Abstract
The integration sites of viral vectors used in human gene therapy can have important consequences for safety and efficacy. However, an extensive evaluation of adeno-associated virus (AAV) vector integration sites has not been completed, despite the ongoing use of AAV vectors in clinical trials. Here we have used a shuttle vector system to isolate and analyze 977 unique AAV vector-chromosome integration junctions from normal human fibroblasts and describe their genomic distribution. We found a significant preference for integrating within CpG islands and the first 1 kb of genes, but only a slight overall preference for transcribed sequences. Integration sites were clustered throughout the genome, including a major preference for integration in ribosomal DNA repeats, and 13 other hotspots that contained three or more proviruses within a 500-kb window. Both junctions were localized from 323 proviruses, allowing us to characterize the chromosomal deletions, insertions, and translocations associated with vector integration. These studies establish a profile of insertional mutagenesis for AAV vectors and provide unique insight into the chromosomal distribution of DNA strand breaks that may facilitate integration.
Successful gene therapy often requires the long-term transgene expression provided by integrating viral vectors. However, integration can cause insertional mutagenesis and oncogene activation, as demonstrated by two X-linked severe combined immune deficiency patients who developed leukemia after treatment with a retroviral vector that integrated near the LMO2 proto-oncogene (14). Large-scale analyses of integration sites can shed light on the process of insertional mutagenesis, and studies of murine leukemia virus, human immunodeficiency virus, avian retroviruses, and vectors based on them have demonstrated distinct integration patterns (16, 27, 33, 40, 46). Of particular importance is the relationship of integration sites to genes, as this may determine the likelihood of oncogene activation. In this regard, human immunodeficiency virus vectors have been shown to integrate preferentially throughout transcription units, while murine leukemia virus vectors integrate preferentially near transcription start sites (40, 46).
Vectors based on adeno-associated virus (AAV) have a linear, single-stranded DNA genome containing a transgene cassette flanked by viral inverted terminal repeats (ITRs). Transduction occurs by multiple pathways, including integration into the host genome (24), expression from linear and circular episomal forms (6, 30), and homologous recombination with chromosomal sequences (37). Definitive evidence for integration has come from sequencing vector-chromosome junctions recovered from human cells (25, 26, 38, 47) and mouse tissues (28, 29), which demonstrated that AAV vectors integrate at nonhomologous chromosomal locations.
Chromosomal sequences surrounding vector proviruses can be deleted or rearranged (25, 26, 29), although it is not known if the AAV vector causes these changes. While integration may occur in only a subset of transduced cells, the large vector doses infused during in vivo gene delivery can lead to substantial numbers of integration events. In mouse models of liver transduction, integrated AAV vector genomes were present at roughly 0.05 copies/cell (32), a value that would correspond to 7.5 × 109 integration events in humans undergoing liver-directed gene therapy with AAV vectors (assuming 1.5 × 1011 cells/liver) (44). The potential consequences of billions of integration events are largely unknown.
Integration requires that the linear AAV vector genome ligate to two chromosomal ends. Unlike retroviral vectors, AAV vectors do not contain an endonuclease to generate chromosomal ends, so they must rely on existing double-strand breaks or nicks. The deletions, insertions, and microhomologies found at AAV vector-chromosome junctions suggest that integration occurs by the nonhomologous end-joining pathway of double-strand break repair (26), and AAV vectors will integrate at a specific double-strand break when it is created in human cells (25). This dependency on host cell factors and chromosomal features allows us to interpret AAV vector integration sites as chromosomal repair events tagged by a provirus.
Here we have performed a large-scale analysis of AAV vector integration sites in normal human cells in the absence of selective pressure, including their relationship to genes, repetitive DNAs, and other chromosomal features. We have characterized the deletions, insertions, and translocations associated with AAV vector integration and the structures of vector proviruses and identified several integration hotspots. Our results establish the profile of insertional mutagenesis associated with AAV vectors, and they suggest that similar integration studies may be a valuable tool for understanding chromosome biology.
MATERIALS AND METHODS
Nucleic acid manipulations.
Plasmids pDG (13) and pA2-TOA (25) have been described. Genomic DNAs were isolated by standard techniques using the Puregene kit (Gentra Systems, Minneapolis, MN). RNA was prepared for microarray studies by using the RNeasy and QIAshredder kits from QIAGEN (Valencia, CA). Plasmid DNAs used for vector production were purified from bacterial pellets by using QIAGEN plasmid maxi kits. Plasmid DNAs containing rescued proviruses were purified and sequenced according to the standard DNA sequencing protocols used in our Genome Center (1, 22). Briefly, the plasmid DNAs were prepared on a QIAGEN Biorobot 3000 utilizing QIAprep 96 Turbo plasmid DNA preparation kit according to the protocols suggested by the manufacturer. The sequencing reactions were carried out with the Big Dye terminator chemistry V3.1 kit (Applied Biosystems, Foster City, CA) using left and right sequencing primers 5′-GATAAG CTG TCA AAC ATG AGA ATT C and 5′-ATCACG AGG CCC TTT CGT CTT CAA G, respectively. Electrophoresis of sequencing reactions was performed on ABI Prism 3700 capillary sequencers and the raw trace data were analyzed and viewed using PHRED/PHRAP/CONSED software tools (8, 9, 12).
Cell culture.
Cells were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium containing 4 g of glucose/liter (Gibco/Invitrogen, Carlsbad, CA), 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin. Primary, normal male human fibroblasts (MHF2) were obtained from the Coriell Institute for Medical Research (Camden, NJ; catalog no. GM05387). 293T cells have been described (7). MHF2 cells were transduced with the AAV2-TOA vector by seeding 6-cm tissue culture dishes with 5 × 105 cells on day 0, replacing the medium and infecting with 2.5 × 1010 genome-containing particles of AAV2-TOA on day 1 (5 × 104 genome-containing particles per cell), and replacing the medium again on day 3. On day 6 cells were detached with trypsin and seeded to one 15-cm dish. On day 10 the cells in a single 15-cm dish were split to three 15-cm dishes. On day 14 genomic DNA was prepared from the 15-cm dishes, except for 3 × 106 cells that were used to seed another set of three 15-cm dishes. This process was repeated every 6 days, and the majority of proviruses were isolated from DNA prepared on the fourth round of this procedure.
Vector preparation.
The serotype 2 AAV vector AAV2-TOA was made by cotransfection of 293T cells with helper plasmid pDG and vector plasmid pA2TOA and purified by benzonase treatment of cell lysates, iodixanol step gradient, heparin affinity column chromatography (HiTrap, Amersham Biosciences, Uppsala Sweden), and HiTrap desalting column as described (48). AAV vector quantification was based on the amount of full-length single-stranded vector genomes detected by alkaline Southern blot analysis (18).
Shuttle vector rescue in bacteria.
Rescue of AAV2-TOA proviruses was done as described (25) with the following modifications: 20 μg of genomic DNA containing integrated proviruses was digested with 80 units of MfeI, AvrII, or NcoI, extracted with phenol and chloroform, and precipitated with ethanol. DNA fragments were resuspended in 355 μl of H2O and brought to 400 μl with 40 μl of 10x ligation buffer and 5 μl of T4 DNA ligase (400 U/μl, New England Biolabs, Beverly, MA). Ligations were incubated at 15°C overnight to circularize fragments, heat inactivated by incubation at 65°C for 20 min, brought to 50 mM NaCl, digested further with 80 units of DpnI for an additional 2 h to remove bacterial DNA, extracted with phenol and chloroform, and precipitated with ethanol.
The DNA pellets were resuspended in 5 μl of H2O, and Escherichia coli strain DH10B (15) was transformed by electroporation with ∼4 μg (1 μl) of DNA at a time. Transformed bacteria were grown on agar containing 50 μg/ml ampicillin and colonies were replated to agar containing 12.5 μg/ml tetracycline. Bacteria resistant to both ampicillin and tetracycline were grown in 96-well culture dishes in freezing medium and then frozen at −80°C for future sequencing. Freezing medium contains 10 g tryptone, 5 g yeast extract, 10 g NaCl, 6.3 g K2HPO4, 1.8 g KH2PO4. 0.5 g sodium citrate, 0.9 g (NH4)2SO4, and 44 ml glycerol per liter of H2O, brought to 10 μM MgSO4 and supplemented with ampicillin after autoclaving.
Microarray analysis of gene expression levels.
We seeded 5 × 105 MHF2 cells in six 6-cm dishes on day 0. On day 1 fresh medium was added to the dishes, and three dishes received 2.5 × 1010 genome-containing particles of AAV2-TOA. On day 3 RNA was harvested from confluent 6-cm dishes and all six samples were processed independently. Labeling of 5 μg of total RNA was performed as described by Affymetrix (Santa Clara, CA); 15 μg of cRNA was used per Affymetrix HG-U133 Plus 2.0 array, which analyzes 47,400 transcripts and variants. Only the subset of probes that identify specific RefSeq gene transcripts (13,069) were used in our analysis. Probe sets that hybridized with more than one gene were excluded. Gene expression levels from all three uninfected cell samples were averaged and compared to those from all three infected cell samples. Where multiple probe sets reflect the transcription level of a single RefSeq gene, the average transcription level was used in rankings.
Database searches and comparisons with genomic features.
DNA sequences were processed with computer programs interpreted by the PERL programming language. Sequences were truncated at bp 500, and expected vector-derived sequences were trimmed. The resulting junction sequences were aligned to build 35 of the human genome and three additional files containing AAV2-TOA vector sequence, nonvector sequence from plasmid pA2TOA, and the 43-kb human ribosomal DNA (rDNA) repeat (GenBank accession no. U13369) (10) using a stand-alone version of BLAT (21) that generates a BLAST alignment score.
The input script was as follows: blat chromosome_file query_file −out=blast8-ooc=11.ooc output_file. An additional 95% homology requirement and BLAST score of >100 were used to establish genomic positions. Alignments were sorted by BLAST score, and those with the five highest scores were saved for further processing. The average match length for all sequences was 383 bp. Nucleotide insertions were defined as sequence preceding the alignment with the highest BLAST score when the alignment did not start at position number 1 of the sequence query. Additional PERL programs were used to remove duplicate junction sequences, compare localized integration sites to various chromosomal features using tables available from the University of California-San Francisco database (20), and determine the positions of restriction enzyme sites in the human genome.
We produced a randomly localized set of genomic positions by generating random numbers between 1 and 5,941,037,819 (the size of the build 35 diploid male genome with chromosomes laid end to end) with the PERL “rand” function. The buffer size had to be increased from 15 to 31 bits to avoid generating duplicate numbers. These random numbers were converted to chromosomal positions by splitting the numeric range of the diploid genome into separate chromosomes with each starting at base pair 1 of the p arm and extending the entire length of the chromosome. These chromosomal positions were used to extract 383 bp of sequence from build 35 of the human genome at each randomly determined position, and the resulting files were aligned with the genome using BLAT as described above. About 7% of these extracted sequences corresponded to gapped or repetitive sequence in the human genome, could not be reliably localized, and were discarded. A set of 10,000 localized positions was used as a control data set (calculated random integration events) for comparison with AAV vector integration site positions. To analyze clustering and hotspots, we used similar sets of 499 and 670 random genomic positions as size-matched controls
To identify oncogenes, we searched several databases, including Entrez Gene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = gene), OMIM (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = OMIM), the Tumor Gene Database (http://www.tumor-gene.org/TGDB/tgdb.html), and the Retrovirus Tagged Cancer Gene Database (http://rtcgd.ncifcrf.gov/).
Statistical analysis.
In all cases statistical significance was determined using the χ2 test to compare AAV vector integration site frequencies with those of randomly generated genomic positions. P values were determined using tables, and those less than 0.01 were considered significant.
RESULTS
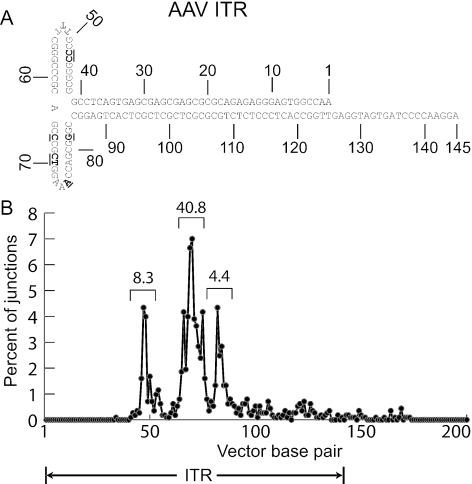
We infected normal human fibroblasts with the AAV shuttle vector AAV2-TOA containing a bacterial replication origin and antibiotic resistance genes, then rescued integrated proviruses and flanking chromosomal DNA in E. coli. as bacterial plasmids. Of the 3,264 sequences obtained, 1,691 had flanking junction DNA that could be localized to build 35 of the human genome, rDNA, vector, or nonvector plasmid sequences, with an average alignment of 383 bp per query. After discarding presumed duplicates (junctions at the exact same nucleotide position), and eliminating sequence reads with BLAST scores <100, a total of 977 unique integration junctions were localized to the human genome (Table 1). Both left and right junctions were obtained from 323 proviruses. Of the flanking sequences, 9.3% were from the vector genome or plasmid backbone, which may represent vector-vector recombination events, foldback priming of DNA synthesis at ITRs, or packaging of plasmid sequences into virions, as observed previously (26, 28, 38). None of the vector proviruses contained intact ITRs, and distinct preferences were noted for junction bases within the ITR secondary structure (Fig. 1).
TABLE 1.
Summary of sequencing and localization
| Parameter | No. |
|---|---|
| Sequences obtained | 3,264 |
| Sequences localized | 1,691 |
| BLAST score >100 | 1,645 |
| Duplicatesa | 519 |
| Vector sequence at junctionb | 97 |
| Plasmid sequence at junctionc | 13 |
| rDNA sequence at junctiond | 58 |
| Junction sequences localized to human genome | 977 |
| Proviruses with only one end localized | 331 |
| Proviruses with both ends localized | 323 |
| Ends localize to the same chromosome | 307 |
| Ends localize to different chromosomes | 16 |
| Unique localized provirusese | 670 |
Proviruses with junctions occurring at the exact same base as another sequenced provirus.
Provirus sequence was joined to other sequences derived from the AAV vector genome.
Provirus sequence was joined to nonvector sequences derived from the AAV vector plasmid.
DNA sequences are not localized to specific chromosomal sites.
Counts proviruses with both ends sequenced once if localized to the same chromosome and twice if localized to different chromosomes, and assumes all proviruses with one end localized were distinct.
FIG. 1.
Location of junction sites in the AAV vector proviruses. In the top panel, the nucleotide sequence of an AAV vector ITR in the flop orientation is shown numbered from 1 to 145 beginning at the 3′ end. The locations of the most common vector-chromosome junctions are indicated by underlined, bold type. In the bottom panel, the percent of vector-chromosome junctions found at specific base pairs in the AAV ITR and adjoining vector sequence is shown. The percent found in three observed peaks is also indicated.
The chromosomal features of vector integration sites are shown in Table 2. As a control, we generated a set of sequences localized to random positions in the human genome that were processed the same way as our provirus sequence reads (see Materials and Methods). Compared to this control set, AAV vectors preferentially integrated in CpG islands, segmental duplications, ribosomal repeats, and satellite DNA, and there were fewer integrants than expected in long terminal repeat elements. The preference for satellite DNA may be an artifact, as most satellite sequences are not localized and would therefore not be included in our random set. There was a modest preference for integrations in transcription units that was not as statistically significant (P < 0.05).
TABLE 2.
Genomic features of integration sites
| Genomic featurec | % of sites
|
Pa | |
|---|---|---|---|
| AAV vector (n = 670) | Random (n = 10,000) | ||
| Transcription units | 38.81 | 34.76 | <0.05 |
| CpG islands | 4.03 | 0.84 | <0.001 |
| Segmental duplications | 8.21 | 5.00 | <0.001 |
| Ribosomal DNA repeats | 7.97 | 0.29b | <0.001 |
| Repeats | |||
| SINE | 13.58 | 13.80 | |
| Alu | 11.94 | 11.26 | |
| MIR | 1.64 | 2.54 | |
| DNA elements | 3.13 | 3.04 | |
| Long terminal repeat elements | 5.67 | 8.60 | <0.01 |
| LINE | 18.51 | 20.39 | |
| Satellite | 4.03 | 0.34 | <0.001 |
| Alpha | 1.34 | 0.20 | <0.001 |
| Beta | 2.69 | 0.14 | <0.001 |
P values of <0.05 are not shown and were not considered statistically significant.
Percent of rDNA in the diploid human genome, assuming 400 rDNA repeats of 43 kb.
SINE, short interspersed nucleotide element; MIR, mammalian interspersed repetitive element; LINE, long interspersed nucleotide element.
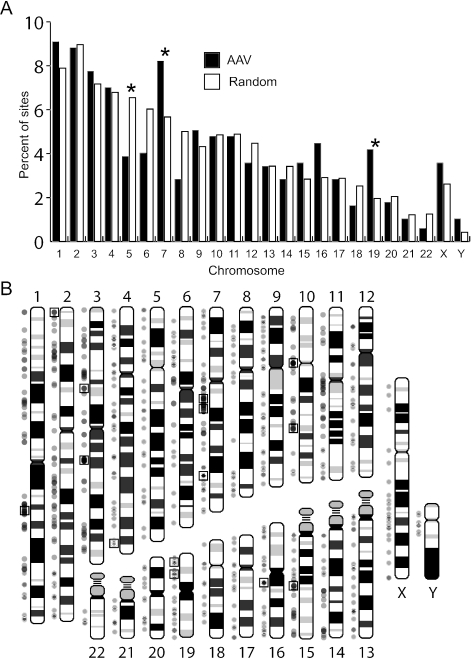
Vector proviruses were found in all human chromosomes (Fig. 2A). Relative to the set of calculated random integration events, some chromosomes had statistically significant differences in vector integration frequencies. Chromosome 5 lacked integration events compared to controls (P < 0.01). Integration hotspots, defined as ≥3 independent proviruses within 500 kb (Fig. 2B), may explain why some chromosomes had statistically significant increases in integration frequencies. Chromosomes 7 and 19 had more vector integrations than expected, with three and two hotspots, respectively. A list of all the hotspots meeting these criteria is shown in Table 3, and in many cases, the hotspot size was significantly less than 500 kb.
FIG. 2.
Chromosomal distribution of integration sites. (A) Localized AAV vector integration sites (n = 670) and a calculated set of random sites (n = 10,000) are graphed as a percentage of all integrants in each chromosome. Only one integration junction was included for AAV vector proviruses where both ends were localized to the same chromosome. Asterisks mark comparisons with P values of <0.01. (B) A human chromosome ideogram is shown with AAV vector integration sites (dots to the left of each chromosome) and hotspots where at least three integrants were found within 500 kb (boxed dots; see Table 3). Each dot represents a unique AAV vector integrant (n = 670) and is 33% opaque to display multiple overlapping integrants. Ribosomal DNA repeats present on the p arm of chromosomes 13, 14, 15, 21, and 22 contained a significant number of AAV vector integrants that are described separately in Fig. 3.
TABLE 3.
AAV vector integration hotspots
| Hotspota | Cytogenetic position | Left boundary | Size (bp) | No. of provirusesb | RefSeq gene(s)c |
|---|---|---|---|---|---|
| chr1HS1d | 1q23.3 | 158232281 | 19,572 | 3-4 | None |
| chr2HS1 | 2p25.3 | 1777992 | 20 | 3 | MYT1L |
| chr3HS1 | 3p14.2 | 61218533 | 479,629 | 3 | PTPRG |
| chr3HS2 | 3q13.31 | 117050246 | 1,857,520 | 11-13 | LSAMP |
| chr7HS1 | 7q11.22 | 69641166 | 265,215 | 3 | AUTS2 |
| chr7HS2d | 7q11.23 | 74755281 | 41,382 | 3 | PMS2L3 |
| chr7HS3 | 7q32.3 | 130042139 | 458,211 | 6-7 | MKLN1 |
| chr10HS1 | 10q11.1-11.2 | 41684847 | 304,399 | 6-9 | None |
| chr10HS2 | 10q23.32 | 93319335 | 351,497 | 4 | PPP1R3C, TNKS2, C10orf13 |
| chr15HS1 | 15q22.2 | 58044788 | 457,399 | 3-4 | FOXB1, ANXA2 |
| chr16HS1d | 16q11.2 | 44949556 | 37,117 | 5-6 | None |
| chr19HS1 | 19p13.3 | 4362420 | 128,182 | 3 | CHAF1A, UBXD1, HDGF2, LRG1 |
| chr19HS2 | 19p13.13 | 13725980 | 54,715 | 3 | MGC10471 |
At least three independent proviruses within 500 kb.
Lower numbers assume that rescued proviruses with only one end localized are duplicates of rescued proviruses where only the other end was localized.
Protein function is described in Table S1 in the supplemental material.
Did not meet hotspot criteria when second-best BLAST score required to be <90% of best score.
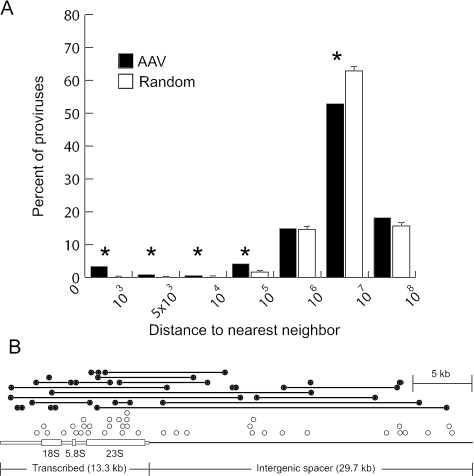
This tendency to integrate in clusters was further quantified by measuring the distances between neighboring integration sites (using data from the left sequencing primer only, to avoid counting sites identified by sequencing opposite ends of the same provirus). Of 499 unique sites, 3.41% or 9.02% of neighboring vector integration sites were within 1 kb or 100 kb, respectively, compared to 0.2% or 1.87%, respectively, of neighboring calculated random integrants (Fig. 3A). A major hotspot with almost 8% of all vector integrations occurred in rDNA repeats (Fig. 3B), which are not localized to the human genome sequence (Table 2). This 43-kb repeat is present at an estimated 400 copies in a diploid human genome (4) and constitutes approximately 0.29% of human DNA.
FIG. 3.
Clustering of AAV vector integration sites and localization within the human ribosomal DNA. (A) The distance between each unique AAV vector integration site identified with one sequencing primer (n = 499) and its nearest neighbor was determined and binned by size, and the percentage of proviruses within each bin was plotted. We used only left junctions in this analysis, to ensure that two different ends of the same provirus were not scored as neighbors. As a control, we performed a similar analysis on three size-matched sets of randomly distributed sites (n = 499), and plotted means with standard deviations. Significant differences (P < 0.01) are marked with asterisks. Each bar represents the number of clones with a nearest neighbor within the distance bounded by the values on the x axis. (B) Each AAV vector integration junction localized to the 43-kb ribosomal DNA repeat unit is shown as a circle above a scale diagram of the repeat. Solid circles connected by lines represent the junctions of proviruses where both ends were localized (n = 21). Open circles represent proviruses where only one end was localized (n = 37). The 13.3-kb transcribed region is drawn with open boxes, where the thick regions represent the 18S, 5.8S, and 23S rRNAs. The 29.7-kb intergenic spacer is depicted as a single bold line.
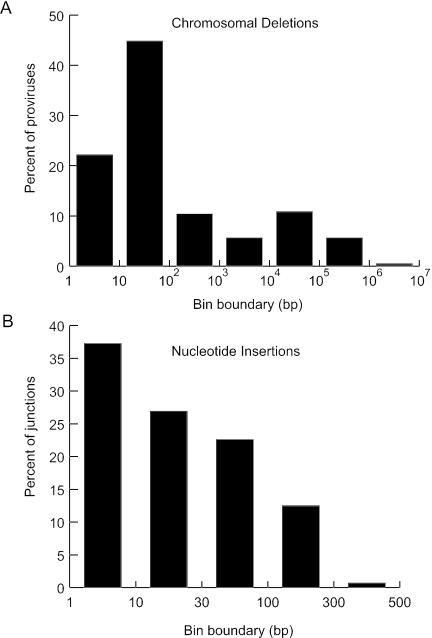
Of the 307 proviruses with both ends localized to the same chromosome, 70% had deletions of genomic DNA ranging in size up to ∼106 bp (Fig. 4A) and 35% of junctions had insertions of DNA that also varied in size (Fig. 4B). Sixteen proviruses had left and right junctions where the best alignment scores were on different chromosomes (Table 4). As these represent possible chromosomal translocations, we used additional criteria to establish their validity. In 11 of 16 proviruses with mismatched ends (MM6 to MM16), the second-best BLAST score was >90% of the best score for at least one end, raising the possibility that there may have been a localization error due to sequence repeats. Many of these junctions mapped to pericentromeric chromosomal regions rich in alpha satellite DNA. The remaining five proviruses had BLAST scores for both ends that were significantly above those of other possible alignments, and three of these had scores over 500 for both ends (MM1 to MM3). Even if one conservatively assumes that only the three translocations meeting the most rigorous criteria are real, this represents nearly 1% of all vector integrations (3 of 323).
FIG. 4.
Deletions and insertions found at integration sites. (A) The sizes of chromosomal deletions associated with AAV vector proviruses (n = 212) were sorted into bins and the percentage of proviruses in each deletion size range bounded by values shown on the x axis was plotted. (B) The sizes of insertions found at AAV vector junctions (n = 416) were sorted into bins and the percentage of proviruses with insertions in each size range bounded by values shown on the x axis was plotted. Junctions linked to vector or plasmid sequence were not included.
TABLE 4.
Vector proviruses where left and right junctions mapped to different chromosomes
| Provirusa | Left-end position
|
Right-end position
|
Gene(s) disrupted
|
|||
|---|---|---|---|---|---|---|
| Cytogenetic | Nucleotide | Cytogenetic | Nucleotide | Left end | Right end | |
| MM1 | 2p16.1 | 59458741 | 5p13.1 | 39681607 | None | None |
| MM2 | 5p15.33 | 734426 | 19p13.11 | 17174853 | TPPP | MYO9B |
| MM3 | 2p25.1 | 11338257 | 1p36.13 | 15759613 | ROCK2 | None |
| MM4 | 19q13.32 | 52662917 | 13q12.11 | 18508827 | SLC8A2 | None |
| MM5 | 7q31.33 | 123752006 | 9p21.3 | 20895739 | None | KIAA1797 |
| MM6 | 19q13.12 | 41437558 | 1p34.1 | 46419046 | None | None |
| MM7 | 10q11.1 | 41684847 | 4p11 | 49483305 | None | None |
| MM8 | 19q13.43 | 63520866 | Xq28 | 154823429 | None | None |
| MM9 | 7q32.3 | 130045779 | 1q23.3 | 158251853 | None | None |
| MM10 | 4p11 | 48982719 | Yq11.1 | 12291893 | None | None |
| MM11 | 16p11.1 | 35077068 | 2q21.2 | 132802318 | None | None |
| MM12 | 8p22 | 15955762 | Yp11.2 | 6508259 | None | None |
| MM13 | 11p11.12 | 510076067 | 3p24.1 | 28703962 | None | None |
| MM14 | 18q21.32 | 55076383 | 8q11.22 | 52569047 | None | None |
| MM15 | 16p13.3 | 1639671 | 14q24.1 | 68454983 | CRAMP1L | ACTN1 |
| MM16 | 15q11.2 | 18360887 | 10q11.21 | 41989246 | None | None |
MM1 to MM3: BLAST score >500, second hit <90% of first hit. MM1 to MM5: BLAST score >100, second hit <90% of first hit.
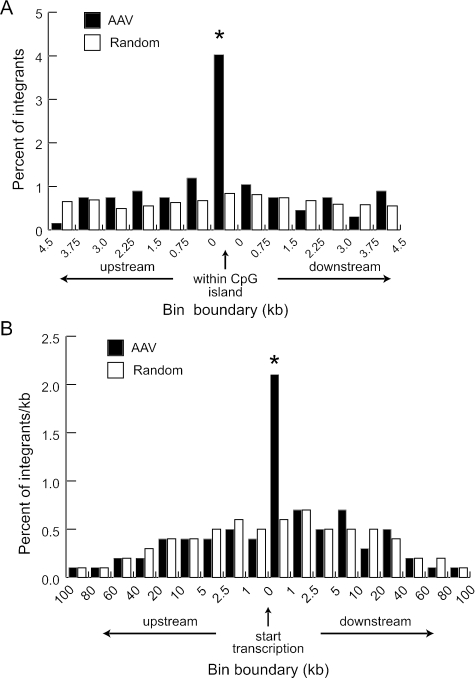
Additional analyses were performed to assess the relationship of integration sites to transcription. CpG islands are frequently found near promoter regions and may regulate gene transcription (3, 23). AAV vectors had a 4.8-fold preference for integration in CpG islands, which did not extend into surrounding regions (Fig. 5A). We also analyzed the distribution of integration sites within transcription units and found a 3.6-fold preference for integrating within 1 kb downstream of the start site, but nowhere else within 100 kb (Fig. 5B). Interestingly, the percentage of calculated random integration events increased slightly near transcription start sites, perhaps reflecting the density of genes in the human genome. Another possible bias comes from the distribution of restriction enzyme sites in the human genome. We analyzed the recognition site positions for each enzyme used in this study relative to CpG islands and transcription starts (Fig. S1 and S2 in the supplemental material). While there were variations in enzyme site frequencies around these genomic elements, they did not correlate with the biases seen for AAV vector integration sites.
FIG. 5.
Integration in CpG islands and at transcription start sites. (A) Localized AAV vector integration sites (n = 670) and a calculated set of random sites (n = 10,000) were mapped relative to those of CpG islands (identified as GC content ≥50%, length >200 bp, and ratio of observed to expected number of CpG dinucleotides >0.6). Integration sites within CpG islands or in twelve 0.75-kb windows flanking each island (the average size of a CpG island is 764 bp) were binned and plotted as the percentage of all sites. Significant differences (P < 0.01) are marked with an asterisk. (B) AAV vector integration sites (n = 670) and a calculated set of random sites (n = 10,000) were mapped relative to the transcription start sites of RefSeq genes, binned into windows of increasing sequence size, and plotted as a percentage of all integrants per kb (to account for the different window sizes). Significant differences (P < 0.01) are marked with an asterisk.
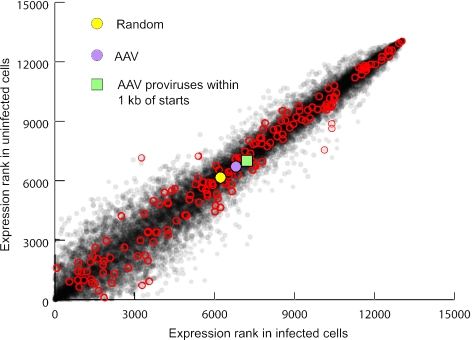
Microarray analysis was performed to determine if integrations occurred preferentially in expressed genes. We ranked 13,069 RefSeq genes by expression level and plotted the ranks from uninfected and infected fibroblasts, highlighting those where vectors integrated (Fig. 6). There was little overall impact of AAV vector infection on gene expression, consistent with an earlier study (41). Two hundred sixty proviruses integrated within the transcribed region of a gene, and the average expression rank of those found on the chip (n = 195) was 1.09-fold above that of genes with calculated random integrants in both infected and uninfected cells. The average expression rank of the genes where integrations occurred within 1 kb of transcription starts (14 found on the chip) was 1.14- to 1.16-fold above that of genes with calculated random integrants. These results suggest that gene expression did not have a major impact on integration. A more significant effect of transcription was observed for the rDNA genes transcribed by RNA polymerase I. Of the 58 proviruses found in rDNA repeats, the integration frequency was 3.7-fold higher in transcribed versus nontranscribed regions (2.71 and 0.74 proviruses/kb, respectively) (Fig. 3B).
FIG. 6.
RefSeq gene expression levels and AAV vector integration. RNA samples from uninfected normal human fibroblasts (y axis) and those infected with AAV2-TOA (x axis) were hybridized to the human U133A Plus 2.0 gene chip array (Affymetrix) and the expression levels of all RefSeq genes assayed in the array were ranked and plotted (light gray dots). RefSeq genes containing AAV vector integrants are circled in red. The average expression rank of RefSeq genes containing a calculated random integrant (n = 2,930; yellow circle), those containing AAV vector integrants (n = 195; purple circle), and those containing AAV vector integrants within 1 kb of transcription start sites (n = 14; green square) are shown.
DISCUSSION
In this report we have used an AAV shuttle vector system to rescue integrated proviruses as bacterial plasmids. This allowed us to recover hundreds of integration events from normal human cells without selection in a scalable process, with automated plasmid purification and junction sequencing. Our approach has several advantages compared to more commonly used PCR-based methods. First, the size of junction sequences is not limited by the efficiency of PCR amplification, and the quality of sequence obtained from plasmids is high, so we were able to generate long sequence reads (average match length of 383 bp). This allows more accurate localization of junctions containing repetitive sequences, and if necessary additional portions of the plasmid can be sequenced. In contrast, the PCR methods used to sequence murine leukemia virus and human immunodeficiency virus integration junctions (40, 46) generated sequences with average match lengths of 98 and 191 bp, respectively, when aligned by our criteria. Second, the shuttle vector allows one to recover both junctions from a provirus, which in the case of AAV is crucial for determining the chromosomal structure at the integration site. Third, while both approaches may be biased by the genomic distribution of restriction sites used, PCR is also biased by parameters affecting amplification. The major drawback to the shuttle vector approach is that the vector must include a selectable bacterial marker and replication origin.
An important issue regarding insertional mutagenesis is its relationship to chromosomal gene transcription. We found that AAV vectors had only a modest preference for integration within transcription units (38.81% versus 34.76% for calculated random integrants), which was less than that reported for AAV vector integration in mouse livers, where 72% of integrants were in genes (29). This could reflect differences in the etiology of chromosomal breaks used for integration, where relatively low hepatocyte proliferation rates may decrease the impact of DNA replication on integration sites and increase the impact of transcription. AAV vectors had a more dramatic preference for integration at transcription start sites, with a >3-fold enrichment within the first kb transcribed, but these accounted for just 2.1% of all integrants due to the small window size. We found only a modest effect of transcription, with the average expression rank of genes containing proviruses at any transcribed position or within 1 kb of the start site only 1.09 to 1.16-fold greater than that of genes with calculated random integration events.
There was also a significant preference for integration in CpG islands (4.03% versus 0.84% for calculated random integrants), which typically act as transcriptional control elements for nearby genes (3, 23). Surprisingly, some of our results were similar to those of murine leukemia virus vectors, where integrations also occurred preferentially near transcription start sites and CpG islands, with only a slight overall preference for genes (46). Thus, despite the distinct life cycles of these viruses, they may share aspects of integration site selection.
AAV vectors do not contain an endonuclease, so integration must occur at chromosomal sites where free DNA ends form. This can take the form of a double-strand break (25) or perhaps a nick that is converted to a double-strand break during DNA replication. The preference for transcriptional start sites, CpG islands, and segmental duplications suggests that these regions may be prone to DNA damage that leads to breaks. In the case of transcription start sites, this damage could be due to local unwinding that initiates transcription and exposes bases. CpG islands can have altered chromatin structure and hypersensitivity to nucleases (42, 45), may act as replication origins (5), and, when methylated, can be mutagenic due to deamination of 5-methylcytosine (34), all of which could increase the likelihood of strand breakage. Segmental duplications are recombinogenic areas of the human genome (39) that may also recombine with AAV vector DNA by similar mechanisms. By the same reasoning, long terminal repeats may be relatively protected from DNA damage, as they were underrepresented sites of AAV vector integration.
The integration hotspots we observed may also be damage-prone areas of the genome. The major hotspot in rDNA could reflect unique aspects of these repeats, which are frequently involved in recombination events with distinct mechanisms in transcribed and nontranscribed regions (11) that may account for the distribution of vector integrations we observed. In crustaceans and insects, rDNA can be a preferred site of transposon insertions (19, 35). Other hotspots correlated with known areas of genomic instability. Hotspots chr7HS1 and chr7HS2 both flank the region where deletions occur in Williams-Beuren syndrome (2), and hotspot chr3HS1 lies near the common fragile site FRA3B (17), an area where DNA gaps and breaks may form and a known integration site for plasmids and papillomavirus (36, 43). The study of AAV vector integration hotspots may lead to new insights into chromosome biology, as the integrated proviruses serve as tags for chromosomal damage at the nucleotide level. Large-scale integration surveys done in specific cell types or under different conditions may help us understand how chromosome structures change during differentiation or genotoxic stress.
Our findings help to define the spectrum of insertional mutagenesis associated with AAV vectors, with implications for their use in gene therapy. Overall, we observed a broad distribution of integrations throughout the human genome, with significant clustering in several hotspots. The effects of integrating in the rDNA repeat hotspot are not known, but given that these genes are already highly expressed and present in multiple copies, there should be minimal phenotypic effects. A major concern at other hotspots is the potential for activating oncogenes by introducing promoters and/or enhancers, and although wild-type AAV has never been shown to cause cancer, the transcriptional control elements are different in vectors. The hotspots we identified did not include known oncogenes, but given the broad distribution of integration sites, one must assume that a provirus could integrate near any gene.
The chromosomal changes associated with integration can be significant, as large deletions and even translocations were observed. Since DNA damage present at these sites presumably exposed free DNA ends prior to integration, a key remaining question is whether the same chromosomal effects would have occurred in the absence of AAV. For example, would repair of damaged chromosomes produce the same deletion sizes or translocations without vector integration? Our study also underscores the variability that occurs in proviral structures. All proviruses were deleted to some degree at their terminal repeats, and many sequence reads identified vector genomes joined to other vector or plasmid sequences that could affect transgene expression. Our results complement a recent report of AAV vector integration in murine hepatocytes with similar findings, albeit with a greater preference for transcribed genes (31). Further research will be required to determine what types of integration events might occur in clinical trials, including studies with cells from different tissues and preclinical animal models.
Supplementary Material
Acknowledgments
We thank Maynard Olson and the University of Washington Genome Center for helpful advice and sequencing and the Center for Expression Arrays for microarray analysis.
This work was supported by grants from the U.S. National Institutes of Health and the Child Health Research Center at Children's Hospital, Seattle, Wash.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Anonymous. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 2.Bayes, M., L. F. Magano, N. Rivera, R. Flores, and L. A. Perez Jurado. 2003. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum Genet. 73:131-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird, A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209-213. [DOI] [PubMed] [Google Scholar]
- 4.Bross, K., and W. Krone. 1972. On the number of ribosomal RNA genes in man. Humangenetik 14:137-141. [DOI] [PubMed] [Google Scholar]
- 5.Delgado, S., M. Gomez, A. Bird, and F. Antequera. 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 17:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, I. L., and J. E. Sylvester. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320-328. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez, I. L., and J. E. Sylvester. 2001. Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73:255-263. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 13.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 14.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 16.Hematti, P., B. K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, and B. Calmels. 2004. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huebner, K., and C. M. Croce. 2001. FRA3B and other common fragile sites: the weakest links. Nat Rev. Cancer 1:214-221. [DOI] [PubMed] [Google Scholar]
- 18.Inoue, N., and D. W. Russell. 1998. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J. Virol. 72:7024-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakubczak, J. L., W. D. Burke, and T. H. Eickbush. 1991. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl. Acad. Sci. USA 88:3295-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karolchik, D., R. Baertsch, M. Diekhans, T. S. Furey, A. Hinrichs, Y. T. Lu, K. M. Roskin, M. Schwartz, C. W. Sugnet, D. J. Thomas, R. J. Weber, D. Haussler, and W. J. Kent. 2003. The University of California-San Francisco Genome Browser Database. Nucleic Acids Res. 31:51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent, W. J. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 23.Larsen, F., G. Gundersen, R. Lopez, and H. Prydz. 1992. CpG islands as gene markers in the human genome. Genomics 13:1095-1107. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin, S. K., P. Collis, P. L. Hermonat, and N. Muzyczka. 1988. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 62:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767-773. [DOI] [PubMed] [Google Scholar]
- 26.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30:147-148. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 73:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai, H., E. Montini, S. Fuess, T. A. Storm, M. Grompe, and M. A. Kay. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297-302. [DOI] [PubMed] [Google Scholar]
- 30.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai, H., X. Wu, S. Fuess, T. A. Storm, D. Munroe, E. Montini, S. Burgess, M. Grompe, and M. A. Kay. 2005. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol. 79:3606-3614. [DOI] [PMC free article] [PubMed]
- 32.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill, J. P., and B. A. Finette. 1998. Transition mutations at CpG dinucleotides are the most frequent in vivo spontaneous single-based substitution mutation in the human HPRT gene. Environ. Mol. Mutagen. 32:188-191. [DOI] [PubMed] [Google Scholar]
- 35.Penton, E. H., B. W. Sullender, and T. J. Crease. 2002. Pokey, a new DNA transposon in Daphnia (Cladocera: Crustacea). J. Mol. Evol. 55:664-673. [DOI] [PubMed] [Google Scholar]
- 36.Rassool, F. V., T. W. McKeithan, M. E. Neilly, E. van Melle, R. Espinosa, 3rd, and M. M. Le Beau. 1991. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc. Natl. Acad. Sci. USA 88:6657-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, D. W., and R. K. Hirata. 1998. Human gene targeting by viral vectors. Nat. Genet. 18:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutledge, E. A., and D. W. Russell. 1997. Adeno-associated virus vector integration junctions. J. Virol. 71:8429-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samonte, R. V., and E. E. Eichler. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 40.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 41.Stilwell, J. L., and R. J. Samulski. 2004. Role of viral vectors and virion shells in cellular gene expression. Mol. Ther. 9:337-346. [DOI] [PubMed] [Google Scholar]
- 42.Tazi, J., and A. Bird. 1990. Alternative chromatin structure at CpG islands. Cell 60:909-920. [DOI] [PubMed] [Google Scholar]
- 43.Wilke, C. M., B. K. Hall, A. Hoge, W. Paradee, D. I. Smith, and T. W. Glover. 1996. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 5:187-195. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, Z. E., A. Rostami-Hodjegan, J. L. Burn, A. Tooley, J. Boyle, S. W. Ellis, and G. T. Tucker. 2003. Inter-individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. Br. J. Clin. Pharmacol. 56:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf, S. F., and B. R. Migeon. 1985. Clusters of CpG dinucleotides implicated by nuclease hypersensitivity as control elements of housekeeping genes. Nature 314:467-469. [DOI] [PubMed] [Google Scholar]
- 46.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 47.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.