Abstract
We aimed to identify cross-clade human immunodeficiency virus type 1 (HIV-1) specific T-cell responses among 10 HLA-typed individuals who were infected with non-B HIV-1 strains (A, AG, C, D, G, or F) and to correlate these responses with genetic variation in documented T-cell epitopes. T-cell reactivity was tested against peptide pools spanning clade B Gag, Pol, Nef, Rev, and Tat consensus, with Gag and Nef providing the highest responses. Nine individuals who responded to clade B Gag demonstrated cross-reactive T-cell responses against clade A and C Gag pools, while six of seven responders to Nef-B reacted to clade A and C Nef pools. An inverse correlation between the height of the T-cell responses and the sequence divergence of the HLA class I-restricted epitopes was identified when we compared autologous Gag and Nef sequences with the reactive consensus pools. This could be explained for the Gag sequences through observed variations in the HLA anchor residues. Through mapping of 30 amino acid cross-clade-reactive regions using Gag-B pools, we were able to link 58% (14/24) of the T-cell responses to regions containing previously described HLA class I-restricted epitopes. Forty-two percent (10/24) of the responses were directed to regions containing new epitopes, for which predicted HLA class I motifs could be recognized in 70% (7/10) of individuals. We demonstrate here that cross-clade T-cell responses are frequently induced in individuals infected with distinct HIV-1 clades, suggesting that interclade variation outside of HLA anchor residues may have less impact on vaccine-induced T-cell reactivity than previously thought.
Human immunodeficiency virus type I (HIV-1) is highly variable and can be subdivided into several distinct clades of virus (A to G) and recombinants of clades (51, 55). This high degree of variation is predicted to have a direct impact on T-cell recognition of variable epitopes and thereby to hamper the design of HIV-1 vaccines aimed at inducing strong cross-clade T-cell responses (46).
It has been well documented that the induction of HLA-restricted cytotoxic-T-lymphocyte (CTL) responses is crucial in controlling HIV-1 replication, as well as being key to slowing down disease progression (38, 40). Vaccine strategies capable of eliciting simian immunodeficiency virus specific CD8+ CTL responses in nonhuman primates have been demonstrated to limit viral replication, as well as to prevent the onset of disease (4, 7, 56). These studies and others collectively suggest that vaccines aimed at inducing strong CTL responses would be effective at preventing infection or limiting disease progression (57). However, viral escape from CTL immune surveillance through the alteration of recognized CTL epitopes has been reported in natural HIV-1/simian immunodeficiency virus infection (3, 26, 32, 34, 53; reviewed in reference 35), as well as in vaccinated nonhuman primates (5, 6).
T-cell responses raised against one HIV-1 clade have been reported to recognize HIV antigens derived from other clades (12, 15, 18, 22, 41, 44), and clade B-based HIV-1 vaccines can elicit cross-clade T-cell responses (27). The characterization of cross-clade specific T-cell responses and the respective roles of sequence variation in HIV-1 antigens and class I haplotypes mediating responses can help further identify the correlatives of cross-clade T-cell responses. In the present study, we analyzed the cross-clade specific T-cell responses in 10 individuals infected with different HIV-1 non-B clades (A, AG, C, D, G, or F). We defined the hierarchy in cross-clade T-cell responses to different clade B peptide pools and compared the top-responding clade B peptides with those representing clades A and C. Through autologous sequence analysis, we revealed an inverse correlation between the strength of the T-cell responses and the degree of viral diversity within known CTL epitopes (9), which for Gag could be associated with HLA anchor residues. Finally, we fine mapped cross-clade Gag T-cell responses to 30- to 40-amino-acid (aa)-long stretches in which we could predict epitopes mediating the cross-clade reactivity by using the Los Alamos Immunology Database and HLA motif scans.
MATERIALS AND METHODS
Samples.
Subjects were selected from the outpatient clinic from the Academic Medical Centre, Amsterdam, The Netherlands. Nine out of 10 individuals tested originated from Africa, the other from South America (Table 1), and all were predicted to have been infected in their home countries. Serum and peripheral blood mononuclear cell (PBMC) samples were obtained and stored frozen prior to analysis. On the sampling date, three patients were on therapy and seven were therapy naive. Their ages ranged between 24 and 50 (mean, 34.5) years, and no patients had clinical signs of AIDS. Viral loads were determined by the NucliSens assay or the HIV-1 RNA QT NASBA assay (Organon Teknika B.V., Boxtel, The Netherlands). Viral loads varied from below the detection limit (log 2 copies/ml) to log 6 copies/ml. Patient HLA typing was commercially determined by the Tissue Typing Laboratory of the University of Pennsylvania Medical Center using standard PCR techniques.
TABLE 1.
Characteristics of subjects included in the study
| Patient | Origin | Age (yr) | Viral load (copies/ml) | Therapy | HIV-1 subtyping
|
HLA typing
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTRb | Gag | Pol | Nef | Env | HLA-A | HLA-B | HLA-Cw | |||||
| M12653 | Rwanda | 26 | 103 | AZT,ddCa | A | A | A | A | A | A36, A74 | B53, B58 | W4, W6 |
| M12453 | Ghana | 34 | 102 | AZT | G | A | G | AG | A | A2, A30 | B27, B50 | W2, W6 |
| M11306 | Kenya | 33 | 104 | C | C | C | C | ND | A23, A23 | B53, B58 | W4, W7 | |
| M12003 | Ethiopia | 36 | 104 | C | C | C | C | ND | A1, A30 | B7, B41 | W7, W17 | |
| M11814 | DRCa | 24 | 105 | D | D | D | F | F | A23, A24 | B35, B58 | W4, W7 | |
| M12020 | DRC | 36 | 104 | AZT | D | D | D | NDd | D | A23, A34 | B44, B53 | W4, W6 |
| M12259 | DRC | 50 | 105 | F | D | D | ND | F | A1, A1 | B8, B55 | W3, W7 | |
| M12115 | Tanzania | 43 | 106 | D | D | D | ND | ND | A1, A24 | B8, B35 | W4, W7 | |
| M12101 | Brazil | 29 | 102 | F | F | F | F | F | A3, A24 | B52, B58 | W3, W6 | |
| M13371 | Nigeria | 34 | 102 | G | G | G | G | ND | A2, A36 | B45, B58 | W3, W6 | |
DRC, Democratic Republic of the Congo.
LTR, long terminal repeat.
AZT, zidovudine; ddC, dideoxycytosine.
ND, not done.
PCR amplification, sequencing, and subtyping.
Partial sequencing and subtyping of Gag, long terminal repeat, Pol, and Env was previously reported by de Baar et al. (20). Complete Nef and Gag sequences were obtained from serum, and nucleic acids were isolated using the silica-based Boom method (14). Reverse transcription and PCRs were performed using previously published methods with modifications (17). Nef sequences were reverse transcribed using the primer 3′ R-M (5′ ACTYAAGGCAAGCTTTATTGAG 3′) or 3′ RMnew (5′ ATTGAGGCTTAAGCAGTGGGTT 3′), followed by primary PCR using the Outer-5-1e primer as previously published (35) for 40 cycles. Secondary PCRs were performed with Inner-5-1e and Outer-3-3e for 25 cycles. Gag sequences were reverse transcribed using SK39 (5′ TTTGGTCCTTGTCTTATGTCCAGAATGC 3′) and prot1 (5′ GCAAATACTGGAGTATTGTATGGATTTTCAGG 3′). Gag primary PCRs were performed using 1-SD (5′ GGCGGCCGCTCTCTCGACGCAGGACTCG3′) plus SK39 and SK145 (5′ AGTGGGGGGACATCAAGCAGCCATGCAAAT 3′) plus prot1 (both for 40 cycles). Secondary PCRs were performed with 3′GAGAE-3 (5′ ACTATTTTATTTAATCCCAGGAT 3′) plus 5′ LOUW-1-GAG (5′ TTGACTAGCGGAGGCTAGAA 3′) and TFS-R (5′ GTTGACAGGTGTAGGTCCTAC 3′) plus FGS011 (5′ AGAGAACCAAGGGGAACTGACATAGCA 3′) (both for 25 cycles). For some samples, a tertiary PCR was performed using 5′Gag-1 (5′ CATGCGAGAGCGTCAGTATTAAGCGG 3′) plus 3′ GAG-3 (5′ ATGCTTAAGCTTCTACTACTTTTACCCATGC 3′) primers with 25 cycles. Positive PCR products were cloned into the TOPO II vector (Invitrogen, Carlsbad, CA) and sequenced using the BigDye Terminator Cycle Sequencing kit (ABI, Foster City, CA) and analyzed using an ABI 377 automated sequencer (ABI). Nef and Gag sequences were analyzed using BioEdit 7.0 (T. Hall, Ibis Therapeutics, CA), Textpad (Helios Software Solutions), and neighbor-joining methods in the Mega3 software (39).
Sequence and epitope analysis.
The genetic distances between patient-specific and peptide amino acid HIV-1 sequences were calculated using Mega3 and are referred to in p-distance values. This distance is the proportion (p) of amino acid sites at which the two compared sequences are different. It is obtained by dividing the number of differences in amino acids by the total number of amino acids compared. Documented HLA-specific CTLs were analyzed using the Los Alamos Immunology Database (9). The analysis of HLA class I anchor motifs was done using the currently available data at the Syfpeithi network (54; http://syfpeithi.de/). The sequence analysis of amino acids external to documented epitopes (see Table 3) was performed using a script analysis (C. Kesmir, Theoretical Biology and Bioinformatics, UU, The Netherlands). This script randomly picks up amino acid positions in a given sequence and calculates the p distance to the reference sequence. For this analysis, we excluded the regions that contain documented CTL epitopes per given HIV-1 region (Gag or Nef). We then calculated the p distance in randomly picked amino acids equal to the total length of combined documented epitopes. To rule out any sampling bias, the procedure was repeated 500 times. Sequence analysis for HLA-specific motifs was performed using the Epitope Location Finder (ELF) (http://hiv-web.lanl.gov/ALABAMA/epitope_analyzer.html). ELF performs various analyses of HIV peptide sequences with the aim to identify optimal CTL epitopes using current knowledge of HLA serotypes, genotypes, and known HLA anchor residues. We subsequently verified whether the predicted HLA-binding motifs corresponded to previously documented epitopes and/or haplotypes in the Los Alamos Immunology Database. We included only ELF results that contained unique combinations of primary anchor and N-terminal residues. We excluded ELF results for HLA-C loci, since the roles of the HLA-C CTL responses in HIV infection are still relatively poorly understood (33).
TABLE 3.
Correlations between cross-clade T-cell responses and sequence diversity in patient-specific HLA class I documented Gag and Nef CTL epitopes
| Type of cross-clade T-cell responses (clades A, B, and C) | Parameter | p distance epitopes | p distance no HLA anchors | p distance HLA anchors −1 | p distance external to CTL epitopes |
|---|---|---|---|---|---|
| Gag | Spearman r | −0,543 | −0,209 | −0,703 | −0.226 |
| 95% confidence interval | (−0.839-<0.001) | (−0.676-0.377) | (−0.902-−0.259) | (−0.685-0.361) | |
| P value (two tailed) | 0.045 | 0.472 | 0.005 | 0.436 | |
| Nef | Spearman r | −0,864 | −0,848 | −0,747 | 0,252 |
| 95% confidence interval | −0.963-−0.563 | −0.958-−0.520 | −0.928-−0.286 | (−0.393-0.731) | |
| P value (two tailed) | 0.001 | 0.001 | 0.007 | 0.430 |
Peptides.
Synthetic peptides were custom ordered from SynPep Corporation (Dublin, CA). Peptide sequences were based on those of the HIV isolates most closely related to the clade B consensus sequences (Los Alamos National Laboratory, Los Alamos, NM [http://hiv-web.lanl.gov]), namely, Gag-A, 90cf4071; Gag-B, CAM-1; Gag-C, C-96xm751.3; Nef-A, Se8891; Nef-B, JRFL; Nef-C, In.21068; Pol-1/2-B, JRFL; Rev-B, JRFL; and Tat-B, JRFL. The peptides were 15-mers overlapping by 11 aa unless otherwise specified. Given the size of the Pol protein, Pol peptides were divided into two pools, Pol-1 and Pol-2, representing each half of the protein. The Gag and Nef amino acid sequences of clades A, B, and C were obtained from primary virus isolate sequences in the Los Alamos HIV-1 database whose sequences were as close as possible to the consensus sequence in that clade type. They were dissolved in straight dimethyl sulfoxide (DMSO) at 20 to 50 mg/ml and stored in small aliquots at −70°C. Peptide pools were composed of an equal volume of each peptide, with up to 120 peptides per pool. The final stock concentration for each peptide in a pool was 0.4 mg/ml.
IFN-γ enzyme-linked immunospot (ELISPOT) assay.
Sterile 96-well microtiter plates with well bottoms of polyvinylidene difluoride (MAIP S45; Millipore, Bedford, MA) were coated overnight at 4°C with 100 μl/well of mouse anti-human gamma interferon (IFN-γ) monoclonal antibody clone 1-D1K (MabTech, Stockholm, Sweden) diluted to 10 μg/ml in sterile Dulbecco's phosphate-buffered saline (PBS) (Gibco-BRL, Grand Island, NY). The coated plates were washed four times and blocked for 2 h in a humidified CO2 incubator at 37°C with 200 μl/well of complete RPMI-1640 medium complemented with 10% fetal bovine serum (R-10; Gibco-BRL, Grand Island, NY). The blocking buffer was removed, and 100 μl/well of PBMCs diluted in R10 was added to result in 2 × 105 and 1 × 105 cells/well. Peptide pools were diluted in R10 and added at 25 μl/well, and the final concentration for each peptide in the pools was 2 to 3 μg/ml. Peptide-free DMSO diluent matching the DMSO concentration in the peptide solutions was used as a negative control (mock antigen). The plates were incubated overnight in a humidified CO2 incubator at 37°C and then washed seven times with PBS containing 0.05% Tween 20 (Sigma, St. Louis, MO). Biotinylated anti-human IFN-γ monoclonal antibody clone 7-B6-1 (MabTech) was diluted to 1 μg/ml in assay diluent consisting of PBS and 0.5% bovine serum albumin (Sigma). Diluted antibody was added to the plates at 100 μl/well, and the plates were incubated for 2 to 4 h at room temperature. The plates were washed four times with PBS-Tween, and 100 μl/well of alkaline phosphatase-conjugated anti-biotin monoclonal antibody (Vector Laboratories, Burlingame, CA) at 1:750 in assay diluent was added to each well. The plates were incubated for 2 h at room temperature and washed four times with PBS-Tween. To develop the spots, 100 μl/well of precipitating alkaline phosphatase substrate nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Pierce, Rockford, IL) was added to each well, and the plates were incubated at room temperature until spots became visible (usually 5 to 10 min). The spots were counted using a digital imager and automated spot-counting software (AutoImmun Diagnostika, Germany). The number of spots per well was normalized to per 106 cells and averaged for each sample and antigen to give a final number of spot-forming cells per 106 cells. Responses were considered positive when they were four times higher than the mock and above 55 spot-forming units (SFU)/106 PBMCs. This criterion was previously determined by statistical assay validation to result in a <1.0% false-positive rate (J. W. Shiver, unpublished observations).
Statistical analysis.
Statistical analyses were performed using GraphPad 3.02 (GraphPad Software, San Diego, Calif.). Means and medians are presented as arithmetic means. All data were analyzed by the use of nonparametric statistics. Kruskal-Wallis nonparametric analyses of variance were used for significant differences between clade-specific responses. Correlations were determined using Spearman's rank correlations. All tests were two tailed and were considered significant if the P value was <0.05.
Nucleotide sequence accession numbers.
The sequences described here have been submitted to GenBank and assigned accession numbers AY887091 through AY887103.
RESULTS
Identification of domains recognized by cross-clade T cells using clade B peptide pools.
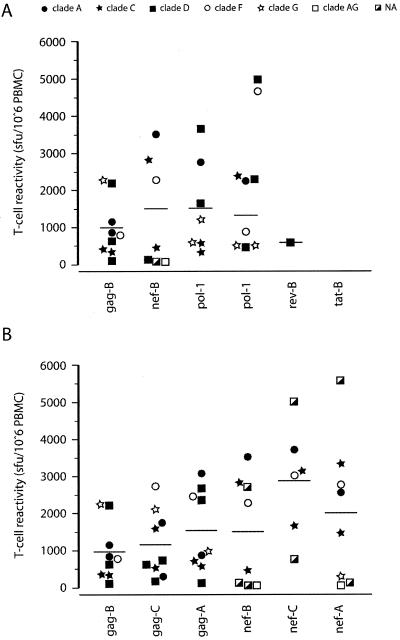
PBMCs from 10 patients were stimulated with clade B pools (Gag-B, Nef-B, Pol-1-B, Pol-2-B, Tat-B, and Rev-B) to identify the localization of epitopes recognized by cross-reactive T cells. T-cell responses were measured using the IFN-γ ELISPOT assay as the readout (Fig. 1A). Nine of the 10 patients responded to Gag-B peptides with responses ranging from 120 to 2,243 SFU/106 PBMCs (mean, 971.6; median, 797), while Nef-B and both Pol-B peptide pools (Pol-1 and Pol-2) were recognized by eight and seven patients, respectively. Reponses to Nef-B peptides ranged from 57 to 3,510 SFU/106 PBMCs (mean, 1,502; median, 1,365), while responses to the two Pol-B pools ranged from 327 to 3,660 SFU/106 PBMCs (Pol-1 mean, 1,533; median, 1,210) and 467 to 2,363 SFU/106 PBMCs (Pol-2 mean, 1,320; median, 870). Rev was positive in only one patient (580 SFU/106 PBMCs), while none of the patients responded to the Tat peptide pool.
FIG. 1.
Cross-clade anti-HIV-1 T-cell responses to clade A, B, and C 15-mer peptide pools. Cross-clade T-cell reactivity was assessed using (A) clade B Gag, Nef, Pol-1, Pol-2, Rev, and Tat and (B) clade A and C Gag and Nef. Responses are shown as the number of IFN-γ-expressing cells per million PBMCs. Serum-derived Gag, Nef, and Pol sequences were subtyped and are indicated by different symbols. For two patients, no subtyping data were available (NA). The median responses are indicated for each group by a horizontal line. Cross-clade B Gag and Nef responses are replotted in panel B.
Comparison of T-cell reactivity to clade A, B, and C Gag and Nef peptide pools.
Since clade B Nef and Gag peptide pools were most frequently recognized by PBMCs of our cohort of non-B-infected individuals, we evaluated Gag and Nef T-cell responses of each donor to clade A and C peptide pools. All patients who recognized Gag-B peptides (nine of nine) reacted to both Gag-A and -C peptide pools (Fig. 1B), while six of seven patients who recognized Nef-B peptides recognized both Nef-A and Nef-C peptides. One patient (M12453-A; the suffix indicates the patient's Gag subtype) recognized only the Nef-A and Nef-B pools. Patient M13371-G recognized only Nef-A peptides, while patient M12115-D recognized only the Nef-B peptide pool. All patients demonstrated cross-clade T-cell responses against a minimum of two to a maximum of four different “heterologous” peptide pools.
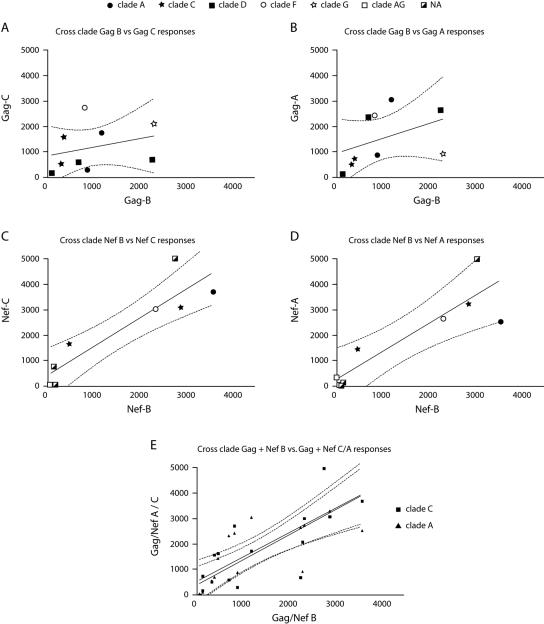
The mean T-cell responses to the Gag-A and Gag-C peptide pools, as well as the Nef-A and Nef-C peptide pools, were higher than those to clade B Gag and Nef pools (Fig. 1B), although no statistically significant interclade differences were detected between the different Gag and Nef pools (Gag, P = 0.398, and Nef, P = 0.201). Due to observed correlations between T-cell responses to homologous and heterologous peptide pools for different viral proteins (16), we compared the magnitudes of clade B Gag or Nef responses to those detected against corresponding A or C clade pools for each individual (Fig. 2). Statistically significant correlations were observed between responses to Gag-B and Gag-A, Nef-B and Nef-A, and Nef-B and Nef-C peptide pools (Fig. 2 and Table 2). A highly significant correlation was identified between clade B and subtypes A and C (P < 0.0001 and P < 0.001, respectively) for the combined Nef and Gag responses shown in Fig. 2E. In the four patients that were tested with both homologous and heterologous clade-derived peptide pools, we found that the highest responses were elicited by the heterologous peptide pools. Patient M12653-A demonstrated the highest response against the Nef-C peptide pool, patient M12453-A against Pol-1-B, patient M11306-C against Nef-A, and patient M12003-C against Pol-2-B. No significant correlations were identified between the T-cell responses and any of the measured clinical parameters, including viral load, age, or therapy status of the patients (data not shown).
FIG. 2.
Correlation between clade B and clade A/C T-cell responses. Clade B T-cell responses were plotted against clade C and A responses for Gag (A and B) and Nef (C and D). Gag and Nef responses were combined for clades A and C and replotted in panel E. Responses are shown as the number of IFN-γ-expressing cells per million PBMCs. Gag and Nef subtypes are indicated with different symbols. For two patients, no subtyping for Nef sequences was available (NA). Correlations were made using Spearman's rank test and are shown together with 95% confidence intervals (dotted lines). Correlation results are summarized in Table 3.
TABLE 2.
Correlation between cross-clade Gag and Nef T-cell responses
| Clade | Parameter | Gag-A | Gag-C | Nef-A | Nef-C | Gag + Nef-A | Gag + Nef-C |
|---|---|---|---|---|---|---|---|
| Gag-B | Spearman r | 0.733 | 0.567 | ||||
| 95% confidence interval | −0.526-1.74 | −0.633-1.34 | |||||
| P value (two tailed) | 0.031 | 0.121 | |||||
| Nef-B | Spearman r | 0.7667 | 0.9222 | ||||
| 95% confidence interval | 0.470-1.75 | 0.620-1.67 | |||||
| P value (two tailed) | 0.0214 | 0.0022 | |||||
| Nef-B + Gag-B | Spearman r | 0.865 | 0.782 | ||||
| 95% confidence interval | 0.636-0.954 | 0.437-0.927 | |||||
| P value (two tailed) | P < 0.0001 | P < 0.001 |
Cross-clade T-cell reactivity related to homology of peptides with autologous amino acid sequences.
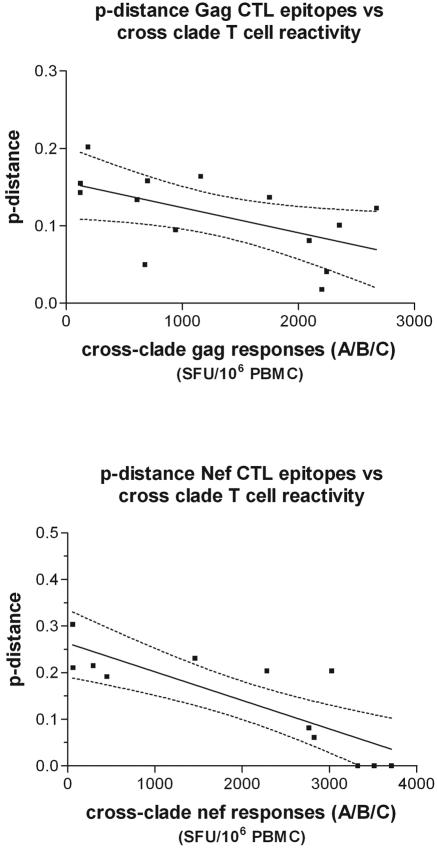
We next studied whether the Gag and Nef cross-clade T-cell responses were influenced by the genetic divergence of patient Gag and Nef sequences from the HIV-1 sequences on which the peptide pools were based (clades A, B, and C). We analyzed the serum-derived Gag sequences of five patients and the Nef sequences of six patients (Gag and Nef, M12653, M11306, M12003, M11814, and M13371; Nef, M12453). We determined the p distance between autologous patient amino acid sequences and amino acid sequences in the different peptide pools for all the documented CTL epitopes combined based on the patient's HLA type. For Gag, we found that per patient, an average of 10 epitopes (range, 6 to 15) were previously documented (9), whereas for Nef, 4.5 (range, 1 to 8) epitopes matched HLA class I restriction in our patient selection. Within the previously documented Gag epitopes, 28% were HLA-A restricted whereas 72% were HLA-B restricted. For Nef, 51.9% of the epitopes were HLA-A restricted and 48.1% were HLA-B restricted. When we analyzed the previously documented epitopes, we found for both Nef and Gag a significant inverse correlation between the p distance of the autologous “epitopes” to peptide pool “epitopes” and T-cell responsiveness (Gag, P = 0.045; Nef, P = 0.001) (Fig. 3 and Table 3). In other words, this demonstrates that the greater the distance of the peptide sequences from the autologous HIV-1 sequences, the lower the T-cell response tends to be. To further analyze whether the correlation we observed was due to the specific residues within or outside HLA class I anchor motifs, we recalculated the p distance between the autologous “epitopes” and peptide “epitopes” excluding the HLA anchor amino acid residues. To correct for the exclusion of the HLA anchor residues, we performed the analysis after removal of the same number of residues 1 aa downstream of the HLA anchor residues (54). The inverse correlation between T-cell responses and the p distance between autologous and peptide CTL epitopes remained significant for Nef, but not for Gag, when the HLA anchor residues were removed from the analysis (Gag, P = 0.472; Nef, P = 0.001), while both remained significant when the adjacent amino acid was removed from the analysis (Gag, P = 0.005; Nef, P = 0.007) (Table 3). This implies that the relevance of variation within Gag epitopes between peptide pools and autologous sequences is mainly due to HLA anchor residues. As a final validation to see whether the observed correlation was due to specific variation in CTL epitopes and not due to stochastic events, we analyzed the p distances for an equal number of randomly picked amino acids exterior to the known epitopes and repeated this 500 times. The average p-distance value was compared with the “epitope” p distances. No significant correlation was found between the variation outside of CTL epitopes and the height of T-cell responses for both Gag and Nef (Gag, P = 0.436; Nef, P = 0.430) (Table 3).
FIG. 3.
Correlation between T-cell responses and genetic distance. The genetic distance (p distance) between documented CTL epitopes for the patient-specific HLA-type was correlated with T-cell responses for Gag (top) and Nef (bottom). Correlations were made using Spearman's rank test and are shown together with 95% confidence intervals (dashed lines). All correlation results are summarized in Table 3.
Expansion of profile of cross-clade Gag responses.
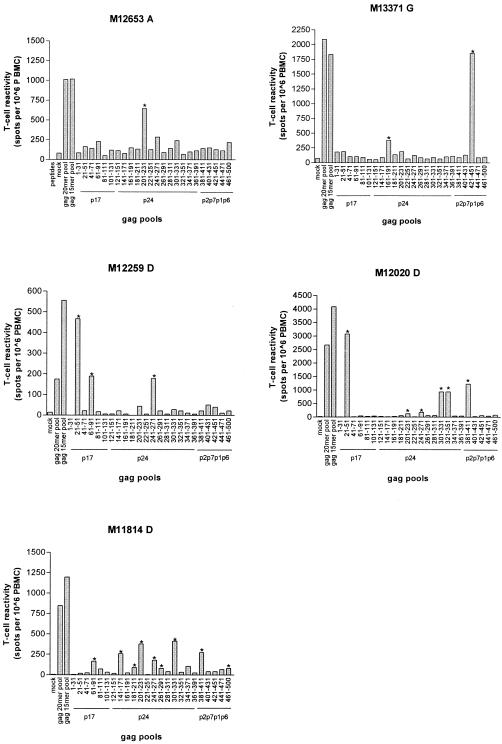
To better define the regions and/or epitopes recognized in cross-clade Gag-B-specific T-cell responses, we mapped the responses of five patients (M13371-G, M12653-A, M11814-D, M12259-D, and M12020-D) demonstrating strong cross-clade reactivity as shown in Fig. 1. Patient PBMCs were tested for reactivity against 23 pools of five peptides each (pool aa 1-31 to aa 441- 471) and one pool of seven peptides (pool aa 461-500). Collectively, these pools spanned the entire Gag region, with each pool of five peptides spanning 30 aa and the pool of seven peptides spanning 40 aa in length. As shown in Fig. 4, the number of positive pools ranged from 1 pool (patient M12653-A) to 12 pools (patient M11814-D), with a mean pool recognition of five.
FIG. 4.
Mapping of cross-clade Gag-B T-cell responses. For five patients, cross-clade Gag-B T-cell responses were mapped using peptide pools stretching 30 aa or 40 aa (only pool aa 461 to 500). The Gag sequence numbering is indicated on the x axis, together with the major Gag regions. Positive responses to Gag pools are indicated by asterisks. All peptide pools are based on strain CAM-1.
T-cell responses were quantitatively (67% [16/24] of the positive peptide pools) and qualitatively (4/5 of the dominant responses) dominated by reactivity to p24. A total of 15 different Gag pools were recognized, with positive responses ranging between 67 and 3,077 SFU/106 PBMCs, with a mean response per patient of 181 to 1,118 SFU/106 PBMCs. Three of five patients reacted to the pools spanning aa 241-271 and aa 201-231 (both p24), while two of the five patients reacted to the pools spanning aa 21-51 and aa 61-91 (both p17). All other cross-clade pools were positive in only one subject. Patient M11814-D had two comparably dominant Gag T-cell responses (Gag pools aa 201-231 and aa 301-331), while other patients had clear dominant responses to just one pool (Fig. 4). Unfortunately, however, due to the lack of cells, we were unable to map the exact epitopes and their restricting alleles. However, to further investigate whether the cross-clade-reactive regions were indicative of previously documented epitopes and haplotypes, we searched the Los Alamos database to identify whether known CTL epitopes could be located within the 30 amino acids spanning Gag pools (Table 4). Of the 24 positive responses, we predicted epitopes restricted by patient-specific HLA class I alleles in 58% (14/24) of positive T-cell responses. When we compared serum-derived Gag sequences, 12 out of 14 (86%) documented epitopes contained no amino acid variation on HLA anchor motifs (Table 4), corroborating the notion that these residues are critical for CTL recognition and that variation at these residues is rare. Six of those 14 responses (40%) mapped to CTL epitopes or peptides previously identified as being recognized either by non-clade B-infected patients or in a cross-clade manner (9). One peptide pool (aa 381 to 411, positive in patients M12020-D and M11814-D) was recently demonstrated to contain a major responsive peptide for clade C-infected patients (48). For 10 of the 24 (42%) positive T-cell responses, no CTL epitopes have been documented that match the patient-specific HLA class I alleles. We scanned these peptide sequences for any known HLA motifs specific for the patient HLA type using ELF (http://hiv-web.lanl.gov/ALABAMA/epitope_analyzer.html). Using ELF,we were able to locate patient-specific HLA motifs in seven of nine “undocumented” positive Gag pools, indicating previously unknown cross-clade-reactive epitopes. Of the seven “predicted” epitopes, six matched documented CTL epitopes but for different HLA class I restrictions, and three “predicted” epitopes matched reported cross-clade-reactive epitopes. Collectively, these results demonstrate that cross-clade T-cell responses are readily detectable and are predominantly Gag and Nef specific. Detectable cross-clade T-cell responses using Gag and Nef peptide pools were inversely related to amino acid sequence variation, while through mapping of cross-clade Gag responses, new undocumented immunoresponsive regions/CTL epitopes or their restricting HLA types became evident.
TABLE 4.
Fine mapping of cross-clade-reactive Gag regions
| Patient | Cross-reactive Gag-B poola(Gag region) | Documented epitopes that match patient HLAb
|
Predicted epitope (ELF) according to patient HLAb
|
Commentsb,c | ||
|---|---|---|---|---|---|---|
| Peptide sequence Epitope sequence Patient sequence | HLA | Peptide sequence Epitope sequence | HLA | |||
| M11814-D | 301-331 (p24) | YKTLRAEQASQEVKNWMTETLLVQNANPDCK | CW4 | |||
| QASQEVKNW | ||||||
| ---------d | ||||||
| 201-231 (p24) | No documented epitope | LKETINEEAAEWDRLHPVHAGPIAPGQMREP | ||||
| ETINEEAAEW | B58 | K: B25 | ||||
| HPVHAGPI | B35 | K: B55 | ||||
| GPIAPGQM | B35 | |||||
| 381-411 (p24) | GNFRNQRKTVKCFNCGKVGHIAKNCRAPRKK | No predicted epitope | K (Subtype C) | |||
| No documented epitope | ||||||
| 141-171 (p24) | QMVHQAISPRTLNAWVKVVEEKAFSPEVIPM | B58 | K, D | |||
| ISPRTLNAW | ||||||
| L--------d | ||||||
| 241-271 (p24) | STLQEQIGWMTNNPPIPVGEIYKRWIILGLN | B35 | ||||
| PPIPVGEIY | ||||||
| --------d | ||||||
| STLQEQIGW | B58 | |||||
| -------A-d | ||||||
| EIYKRWIIL | A24 | |||||
| ---------d | ||||||
| 61-91 (p17) | No documented epitope | LGQLQPSLQTGSEELRSLYNTVATLYCVHQK | ||||
| LYNTVATL | A24 | K, CC, cHLA | ||||
| 341-371 (p24) | No documented epitope | ATLEEMMTACQGVGGPGHKARVLAEAMSQVT | ||||
| GPGHKARVL | B35 | K: B7 | ||||
| 181-211 (p24) | -TPQDLNTMLNTVGGHQAAMQMLKETINEEAAE | B58 | K, D, CC | |||
| ATPQDLNTMLNT | ||||||
| ------------ | ||||||
| 461-500 (p2p1) | No documented epitope | ESFRFGEEKTTPSQKQEPIDKELYPLASLRSLFGNDPSSQ | ||||
| TPSQKQEPI | B35 | K: B7 | ||||
| EPIDKELYPL | B35 | |||||
| LYPLASLRSL | A24 | |||||
| YPLASLRSLF | B35 | K: B7 | ||||
| LASLRSLF | B58 | |||||
| 261-291 (p24) | IYKRWIILGLNKIVRMYSPTSILDIRQGPKE | A24 | ||||
| IYKRWIILGL | ||||||
| ----------d | ||||||
| 81-111 (p17) | No documented epitope | TVATLYCVHQKIDVKDTKEALEKIEEEQNKS | ||||
| LYCVHQKI | A24 | K: A11, cHLA | ||||
| 441-471 (p1p6) | No documented epitope | HKGRPGNFLQSRPEPTAPPEESFRFGEEKTT | ||||
| EPTAPPEESF | B35, B58 | |||||
| M13371-G | 421-451 (p24) | HQMKDCNER--QANFLGKIWPSHKGRPGNFLQS | A2 | K | ||
| HQMKDCNER--QAN | ||||||
| -------T-QG--- | ||||||
| 161-191 (p24) | EKAFSPEVIPMFSALSEGATPQDLNTMLNTV | B58 | K, D, CC | |||
| KAFSPEVIPMF | ||||||
| -----------d | ||||||
| TPQDLNQMLNTV | B58 | K, D, CC | ||||
| ------T-----d | ||||||
| M12653-A | 201-231 (p24) | LKETINEEAAEWDRLHPVHAGPIAPGQMREP | B53 | |||
| ETINEEAAEW | ||||||
| D---------d | ||||||
| M12259-D | 21-51 (p17) | LRPGGKKKYKLKHIVWASRELERFAVNPGLL | B8 | |||
| GGKKKYKL | ||||||
| -------M | ||||||
| 61-91 (p17) | LGQLQPSLQTGSEELRSLYNTVATLYCVHQK | A1 | K, D, CC | |||
| GSEELRSLY | ||||||
| -TEGIK--H | ||||||
| ELRSLYNTV | B8 | |||||
| GIK--H--- | ||||||
| 241-271 (p24) | STLQEQIGWMTNNPPIPVGEIYKRWIILGLN | B8 | ||||
| GEIYKRWII | ||||||
| ---------d | ||||||
| M12020-D | 21-51 (p17) | LRPGGKKKYKLKHIVWASRELERFAVNPGLL | Cw4 | K, D | ||
| RPGGKKKYKL | ||||||
| ----N-----d | ||||||
| 381-411 (p24) | GNFRNQRKTVKCFNCGKVGHIAKNCRAPRKK | No predicted epitope | K (Subtype C) | |||
| No documented epitope | ||||||
| 301-331 (p24) | YKTLRAEQASQEVKNWMTETLLVQNANPDCK | Cw4 | ||||
| QASQDVKNW | B53 | |||||
| ----E----d | ||||||
| AEQASQDVKNW | B44 | |||||
| ------E----d | ||||||
| 241-271 (p24) | STLQEQIGWMTNNPPIPVGEIYKRWIILGLN | STLQEQIGWMTNNPPIPVGEIYKRWIILGLN | ||||
| No documented epitope | PPIPVGEIY | B53 | K, D, CC, cHLA, V | |||
| 201-231 (p24) | LKETINEEAAEWDRLHPVHAGPIAPGQMREP | B53 | K, CC | |||
| ETINEEAAEW | ||||||
| ----------d | ||||||
| 461-500 (p15) | No documented epitope | ESFRFGEEKTTPSQKQEPIDKELYPLASLRSLFGNDPSSQ | ||||
| TPSQKQEPI | B53 | K: B7 | ||||
| QEPIDKELY | B44 | |||||
| EPIDKELYPL | B53 | |||||
| YPLASLRSL | B53 | K: B7 | ||||
Pools are ranked according to height of cross-clade T-cell responses (Fig. 4).
As listed in the Los Alamos Immunology Database. HLA anchor motifs specific for the patient haplotype are given in boldface.
Characteristics of documented or predicted epitopes and documented restricting haplotype (if different from predicted epitope) are as follows: K, known epitope/peptide in non-clade B setting; D, known dominant epitope in non-clade B setting; CC, Known cross-clade-responding epitope; cHLA, Epitope known to be conserved across subtypes and commonly HLA restricted; V, epitope included in polyepitope CTL vaccine.
Serum-derived Gag sequences which contain no amino acid variation on HLA anchor motifs.
DISCUSSION
One of the key features of an effective HIV-1 vaccine must be its ability to elicit and maintain adequate cellular immune responses, preferably across clade-specific boundaries (46). We have identified and characterized in the present study HIV-1 cross-clade T-cell activity in 9 out of 10 non-clade B-infected individuals. Gag and Nef were most frequently recognized, which is generally in agreement with previous reports (12, 15, 18, 22, 41, 44). T-cell responses were assessed using peptide pools based on primary isolates that most closely resembled the consensus sequences of the particular clades. In this way, the antigen represents a naturally occurring HIV-1 strain but at the same time reflects the sequences predominating in a specific clade. The observation that in certain patients T-cell responses to heterologous clades were higher than responses to homologous clades corresponds to a recent study by Sebado and coworkers in a comparable-size patient cohort. They described a heterologous peptide pool (clade C strain) as being more suitable for the detection of T-cell responses than consensus B-based peptide pools. Putatively, the generation of consensus sequences will erase functional HLA anchor residues and thus explain the distinction seen between the peptide pools. However, in a large recently conducted study (250 patients) (16), intraclade responses were found to be higher than interclade responses using the same reagents, leading to a probable dilution of the aforementioned phenomenon. These results prompted us to analyze amino acid variation in reported CTL epitopes. The T-cell responses observed in HIV-infected patients reflect mostly, if not all, CD8+ cytotoxic-T-cell reactivity (16). We identified a significant inverse correlation between T-cell responses and the amino acid divergence of the predicted class I epitopes and the peptides utilized. We observed that the correlation for cross-clade Gag responses was explained by the discrepancy at HLA anchor residues, suggesting that variation in Gag is allowed at particular HLA anchor residues that are important for efficient antigen presentation while this variation comes at little cost to viral fitness (37). This phenomenon is suggestive of a history of HLA-associated CTL pressure and selection (38, 47), and the results argue that if Gag is included in an HIV-1 vaccine, as is usually or likely the case, successful immunogenicity may be predictable in a given population on the basis of HLA (super) motifs most frequently present in the vaccine target population. It has been shown recently for clade C viruses that various HLA alleles can show promiscuity at the same epitopes (43). Several reasons can be proposed for why the significance of HLA anchor residues in Nef is not detected. Currently, twice as many epitopes have been defined in Gag as in Nef (9), which is also reflected in our patient population. As a result, a proportionally bigger data set of Nef epitopes is required to achieve statistical power. It has also been shown recently that substantially greater selection pressure is imposed by HLA-B than by HLA-A on HIV-1 (38), which could help explain the dichotomy found between Gag and Nef, since in our analysis, a greater proportion of Gag epitopes than Nef epitopes were restricted by the HLA-B haplotypes (72% versus 48%). Lastly, the Nef protein is known to accept a higher degree of variation with a less pronounced effect on protein function (2), thereby helping to explain why we did not observe a significant effect mediated by the HLA anchor motifs alone, although variation in cross-clade T-cell responses could be linked to alterations in predicted class I-restricted Nef epitopes. However, these results need to be evaluated in larger patient populations infected with different non-B clades, preferably with a higher heterogeneity of HLA class I genes. This will allow the assessment of whether the observed effect of variation on HLA anchor is HIV-1 antigen, clade, and/or haplotype specific.
Two major points must be taken into consideration when evaluating the cross-clade reactivity data. First, although the IFN-γ ELISPOT is currently the most frequently used technique for determining CD8+ T-cell responses, it does not necessarily provide a clear correlate of protection. In the past, positive correlations (10, 25, 42, 49), negative correlations (50), and no correlations (1, 19, 31) have been identified for both the height and diversity of T-cell responses and clinical parameters, such as viral load and CD4+ counts. Moreover, these were weak correlations or true only for single gene regions (49). New markers therefore need to be constantly evaluated for possible use as immune correlates (52). Recently, it was shown that long-term-nonprogressing HIV-1 patients maintain HIV-1-specific CD8+ T-cell populations that elicit multiple functional (MIP-1β, tumor necrosis factor α, interleukin-2, and CD107a) abilities (11; M. Betts personal communication), and these polyfunctional readouts may be more precise in defining true cross-clade and protective T-cell responses. Secondly, although we were able to narrow down the Gag-specific cross-clade T-cell responses to specific Gag domains in this study, using peptide pools with overlapping peptides is known to underestimate the true T-cell responses (8, 24). The use of optimal-size epitopes will allow a more accurate analysis of cross-clade T-cell responses that are epitope specific (8).
From the mapping of Gag responses in four individuals using 30 amino acid peptide pools, we conclude that Gag p24 most frequently elicits cross-clade T-cell responses and that nearly all autologous Gag sequences in documented HLA class epitopes in the peptide pools were conserved and likely mediating the responses. Nonetheless, further analyses (e.g., fine mapping with truncated peptides) are needed to specifically identify the recognized epitopes and reveal their HLA restrictions. The majority of positive T-cell responses, however, could be linked to previously documented epitopes, half of which are known to be cross-clade responsive. In addition, we could identify additional epitopes that are in part responsible for the cross-clade T-cell responses observed in our study by using HLA motif-finding algorithms. Although it is known that HLA motif prediction can miss in vivo-responsive CTL epitopes (23), a bioinformatics approach has proven to correlate with in vivo findings (13, 21, 45, 58). In total, 25% (6/24) of the positive Gag pools in our study could be linked with previously documented cross-clade-responding epitopes in non-clade B-infected patients. For instance, the peptide pools aa 161 to 191 and aa 181 to 211 contain the epitope TL9 (TPQDLNTML), which has been associated with dominant recognition by clade C-infected individuals (48) and has been shown to be targeted by different ethnicities in clade B settings (29). This epitope, among others, has been proposed (28, 29) to be incorporated into HIV-1 vaccines and is currently in phase III trials (36) (Table 4). These responses may not necessarily be dominant responses (as were detected in our cohort), but it is known that subdominant Gag T-cell responses might be equally effective in controlling viremia (30).
In summary, we present evidence of broad cross-clade HIV-1 T-cell responses that are focused on Gag and Nef. We show, using the current data on CTL domains, that the magnitudes of the T-cell responses correlate with sequence diversity in CTL epitopes, and in the case of Gag, the magnitude of the T-cell response was directly linked to the homology between HLA anchor residues in the infecting virus and those in the peptides used in the experimental setting. A recent study analyzing a large cohort of individuals encompassing different ethnicities showed that T-cell responses are highly focused on specific regions in Gag and Nef (29). Moreover, more than 90% of the examined population mounted T-cell responses to relatively small stretches in these areas, which in turn may serve as suitable candidates for vaccine constructs. Analyses of the HLA anchor motifs of the predominant HLA class I haplotypes in the vaccine target population will putatively predict T-cell responses and help in the design of future immunogens.
Acknowledgments
This work was funded by AIDSFONDS grant 6002.
We thank the patients of the Academic Medical Centre clinic and Amsterdam Cohort Studies, Michel de Baar and Can Kesmir for their excellent scientific contributions, and Wim van Est for the artwork.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, A., S. Pillai, H. Ng, R. Lubong, D. D. Richman, B. D. Jamieson, Y. Ding, M. J. McElrath, J. C. Guatelli, and O. O. Yang. 2003. Broadly increased sensitivity to cytotoxic T lymphocytes resulting from Nef epitope escape mutations. J. Immunol. 171:3999-4005. [DOI] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 8.Beattie, T., R. Kaul, T. Rostron, T. Dong, P. Easterbrook, W. Jaoko, J. Kimani, F. Plummer, A. McMichael, and S. Rowland-Jones. 2004. Screening for HIV-specific T-cell responses using overlapping 15-mer peptide pools or optimized epitopes. AIDS 18:1595-1598. [DOI] [PubMed] [Google Scholar]
- 9.Bette, T., M. Korber, C. Brander, B. F. Haynes, R. Koup, J. P. Moore, B. D. Walker, and D. I. Watkins. 2003. HIV immunology and HIV/SIV vaccine databases 2003. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 10.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 102:4512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. Luo, N. N′Gandu, H. Sheppard, B. H. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bond, K. B., B. Sriwanthana, T. W. Hodge, A. S. De Groot, T. D. Mastro, N. L. Young, N. Promadej, J. D. Altman, K. Limpakarnjanarat, and J. M. McNicholl. 2001. An HLA-directed molecular and bioinformatics approach identifies new HLA-A11 HIV-1 subtype E cytotoxic T lymphocyte epitopes in HIV-1-infected Thais. AIDS Res. Hum. Retrovir. 17:703-717. [DOI] [PubMed] [Google Scholar]
- 14.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao, H., P. Kanki, J. L. Sankale, A. Dieng-Sarr, G. P. Mazzara, S. A. Kalams, B. Korber, S. Mboup, and B. D. Walker. 1997. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 71:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coplan, P. M., S. B. Gupta, S. A. Dubey, P. Pitisuttithum, A. Nikas, B. Mbewe, E. Vardas, M. Schechter, E. G. Kallas, D. C. Freed, T. M. Fu, C. T. Mast, P. Puthavathana, J. Kublin, C. K. Brown, J. Chisi, R. Pendame, S. J. Thaler, G. Gray, J. McIntyre, W. L. Straus, J. H. Condra, D. V. Mehrotra, H. A. Guess, E. A. Emini, and J. W. Shiver. 2005. Cross-reactivity of anti-HIV-1 T cell immune responses among the major HIV-1 clades in HIV-1-positive individuals from 4 continents. J. Infect. Dis. 191:1427-1434. [DOI] [PubMed] [Google Scholar]
- 17.Cornelissen, M., B. R. van den, F. Zorgdrager, V. Lukashov, and J. Goudsmit. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Currier, J. R., M. deSouza, P. Chanbancherd, W. Bernstein, D. L. Birx, and J. H. Cox. 2002. Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles. J. Virol. 76:4971-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalod, M., M. Dupuis, J. C. Deschemin, D. Sicard, D. Salmon, J. F. Delfraissy, A. Venet, M. Sinet, and J. G. Guillet. 1999. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8+ responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J. Virol. 73:7108-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Baar, M. P., A. de Ronde, B. Berkhout, M. Cornelissen, K. H. Van Der Horn, A. M. Van Der Schoot, F. de Wolf, V. V. Lukashov, and J. Goudsmit. 2000. Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer-binding site. AIDS Res. Hum. Retrovir. 16:499-504. [DOI] [PubMed] [Google Scholar]
- 21.De Groot, A. S., B. Jesdale, W. Martin, A. C. Saint, H. Sbai, A. Bosma, J. Lieberman, G. Skowron, F. Mansourati, and K. H. Mayer. 2003. Mapping cross-clade HIV-1 vaccine epitopes using a bioinformatics approach. Vaccine 21:4486-4504. [DOI] [PubMed] [Google Scholar]
- 22.Dorrell, L., T. Dong, G. S. Ogg, S. Lister, S. McAdam, T. Rostron, C. Conlon, A. J. McMichael, and S. L. Rowland-Jones. 1999. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J. Virol. 73:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorrell, L., B. E. Willcox, E. Y. Jones, G. Gillespie, H. Njai, S. Sabally, A. Jaye, K. DeGleria, T. Rostron, E. Lepin, A. McMichael, H. Whittle, and S. Rowland-Jones. 2001. Cytotoxic T lymphocytes recognize structurally diverse, clade-specific and cross-reactive peptides in human immunodeficiency virus type-1 gag through HLA-B53. Eur. J. Immunol. 31:1747-1756. [DOI] [PubMed] [Google Scholar]
- 24.Draenert, R., M. Altfeld, C. Brander, N. Basgoz, C. Corcoran, A. G. Wurcel, D. R. Stone, S. A. Kalams, A. Trocha, M. M. Addo, P. J. Goulder, and B. D. Walker. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19-29. [DOI] [PubMed] [Google Scholar]
- 25.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 76:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari, G., W. Humphrey, M. J. McElrath, J. L. Excler, A. M. Duliege, M. L. Clements, L. C. Corey, D. P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrari, G., D. D. Kostyu, J. Cox, D. V. Dawson, J. Flores, K. J. Weinhold, and S. Osmanov. 2000. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res. Hum. Retrovir. 16:1433-1443. [DOI] [PubMed] [Google Scholar]
- 29.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallimore, A., J. Hombach, T. Dumrese, H. G. Rammensee, R. M. Zinkernagel, and H. Hengartner. 1998. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur. J. Immunol. 28:3301-3311. [DOI] [PubMed] [Google Scholar]
- 31.Gea-Banacloche, J. C., S. A. Migueles, L. Martino, W. L. Shupert, A. C. McNeil, M. S. Sabbaghian, L. Ehler, C. Prussin, R. Stevens, L. Lambert, J. Altman, C. W. Hallahan, J. C. deq Uiros, and M. Connors. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082-1092. [DOI] [PubMed] [Google Scholar]
- 32.Geels, M. J., M. Cornelissen, H. Schuitemaker, K. Anderson, D. Kwa, J. Maas, J. T. Dekker, E. Baan, F. Zorgdrager, R. van den Burg, M. van Beelen, V. V. Lukashov, T. M. Fu, W. A. Paxton, L. van der Hoek, S. A. Dubey, J. W. Shiver, and J. Goudsmit. 2003. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J. Virol. 77:12430-12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulder, P. J., M. Bunce, G. Luzzi, R. E. Phillips, and A. J. McMichael. 1997. Potential underestimation of HLA-C-restricted cytotoxic T-lymphocyte responses. AIDS 11:1884-1886. [DOI] [PubMed] [Google Scholar]
- 34.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 35.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630-640. [DOI] [PubMed] [Google Scholar]
- 36.Hanke, T., and A. J. McMichael. 2000. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat. Med. 6:951-955. [DOI] [PubMed] [Google Scholar]
- 37.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 40.Letvin, N. L., and B. D. Walker. 2003. Immunopathogenesis and immunotherapy in AIDS virus infections. Nat. Med. 9:861-866. [DOI] [PubMed] [Google Scholar]
- 41.Lynch, J. A., M. deSouza, M. D. Robb, L. Markowitz, S. Nitayaphan, C. V. Sapan, D. L. Mann, D. L. Birx, and J. H. Cox. 1998. Cross-clade cytotoxic T cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J. Infect. Dis. 178:1040-1046. [DOI] [PubMed] [Google Scholar]
- 42.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 78:3233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masemola, A. M., T. N. Mashishi, G. Khoury, H. Bredell, M. Paximadis, T. Mathebula, D. Barkhan, A. Puren, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J. Immunol. 173:4607-4617. [DOI] [PubMed] [Google Scholar]
- 44.McAdam, S., P. Kaleebu, P. Krausa, P. Goulder, N. French, B. Collin, T. Blanchard, J. Whitworth, A. McMichael, and F. Gotch. 1998. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS 12:571-579. [DOI] [PubMed] [Google Scholar]
- 45.McKinney, D. M., R. Skvoretz, B. D. Livingston, C. C. Wilson, M. Anders, R. W. Chesnut, A. Sette, M. Essex, V. Novitsky, and M. J. Newman. 2004. Recognition of variant HIV-1 epitopes from diverse viral subtypes by vaccine-induced CTL. J. Immunol. 173:1941-1950. [DOI] [PubMed] [Google Scholar]
- 46.McMichael, A., M. Mwau, and T. Hanke. 2002. HIV T cell vaccines, the importance of clades. Vaccine 20:1918-1921. [DOI] [PubMed] [Google Scholar]
- 47.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 48.Novitsky, V., H. Cao, N. Rybak, P. Gilbert, M. F. McLane, S. Gaolekwe, T. Peter, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2002. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J. Virol. 76:10155-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogg, G. S., S. Kostense, M. R. Klein, S. Jurriaans, D. Hamann, A. J. McMichael, and F. Miedema. 1999. Longitudinal phenotypic analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes: correlation with disease progression. J. Virol. 73:9153-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 52.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10:806-810. [DOI] [PubMed] [Google Scholar]
- 53.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rammensee, H., J. Bachmann, N. P. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 55.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 56.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 57.van Ballegooijen, M., J. A. Bogaards, G. J. Weverling, M. C. Boerlijst, and J. Goudsmit. 2003. AIDS vaccines that allow HIV-1 to infect and escape immunologic control: a mathematic analysis of mass vaccination. J. Acquir. Immune Defic. Syndr. 34:214-220. [DOI] [PubMed] [Google Scholar]
- 58.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]