Abstract
APOBEC3G/CEM15 (hA3G) is a novel host factor that confers resistance to lentiviral infection under experimental conditions. Human immunodeficiency virus (HIV) type 1, however, produces viral infectivity factor (Vif) that targets hA3G for proteolysis, thereby escaping this defense system. To examine hA3G's contribution to the protection against HIV disease progression in humans, we quantified hA3G mRNA levels in peripheral blood mononuclear cells from 6 HIV-uninfected and 25 HIV-infected subjects; the latter group included 8 long-term nonprogressors (LTNPs) and 17 progressors. None of the HIV-infected subjects were receiving antiretroviral therapy. We found a striking inverse correlation between hA3G mRNA levels and HIV viral loads (P ≤ 0.00009) and a highly significant positive correlation between hA3G mRNA levels and CD4 cell counts (P ≤ 0.00012) in these patients. Furthermore, we discovered that the order of hA3G mRNA levels is LTNPs > HIV-uninfected subjects > progressors.
Approximately 5% of human immunodeficiency virus (HIV)-infected individuals, called long-term nonprogressors (LTNPs), have a low rate of disease progression in the absence of therapeutic interventions, are usually asymptomatic, and maintain high CD4 cell counts and low HIV viral loads. Potential mechanisms for long-term survival include virological, immunological, and genetic elements (2, 3, 6, 7, 10-13). APOBEC3G/CEM15 (hA3G) is a newly discovered host factor that is involved in restriction of HIV-1 infectivity and belongs to the family of APOBEC-1-related proteins, which carry out deamination of cytidine/deoxycytidine (8, 9, 14, 18, 19). hA3G may inhibit HIV-1 infectivity by mediating dC-to-dU mutation on minus-strand DNA during reverse transcription and corresponding creation of templates for dG-to-dA transition during plus-strand synthesis (19). Indeed, several previous publications have identified G-to-A hypermutation in subjects with HIV infection (1, 5, 15-17). Notably, in some LTNPs, viral sequence defects due to G-to-A hypermutations have been noted (4). We hypothesized that increased expression of hA3G may be associated with control of HIV disease progression and tested the hypothesis by quantifying hA3G mRNA levels in peripheral blood mononuclear cells (PBMCs) of HIV-infected and -uninfected subjects.
We established a real-time PCR assay to quantify hA3G mRNA levels in human PBMCs. Each subject gave written informed consent as approved by the Research Subject Review Board of the University of Rochester. Human subject study regulations and guidelines were strictly followed. PBMCs were obtained from consenting human subjects and cryopreserved. Prior to RNA isolation, cryopreserved PBMCs were thawed, washed with phosphate-buffered saline, and stimulated with 1 μg each of anti-CD3 and anti-CD28 antibodies for 18 to 20 h. Aliquots of 2 × 106 to 5 ×106 cells were resuspended in 1 ml of TriReagent (MRC), and total cellular RNA was isolated according to standard protocols. Poly(A)+ RNA was isolated using the MicroPoly (A) Purist kit (Ambion) and stored in RNase-free water (Ambion) at −80°C. Purified poly(A)+ RNA was quantified by measuring the optical density at 260 nm (OD260) and the OD280, and all RNAs were found to have a OD260/OD280 ratio of 1.95 or greater. hA3G gene expression was examined by using TaqMan chemistry with probes and primers designed to uniquely amplify h3G/APOBECEG (NM_0218220). The primers used were as follows: forward, 5′CGCAGCCTGTGTCAGAAAAG3′ (nucleotides 637 to 657); reverse, 5′CCAACAGTGCTGAAATTCGTCATA3′ (nucleotides 714 to 691); and probe, FAM-5′GTGCCACCATGAAGA3′-BHQ1 (nucleotides 668 to 682) (where FAM is 6-carboxyfluorescein and BHQ1 is black-hole quencher 1). The following dyes, in combination, for probe generation were used for detection and data normalization: FAM (for the genes of interest), hexachloro- 6-carboxyfluorescein (for normalized genes, see below), BHQ1 (nonfluorescent quencher), and carboxy-X-rhodamine. Validation experiments were performed to determine the specificity and efficiency of the primers and probes designed to selectively amplify hA3G mRNA over closely related APOBEC3B (hA3B) and APOBEC3F (hA3F) (18). A commercially available primer/probe combination was used to quantify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalizing control sequence for the number of cell equivalents in poly(A)+ mRNA starting material used for the quantification of hA3G mRNA. Following probe and primer optimization, all reverse transcriptase, first-strand cDNA products were diluted and used in a 10-μl PCR mixture containing 5 μl of ABI 2× Universal Master Mix, 1.25 μl each of forward and reverse primers (final stock concentrations ranging from 200 to 900 nM depending on the primer set), 1 μl of probe (stock ranging from 50 to 200 nM), and RNase/DNase-free water. All reactions were run in an ABI 7900, with 1 cycle at 50°C (2 min) followed by 95°C (10 min) and 40 cycles of 95°C (15 s) followed by 60°C (1 min). Data were collected and analyzed using Sequence Detection software (ABI, Foster City, CA), and relative quantitation was determined using the comparative threshold cycle method performed with Microsoft Excel (see ABI user bulletin no. 2, Relative Gene Expression Quantitation [http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf]).
Figure 1 shows sample results of this assay. Real-time PCR assays were performed using samples from a subject with HIV infection, a positive control (CEM15 control, a plasmid carrying hA3G cDNA), and a negative control (APOBEC3F, a plasmid carrying hA3F cDNA) (Fig. 1a), and GAPDH from the human sample in a separate reaction (Fig. 1b) as a control for cell number that was used to normalize the hA3G quantification. Each sample was tested in duplicate. These results indicated that hA3G quantification was within the reliable detection limits of the assay. Importantly, GAPDH mRNA was expressed at similar levels in each patient sample (Fig. 1b).
FIG. 1.
Results of real-time PCR assays using a sample from an HIV-infected subject, a CEM15 positive control, and an APOBEC3F negative control (a) and GAPDH from the same patient and another human sample in a separate reaction as a control for cell number (b). ΔRn, change in the amount of fluorescence.
Using this method, we studied 6 HIV-uninfected and 25 antiretroviral antibody-naïve chronically HIV-infected subjects, including 8 LTNPs whose average viral load was 1.8 × 103 (±1.1 × 103) copies/ml and whose average CD4 cell count was 755 (±284)/μl and 17 progressors whose average viral load was 1.5 × 105 (±2.5 × 105) copies/ml and whose average CD4 cell count was 324 (±208)/μl (Table 1). HIV-1 RNA levels were quantified using the Amplicor HIV-1 Monitor assay (Roche Molecular Systems, Branchburg, NJ), which has a detection limit of 50 HIV-1 RNA copies/ml. The CD4 cell counts and percentages were determined using whole blood and the MultiSet program (Becton Dickinson, San Jose, CA) by flow cytometer techniques in a Clinical Laboratory Improvement Amendments-certified laboratory. PBMCs from these subjects were stimulated, and samples were coded and sent to another lab for poly(A)+ mRNA extraction. The samples were recoded and sent for cDNA synthesis and real-time PCR assays. The amounts of hA3G mRNA were standardized against the GAPDH levels in each sample and calculated as copies of mRNA/μg cDNA. The hA3G mRNA levels in each subject were determined, and the average values (± standard deviations) were 132 (±23) copies/mg cDNA in HIV-uninfected subjects, 189 (±59) copies/μg cDNA in LTNPs, and 105 (±15) copies/μg cDNA in progressors. In all HIV-infected subjects, the average value was 132 (±53) copies/μg cDNA (Table 1). By the Mann-Whitney U test, the hA3G mRNA levels in LTNPs were significantly higher than those in progressors (P ≤ 0.001) and those in HIV-uninfected controls (P ≤ 0.020). In addition, the hA3G levels in HIV-uninfected controls were also higher than those in progressors (P ≤ 0.008).
TABLE 1.
CEM15 mRNA levels in HIV-infected and -uninfected study subjects
| Subject group or patient no. | Viremia (copies/ml) | CD4 count/μl | Yr of HIV infection | CEM15 mRNA copies/μg of cDNA |
|---|---|---|---|---|
| HIV-uninfecteda | ||||
| Mean | 132 | |||
| SD | 23 | |||
| HIV-infected | ||||
| LTNPs | ||||
| 1 | 5.0E+01 | 1,320 | 8 | 321 |
| 2 | 8.1E+02 | 492 | 18 | 173 |
| 3 | 1.3E+03 | 591 | 9 | 189 |
| 4 | 1.7E+03 | 737 | 12 | 114 |
| 5 | 2.2E+03 | 648 | 16 | 175 |
| 6 | 2.6E+03 | 478 | 15 | 204 |
| 7 | 3.0E+03 | 1,000 | 18 | 161 |
| 8 | 3.1E+03 | 775 | 14 | 176 |
| Mean | 1.8E+03 | 755 | 189 | |
| SD | 1.1E+03 | 284 | 59 | |
| Progressors | ||||
| 9 | 2.4E+03 | 637 | 2 | 133 |
| 10 | 3.0E+03 | 237 | 19 | 103 |
| 11 | 5.3E+03 | 247 | 12 | 98 |
| 12 | 5.3E+03 | 600 | 4 | 93 |
| 13 | 5.8E+03 | 418 | NAb | 99 |
| 14 | 1.6E+04 | 462 | 1 | 93 |
| 15 | 2.1E+04 | 582 | 3 | 121 |
| 16 | 2.3E+04 | 263 | 17 | 100 |
| 17 | 2.4E+04 | 166 | NA | 98 |
| 18 | 2.7E+04 | 177 | 6 | 120 |
| 19 | 3.7E+04 | 516 | 3 | 125 |
| 20 | 7.3E+04 | 209 | NA | 98 |
| 21 | 1.4E+05 | 599 | 10 | 94 |
| 22 | 2.1E+05 | 17 | 7 | 135 |
| 23 | 4.1E+05 | 11 | 11 | 98 |
| 24 | 7.5E+05 | 211 | 12 | 92 |
| 25 | 7.5E+05 | 157 | 10 | 89 |
| Mean | 1.5E+05 | 324 | 105 | |
| SD | 2.5E+05 | 208 | 15 | |
| LTNPs + progressors | ||||
| Mean | 1.0E+05 | 462 | 132 | |
| SD | 2.2E+05 | 307 | 53 |
n = 6.
NA, not available.
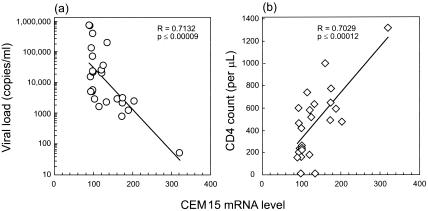
To determine if the augmented hA3G gene expression had any functional implications, we performed a rank correlation test between hA3G mRNA levels and HIV viremia and CD4 cell counts in data from the 25 HIV-infected individuals and found a striking inverse correlation between hA3G mRNA levels and viral loads (R = −0.7132, P ≤ 0.00009) (Fig. 2a) and a highly significant positive correlation between hA3G mRNA levels and CD4 cell counts (R = 0.7029, P ≤ 0.00012) (Fig. 2b). Moreover, these correlations remain even after removing the one LTNP (subject 1) who has the highest hA3G value (R = −0.5988, P ≤0.0022 [for viral load]; R = 0.4962, P ≤ 0.014 [for CD4 cell count]).
FIG. 2.
Results of rank correlation test between hA3G mRNA levels and HIV viremia and CD4 cell counts for the 25 HIV-infected individuals.
Although it has been shown that hA3G contributes to the control of HIV and simian immunodeficiency virus replication in cell cultures and animal experiments (9, 14), our results are the first to demonstrate correlations between hA3G mRNA levels and HIV viral load and CD4 cell count, both of which are predictors of HIV disease progression in patients who have not received antiretroviral drugs or other forms of therapeutic intervention. In addition, we have found that LTNPs have significantly higher hA3G mRNA levels than do HIV-uninfected controls or the progressors, whose hA3G mRNA levels are significantly lower that of HIV-uninfected controls.
The lower rate of HIV disease progression in LTNP patients is likely to be multifactorial. However, our data suggest that increased hA3G gene expression may provide a competitive advantage over time and that mutations in the HIV genome may accumulate to the point of debilitating the virus and suppressing viremia. In summary, we have provided evidence that enhanced hA3G gene expression in humans may provide protective effects against HIV disease progression by reducing viral burden and increasing CD4+ T-cell count over time, and the increased hA3G levels may partly contribute to the slow disease progression in LTNPs.
REFERENCES
- 1.Borman, A. M., C. Quillent, P. Charneau, K. M. Kean, and F. Clavel. 1995. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G→A hypermutation during negative-strand viral DNA synthesis. Virology 208:601-609. [DOI] [PubMed] [Google Scholar]
- 2.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 3.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 4.Huang, Y., L. Zhang, and D. D. Ho. 1998. Characterization of gag and pol sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology 240:36-49. [DOI] [PubMed] [Google Scholar]
- 5.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 7.Lum, J. J., O. J. Cohen, Z. Nie, J. G. Weaver, T. S. Gomez, X. J. Yao, D. Lynch, A. A. Pilon, N. Hawley, J. E. Kim, Z. Chen, M. Montpetit, J. Sanchez-Dardon, E. A. Cohen, and A. D. Badley. 2003. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J. Clin. Investig. 111:1547-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 9.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 12.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10:806-810. [DOI] [PubMed] [Google Scholar]
- 13.Rinaldo, C., X.-L. Huang, Z. Fan, M. Ding, L. Beltz, A. Logar, D. Panicali, G. Mazzara, J. Liebmann, M. Cottrill, and P. Gupta. 1995. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J. Virology 69:5838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 15.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83:801-805. [DOI] [PubMed] [Google Scholar]
- 16.Vartanian, J.-P., A. Meyerhans, B. Åsjö, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vartanian, J. P., A. Meyerhans, M. Sala, and S. Wain-Hobson. 1994. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc. Natl. Acad. Sci. USA 91:3092-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207-216. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]