plasmid replication as shown by a DpnI-resistant fragment (Fig. 7A, lane 1). When either pAcLEF-3 (Fig. 7A, lane 2) or pAcP143 (Fig. 7A, lane 3) was absent from the transfection mixture, no plasmid replication occurred. Replication did occur in the presence of LEF-3 and the fusion protein of the N-terminal 56 amino acids from LEF-3 fused in-frame with the P143 open reading frame [pAcLEF3(1-55)P143] (Fig. 7A, lane 4), clearly demonstrating that fusing an additional 56 amino acids to the N terminus of P143 did not disrupt its function during DNA replication. However, when full-length LEF-3 was absent from the plasmid library, pAcLEF3(1-55)P143 on its own did not support plasmid replication (Fig. 7A, lane 5).
We confirmed that pAcLEF3(1-55)P143 expressed a P143 fusion protein which localized to the nucleus by immunofluorescence microscopy. In the absence of the LEF-3 NLS domain, P143 displayed as a bright red ring surrounding the nucleus, while P143 fused to the LEF-3 NLS localized completely in the nucleus (Fig. 7B). These results were also confirmed by biochemical fractionation of transfected cells. P143 alone was found in the cytoplasmic fraction, while P143 fused with the LEF-3 NLS was found in the nuclear fraction (Fig. 7C). Together, these data suggest that simply transporting P143 into the nucleus is not sufficient for viral DNA replication in the absence of LEF-3. We conclude that although one role for LEF-3 during virus infection is to transport P143 to the nucleus, LEF-3 performs other essential functions once inside the nucleus.
DISCUSSION
AcMNPV genome replication occurs in the nucleus of infected cells, requiring proper transport and assembly of the viral proteins essential for DNA replication to this intracellular compartment. In this study, we have identified a region of LEF-3 that is required for nuclear import of LEF-3. A variety of deletion mutants, all expressing the region including amino acids 5 to 56, produced LEF-3-related polypeptides that were localized to the nucleus. When these residues were fused in-frame with the GFP reporter gene, GFP was transported to the nucleus. Finally, when LEF-3 residues 5 to 56 were fused in-frame with a GFP-P143 fusion protein, a protein predicted to be 180 kDa, the GFP fluorescence was also localized to the nucleus. These results were confirmed by biochemical fractionation of transfected cells. All these data are consistent with the hypothesis that residues 5 to 56 of LEF-3 contain a nuclear localization signal domain and that this domain can function in a variety of configurations to actively transport large proteins into the nucleus of insect cells.
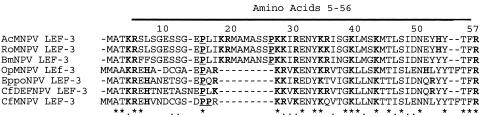
The LEF-3 5 to 56 domain includes a number of basic residues that are conserved among different species of the group I NPVs (Fig. 8). Three closely related species (AcMNPV, Rachiplusia ou MNPV [RoMNPV], and Bombyx mori NPV [BmNPV]) appear to have an insert in this region consisting of nine amino acids (eight are identical in all these viruses), including flanking basic amino acids and a proline, compared with other group I NPVs. We have previously shown that Christoneura fumifurana MNPV (CfMNPV) LEF-3 self-localizes to the nucleus of infected cells (2), suggesting that this nine-amino-acid region is not essential for nuclear localization. Since all of our constructs in the vector pHSEH are His tagged, there is a string of eight basic amino acids fused immediately upstream of any sequences cloned into this vector. These amino acids potentially could mimic basic residues in an NLS, making it difficult to determine whether the first four amino acids of LEF-3 are essential for nuclear localization. However, it is clear that the His tag on its own cannot function as an NLS, since constructs of GFP or P143 in this vector remain cytoplasmic.
FIG. 8.
Amino acid sequence similarity between baculovirus group I NPV LEF-3 proteins. The N-terminal 57 amino acids of AcMNPV LEF-3 were aligned with other group I baculoviruses by ClustalX (25). The presence of identical (*) and conserved (.) amino acids is shown in the bottom row. Basic amino acids (K, R, and H) are highlighted in bold, and prolines are shown in bold and underlined. The numbers along the top refer to the amino acid number of AcMNPV LEF-3. The addition of residues 5 to 56 of AcMNPV LEF-3 was sufficient to localize GFP or P143 to the nucleus.
The conservation of residues in the N-terminal 56 residues of group I NPV LEF-3s argues that the LEF-3s expressed by these viruses use a common host mechanism for nuclear import. It is likely that they interact with similar importin proteins from their insect hosts (for a review, see reference 20). The group II NPVs have a greater diversity of sequence in their N-terminal regions, suggesting that there may be more diversity in the nuclear import systems in their insect hosts.
At least three types of NLS have been described (9). The classical NLS has been generalized to either a four-residue pattern (called ′pat4′) composed of 4 basic amino acids (K or R), or composed of three basic amino acids (K or R) and either H or P, or a seven-residue pattern (called pat7′) that starts with P and is followed within three residues by a basic segment containing three K or R residues out of four. The group I NPVs all have pat7-type patterns (PKKIREN, PPRKRVK, or PRKRVKE) but LEF-3 from Orgyia pseudotsugata MNPV (OpMNPV) and CfMNPV also carry a predicted bipartite pattern of two basic residues, a 10-residue spacer, and another basic region consisting of at least three basic residues out of five residues (e.g., CfMNPV residues 5 to 21 are KREHVNDCGSDPPRKRVK). The NLSs of other baculovirus proteins, including polyhedrin (3, 13), IE-1 (23), and PP31 (1a), have been associated with basic residues of sequence KRKK or KVNRR. The LEF-3 NLS functions as a single domain, most similar to the polyhedrin NLS. We are currently investigating the specific amino acid residues that would define the group I LEF-3 NLS.
We have also identified a region of LEF-3, between residues 83 and 125, that is essential for interaction with P143 in trans. LEF-3 interaction with P143 is essential for the nuclear localization of P143 (27), where it presumably functions as a helicase during viral DNA replication (18). Although we have recently shown that LEF-3 and P143 are bound to viral DNA in vivo (12), it was not known whether LEF-3 plays a functional role during the replication process. Both LEF-3 and P143 are DNA binding proteins. LEF-3 has single-stranded DNA binding activity (8), while P143 can bind double-stranded DNA (15). The P143 interaction domain of LEF-3 has been investigated by a yeast two-hybrid analysis (4), where it was demonstrated that LEF-3 amino acids 1 to 165 but not 1 to 77 were capable of interacting with P143. Our data are consistent with those results and narrow down the interacting region to between amino acids 83 and 125. This region is well conserved within the group I NPVs, particularly between two absolutely conserved cysteine (AcMNPV amino acids 82 and 106) residues that may be important for disulfide bond formation in the tertiary structure of LEF-3.
We demonstrated by both immunofluorescence and biochemical fractionation that a protein consisting of the LEF-3 NLS domain fused in-frame with P143 was localized to the nucleus. In addition, this construct could substitute for authenticate P143 in supporting DNA replication in transient assays but only in the presence of intact LEF-3. These results confirm previous experiments where fusing GFP to the CfMNPV P143 protein did not inhibit the ability of P143 to support DNA replication (2). From these results, we conclude that the presence of P143 in the nucleus is not sufficient to support DNA replication, in the absence of LEF-3. LEF-3 must function directly in the replication process, likely in a complex with the P143 helicase on replicating DNA, as recently suggested by chromatin immunoprecipitation experiments (12).
Replication factor A is the host single-stranded DNA-binding protein complex required for eukaryotic DNA replication (11, 26). Our results show that the lepidopteran equivalent of replication factor A cannot substitute for LEF-3. A second baculovirus single-stranded DNA-binding protein has been identified and characterized (dbp, AcORF25) (21). This gene, unlike lef-3, is conserved in all lepidopteran baculoviruses (16). DBP has not been demonstrated to be essential for plasmid DNA replication in transient assays, so its role in viral DNA replication is unknown. However, it has been suggested that DBP is a true single-stranded DNA-binding protein, based on amino acid sequence homologies and its ability to destabilize duplex DNA (21). We are currently investigating whether DBP can substitute for LEF-3 in transient replication assays when P143 is present in the nucleus.
LEF-3 also interacts with a viral alkaline nuclease (22), and it has been suggested that an alkaline nuclease-LEF-3 complex might be involved in repair and recombination of baculovirus genomes during replication. Because little is known about the mechanism of baculovirus replication, it will be of great interest to explore the functional role of other domains within LEF-3.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and Genome Canada through the Ontario Genomics Institute.
REFERENCES
- 1.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. Volkman. 2000. Family Baculoviridae, p. 195-202. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Ca.
- 1a.Broussard, D. R., L. A. Guarino, and D. L. Jarvis. 1996. Mapping functional domains in AcMNPV pp31. Virology 222:318-331. [DOI] [PubMed] [Google Scholar]
- 2.Chen, T., D. Sahri, and E. B. Carstens. 2004. Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication. J. Virol. 78:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eason, J. E., R. H. Hice, J. J. Johnson, and B. A. Federici. 1998. Effects of substituting granulin or a granulin-polyhedrin chimera for polyhedrin on virion occlusion and polyhedral morphology in Autographa californica multinucleocapsid nuclear polyhedrosis virus. J. Virol. 72:6237-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans, J. T., G. S. Rosenblatt, D. J. Leisy, and G. F. Rohrmann. 1999. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143). J. Gen. Virol. 80:493-500. [DOI] [PubMed] [Google Scholar]
- 5.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-170. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 6.Friesen, P. D., and L. K. Miller. 2001. Insect viruses, p. 599-628. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 7.Gordon, J. D., and E. B. Carstens. 1984. Phenotypic characterization and physical mapping of a temperature-sensitive mutant of Autographa californica nuclear polyhedrosis virus defective in DNA synthesis. Virology 138:69-81. [DOI] [PubMed] [Google Scholar]
- 8.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks, G. R., and N. V. Raikhel. 1995. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11:155-188. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 12.Ito, E., D. Sahri, R. Knippers, and E. B. Carstens. 2004. Baculovirus proteins IE-1, LEF-3, and P143 interact with DNA in vivo: a formaldehyde cross-linking study. Virology 329:337-347. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis, D. L., D. A. Bohlmeyer, and A. Garcia, Jr. 1991. Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology 185:795-810. [DOI] [PubMed] [Google Scholar]
- 14.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laufs, S., A. Lu, L. K. Arrell, and E. B. Carstens. 1997. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology 228:98-106. [DOI] [PubMed] [Google Scholar]
- 16.Lauzon, H. A. M., C. J. Lucarotti, P. J. Krell, Q. L. Feng, A. Retnakaran, and B. M. Arif. 2004. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 78:7023-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y., A. L. Passarelli, and L. K. Miller. 1993. Identification, sequence, and transcriptional mapping of lef-3, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:5260-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 19.Lu, A., P. J. Krell, J. M. Vlak, and G. F. Rohrmann. 1997. Baculovirus DNA replication, p. 171-191. In L. K. Miller (ed.), The baculoviruses. Plenum, New York, N.Y.
- 20.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikhailov, V. S., A. L. Mikhailova, M. Iwanaga, S. Gomi, and S. Maeda. 1998. Bombyx mori nucleopolyhedrovirus encodes a DNA-binding protein capable of destabilizing duplex DNA. J. Virol. 72:3107-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2003. Baculovirus alkaline nuclease possesses a 5′→3′ exonuclease activity and associates with the DNA-binding protein LEF-3. J. Virol. 77:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson, V. A., J. A. Wetter, and P. D. Friesen. 2002. Baculovirus transregulator IE1 requires a dimeric nuclear localization element for nuclear import and promoter activation. J. Virol. 76:9505-9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 27.Wu, Y., and E. B. Carstens. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32-40. [DOI] [PubMed] [Google Scholar]