Abstract
The mechanisms involved in the emergence of virulent mammalian viruses were investigated in the adult immunodeficient SCID mouse infected by the attenuated prototype strain of the parvovirus Minute Virus of Mice (MVMp). Cloned MVMp intravenously inoculated in mice consistently evolved during weeks of subclinical infection to variants showing altered plaque phenotypes. All the isolated large-plaque variants spread systemically from the oronasal cavity and replicated in major organs (brain, kidney, liver), in sharp contrast to the absolute inability of the MVMp and small-plaque variants to productively invade SCID organs by this natural route of infection. The virulent variants retained the MVMp capacity to infect mouse fibroblasts, consistent with the lack of genetic changes across the 220-to-335 amino acid sequence of VP2, a capsid domain containing main determinants of MVM tropism. However, the capsid of the virulent variants shared a lower affinity than the wild type for a primary receptor used in the cytotoxic infection. The capsid gene of a virulent variant engineered in the MVMp background endowed the recombinant virus with a large-plaque phenotype, lower affinity for the receptor, and productive invasiveness by the oronasal route in SCID mice, eventually leading to 100% mortality. In the analysis of virulence in mice, both MVMp and the recombinant virus similarly gained the bloodstream 1 to 2 days postoronasal inoculation and remained infectious when adsorbed to blood cells in vitro. However, the wild-type MVMp was cleared from circulation a few days afterwards, in contrast to the viremia of the recombinant virus, which was sustained for life. Significantly, attachment to an abundant receptor of primary mouse kidney epithelial cells by both viruses could be quantitatively competed by wild-type MVMp capsids, indicating that virulence is not due to an extended receptor usage in target tissues. We conclude that the selection of capsid-receptor interactions of low affinity, which favors systemic infection, is a major evolutionary process in the adaptation of parvoviruses to new hosts and in the cause of disease.
Understanding the molecular mechanisms underlying the emergence of pathogenic viruses is crucial for their control and prevention. Among the multiple factors producing emerging microbial threat (62), one major factor is inherent viral evolutionary capacity (reviewed in reference 23), which for many RNA virus is rapid due to high mutation rates and large population sizes (22, 25), whereas double-stranded DNA viruses replicating by polymerases with proofreading activities apparently evolve at a much lower rate (24, 40). However, accumulating experimental evidence supports the idea that single-stranded DNA viruses can also evolve rapidly, leading to heterogeneous populations demonstrable in natural hosts (28, 32, 35, 41, 48, 87). Evolution in viral populations is usually evidenced by sequence comparison among different spatial or temporal virus isolates sampled in nature, but the development of experimental models of virulence evolution, such as adaptation to their hosts, is critical for dealing with emerging viruses (26). Indeed, the evolutionary dynamics of virus populations, including important human pathogens (54, 88), have been monitored in experimental and often immunocompromised hosts, which may act as a reservoir of persistent viruses (44, 85).
More-detailed information on the molecular determinants of the multiple factors involved in virus host range and pathogenicity is obtainable for genetically simple viruses. In the Parvoviridae, a family of nonenveloped icosahedral viruses with a 5-kb single-stranded DNA genome organized into two overlapping transcription units (19, 59), host range, tropism, and fitness determinants were mapped in the early promoter (21) and in the NS1 and NS2 nonstructural proteins (30, 49, 69, 82), though most determinants have been mapped to the capsid gene encoding the VP1 and VP2 structural proteins. For example, a region of the capsid gene determined the host range of porcine parvovirus (PPV) (7, 82), and a few residues of the VP2 protein influenced in vivo replication of the Aleutian mink disease virus (8, 27) or conferred the tropism of the canine parvovirus (CPV) toward canine and feline cells (37). Interestingly, specific recognition, at the cell surface, of the transferrin receptor dictates the host range of the CPV and feline parvovirus (FPV) in nature (36, 63) and underlies the emergence of new isolates during the rapid evolution of these viruses (75, 81).
The two best-characterized strains of the parvovirus Minute Virus of Mice (MVM), which show different tropism and pathogenicity characteristics even though they share 97% homology in their nucleotide sequences (3), comprise an interesting simple viral model to precisely define molecular bases of parvovirus virulence. The immunosuppressive strain (MVMi) was isolated from the EL-4(G-) lymphoma (10), whereas the prototype strain, parvovirus MVM (MVMp), was isolated from an adenovirus stock (20), grown in mouse embryo culture, and plaque purified in mouse fibroblasts without forced adaptive passages (reviewed in reference 86). In vitro, MVMp productively infects mouse fibroblasts such as the A9 cell line (19), and MVMi infects mouse T-lymphoid cell lines (57) and primary hemopoietic precursors (71). The tropism and host range determinants of MVMi and MVMp localized mainly in a region of the genome contained within the capsid gene (1, 16, 30) that for the acquisition of fibrotropism was mapped to a 237-nucleotide (nt) sequence called the “allotropic determinant” (30), of which two amino acid (residues 317 and 321) played the major role (5), via the VP2 major capsid protein (56). In the MVM capsid structure (2), these two VP2 residues localize nearby some of the important amino acids determining parvovirus CPV and PPV host range (33, 37, 82). In the nonpermissive cells the infection of the incoming MVM virion is restricted prior to transcription and gene expression (1, 29), though both virus strains compete for specific binding sites (79) which are highly abundant in permissive mouse fibroblasts (45). Therefore, it was suggested that the MVM strains use a common cell surface receptor for attachment and that target cell specificity would be mediated by the subsequent interaction of the allotropic determinant with unknown intracellular host factors (79), which may alter nuclear decapsidation or complete genome release (65).
At the organism level, the virulence of the MVM strains was studied at three developmental stages of the mouse host. (i) In intranasal inoculations of the newborn, MVMi induced a runting syndrome at a low viral dose (43) and a lethal infection in some inbred strains at a high dose with renal papillary hemorrhage and replication in endothelia (13), neuroblasts (66), and hemopoietic precursors (73), whereas MVMp infection was asymptomatic and the virus replicated at a low titer in several organs, with the exception of the high titers found in intestine (12). Significantly, a recombinant MVMp virus carrying the MVMi allotropic determinant caused a lethal infection of the newborn and replicated in the same target cells as MVMi (12). (ii) In the developing embryo, the MVM strains infected a broad set of cell types that partly overlapped but in which the tropism of MVMp for fibroblasts and of MVMi for endothelium, and the higher pathogenicity of MVMi, were conserved (42). (iii) In adult mice with severe combined immunodeficiency (SCID), MVMi infection led to an acute lethal leukopenia (74) and the suppression of long-term repopulating hemopoietic stem cells (72). The SCID mouse as a model to gain insights into hallmarks of MVMi biology has been recently validated in the natural selection of pathogenic mutants resistant to neutralizing anticapsid antibodies (48) and of viruses with increased fitness by enhanced NS2-mediated sequestration of the cellular CRM1 nuclear export receptor (49).
In this study, the MVM evolutionary potential was exploited for the analysis of parvovirus virulence through attempting the adaptation of MVMp to adult SCID mice. In this model of experimental evolution, we show that the apathogenic MVMp consistently evolves, after weeks of subclinical infection, to virulent variants that have acquired the capacity to cause a systemic lethal disease by the natural oronasal route. The virulent variants remained fibrotropics, in agreement with the lack of genetic changes in the main capsid determinant of MVM tropism. The viral emergence was not due to the access to a different receptor. Instead, the capsid of the virulent variants exhibited a lowered avidity for a primary receptor used in the MVMp productive infection. The study provides evidence for the crucial role that the tightness of capsid-receptor recognition plays in parvovirus virulence at the organism level, a new concept dramatically exemplified by the MVMp adaptation to SCID mice, in which the virus evolves from being asymptomatic to inducing a lethal disease.
MATERIALS AND METHODS
Viruses.
The stocks of the prototype strain of the parvovirus Minute Virus of Mice (MVMp) (20) were prepared by transfecting the pMM984 molecular clone (58) in the human transformed NB324K cells, purified devoid of empty particles, and sterile filtered (0.22 μm) as previously described (52). The MVMp variants arising in mice were plaque isolated from NB324K cell monolayers inoculated with freshly homogenized organs, and stocks were prepared similarly from cell cultures inoculated at low multiplicity of infection (MOI) to minimize secondary mutations.
The recombinant WT-VP3B virus was constructed by exchanging the Hind III-XbaI (nt 2650 to 4342) restriction fragment of pMM984 encompassing 89% of the coding sequence of the VP2 protein (3) for the corresponding fragment of the 3B virus genome, gel purified from restriction enzyme-digested DNA replicative intermediates that had been isolated from infected NB324K cells (57). The chimeric plasmid was transfected and amplified in the Escherichia coli strain JC8111 (9), and viral stocks were prepared and purified from electroporated NB324K cells following described methods (69). Except for the particular comparative assay shown (see Fig. 2), the virus titers in all the experiments described in this report were determined by a PFU assay on NB324K monolayers (80). The specific infectivity of the viral stocks prepared for MVMp and the isolated variants was close to 1 PFU/500 viral particles as determined by hemagglutination and VP protein staining (see below).
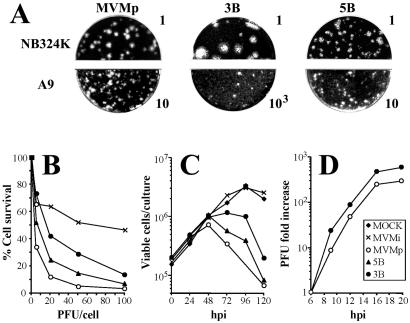
FIG. 2.
Features of the isolated MVMp variants in culture. (A) Plaque phenotype of MVMp and of the 3B and 5B virus variants in two fibroblast cell lines. Numbers are arbitrary units to designate the infectious titers (PFU) of the inocula. (B) Killing of mouse A9 cells. Monolayers of A9 cells were inoculated at 37°C with MVMp viruses at the indicated MOI normalized by a PFU assay in NB324K cells, and viable cells were determined as countable colonies 10 days afterwards. Represented is the average from five independent experiments. (C) Viral progression. Cultures of 3 × 105 A9 cells were inoculated at an MOI of 0.1, and the numbers of viable cells were counted by trypan blue exclusion as a function of time. The result is the average of two determinations. (D) Virus yield in a one-step growth assay. Cultures of highly synchronized A9 cells (19) were inoculated at an MOI of 20, and the total infectious virus levels were determined as PFU as a function of time. The figure shows the average of three independent experiments.
Mice.
The severe combined immunodeficient C.B-17 inbred strain of SCID mice (11), originally obtained from Jackson Laboratories (Bar Harbor, ME), was handled and bred in our animal facility under the conditions previously described (74). In brief, mice were allowed autoclaved food and water ad libitum and housed under sterile conditions in microisolators under a 9 a.m.-to-9 p.m. light-dark cycle. Female mice 6 to 8 weeks old were inoculated either intravenously in the tail vein with a single viral dose of 0.1 ml of phosphate-buffered saline (PBS) or oronasally with 0.01 ml of PBS. Mice were inspected for clinical signs twice per week, and the number of white blood cells was determined when indicated as described previously (74).
Monitoring viral multiplication in mice.
Animals were euthanized and the excised organs washed in PBS, containing 0.9 mM CaCl2 and 0.5 mM MgCl2, weighed, and quick frozen in dry ice. The organs were diluted in PBS to a 10% wt/vol ratio and rapidly homogenized (UltraTurrax T25; IKA-Labortechnik). To determine infectious virus in the organs, homogenized samples centrifuged to remove debris were serially diluted in PBS and inoculated onto monolayers of NB324K cells for plaque assays. Viral DNA synthesis in mice was determined by low-molecular-weight DNA extraction (57) and Southern blotting as described previously (74). When indicated, the plaques arising from the organs were probed to determine whether they were of MVM origin. For this purpose, nitrocellulose lifts were taken at 6 days postinfection (dpi) from the monolayers and hybridized to an MVM DNA probe at high stringency.
For the determination of MVM viremia, a method was developed for efficient isolation of virions associated with circulating peripheral blood cells. Cells from 10 to 50 μl of blood obtained from the tail vein were washed three times in 1.5 ml of PBS and lysed by digestion with 0.2% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS) for 15 min at 37°C, and low-molecular-weight products were removed by spun-column chromatography (2,000 × g, 2 min) in Sephadex G-50-80. This procedure allowed 50 to 80% recoveries in the excluded fraction of infectious MVMp bound for 30 min at 37°C to mouse erythrocytes in control tests in vitro, while the freeze-thaw extraction cycles commonly used for infected cell lines yielded about 1% of the bound virus. Viral genomes in the excluded fraction equivalent to 1 μl of blood were extracted by Hirt's procedure (57) and measured by a semiquantitative PCR of 25 cycles with the oligonucleotides VVP6 (nt 3444 to 3460) and VPSEQO (nt 4706 to 4688) in a Perkin-Elmer DNA Cetus (Gene Amp PCR system 9600) or Bio-Rad gene cycler as described previously (48). The amplified 1.25-kbp DNA fragment was resolved by 1% agarose gel electrophoresis.
Cell lines and primary kidney cultures.
The A9 mouse fibroblast cell line (a variant of mouse L cells) (46) and the NB324K simian virus 40-transformed human newborn kidney cell line (76), described as permissive hosts for the productive infection of MVM strains (80), were maintained with a minimal number of passages in Dulbecco's Modified Eagle Medium supplemented with 5% heat-inactivated fetal calf serum (Gibco BRL). When indicated, A9 cells were synchronized by isoleucine deprivation and aphidicolin treatment as described previously (19). To obtain primary epithelial cells from mouse kidneys, the method of culturing in d-valine as a selective agent (31, 50) was used. Kidneys were excised from 2- to 3-week-old SCID mice, the capsule was aseptically removed, and the organ was carefully minced and dispersed to cell suspensions by gentle treatment with a Dounce homogenizer and collagenase and seeded in dishes with MEM-D-Val selective medium (Gibco) supplemented with 15% fetal calf serum, and cultures were grown for 2 weeks before use.
The effect of one round of MVM infection on cell proliferative capacity was measured by a previously described clonogenic assay (17, 67) with minor modifications. Cell monolayers seeded at a density of 5,000 cells/cm2 the day before, or in suspension at a concentration of 105 cells/ml, were infected at increasing MOIs (0.2 to 100 PFU/cell) and plated at 4 h postinfection at various densities (2 × 102 to 2 × 104 cells per 60-mm-diameter dish) in triplicate to obtain a number of statistically representative cellular clones. Cells were incubated for 10 days in medium supplemented with a neutralizing dilution of an MVM capsid antiserum to block reinfections, and arising colonies were fixed in absolute methanol and stained with 1% crystal violet. Survival is expressed as the percentage of colonies in the infected versus the uninfected cultures normalized for the number of plated cells.
Binding assays.
Cells detached from the plates by controlled trypsin digestion (A9) or EDTA treatment (primary epithelial kidney cells) were incubated for 30 min at 37°C in medium with serum to restore surface properties, washed with PBS three times, and incubated with [35S]methionine-cysteine radiolabeled MVM particles purified as described previously (52, 53). Typical binding assays were performed with 1 × 106 to 2 × 106/ml cells incubated with 15,000 cpm of 35S-labeled capsid in 100 μl of PBS at 4°C to minimize virus uptake into cells for the time indicated in the figure legends. Upon binding, cells were extensively washed in PBS to remove unbound capsids and centrifuged and the radioactivity associated with the cell pellets was determined by liquid scintillation counting. In the study of cytotoxic interactions of infectious particles to cells, 105/ml A9 cells in 150 μl of PBS were incubated with purified viruses (MOI of 100) at 4°C for the indicated time periods and cell survival was determined by a clonogenic assay as above.
The specificity of the attachment of labeled capsids and infectious virions to cells was probed by competing with graded amounts of unlabeled empty capsids. The specific activity of labeled capsids (1 × 105 to 3 × 105 cpm/μg) and the stoichiometry in the competitions were carefully adjusted for each experiment by measuring the number of particles by VP protein staining with Coomassie blue in 10% SDS-polyacrylamide gel electrophoresis, using bovine serum albumin protein (Sigma) as the standard, and values were further validated in standard hemagglutination with mouse erythrocytes. Wild-type (wt) and MVMp variant capsids showed similar levels of hemagglutination activity (2 h at 4°C), with titers in the range of 2 × 103 to 6 × 103 hemagglutination units/μg of VP protein.
RESULTS
A heterogeneous MVMp plaque phenotype develops in SCID mice weeks postinfection.
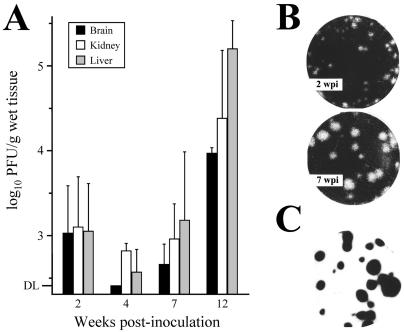
To explore the capacity of a DNA virus to gain virulence in a single host and the mechanisms involved, the apathogenic prototype strain of the parvovirus Minute Virus of Mice (MVMp) was intravenously inoculated in adult SCID mice and viral features were monitored for up to 12 weeks postinfection (wpi). The titers of infectious virus in three major organs (brain, kidney, and liver) of mice inoculated with a viral dose of 107 PFU remained at low levels in all the organs at 2 wpi and dropped to levels close to the detection limit of the plaque assay by the fourth wpi (Fig. 1A). Remarkably, by 7 wpi the infectious titers in the organs began to rise, and at 12 wpi titers had increased to 104 to 105 PFU/g. Similar results were obtained with inoculations at doses of 106 or 108 PFU/mouse (n = 3 per dose), though the time period prior to the titer increase varied accordingly (not shown). MVMp multiplication in mouse organs up to 12 wpi (n = 9) was not associated with any overt symptom, such as the characteristic pathological signs and leukopenic syndrome that develop by 6 wpi in SCID mice infected with the hematotropic MVMi strain (49, 74). This is in agreement with the inability of MVMp to infect primary bone marrow precursors in vitro (71). Interestingly, the rise in infectious titers in the organs in the weeks postinfection correlated with the emergence of an increasing proportion of viruses forming plaques of larger size (Fig. 1B), a consistent phenomenon observed in all of the inoculated animals. Viral plaques, regardless of size, corresponded to MVM as judged by the hybridization under stringent conditions to a radioactive probe of the MVM genome (Fig. 1C).
FIG. 1.
Emergence of MVMp variants in SCID mice. (A) Infectious titers in three major organs of SCID mice inoculated intravenously with MVMp (107 PFU/mouse), as determined by plaque assay in the NB324K cell line. The mean values and standard deviation from at least three mice per point from two independent inoculations are shown. DL, detection limit of the assay (250 PFU/g of wet tissue). (B) Examples of the heterogeneous plaque size distribution of MVMp variants harvested from mouse organs at two postinfection times. (C) Plaques of all sizes hybridize to a 32P-labeled DNA probe of MVM genome after overnight exposure.
In vivo-isolated MVMp variants show altered plaque-forming capacity in fibroblasts.
To gain insights into the phenotypic features of the viruses growing in MVMp-infected mice, a collection of plaque-isolated virus clones randomly sampled at 12 wpi from different organs of several animals was established for further analysis (Table 1). Features of the infection of these virus clones were first characterized by plaque assay in two reference cell lines (A9 and NB324K) in which the MVM strains showed distinct plaque-forming capacity (5, 80). Most isolated clones formed plaques larger than the wt in human NB324K cells but showed a poor plaque-forming capacity in A9 mouse fibroblasts. Indeed, as shown in Table 1, a tight correspondence between plaque size in NB324K and the ratio of NB324K/A9 plaque titer was noticed in the collection of viruses tested. This is exemplified in Fig. 2A for two clones isolated from the brains of mouse 3 (variant 3B) and mouse 5 (variant 5B). The 5B variant showed a plaque-forming capacity comparable to that of the wt in both cell types, whereas the 3B variant formed larger plaques in NB324K cells and its plaque-forming capacity in A9 cells was about 50 times lower than that of the wt. Interestingly, the 3B and the other large-plaque variants damaged the A9 monolayers reducing cell density when inoculated at high MOI (not shown).
TABLE 1.
Plaque phenotype of MVMp variants isolated from SCID mice
| MVMp varianta | NB324K/A9 Plaque titer ratio
|
NB324K plaque size (mm)b | |
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| 1B | 54 | 128 | 1.90 ± 0.66 |
| 2L | 94 | 138 | 1.93 ± 0.83 |
| 3B | 740 | 1209 | 2.51 ± 0.85 |
| 3L | NDc | 551 | 2.01 ± 0.93 |
| 4B | 1560 | 468 | 1.94 ± 0.79 |
| 4K | ND | 1132 | 3.02 ± 0.92 |
| 4L | 1163 | 1475 | 2.63 ± 0.99 |
| 5B | ND | 27 | 1.15 ± 0.30 |
| MVMp | 20 | 19 | 1.23 ± 0.50 |
Numbers indicate different mice. B, brain; K, kidney; L, liver.
Values are the mean and standard error (n = 30).
ND, not determined.
To study the bases of the poor plaque formation by most MVMp variants in A9 cells, several parameters of the interaction with this cell type of the 3B and 5B representative variants were analyzed. The capacity of these two variants to suppress A9 viability as measured by a clonogenic assay was important though lower than that of the wt (Fig. 2B) and thus significantly higher than the value obtained for the infection with the MVMi strain, which had been previously described to be restricted in this cell type (80). Correspondingly, both variants progressed in A9 cultures inoculated at low MOI, as judged by the decay of viable cells, in contrast with the continuous growth of mock- and MVMi-inoculated cultures (Fig. 2C). Moreover, the yield of 3B infectious virus in a one-step multiplication curve was comparable to that of the wt (Fig. 2D). Thus, the cytotoxicity and virus production tests manifested a nonrestricted multiplication of the 3B variant in A9 fibroblasts, in spite of its poor plaque-forming capacity in this cell type.
Virulent MVMp variants emerge in SCID mice.
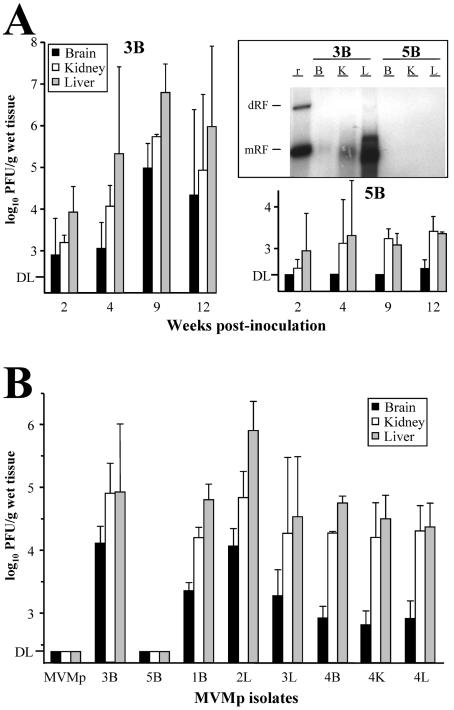
To investigate whether the different plaque phenotypes reflect a pertinent biological property, the capacity of the 3B and 5B variants to spread and multiply in three major organs was monitored following reinoculation into SCID mice. The infectious titer of the 5B variant inoculated intravenously (Fig. 3A, right panel) remained at low levels (kidney and liver) or below the detectable level (brain) in these mouse organs up to 12 wpi. In sharp contrast, the presence of the 3B variant was demonstrable at moderate infectious titer in all three organs by 2 wpi, and the titer increased by several orders of magnitude within a few weeks (Fig. 3A, left panel). Consistent with this observation, the accumulation of viral DNA replicative intermediates analyzed at 9 wpi was high in the liver of 3B inoculated mice, moderate in the kidney, and low in the brain, whereas 5B replicative forms were undetectable by conventional Southern blotting in any of the organs (Fig. 3A, middle panel). These analyses probed the capacity of the 3B virus to multiply in mouse organs.
FIG. 3.
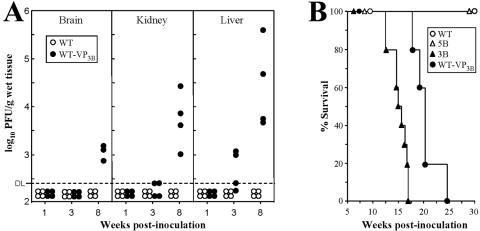
Virulence of isolated MVMp variants in SCID mice. (A) Multiplication of two MVMp variants reinoculated in SCID mice. The figure shows the infectious titers in the organs of SCID mice at the indicated times postintravenous inoculation (107 PFU/mouse) with the 3B and 5B variants. Upper right panel: Southern blot of low-molecular-weight DNA extracted from mouse organs, by the Hirt procedure, at 9 wpi and hybridized with a 32P-labeled MVM probe. DNA extracted from 5 mg of organ was loaded per gel slot, and membranes were exposed for 40 h for autoradiography. B, brain; K, kidney; L, liver; r, replicative intermediate markers. The positions of the dimer replicative form (dRF) and monomeric replicative form (mRF) are indicated. (B) Infectious titers in major organs of SCID mice oronasally inoculated (107 PFU/mouse) with the indicated purified viruses. The data for the wt and 5B viruses were obtained at 12 wpi and for the rest of variants at 8 wpi. Values are the mean and standard deviation for at least 3 mice per point from two independent inoculations. DL, detection limit of the assay.
The virulence of the MVMp variants showing characteristic plaque phenotypes (see Table 1) was analyzed by the natural oronasal route of MVM infection. The wt and the 5B small-plaque variant were undetectable in any of the mouse organs up to 12 wpi (Fig. 3B). However, variant 3B, as well as all the other large-plaque variants, spread from the oronasal route and reached variable, but high, infectious titers in the three organs by 8 wpi. As before, these infectious titers correlated with the synthesis of viral DNA replicative intermediates demonstrable in blot analysis (not shown). These results demonstrate the emergence in MVMp-infected SCID mice of virulent variants with the capacity to infect mice by the oronasal route and replicate in major organs distant from the inoculation site.
The capsid of virulent variants binds with lower affinity than MVMp to the receptor used for the cytotoxic infection.
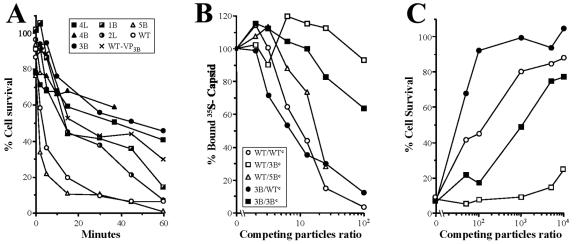
In the absence of molecular data accounting for virulence and plaque formation, the slightly different slopes of A9 killing (Fig. 2B and C) suggested that the initial virus-cell interaction might be altered in the isolated MVMp variants. We investigated this possibility by analyzing features of the virus-cell interaction at 4°C to avoid postbinding effects. As the expression of the cytotoxic NS1 nonstructural protein (15, 69) is a reliable test for the onset of MVM productive infection, the kinetics of cell interaction leading to the cytotoxic infection of representative MVMp variants was examined first. As shown in Fig. 4A, the survival curves of A9 cells indicated a rapid binding of the MVMp and 5B variant, as most infectious virus was bound by 15 min. In contrast, the 3B and the other large-plaque variants tested (4L, 4B, 2L, and 1B) interacted, without exception, with a much lower kinetics, as only about 50% of the cells were killed after 30 min of interaction. These characteristic cytotoxic interactions indicated a distinctly lower avidity of the MVMp variants for A9 cells.
FIG. 4.
Capsid affinity of MVMp variants to a primary fibroblast receptor. (A) Kinetics of cytotoxic interaction of MVMp variants to A9 cells. Purified viruses (MOI of 100) were allowed to bind at 4°C to A9 cells in suspension for the indicated time periods, and levels of surviving cells were determined by clonogenic assays. Representative results from at least two independent experiments are shown as percentage of survival with respect to the plating efficiency of the mock-infected cells. (B) MVMp and virus variants compete for A9 binding. 35S-labeled MVMp (blank symbols) or 3B (filled symbols) capsids were allowed to bind for 1 h at 4°C in the presence of competitor unlabeled MVMp (circles), 3B (squares), or 5B (triangles) capsids. Represented is the percentage of 35S radioactivity bound to cells (100% binding was approximately 3,000 cpm). (C) The 3B virulent variant shares receptor with MVMp for A9 infection. MVMp (blank symbols) or 3B (filled symbols) infectious viruses binding to A9 cells (MOI 100) for 1 h at 4°C were incubated with the indicated ratios of purified competitor MVMp (circles) or 3B (squares) capsids, and levels of surviving cells were determined by a clonogenic assay. The result is the average from three independent determinations.
To explore whether the MVMp variants share the receptor with the wt for binding to A9 susceptible fibroblasts, competition experiments between viral particles were attempted. The binding of 35S-labeled wt capsid was efficiently competed by increasing amounts of the homologous nonlabeled capsid (Fig. 4B). The 5B capsid also competed the binding efficiently when added at ratios higher than 12, in sharp contrast to the lack of competition capacity of the 3B capsid even at the highest assayed ratio (100-fold). In the reciprocal test, however, the binding of 35S-labeled 3B capsid to A9 cells was competed by the wt capsid much more efficiently than by the homologous 3B capsid (Fig. 4B). These competition rates supported the notion that the MVMp viruses share a receptor for the initial binding to A9 cells, though the 3B interaction with this receptor occurs with a significantly lower affinity.
To determine whether the different binding affinities are to a nonproductive attachment factor or involve the receptor used in the MVMp infection, the cytotoxic interaction of wt and 3B viruses was competed with increasing amounts of the respective homologous and heterologous empty capsids. According to the number of surviving viable cells shown in Fig. 4C, the infection of the 3B variant was completely competed by the wt capsid by use of a particle ratio of 102-fold excess, but a similar level of competition of the MVMp infection by the homologous capsid required at least a 103-fold excess. Instead, the 3B capsid significantly competed the infection of the homologous 3B virus only at very high particle ratios, close to 104-fold excess, and moreover its capacity to compete out the MVMp infection was poor at even the highest particle ratio. These data, which are consistent with the binding affinities shown in Fig. 4B, led us to conclude that MVMp and the 3B virulent variant use the same primary receptor to initiate the cytotoxic infection of A9 fibroblasts but that the interaction of 3B with this receptor occurs with lower affinity.
The VP gene determines MVMp plaque morphology, affinity of capsid-receptor interaction, and virulence in SCID mice.
To study the genetic relationship between capsid affinity and virulence in mice, a chimeric genome was constructed, replacing most coding sequence of the VP gene in the MVMp wt genome with the equivalent region of the 3B genome (see Materials and Methods), and the phenotypic properties of the resulting recombinant virus (named WT-VP3B) were studied in vitro and in vivo. The plaque-forming capacity of the recombinant virus in A9 and NB324K cells was similar to that of the parental 3B variant, namely, large plaques in NB324K (2.6 ± 0.6 mm) and around a 1,000-fold higher NB324K/A9 ratio (not shown). As found for large-plaque variants, the plaque phenotype of the WT-VP3B recombinant virus correlated with slower kinetics in the cytotoxic interaction with A9 cells (Fig. 4A). These experiments suggested that the affinity of the capsid for the receptor is a major factor in MVMp plaque phenotype.
The virulence of the MVMp-VP3B virus in comparison with the wt was determined by the oronasal route in SCID mice. Consistent with the experiments shown in Fig. 3B, the titer of wt infectious virus in the major organs remained below detectable levels up to 8 wpi (Fig. 5A). In contrast, infectious WT-VP3B virus was recovered from the liver of some animals as early as 3 wpi and from the three organs in all the animals by 8 wpi (Fig. 5A) at titers slightly lower than, but paralleling, the results obtained for the parental 3B variant. As described above for MVMp, the WT-VP3B infection was not associated with the leukopenic syndrome induced by the MVMi strain since 6 wpi in SCID mice (74). These results indicate that the 3B capsid facilitates systemic infection from the oronasal route.
FIG. 5.
The MVMp-derived recombinant WT-VP3B virus is virulent when inoculated by the oronasal route. (A) Virus titers in organs of adult SCID mice after intranasal inoculation with purified recombinant WT-VP3B (107 PFU/mouse) or the wt strain (108 PFU/mouse). Each point corresponds to the infectious titer found in the organ of one individual mouse (n = 12 per virus). DL, detection limit of the assay. (B) Lethality of recombinant and variant MVMp viruses in SCID mice. Adult SCID mice were inoculated by the oronasal route with 107 PFU/mouse of purified WT-VP3B (n = 5), 3B (n = 10), or 5B (n = 10) viruses or with 108 PFU/mouse of the wt MVMp (n = 6) and scored for survival for 30 weeks.
In a further long-range study, the virulence of several MVMp viruses was monitored as a function of SCID mouse survival (Fig. 5B). Remarkably, intranasal infection with the recombinant WT-VP3B and the 3B variant that forms large plaques eventually led to overt pathological signs (hunched posture and ruffled fur) and to a lethal outcome in all of the inoculated animals by 25 wpi. The pathological basis of this lethality remains unstudied. Note that the course of disease was slightly delayed for the recombinant versus the 3B parental virus but significantly delayed with respect to the 8 wpi lethality observed for infections with the MVMi strain (49, 74). However, SCID mice inoculated with the small-plaque 5B variant, as well as with a higher dose of the wt, survived without clinical symptoms for months. This study assigns the capsid gene the role as the main determinant of MVMp virulence by the oronasal route.
In an attempt to localize the amino acid residues involved in MVMp virulence in SCID, the nucleotide sequence of a region of MVM genome (nt 3450 to 3800), corresponding to the amino acid residues 220 to 335 of VP2 (3), was determined in the entire collection of isolated MVMp variants (Fig. 3B and Table 1). This region encompasses the allotropic determinant governing the tropism of MVM strains (1, 5, 16, 30), as well as other main VP residues controlling CPV or PPV host range (7, 33, 37, 82) and adaptation in culture (4). DNA isolated from the respective viral stocks by a modified Hirt's procedure (57) was amplified by PCR and sequenced following a previously described methodology (48). None of the virus isolates, regardless of phenotypic features and virulence, showed any nucleotide change with respect to the updated reported sequence of the wild-type MVMp genome (reference 5 and data not shown). Thus, we had to conclude that the genetic basis of MVMp virulence in SCID is not directly related to the tropism determinants of MVM and other evolutionary close parvoviruses.
Analysis of MVMp attenuation in SCID mice: viremia and interaction with primary cells.
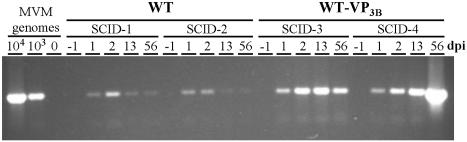
To study, at the organism level, the basis of the virulence gained by the WT-VP3B virus with respect to the attenuated wt, the capacities of both viruses to cause viremia during natural infection were compared. Blood samples were collected from the tail vein at several dpi from two oronasally inoculated adult SCID mice per virus (108 PFU/mouse) and monitored for MVM viremia. As previously reported (34), MVM viruses were found associated with blood cells and not in the plasma fraction, and they could be efficiently recovered by an SDS-based extraction method and the number of genomes was estimated by semiquantitative PCR (see Materials and Methods). Blood samples collected from mice prior to virus inoculations were negative for MVM sequences (Fig. 6), but DNA of the WT-VP3B, as well as that of the MVMp viruses, was consistently demonstrated at similar genomic copy numbers (lower than 103/μl) in the cellular fraction of the blood at 1 to 2 dpi. However, a major difference between the two viruses was evident from their rate of clearance from the circulation, as the level of MVMp genomes in blood had dropped by 13 dpi and became barely detectable by 56 dpi in the two analyzed mice (SCID-1 and SCID-2), whereas the number of WT-VP3B genomes increased at 13 dpi and either declined (SCID-3) or increased to a titer higher than 104/μl (SCID-4) at 56 dpi. Parallel studies in vitro suggested that MVMp clearance was not due to inactivation or irreversible binding to mouse erythrocytes, as a similar proportion of wt and recombinant infectious virus, bound to erythrocytes, could be harvested by the SDS treatment mentioned above; moreover, 1 to 5% of 104 PFU of either viruses, bound to 107 erythrocytes, spontaneously released and formed plaques on NB324K cell monolayers (2.5 × 105 cells) after a 1-h incubation (not shown). These studies showed that both viruses invade the bloodstream by the oronasal route, where they remained presumably infectious, but the attenuated MVMp was cleared from circulation within a few dpi, whereas the accumulation of the WT-VP3B virus continued over many weeks, corresponding to its multiplication during this time in major organs of the host mouse (Fig. 5A).
FIG. 6.
Viremia and clearance of the MVMp and WT-VP3B viruses in SCID mice. Genomic copy numbers of the MVMp and WT-VP3B viruses in blood cells obtained from two oronasally inoculated SCID mice (108 PFU/mouse) at the indicated dpi. Low-molecular-weight DNA isolated from an equivalent of 1 μl of blood cells was subjected to semiquantitative PCR amplification of a 1.25-kbp region of MVM genome and loaded in 1% agarose gel. PCR amplification of measured numbers of viral particles (determined by hemagglutination) from the purified viral stocks used to inoculate the mice is shown in the left lanes.
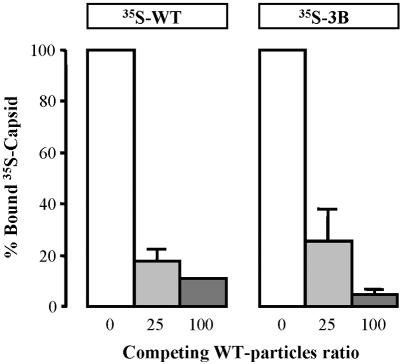
The inability of circulating MVMp to replicate in mouse organs and cause disease prompted us to investigate the interaction of viruses exhibiting different levels of virulence toward primary cells of the kidney, a main target organ for all the isolated virulent variants (Fig. 3). For this purpose, the binding of MVMp and 3B capsids to SCID kidney epithelial cells cultured with selective medium (see Materials and Methods) was analyzed. Epithelial cultures inoculated with the MVMp and 3B viruses (MOI of 5) showed some morphological cytopathic effect by 4 dpi (not shown), denoting permissiveness to viral infection. 35S-labeled MVMp and 3B capsids attached to primary cells at similar percentages (5 to 8%) of different inputs of particles; moreover, this binding was specific, as it could be efficiently competed by an excess of nonlabeled MVMp capsids (Fig. 7). Significantly, binding of the 3B capsid was almost completely inhibited at a 100-fold excess of competitor, a ratio also inhibiting the binding to fully permissive A9 fibroblasts (Fig. 4B). This analysis indicated that the receptor usage of the virulent variant 3B to initiate the infection of primary mouse cells is either shared or included within the receptor usage of MVMp. Attempts to determine the affinity of the interactions and dissociation constants by Scatchard analysis gave inconsistent values between preparations of the primary cultures. In spite of their heterogeneity, the preliminary data suggest a saturable binding of both types of capsid to a receptor represented in the range of 4 × 105 to 8 × 105 binding sites per cell (not shown), as previously reported for established cell lines susceptible to MVM infection (45, 79).
FIG. 7.
Competitive binding of MVMp and 3B capsids to primary cells. The figure shows the specific interaction of 35S-labeled MVMp (wt) and 3B purified capsids to primary epithelial cells isolated from SCID kidney. Values are represented as the percentage of cell-bound radioactivity not competed off by the indicated excess of nonlabeled wt capsid (100% is about 2,000 cpm). The result is (with one exception) the average of three independent determinations, and standard errors are shown.
DISCUSSION
This study explored mechanisms of emergence of virulent DNA viruses in an immunodeficient mammalian host. Adult SCID mice were inoculated with MVMp and monitored for virus evolution and disease. Regarding the components of this experimental model, it is worth mentioning that the MVMp is a fibrotropic strain of laboratory and wild mice that might have been attenuated in culture over time, although it was never subjected to adaptive passages at high MOI (86). Likewise, for the SCID mouse, although it is certainly a limited model for immune competent hosts, the immunodeficiency stage may constitute a valuable scenario for the common parvoviral infections of immunocompromised and newborn hosts (59). Moreover, the SCID mouse has allowed us to isolate virus mutants with important biological features under different selective pressures (48, 49), which may develop in natural persistent infections and long-run adaptive processes. Here, the adaptation of MVMp to SCID led to the emergence of virus variants that were studied further to obtain insights into parvoviral virulence
Emergence of MVMp variants in SCID mice.
The course of MVMp multiplication in intravenously inoculated SCID mice developed with a characteristic rise in infectious titers in the mouse organs since 7 wpi (Fig. 1) and the absence of pathological signs for several weeks postinfection. This pattern is in sharp contrast to the severe leukopenic syndrome found in infections with the virulent MVMi strain by 6 wpi (74), consistent with the lack of MVMp tropism toward lymphohemopoietic cells (71). Unlike MVMi, the emerging variants conserved fibrotropism (Fig. 2) and did not show genetic changes in the allotropic determinant of the capsid (see Results) that contains 10% of the single-nucleotide differences between MVMi and MVMp (3, 5), suggesting that the acquisition of virulence is not due to gaining tropism for hemopoietic cells. Experiments are currently under way to identify the primary cells targeted by MVMp in adult SCID mice.
These virus variants arising in the organs showed, however, an altered plaque phenotype with respect to MVMp, as they formed large plaques in human cells but failed to efficiently plaque in A9 mouse fibroblasts (Table 1). The MVM plaque phenotype may be altered by several determinants of the VP (5, 53) and NS (49, 69) genes, acting at distinct life cycle steps. Here, the virus cycle of large-plaque variants did not show however any major different with respect to the MVMp other than a lower avidity in the primary interaction leading to A9 cell killing (Fig. 2). Furthermore, this interaction could be completely competed by wt capsids (Fig. 4), indicating that the receptor usage by the emerging MVM variants is within the receptor repertoire used by MVMp for A9 infection. However, whether the higher affinity of the MVMp capsid to cells is due to the access to additional binding sites that are not competed for by virulent capsids remains unexplored. Thus, it is likely that the lower avidity of the variant capsids for the A9 cell surface, which also occurs in NB324K cells (to be described elsewhere), facilitates their spreading in monolayers. This may account for the larger plaques in NB324K cells, whereas a less extensive A9 cell lysis (Fig. 2B) would result in diffused plaques not evident by eye inspection.
The adaptation of MVMp to SCID mice was consistently associated with gain of virulence. The emergence of variants showing a dramatic increase in virulence in reinoculated SCID mice indicated that MVMp genetically changed in the mice (Fig. 3). Given the altered binding to cells of the variant capsids, the analysis of virulence was focused in the capsid gene, a region of parvovirus genome where otherwise most determinants of host range and tropism have been mapped (see the introduction). The phenotypic analysis of the WT-VP3B recombinant virus, carrying the VP gene of the 3B virulent variant in the MVMp background, demonstrated that the large-plaque morphology, low affinity of receptor interaction, and intranasal virulence are indeed determined by the capsid gene (Fig. 5). The lack of genetic changes in the VP allotropic determinant sequenced in eight virus isolates (Table 1), together with the high genetic heterogeneity of the MVMi populations replicating in SCID mice (48, 49), indicate that a comprehensive sequence analysis of a large collection of MVMp variants will be required to understand the genetic basis of virulence.
Capsid affinity for the receptor in parvovirus pathogenesis.
The basis of the strikingly different pathogenicity of MVMp versus the isolated virulent variants harboring capsids with low receptor affinity was analyzed at several levels. Mice oronasally inoculated by either MVMp or virulent variants become viremic with similar viral genome copy numbers by 1 to 2 dpi (Fig. 6), indicating that MVMp is not trapped during passage through the respiratory or gastrointestinal route of entry. Moreover, the virus remained infectious and bound to blood cells in vitro and therefore could access target tissues by hematogeneous spreading. In the interaction with primary epithelial kidney cells (Fig. 7), both viruses bound similarly and could be competed with wt capsids, suggesting that, as probed for the A9 mouse fibroblast cell line (Fig. 4B and C), the virulent variants do not access alternative receptors in this target organ. Thus, unlike most viral systems analyzed to date, the virulence in the MVMp-SCID model may not rely on extended receptor usage.
A comprehensive understanding of the connection between the affinity of capsid receptor interaction and MVMp pathogenesis deserves further research. In mouse infections by polyomavirus strains with different levels of pathogenicity (6), a weak interaction with the receptor appeared to facilitate spreading to distal host-susceptible organs, while high-affinity binding retained the nonpathogenic strains within nearby cells, limiting systemic infection. Analogously, in the MVMp-SCID model addressed here, in which virulent variants naturally emerged in an adult mouse host, this hypothesis would imply that the high-affinity binding of the MVMp apathogenic strain to cell sites not competed by the virulent capsids involves nonproductive receptors. Our study indicates, however, that the MVMp reached the bloodstream, where it remained infectious and bound to circulating cells (see above), confining this hypothetical restriction to postviremia interactions.
As an alternative hypothesis, the affinity of recognition of a common receptor by MVMp and the virulent variants in cells of target mouse tissues may be crucial for the initiation of a productive MVM infection. Viruses enter animal cells through complex entry and uncoating programs (78), and, as exemplified for the fusion step in influenza virus (60), the affinity of binding to the receptor may modulate multiple events required for infection. The entry of the genetically simpler members of the Parvoviridae is also a complex and only partly understood process (reviewed in reference 83) involving structural transitions of a metastable capsid to specifically externalize the VP N-terminal sequences at the virion surface (18, 47, 53, 68, 84). It is therefore conceivable that the number and tightness of parvovirus-receptor binding sites affect the capsid configuration at precise subcellular compartments, modulating overall the efficiency of nuclear invasion by the virus genome. In this sense, it may be worth evaluating the concept of differential affinity of the capsid for cell-surface receptors in other parvovirus infections, particularly in the hypothetical involvement of postattachment host factors determining MVMp/MVMi target cell specificity (29, 65, 79) and cell permissiveness to PPV infections (61), and in a pathogenic role of the recognition by CPV/feline parvovirus host range variants of the transferrin receptors (36, 38).
In a number of viral systems, alterations in the recognition of sugars as attachment receptors have been shown to modulate infection and pathogenicity (see, e.g., references 6, 14, 39, 51, 55, 64, 70, and 77). Interestingly, the sensitivity of the infection to enzymes suggested that the sialic part of a surface glycoprotein acts as an MVM receptor in permissive cells (19), and thus the interaction of the viral capsid with this receptor component could modulate the pathways of MVMp infection as hypothesized above. Experiments are under way in this laboratory to address this and other complex issues of MVMp pathogenesis, such as the identification of the host cells in mouse tissues targeted during MVMp replication and evolution, the nature of the primary receptor, and the genetic basis of the virulence.
In summary, this report demonstrates that a change of capsid affinity for the receptor is a major mechanism in the natural selection of virulent viruses in a single immunodeficient mammalian host. Two further conclusions, with potential general interest, could be drawn from this study. (i) Antiviral therapies targeting virus-receptor interactions must be undertaken cautiously, as they may favor severe systemic spreading of otherwise local infections or the selection of virus escape mutants with increased pathogenicity. (ii) The development of gene therapy protocols in which a hematogeneous systemic treatment with the viral vectors is desired might more usefully employ viral coats of low receptor affinity. The SCID mouse may constitute a suitable mammalian model for this kind of trial, since it allows the isolation of virus mutants useful in characterizing long-term pathogenic factors (references 48 and 49 and references therein).
Acknowledgments
We are indebted to Peter Tattersall for providing the pMM984 infectious plasmid clone and thoughtful comments.
This work was supported by grant SAF 2001-1325 CICYT from the Spanish Ministry of Science, by EU-Contract QLK3-CT-2001-01010, by grant 07B/0020/2002 from the Comunidad de Madrid, and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.”
REFERENCES
- 1.Antonietti, J.-P., R. Sahli, P. Beard, and B. Hirt. 1988. Characterization of the cell type-specific determinant in the genome of minute virus of mice. J. Virol. 62:552-557.W [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Astell, C. R., E. M. Gardiner, and P. Tattersall. 1986. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J. Virol. 57:656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badgett, M. R., A. Auer, L. E. Carmichael, C. R. Parrish, and J. J. Bull. 2002. Evolutionary dynamics of viral attenuation. J. Virol. 76:10524-10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball-Goodrich, L. J., and P. Tattersall. 1992. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J. Virol. 66:3415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, P. H., C. Cui, T. Stehle, S. C. Harrison, J. A. DeCaprio, and T. L. Benjamin. 1999. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 73:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergeron, J., B. Hébert, and P. Tijssen. 1996. Genome organization of the kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 70:2508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom, M. E., D. B. Bradley, W. Wei, S. Perryman, and J. B. Wolfinbarger. 1993. Characterization of chimeric full-length molecular clones of aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J. Virol. 67:5976-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissy, R., and C. R. Astell. 1985. An Escherichia coli recBC sbc BrecF host permits the deletion-resistant propagation of plasmid clones containing the 5′-terminal palindrome of minute virus of mice. Gene 35:179-185. [DOI] [PubMed] [Google Scholar]
- 10.Bonnard, G. D., E. K. Manders, D. A. Campbell, R. B. Herberman, and M. J. Collins. 1976. Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. J. Exp. Med. 143:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301:527-530. [DOI] [PubMed] [Google Scholar]
- 12.Brownstein, D. G., A. L. Smith, E. A. Johnson, D. J. Pintel, L. K. Naeger, and P. Tattersall. 1992. The pathogenesis of infection with minute virus of mice depends on expression of the small nonstructural protein NS2 and on the genotype of the allotropic determinants VP1 and VP2. J. Virol. 66:3118-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brownstein, D. G., A. L. Smith, R. O. Jacoby, E. A. Johnson, G. Hansen, and P. Tattersall. 1991. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype. Lab. Investig. 65:357-363. [PubMed] [Google Scholar]
- 14.Byrnes, A., and D. E. Griffin. 2000. Large-plaque mutants of sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J. Virol. 74:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colomar, M. C., B. Hirt, and P. Beard. 1998. Two segments in the genome of the immunosuppressive minute virus of mice determine the host-cell specificity, control viral DNA replication and affect viral RNA metabolism. J. Gen. Virol. 79:581-586. [DOI] [PubMed] [Google Scholar]
- 17.Cornelis, J. J., N. Spruyt, P. Spegelaere, E. Guetta, T. Darawshi, S. F. Cotmore, J. Tal, and J. Rommelaere. 1988. Sensitization of transformed rat fibroblasts to killing by parvovirus minute virus of mice correlates with an increase in viral gene expression. J. Virol. 62:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., A. M. D'Abramo, C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virions expose the VP1 N terminus and viral genome without particle disassembly. Virology 254:169-181. [DOI] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 20.Crawford, L. V. 1966. A minute virus of mice. Virology 29:605-612. [DOI] [PubMed] [Google Scholar]
- 21.Davis, C., N. Segev-Amzaleg, I. Rotem, M. Mincberg, N. Amir, S. Sivan, I. Gitelman, and J. Tal. 2003. The P4 promoter of the parvovirus minute virus of mice is developmentally regulated in transgenic P4-LacZ mice. Virology 306:268-279. [DOI] [PubMed] [Google Scholar]
- 22.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 23.Domingo, E., R. Webster, and J. Holland (ed.). 1999. Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 24.Drake, J. W. 1991. A constant rate of spontaneous mutations in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert, D. 1998. Experimental evolution of parasites. Science 282:1432-1435. [DOI] [PubMed] [Google Scholar]
- 27.Fox, M. J., M. A. McCrackin Stevenson, and M. E. Bloom. 1999. Replication of aleutian mink disease parvovirus in vivo is influenced by residues in the VP2 protein. J. Virol. 73:8713-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 100:6081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner, E. M., and P. Tattersall. 1988. Evidence that developmentally regulated control of gene expression by a parvoviral allotropic determinant is particle mediated. J. Virol. 62:1713-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardiner, E. M., and P. Tattersall. 1988. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J. Virol. 62:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert, S. F., and B. R. Migeon. 1975. d-Valine as a selective agent for normal human and rodent epithelial cells in culture. Cell 5:11-17. [DOI] [PubMed] [Google Scholar]
- 32.Gottschalck, E., S. Alexandersen, A. Cohn, L. A. Poulsen, M. E. Bloom, and B. Aasted. 1991. Nucleotide sequence analysis of aleutian mink disease parvovirus shows that multiple virus types are present in infected mink. J. Virol. 65:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govindasamy, L., K. Hueffer, C. R. Parrish, and M. Agbandje-McKenna. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 77:12211-12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris, R. E., P. H. Coleman, and P. S. Morahan. 1974. Erythrocyte association and interferon production by minute virus of mice. Proc. Soc. Exp. Biol. Med. 145:1288-1292. [DOI] [PubMed] [Google Scholar]
- 35.Hueffer, K., and C. R. Parrish. 2003. Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 6:392-398. [DOI] [PubMed] [Google Scholar]
- 36.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J.-Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hueffer, K., L. Govindasamy, M. Agbandje-Mckenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77:10099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hueffer, K., L. Palermo, and C. R. Parrish. 2004. Parvovirus infection of cells by using variants of the feline transferrin receptor altering clathrin-mediated endocytosis, membrane domain localization, and capsid-binding domains. J. Virol. 78:5601-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulst, M. M., H. G. P. van Gennip, A. C. Vlot, E. Schooten, and R. J. M. Moormann. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 75:9585-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang, Y. T., B.-Y. Liu, D. M. Coen, and C. B. C. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isnard, M., M. Granier, R. Frutos, B. Reynaud, and M. Peterschmitt. 1998. Quasispecies nature of three maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 79:3091-3099. [DOI] [PubMed] [Google Scholar]
- 42.Itah, R., J. Tal, and C. Davis. 2004. Host cell specificity of minute virus of mice in the developing mouse embryo. J. Virol. 78:9474-9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimsey, P. B., H. D. Engers, B. Hirt, and V. Jongeneel. 1986. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J. Virol. 59:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lednicky, J., A. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linser, P., H. Bruning, and R. W. Armentrout. 1977. Specific binding sites for a parvovirus, minute virus of mice on cultured mouse cells. J. Virol. 24:211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Littlefield, J. W. 1964. Three degrees of guanylic acid-inosinic acid pyrophosphorylase deficiency in mouse fibroblast. Nature 203:1142-1144. [DOI] [PubMed] [Google Scholar]
- 47.Lombardo, E., J. C. Ramírez, J. García, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-Bueno, A., M. Mateu, and J. M. Almendral. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 77:2701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.López-Bueno, A., N. Valle, J. M. Gallego, J. Pérez, and J. M. Almendral. 2004. Enhanced cytoplasmic sequestration of the nuclear export receptor CRM1 by NS2 mutations developed in the host regulates parvovirus fitness. J. Virol. 78:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyerla, T. A., S. K. Gross, and R. H. McCluer. 1986. Glycosphingolipid patterns in primary mouse kidney cultures. J. Cell. Physiol. 19:390-394. [DOI] [PubMed] [Google Scholar]
- 51.Mandl, C. W., S. L. Allison, H. Holzmann, T. Meixner, and F. X. Heinz. 2000. Attenuation of tick-borne encephalitis virus by structural-based site-specific mutagenesis of a putative flavivirus receptor binding site. J. Virol. 74:9601-9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maroto, B., J. C. Ramírez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maroto, B., N. Valle, R. Saffrich, and J. M. Almendral. 2004. Nuclear export of the non-enveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 78:10685-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H.-D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxwell, I. H., A. L. Spitzer, F. Maxwell, and D. J. Pintel. 1995. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J. Virol. 69:5829-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMaster, G. K., P. Beard, H. D. Engers, and B. Hirt. 1981. Characterization of an immunosuppressive parvovirus related to the minute virus of mice. J. Virol. 38:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merchlinsky, M. J., P. J. Tattersall, J. J. Leary, S. F. Cotmore, E. M. Gardiner, and D. C. Ward. 1983. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J. Virol. 47:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication. In D. M. Knipe, and P. M. Howley (ed.), Field's virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 60.Ohuchi, M., R. Ohuchi, T. Sakai, and A. Matsumoto. 2002. Tight binding of influenza virus hemagglutinin to its receptor interferes with fusion pore dilation. J. Virol. 76:12405-12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oraveerakul, K., C.-S. Choi, and T. W. Molitor. 1992. Restriction of porcine parvovirus replication in nonpermissive cells. J. Virol. 66:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ostroff, S. M., J. E. McDade, J. W. LeDuc, and J. M. Hughes. 2005. Emerging and reemerging infectious disease threats, p. 173-192. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, vol. II. Elsevier, Philadelphia, PA. [Google Scholar]
- 63.Parker, J. S. L., W. J. Murphy, D. E. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferring receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porotto, M., M. Murrell, O. Greengard, M. C. Lawrence, J. L. McKimm-Breschkin, and A. Moscona. 2004. Inhibition of parainfluenza virus type 3 and Newcastle disease virus hemagglutinin-neuraminidase receptor binding: effect of receptor avidity and steric hindrance at the inhibitor binding sites. J. Virol. 78:13911-13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Previsani, N., S. Fontana, B. Hirt, and P. Beard. 1997. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J. Virol. 71:7769-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramírez, J. C., A. Fairén, and J. M. Almendral. 1996. Parvovirus minute virus of mice strain i multiplication and pathogenesis the newborn mouse brain are restricted to proliferative areas and to migratory cerebellar young neurons. J. Virol. 70:8109-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez, J. C., J. A. Santarén, and J. M. Almendral. 1995. Transcriptional inhibition of the parvovirus Minute Virus of Mice by constitutive expression of an antisense RNA targeted against the NS-1 transactivator protein. Virology 206:57-68. [DOI] [PubMed] [Google Scholar]
- 68.Reguera, J., A. Carreira, L. Riolobos, J. M. Almendral, and M. G. Mateu. 2004. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA 101:2724-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rubio, M. P., S. Guerra, and J. M. Almendral. 2001. Genome replication and postencapsidation functions mapping to the nonstructural gene restrict the host range of a murine parvovirus in human cells. J. Virol. 75:11573-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sa-Carvalho, D., E. Reider, B. Baxt, R. Rodarte, A. Tanuri, and P. Mason. 1997. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 71:5115-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segovia, J. C., A. Real, J. A. Bueren, and J. M. Almendral. 1991. In vitro myelosuppressive effects of the parvovirus minute virus of mice (MVMi) on hematopoietic stem and committed progenitor cells. Blood 77:980-988. [PubMed] [Google Scholar]
- 72.Segovia, J. C., G. Guenechea, J. M. Gallego, J. M. Almendral, and J. A. Bueren. 2003. Parvovirus infection suppresses long-term repopulating hematopoietic stem cells. J. Virol. 77:8495-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Segovia, J. C., J. A. Bueren, and J. M. Almendral. 1995. Myeloid depression follows infection of susceptible newborn mice with the parvovirus Minute Virus of Mice (strain i). J. Virol. 69:3229-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Segovia, J. C., J. M. Gallego, J. A. Bueren, and J. M. Almendral. 1999. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected with the parvovirus minute virus of mice. J. Virol. 73:1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shein, H. M., and J. F. Enders. 1962. Multiplication and cytopathogenicity of simian vacuolating virus 40 in cultures of human tissues. Proc. Soc. Exp. Biol. Med. 109:495-500. [DOI] [PubMed] [Google Scholar]
- 77.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 78.Smith, A., and A. Helenius. 2004. How viruses enter animal cells. Science 304:237-242. [DOI] [PubMed] [Google Scholar]
- 79.Spalholz, B. A., and P. Tattersall. 1983. Interaction of minute virus of mice with differentiated cells: strain-dependent target cell specificity is mediated by intracellular factors. J. Virol. 46:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus cell interaction of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Truyen, U., A. Gruemberg, S.-F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vasudevacharya, J., and R. W. Compans. 1992. The NS and capsid genes determine the host range of porcine parvovirus. Virology 187:515-524. [DOI] [PubMed] [Google Scholar]
- 83.Vihinen-Ranta, M., and C. R. Parrish. 2004. Pathways of cell infection by parvoviruses and adeno-associated viruses. J. Virol. 78:6709-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Villarreal, L. P., V. P. Defilippis, and K. A. Gottieb. 2000. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 272:1-6. [DOI] [PubMed] [Google Scholar]
- 86.Ward, D. C., and P. J. Tattersall. 1982. Minute virus of mice, p. 313-334. In H. L. Foster, J. D. Small, and J. G. Fox (ed.), The mouse in biochemical research, vol. II. Academic Press, New York, N.Y. [Google Scholar]
- 87.Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne, and J. J. Bull. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285:422-424. [DOI] [PubMed] [Google Scholar]
- 88.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptative evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]