Abstract
Treatment with alpha interferon is a standard therapy for patients with chronic hepatitis B virus (HBV) infections. This treatment can reduce virus load and ameliorate disease symptoms. However, in the majority of cases, alpha interferon therapy fails to resolve the chronic HBV infection. The reason alpha interferon therapy is inefficient at resolving chronic HBV infections is assumed to be because it fails to eliminate covalently closed circular (CCC) HBV DNA from the nuclei of infected hepatocytes. In an attempt to address this issue, the stability of HBV CCC DNA in response to alpha/beta interferon induction was examined in HNF1α-null HBV transgenic mice. Alpha/beta interferon induction by polyinosinic-polycytidylic acid [poly(I-C)] treatment efficiently eliminated encapsidated cytoplasmic HBV replication intermediates while only modestly reducing nuclear HBV CCC DNA. These observations indicate that nuclear HBV CCC DNA is more stable than cytoplasmic replication intermediates in response to alpha/beta interferon induction. Consequently it appears that for therapies to resolve chronic HBV infection efficiently, they will have to target the elimination of the most stable HBV replication intermediate, nuclear HBV CCC DNA.
Hepatitis B virus (HBV) infection is associated with significant morbidity and mortality (4, 26). Currently, it is estimated there are approximately 400 million chronic carriers in the world (26). Reliable therapies for chronic HBV infection are required, as the long-term consequences include cirrhosis of the liver and hepatocellular carcinoma, which have very poor prognoses (26). Chronic HBV infections can be treated with various antiviral drugs such as lamivudine and adefovir (26). These nucleoside analogues specifically inhibit the HBV reverse transcriptase/DNA polymerase activity and inhibit viral replication (26). Unfortunately, treatment with these antiviral agents often leads to the selection of drug-resistant variants, preventing the resolution of the chronic carrier state (3, 26).
In addition to the use of antiviral drugs, chronic HBV infection can be treated with alpha interferon therapy (26). As observed with antiviral therapy, alpha interferon therapy may suppress HBV replication during the treatment phase but it often fails to resolve the chronic HBV infection or the associated disease state (24, 26). The reason these therapies fail to resolve the chronic HBV infection is unclear, but it may be due to the existence of covalently closed circular (CCC) HBV DNA in the nuclei of the infected hepatocytes (34, 40, 44). It is possible that the half-life of HBV CCC DNA is sufficiently long that the current antiviral drugs fail to eliminate this viral replication intermediate during the standard period of therapy (1, 12, 27, 30, 44, 49).
During the initial stages of an HBV infection, the virion enters the hepatocyte and releases the viral nucleocapsid into the cytoplasm (35). The nuclear localization signal sequences in the nucleocapsid presumably delivers the 3.2-kb partially double-stranded genomic DNA located within the nucleocapsid to the nucleus (15). In the nucleus, the partially double-stranded genomic DNA is converted to HBV CCC DNA with the aid of various enzyme activities associated with the nuclear replication machinery (29, 35). HBV CCC DNA subsequently serves as the transcriptional template for the synthesis of the viral transcripts (35).
The HBV 3.5-kb pregenomic RNA is translated to produce both the core protein and the viral polymerase (31, 45). The polymerase protein binds to the epsilon sequence within the pregenomic RNA, and this complex is encapsidated by the viral core protein to generate an immature viral nucleocapsid (22). Subsequently the viral polymerase converts the pregenomic RNA into partially double-stranded genomic DNA, producing a mature nucleocapsid particle that is essentially identical to the nucleocapsid derived from the infecting virion (45). Consequently, the newly synthesized mature nucleocapsid can deliver its viral genomic DNA to the nucleus, resulting in the amplification of nuclear HBV CCC DNA (40).
During natural infection it is estimated that there are up to 50 copies of CCC DNA in the nuclei of infected hepatocytes (1, 12, 29, 48, 49). The amplification of nuclear CCC DNA appears to be limited by the translation of the viral transcripts encoding the surface antigen polypeptides (39). The surface antigen polypeptides are synthesized as transmembrane polypeptides which associate with mature nucleocapsids at the endoplasmic reticulum and bud as virions into the lumen of the endoplasmic reticulum (6, 7, 18). HBV virions are subsequently secreted from the hepatocyte after processing and transport through the endoplasmic reticulum and Golgi apparatus (23, 46).
In this study, the stability of HBV CCC DNA in the hepatocytes of HNF1α-null HBV transgenic mice has been examined in response to the induction of an alpha/beta interferon response. HBV transgenic mice do not normally have significant levels of nuclear CCC DNA despite synthesizing high levels of cytoplasmic replication intermediates (19, 34). However, HBV transgenic mice lacking HNF1α not only synthesize high levels of cytoplasmic replication intermediates but also produce detectable levels of nuclear HBV CCC DNA (34). Induction of an alpha/beta interferon response in HBV transgenic mice by polyinosinic-polycytidylic acid [poly(I-C)] treatment dramatically reduces the level of cytoplasmic replication intermediates (20, 28, 41, 43). This reduction in cytoplasmic replication intermediates requires a functional alpha/beta interferon receptor, demonstrating this effect is mediated primarily by alpha/beta interferon (28). Therefore, it was of interest to determine if poly(I-C) induction of an alpha/beta interferon response in HNF1α-null HBV transgenic mice might reduce the level of cytoplasmic and nuclear viral replication intermediates to a similar or different extent.
From this analysis, it is apparent that cytoplasmic replication intermediates are much more sensitive to elimination by alpha/beta interferon induction than nuclear HBV CCC DNA. These observations support the contention that elimination of nuclear HBV CCC DNA is the major problem in resolving chronic HBV infection. In addition, these findings help to explain why alpha interferon therapy may reduce patients' viral load without necessarily preventing the recurrence of viral biosynthesis after the completion of therapy (26).
MATERIALS AND METHODS
Transgenic mice.
The production and characterization of the HBV transgenic mouse lineage 1.3.32 has been described (19). These HBV transgenic mice contain a single copy of the terminally redundant, 1.3-genome-length copy of the HBVayw genome integrated into the mouse chromosomal DNA. High levels of HBV replication occur in the livers of these mice. The mice used in the breeding experiments were homozygous for the HBV transgene and were maintained on the C57BL/6 genetic background.
The production and characterization of the HNF1α-null mice have been described (32). These mice do not express HNF1α and display hepatic dysfunction, phenylketonuria, renal Fanconi syndrome, and infertility (25, 32). The mice used in the breeding experiments were heterozygous for HNF1α and maintained on the Sv/129 genetic background.
HNF1α-null HBV transgenic mice were generated by mating the HBV transgenic mice with the heterozygous HNF1α+/− mice. The resulting heterozygous HNF1α+/− HBV transgenic F1 mice were subsequently mated with the heterozygous HNF1α+/− mice and the F2 mice were screened for the HBV transgene and HNF1α-null allele by PCR analysis of tail DNA. Tail DNA was prepared by incubating 1 cm of tail in 500 μl of 100 mM Tris hydrochloride (pH 8.0), 200 mM NaCl, 5 mM EDTA, 0.2% (wt/vol) sodium dodecyl sulfate containing 100 μg/ml proteinase K for 16 to 20 h at 55°C. Samples were centrifuged at 14,000 rpm in an Eppendorf 5417C microcentrifuge for 10 min and the supernatant was precipitated with 500 μl of isopropanol. DNA was pelleted by centrifugation at 14,000 rpm in an Eppendorf 5417C microcentrifuge for 10 min and subsequently dissolved in 100 μl of 5 mM Tris hydrochloride (pH 8.0), 1 mM EDTA.
The HBV transgene was identified by PCR analysis using the oligonucleotides TCGATACCTGAACCTTTACCCCGTTGCCCG (oligonucleotide XpHNF4-1, HBV coordinates 1133 to 1159) and TCGAATTGCTGAGAGTCCAAGAGTCCTCTT (oligonucleotide CpHNF4-2, HBV coordinates 1683 to 1658), and 1 μl of tail DNA. The samples were subjected to 30 amplification cycles involving denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension from the primers at 72°C for 2 min. A PCR product of 551 bp indicated the presence of the HBV transgene. The HNF1α wild-type and null alleles were identified by PCR analysis using the oligonucleotides CAGAGCTTGACTAGTGGGATTTGG (oligonucleotide mHNF1α-1, HNF1α promoter plus-strand sequence), ACCCTCTCCAACCATCAGGTAGG (oligonucleotide mHNF1α-2, HNF1α exon 1 minus-strand sequence), and AACTGTTGGGAAGGGCGATCGGTG (oligonucleotide BGAL, β-galactosidase minus-strand sequence), and 1 μl of tail DNA. The samples were subjected to 35 amplification cycles involving denaturation at 96°C for 1 min, annealing at 56°C for 1 min, and extension from the primers at 72°C for 1 min. A PCR product of 276 bp indicated the wild-type HNF1α genotype, whereas a PCR product of 390 bp indicated the mutated HNF1α genotype. The 20-μl reaction conditions used were as described by the manufacturer (PGC Scientifics) and contained 1.5 units of Taq DNA polymerase.
Alpha/beta interferon expression was induced in the HBV transgenic mice essentially as described (41). Mice were injected four times with 200 μg of poly(I-C) in phosphate-buffered saline at 24-hour intervals. Mice were sacrificed 4 to 6 h after the final intraperitoneal injection of poly(I-C). Liver tissue was frozen in liquid nitrogen and stored at −70°C prior to DNA and RNA extraction.
HBV DNA and RNA analysis.
Total DNA and RNA were isolated from the livers of HBV transgenic mice as described (10, 37). DNA (Southern) filter hybridization analyses were performed using 20 μg of HindIII-digested DNA (37). Filters were probed with 32P-labeled HBVayw genomic DNA (16) to detect HBV sequences. RNA (Northern) filter hybridization analyses were performed using 10 μg of total cellular RNA as described (37). Filters were probed with 32P-labeled HBVayw genomic DNA to detect HBV sequences and the mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA to detect the GAPDH transcript used as an internal control (36). Filters were probed with 32P-labeled mouse 2′,5′-oligoadenylate synthase (2OAS) cDNA to detect the 2OAS transcript as an indirect measure of alpha/beta interferon induction (41).
RNase protection assays were performed using the Pharmingen Riboquant kit and riboprobes were synthesized using the Ambion Maxiscript kit as described by the manufacturers. Transcription initiation sites for the 3.5-kb HBV transcripts were examined using 20 μg of total cellular RNA and a 333-nucleotide-long (HBV coordinates 1990 to 1658) 32P-labeled HBV riboprobe. As an internal control for the RNase protection analysis, a 32P-labeled mouse ribosomal protein L32 gene riboprobe spanning 101 nucleotides of exon 3 was utilized (14). All riboprobes contained additional flanking vector sequences that are not protected by HBV transgenic mouse RNA.
Nuclear CCC HBV DNA was prepared from mouse livers by a modification of a previously described procedure (47). Approximately 50 mg of mouse liver was homogenized in a Potter-Elvehjem tissue grinder in 0.5 ml of 50 mM Tris hydrochloride (pH 8.0), 1 mM EDTA, 0.2% (vol/vol) NP-40, 0.15 M NaCl at 4°C. The homogenate was centrifuged for 30 seconds at 14,000 rpm in an Eppendorf 5417C microcentrifuge at 4°C. The nuclear pellet was suspended in 0.2 ml of 10 mM Tris hydrochloride (pH 8.0), 1 mM EDTA at 4°C. Nuclei were lysed by the addition of 0.2 ml of 6% (wt/vol) sodium dodecyl sulfate, 0.15 M NaOH. The lysate was vigorously mixed and incubated at 37°C for 30 min. The alkaline lysate was neutralized by the addition of 0.1 ml of 3 M potassium acetate (pH 5.0) and centrifuged for 2 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge at 25°C. The supernatant was extracted with 0.5 ml of water-saturated phenol and precipitated with ethanol in the presence of 5 μg of glycogen (Sigma). The precipitate was suspended in 25 μl of 10 mM Tris hydrochloride (pH 8.0), 1 mM EDTA, and the isolated HBV DNA was examined by filter hybridization analysis. Filters were probed with 32P-labeled HBVayw genomic DNA (16) and the mouse mitochondrial cytochrome b gene region between coordinates 14243 and 14550 (5) to detect HBV CCC DNA and mitochondrial DNA sequences, respectively. Filter hybridization and RNase protection analyses were quantitated by phosphorimaging using a Packard Cyclone Storage Phosphor System.
HBV antigen analysis.
Hepatitis B virus e antigen (HBeAg) analysis was performed using 2 μl of mouse serum and the HBe enzyme-linked immunosorbent assay as described by the manufacturer (International Immunodiagnostics). The level of antigen was determined in the linear range of the assay.
RESULTS
Effect of poly(I-C) injection on HBeAg synthesis in HBV transgenic mice.
HBV transgenic mice were bred with HNF1α heterozygous mice, and HBV transgenic mice hemizygous for the HBV transgene and heterozygous for the wild-type HNF1α allele or homozygous for the HNF1α-null allele were identified in the F2 generation. For these studies, HBV transgenic mice that were heterozygous for the HNF1α wild-type allele were used as controls and compared with HBV transgenic mice that were homozygous for the HNF1α-null allele.
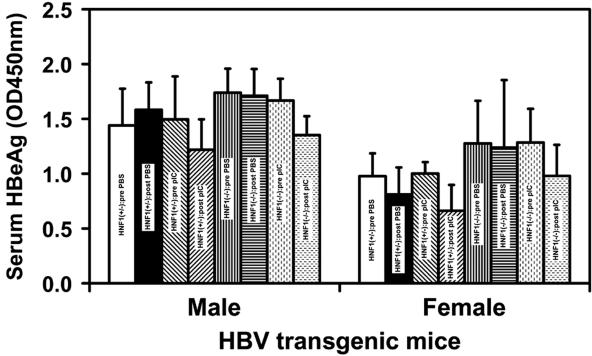
Control HBV transgenic mice heterozygous for the HNF1α wild-type allele and HNF1α-null HBV transgenic mice were injected with poly(I-C) to induce an alpha/beta interferon response (41). Multiple rather than single poly(I-C) injections were employed in an attempt to sustain the alpha/beta interferon induction and to confirm the effect of alpha/beta interferon on HBV transcription (41). Male and female mice of each genotype were assayed for the level of HBeAg in their sera before and after injection with either phosphate-buffered saline or poly(I-C) (Fig. 1). HBV transgenic mice injected with phosphate-buffered saline did not display a significant alteration in the level of serum HBeAg as a consequence of this treatment. In contrast, HBV transgenic mice injected with poly(I-C) displayed a statistically significant decrease of approximately 25% in the level of serum HBeAg as a consequence of this treatment. The decrease in serum HBeAg was similar in HNF1α-expressing and HNF1α-null HBV transgenic mice, suggesting that the induction of alpha/beta interferon and its effect on HBeAg synthesis were similar in both groups of mice. This indicates the absence of HNF1α in these mice did not greatly influence the production and function of alpha/beta interferon, at least, with respect to its effects on HBeAg synthesis. As HBeAg is translated from the 3.5-kb precore RNA (42), these observations suggest that the induction of an alpha/beta interferon response in these HBV transgenic mice may have a modest effect on the synthesis of the 3.5-kb precore RNA.
FIG. 1.
Effect of poly(I-C) injection on serum HBeAg synthesis in HBV transgenic mice. HNF1(+/−), HNF1α-expressing HBV transgenic mice heterozygous for the wild-type HNF1α allele; HNF1(−/−), HNF1α-null HBV transgenic mice. Mice were injected with phosphate-buffered saline (PBS) or poly(I-C) (pIC). Serum HBeAg levels were measured before (pre) and after (post) injection (OD450 nm). The mean HBeAg levels plus standard deviations derived from seven male HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven male HNF1α+/− HBV transgenic mice injected with poly(I-C), four male HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, eight male HNF1α−/− HBV transgenic mice injected with poly(I-C), seven female HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven female HNF1α+/− HBV transgenic mice injected with poly(I-C), eight female HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, and nine female HNF1α−/− HBV transgenic mice injected with poly(I-C) are shown. The levels of HBeAg in each group of HBV transgenic mice injected with poly(I-C) were decreased in a statistically significantly manner as determined by a paired Student t test (P < 0.05).
Effect of poly(I-C) injection on viral transcription in HBV transgenic mice.
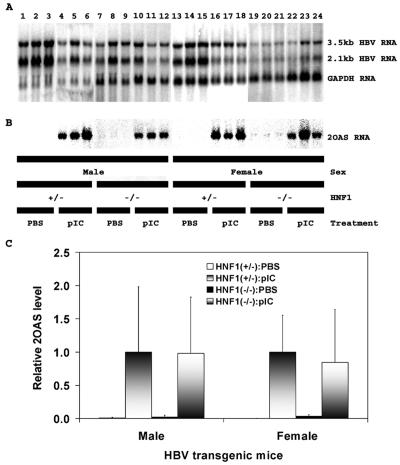
HBV transgenic mice were examined for their steady-state levels of HBV transcripts by analysis of the total liver RNA (Fig. 2). The steady-state levels of the HBV 3.5- and 2.1-kb transcripts in the livers of the HBV transgenic mice with and without HNF1α were not greatly influenced by the induction of alpha/beta interferon mediated by poly(I-C) (Fig. 2A). The HBV transcripts were reduced by less than twofold based on this RNA filter hybridization analysis (Fig. 2A), which is consistent with the modest reduction in serum HBeAg (Fig. 1). This effect of multiple poly(I-C) injections on the steady-state levels of HBV transcripts in these mice is less than previously reported (41) but supports the suggestion that induction of alpha/beta interferon does decrease viral RNA levels.
FIG. 2.
RNA (Northern) filter hybridization analysis of HBV and 2′,5′-oligoadenylate synthase transcripts in the livers of HBV transgenic mice. Groups of three representative mice of each sex and genotype are shown. (A) The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for the quantitation of the HBV 3.5- and 2.1-kb RNAs. The probes used were HBVayw genomic DNA plus glyceraldehyde-3-phosphate dehydrogenase cDNA. (B) The synthesis of the 2′,5′-oligoadenylate synthase (2OAS) transcript was used as an indirect measure of the induction of alpha/beta interferon by poly(I-C) injection. The probe used was the 2′,5′-oligoadenylate synthase cDNA. HNF1(+/−), HNF1α-expressing HBV transgenic mice heterozygous for the wild-type HNF1α allele; HNF1(-/-), HNF1α-null HBV transgenic mice. Mice were injected with phosphate-buffered saline (PBS) or poly(I-C) (pIC). (C) Quantitative analysis of the 2OAS transcript in HBV transgenic mice. The mean 2OAS transcript levels plus standard deviations derived from seven male HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven male HNF1α+/− HBV transgenic mice injected with poly(I-C), four male HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, eight male HNF1α−/− HBV transgenic mice injected with poly(I-C), seven female HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven female HNF1α+/− HBV transgenic mice injected with poly(I-C), eight female HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, and nine female HNF1α−/− HBV transgenic mice injected with poly(I-C) are shown. The levels of the 2OAS transcript in the HNF1α-null HBV transgenic mice are not statistically significantly different from their levels in the control HNF1α-expressing HBV transgenic mice as determined by a Student t test (P > 0.05).
The alpha/beta interferon response induced by poly(I-C) was similar in control HBV transgenic mice heterozygous for the HNF1α wild-type allele and HNF1α-null HBV transgenic mice as measured by the induction of 2′,5′-oligoadenylate synthase gene expression (Fig. 2B and C). Similar levels of 2OAS gene expression were also observed in control and HNF1α-null HBV transgenic mice using an RNase protection assay (A. L. Anderson and A. McLachlan, unpublished data). These results indicate that the absence of HNF1α in the HNF1α-null HBV transgenic mice did not significantly affect the ability of poly(I-C) to induce an alpha/beta interferon response.
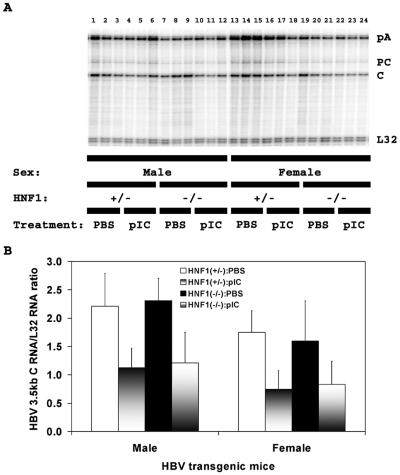
The effect of the induction of alpha/beta interferon on the steady-state levels of the precore and pregenomic HBV 3.5-kbRNAs was examined by RNase protection analysis (Fig. 3). In the case of the precore RNA, induction of alpha/beta interferon by poly(I-C) did not significantly reduce the abundance of this viral transcript, consistent with the modest reduction in serum HBeAg. The pregenomic RNA was reduced approximately twofold in both HNF1α-expressing and HNF1α-null HBV transgenic mice, which was consistent with the effect of induction of alpha/beta interferon on the HBV 3.5-kb RNA observed by RNA filter hybridization analysis (Fig. 2A). Therefore, it appears that the induction of alpha/beta interferon by poly(I-C) reduces HBV transcription to a modest but significant extent, as previously reported (20, 28, 41).
FIG. 3.
RNase protection analysis mapping the transcription initiation sites of the precore (PC) and pregenomic (C) transcripts from the livers of HBV transgenic mice. (A) Groups of three representative mice of each sex and genotype are shown. The 3′ ends of the all the HBV transcripts corresponding to the polyadenylation site (pA) of these RNAs also generated a protected fragment in this analysis. The riboprobes used included the HBVayw sequence spanning nucleotide coordinates 1990 to 1658 and the mouse ribosomal protein L32 gene riboprobe spanning 101 nucleotides of exon 3. The 3.5-kb HBV RNAs protect fragments of 283 (pA), 206 (pC), and 175 (C) nucleotides, respectively. The mouse ribosomal protein L32 RNA protects a fragment of 101 nucleotides, designated L32, when probed with the L32 probe. HNF1(+/−), HNF1α-expressing HBV transgenic mice heterozygous for the wild-type HNF1α allele; HNF1(-/-), HNF1α-null HBV transgenic mice. Mice were injected with phosphate-buffered saline (PBS) or poly(I-C) (pIC). (B) Quantitative analysis of the HBV pregenomic RNA in HBV transgenic mice. The mean HBV pregenomic (C) RNA-to-L32 RNA ratio plus standard deviations derived from seven male HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven male HNF1α+/− HBV transgenic mice injected with poly(I-C), four male HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, eight male HNF1α−/− HBV transgenic mice injected with poly(I-C), seven female HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven female HNF1α+/− HBV transgenic mice injected with poly(I-C), eight female HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, and nine female HNF1α−/− HBV transgenic mice injected with poly(I-C) are shown. The lower HBV pregenomic RNA to L32 RNA ratios in the poly(I-C)-injected HBV transgenic mice are statistically significantly different from their ratios in the control phosphate-buffered saline injected HBV transgenic mice as determined by a Student t test (P < 0.05).
Effect of poly(I-C) injection on cytoplasmic viral replication intermediates in HBV transgenic mice.
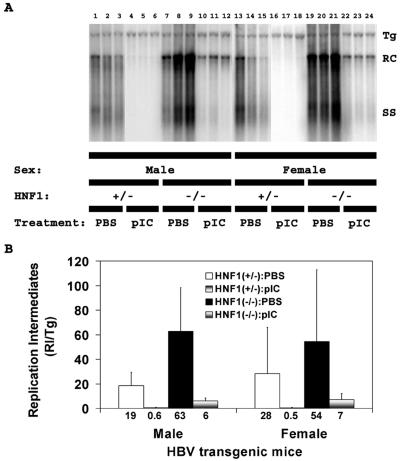
As reported previously, the level of replication intermediates in the livers of HNF1α-null HBV transgenic mice was approximately two- to threefold higher than observed in HNF1α-expressing HBV transgenic mice (Fig. 4) (34). The induction of alpha/beta interferon by poly(I-C) reduced the level of cytoplasmic HBV replication intermediates 30- to 50-fold in the control HNF1α-expressing HBV transgenic mice (Fig. 4). This observation is consistent with previous findings (20, 28) and demonstrates that the level of cytoplasmic viral replication intermediates is affected to a much greater extent than pregenomic RNA abundance in response to alpha/beta interferon induction.
FIG. 4.
DNA (Southern) filter hybridization analysis of HBV DNA replication intermediates in the livers of HBV transgenic mice. (A) Groups of three representative mice of each sex and genotype are shown. The HBV transgene was used as an internal control for the quantitation of the HBV replication intermediates. The probe used was HBVayw genomic DNA. Tg, HBV transgene; RC, HBV relaxed circular replication intermediates; SS, HBV single-stranded replication intermediates; HNF1(+/−), HNF1α-expressing HBV transgenic mice heterozygous for the wild-type HNF1α allele; HNF1(-/-), HNF1α-null HBV transgenic mice. Mice were injected with phosphate-buffered saline (PBS) or poly(I-C) (pIC). (B) Quantitative analysis of the HBV DNA replicative intermediate (RI) levels in HBV transgenic mice. The mean HBV DNA replicative intermediate levels (values for replicative intermediates per transgene are indicated below the bar graph) plus standard deviations derived from seven male HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven male HNF1α+/− HBV transgenic mice injected with poly(I-C), four male HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, eight male HNF1α−/− HBV transgenic mice injected with poly(I-C), seven female HNF1α+/− HBV transgenic mice injected with phosphate-buffered saline, seven female HNF1α+/− HBV transgenic mice injected with poly(I-C), eight female HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, and nine female HNF1α−/− HBV transgenic mice injected with poly(I-C) are shown. The lower levels of the HBV DNA replication intermediates in the poly(I-C)-injected HBV transgenic mice are statistically significantly different from the levels in the control phosphate-buffered saline injected HBV transgenic mice as determined by a Student t test (P < 0.05).
Similar to the control HBV transgenic mice, the induction of alpha/beta interferon by poly(I-C) reduced the level of cytoplasmic HBV replication intermediates in the HNF1α-null HBV transgenic mice, although to a somewhat lesser extent (Fig. 4). The cytoplasmic viral replication intermediates were reduced 10- and 8-fold in male and female HNF1α-null HBV transgenic mice, respectively (Fig. 4). This indicates that although HNF1α is not essential for the inhibition of viral replication by alpha/beta interferon, it appears it may affect either directly or indirectly the magnitude of this inhibition. The reason for this effect is unclear, but it may be related to some of the metabolic changes associated with the loss of HNF1α in the liver (2, 8, 21, 25, 32, 33, 38).
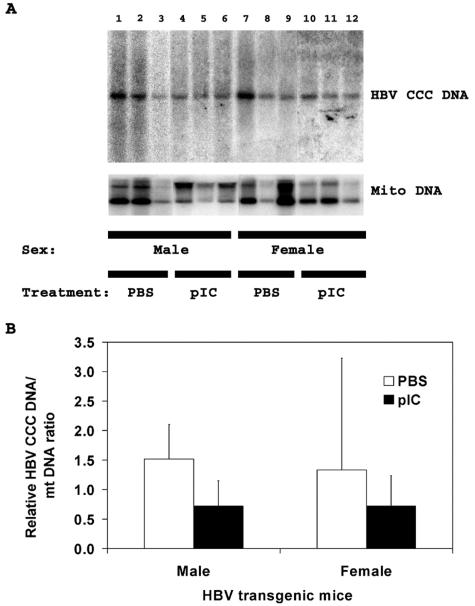
Effect of poly(I-C) injection on nuclear HBV covalently closed circular DNA in HNF1α-null HBV transgenic mice.
As reported previously, HNF1α-expressing HBV transgenic mice synthesize very limited amounts of nuclear HBV CCC DNA whereas HNF1α-null HBV transgenic mice have detectable levels of this viral replication intermediate (34). As the level of cytoplasmic replication intermediates was significantly reduced by the induction of alpha/beta interferon with poly(I-C) in HNF1α-null HBV transgenic mice (Fig. 4), it was of interest to examine the stability of the nuclear HBV CCC DNA in these mice. The level of HBV CCC DNA in the livers of the HNF1α-null HBV transgenic mice was reduced approximately twofold by the induction of alpha/beta interferon in both male and female mice (Fig. 5). This observation clearly indicates that HBV CCC DNA is more stable than the encapsidated cytoplasmic viral replication intermediates. However, it is apparent that the level of HBV CCC DNA does decrease in response to the poly(I-C) injection, and it may have a half-life of approximately 3 days in HNF1α-null HBV transgenic mice after the induction of an alpha/beta interferon response.
FIG. 5.
DNA (Southern) filter hybridization analysis of HBV CCC DNA replication intermediates and mitochondrial DNA in the livers of HNF1α-null HBV transgenic mice. (A) Groups of three representative mice of each sex are shown. The mitochondrial DNA was used as an internal control for the quantitation of the HBV CCC DNA. The probe used was HBVayw genomic DNA and a mouse mitochondrial DNA fragment. HBV CCC DNA, HBV covalently closed circular DNA; Mito DNA, mouse mitochondrial DNA. Mice were injected with phosphate-buffered saline (PBS) or poly(I-C) (pIC). (B) Quantitative analysis of the HBV CCC DNA replication intermediate levels in HNF1α-null HBV transgenic mice. The mean HBV CCC DNA replication intermediate levels plus standard deviations derived from four male HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, eight male HNF1α−/− HBV transgenic mice injected with poly(I-C), eight female HNF1α−/− HBV transgenic mice injected with phosphate-buffered saline, and nine female HNF1α−/− HBV transgenic mice injected with poly(I-C) are shown. The lower levels of the HBV CCC DNA replication intermediates in the poly(I-C)-injected male but not the female HNF1α-null HBV transgenic mice are statistically significantly different from the levels in the control phosphate-buffered saline-injected HNF1α-null HBV transgenic mice as determined by a Student t test (P < 0.05).
DISCUSSION
HBV is a major human pathogen, causing approximately a million deaths annually (9). There are currently no reliable therapies for chronic HBV infection, which is associated with cirrhosis of the liver and hepatocellular carcinoma (26). Chronic HBV patients may be treated with various nucleoside analogues, which selectively inhibit the viral polymerase, or alpha interferon, which reduces viral load by unknown mechanisms that are presumably associated with its known antiviral properties (26). Although these therapies may improve the prognosis for the patient, these treatments generally fail to resolve the chronic HBV infection (26). It is possible that the reason these antiviral therapies fail to resolve chronic HBV infections is their failure to eliminate HBV CCC DNA from the nucleus of infected hepatocytes (44).
HBV CCC DNA is the presumptive transcriptional viral template (17, 35). The first HBV CCC DNA molecule is generated from the genome of the viral particle that initially infects the hepatocyte. This HBV CCC DNA molecule is transcribed, generating the four viral transcripts that are translated to produce the seven viral polypeptides (17, 35). In the cytoplasm, the pregenomic RNA is reverse transcribed within the nucleocapsid to produce a mature capsid particle containing the 3.2-kb partially double-stranded HBV DNA genome (45). The mature capsid particle may either interact with the envelope polypeptides in the membrane of the endoplasmic reticulum to initiate the synthesis of a new viral particle or cycle the viral genome back to the nucleus to amplify the pool of HBV CCC DNA molecules that can serve as transcriptional templates. The fate of the mature capsid is regulated, at least in part, by the level of synthesis of the envelope polypeptides. In the presence of abundant envelope polypeptide synthesis, mature capsids preferentially traffic through the viral biosynthetic pathway (39).
Although some of the factors that regulate the synthesis of HBV CCC DNA are apparent, little is known about the factors that control the loss of this molecule from the nucleus of the infected cell. Despite the possibility that chronic HBV infection is not resolved due to the inherent stability of HBV CCC DNA, there is limited evidence supporting this contention. The stability of HBV CCC DNA in cell culture has been examined when viral replication was inhibited with nucleoside analogs, and it was estimated that the half-life of HBV CCC DNA was approximately 3 days (13). For chimpanzees infected with HBV, it has been estimated that the half-life of HBV CCC DNA during the noncytolytic phase of natural infection is between 9 and 14 days (44). For woodchuck hepatitis virus and duck hepatitis B virus infection, estimates of the stability of the viral CCC DNA have been quite variable. Duck hepatitis B virus CCC DNA was estimated to have a half-life in cell culture of between 3 and 5 days (11), whereas its half-life in vivo was estimated to be between 35 and 57 days (1). Woodchuck hepatitis virus CCC DNA was estimated to have a half-life in cell culture of more than 24 to 36 days (12, 30), and its half-life was estimated to be between 33 and 50 days in vivo, suggesting this molecule was very stable under both conditions (49).
To address further the issue of the stability of HBV CCC DNA in vivo in the absence of infection and its associated cellular immune response, HNF1α-null HBV transgenic mice were injected with poly(I-C) to induce an alpha/beta interferon response (Fig. 2). The potential cytopathic effects of poly(I-C) injections on the liver were monitored by measuring serum alanine aminotransferase levels. HNF1α-null HBV transgenic mice responded to poly(I-C) in a manner similar to the control HBV transgenic mice, and no cytopathic effects were observed in response to alpha/beta interferon induction. HBV pregenomic RNA synthesis was reduced approximately twofold in HBV transgenic mice of both genotypes (Fig. 3). In addition, HBV transgenic mice of both genotypes showed a dramatic reduction in the steady-state levels of viral replication intermediates in their livers in response to the induction of alpha/beta interferon (Fig. 4).
As the HNF1α-null but not the control HBV transgenic mice synthesize measurable levels of HBV CCC DNA (34), it was possible to determine the short-term effect of the induction of alpha/beta interferon on HBV CCC DNA stability in these mice. In contrast to the dramatic reduction in cytoplasmic replication intermediates, HBV CCC DNA levels were not greatly affected by the induction of alpha/beta interferon (Fig. 5). This indicates that nuclear HBV CCC DNA is more stable than encapsidated cytoplasmic viral replication intermediates in the HNF1α-null HBV transgenic mice model of the chronic HBV carrier state. In this model, HBV CCC DNA is not completely stable, as the levels decreased approximately twofold during the 3-day induction of alpha/beta interferon. This suggests that HBV CCC DNA may have a half-life of approximately 3 days in this model system. Obviously, the precise relevance of this observation in the HNF1α-null HBV transgenic mouse to natural infection requires further investigation. However, this half-life is similar to that determined for HBV CCC DNA in cell culture but somewhat shorter than estimated for natural infection of the chimpanzee (13, 44).
In addition, it should be noted that the stability of the HBV CCC DNA was only examined during an initial 3-day period. It is possible that the HBV CCC DNA in the HNF1α-null HBV transgenic mice remaining after 3 days of alpha/beta interferon induction may have a much longer half-life than observed in the initial treatment period. Nevertheless, assuming a half-life for HBV CCC DNA of 3 days, 5 × 1010 hepatocytes in the adult human liver (44), and 10 copies of HBV CCC DNA per cell, the time required to eliminate HBV CCC DNA from a infected liver by alpha interferon therapy can be estimated. Based on these assumptions, it would take 117 days, or 39 half-lives, to reduce the number of HBV CCC DNA molecules in the liver to less than one. As most alpha interferon treatments for chronic HBV are administered for somewhat longer periods of time, in the range of 4 to 6 months (26), their failure to resolve the infection efficiently suggest two possibilities. Either the half-life of HBV CCC DNA is longer than 3 days in natural infection, or low levels of viral replication persist during treatment and HBV CCC DNA synthesis occurs at a level preventing the resolution of the infection. Determining the relative importance of these two possibilities is likely to have important implications for therapies aiming to resolve chronic HBV infections. However, as extending alpha interferon therapy may be beneficial (26), it is possible that the elimination of HBV CCC DNA may be a slow but progressive response to this and possibly other forms of anti-HBV treatment.
Acknowledgments
We thank Luca G. Guidotti and Francis V. Chisari (Scripps Research Institute, La Jolla, CA) for providing the HBV transgenic mice. We are grateful to Susan L. Uprichard (Scripps Research Institute, La Jolla, CA) for plasmids pmGAPDH and pmOAS, containing the mouse glyceraldehyde-3-phosphate dehydrogenase and 2′,5′-oligoadenylate synthase cDNAs, respectively.
This work was supported by Public Health Service grant AI30070 from the National Institutes of Health.
Footnotes
Publication number 17195-CB from the Scripps Research Institute.
REFERENCES
- 1.Addison, W. R., K. A. Walters, W. W. S. Wong, J. S. Wilson, D. Madej, L. D. Jewell, and D. L. J. Tyrrell. 2002. Half-life of the duck hepatitis B virus covalently closed circular DNA pool in vivo following inhibition of viral replication. J. Virol. 76:6356-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama, T. E., J. M. Ward, and F. J. Gonzalez. 2000. Regulation of the liver fatty acid-binding protein gene by hepatocyte nuclear factor 1a (HNF1a)-Alterations in fatty acid homeostasis in HNF1a-deficient mice. J. Biol. Chem. 275:27117-27122. [DOI] [PubMed] [Google Scholar]
- 3.Angus, P., R. Vaughan, S. Xiong, H. L. Yang, W. Delaney, C. Gibbs, C. Brosgart, D. Colledge, R. Edwards, A. Ayres, A. Bartholomeusz, and S. Locarnini. 2003. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology 125:292-297. [DOI] [PubMed] [Google Scholar]
- 4.Beasley, R. P. 1988. Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. J., R. A. Van Etten, C. T. Wright, M. W. Walberg, and D. A. Clayton. 1981. Sequence and genome organization of mouse mitochondrial DNA. Cell 26:167-180. [DOI] [PubMed] [Google Scholar]
- 6.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss, V., and R. Thomssen. 1994. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 68:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, C., T. E. Akiyama, G. Kudo, and F. J. Gonzalez. 2003. Hepatic expression of cytochrome P450s in hepatocyte nuclear factor 1-alpha (HNF1a)-deficient mice. Biochem. Pharmacol. 66:2011-2020. [DOI] [PubMed] [Google Scholar]
- 9.Chisari, F. V. 2000. Viruses, immunity, and cancer: Lessons from hepatitis B. Am. J. Pathol. 156:1118-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Civitico, G. M., and S. A. Locarnini. 1994. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology 203:81-89. [DOI] [PubMed] [Google Scholar]
- 12.Dandri, M., M. R. Burda, H. Will, and J. Petersen. 2000. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology 32:139-146. [DOI] [PubMed] [Google Scholar]
- 13.Delaney, W. E., T. G. Miller, and H. C. Isom. 1999. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (−)-b-2′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob. Agents Chemother. 43:2017-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudov, K. P., and R. P. Perry. 1984. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell 37:457-468. [DOI] [PubMed] [Google Scholar]
- 15.Eckhardt, S. G., D. R. Milich, and A. McLachlan. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 17.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 18.Gerelsaikhan, T., J. E. Tavis, and V. Bruss. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 70:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., A. Morris, H. Mendez, R. Koch, R. H. Silverman, B. R. G. Williams, and F. V. Chisari. 2002. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J. Virol. 76:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiraiwa, H., C. J. Pan, B. C. Lin, T. E. Akiyama, F. J. Gonzalez, and J. Y. Chou. 2001. A molecular link between the common phenotypes of type 1 glycogen storage disease and HNF1a-null mice. J. Biol. Chem. 276:7963-7967. [DOI] [PubMed] [Google Scholar]
- 22.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamimura, T., A. Yoshikawa, F. Ichida, and H. Sasaki. 1981. Electron microscopic studies of Dane particles in hepatocytes with special reference to intracellular development of Dane particles and their relation with HBeAg in serum. Hepatology 1:392-397. [DOI] [PubMed] [Google Scholar]
- 24.Lai, C. L., C. K. Ching, A. K. M. Tung, E. Li, J. Young, A. Hill, B. C. Y. Wong, J. Dent, and P. C. Wu. 1997. Lamivudine is effective in suppressing hepatitis B virus DNA in Chinese hepatitis B surface antigen carriers: A placebo-controlled trial. Hepatology 25:241-244. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y. H., B. Sauer, and F. J. Gonzalez. 1998. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1a knockout mouse. Mol. Cell. Biol. 18:3059-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000-Summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 27.Luscombe, C., J. Pedersen, E. Uren, and S. Locarnini. 1996. Long-term ganciclovir chemotherapy for congenital duck hepatitis B virus infection in vivo: Effect on intrahepatic-viral DNA, RNA, and protein expression. Hepatology 24:766-773. [DOI] [PubMed] [Google Scholar]
- 28.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, R. H., and W. S. Robinson. 1984. Hepatitis B virus DNA forms in nuclear and cytoplasmic fractions of infected human liver. Virology 137:390-399. [DOI] [PubMed] [Google Scholar]
- 30.Moraleda, G., J. Saputelli, C. E. Adrich, D. Averett, L. Condreay, and W. S. Mason. 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J. Virol. 71:9392-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou, J.-H., H. Bao, C. Shih, and S. M. Tahara. 1990. Preferred translation of human hepatitis B virus polymerase from core protein-but not from precore protein-specific transcript. J. Virol. 64:4578-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontoglio, M., J. Barra, M. Hadchouel, A. Doyen, C. Kress, J. P. Bach, C. Babinet, and M. Yaniv. 1996. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84:575-585. [DOI] [PubMed] [Google Scholar]
- 33.Pontoglio, M., D. M. Faust, A. Doyen, M. Yaniv, and M. C. Weiss. 1997. Hepatocyte nuclear factor 1a gene inactivation impairs chromatin remodeling and demethylation of the phenylalanine hydroxylase gene. Mol. Cell. Biol. 17:4948-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raney, A. K., C. M. Eggers, E. F. Kline, L. G. Guidotti, M. Pontoglio, M. Yaniv, and A. McLachlan. 2001. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1a-null hepatitis B virus transgenic mice. J. Virol. 75:2900-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raney, A. K., and A. McLachlan. 1991. The biology of hepatitis B virus, p. 1-37. In A. McLachlan (ed.), Molecular biology of the hepatitis B virus. CRC Press, Boca Raton, Florida.
- 36.Sabath, D. E., H. E. Broome, and M. B. Prystowsky. 1990. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene 91:185-191. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shih, D. Q., M. Bussen, E. Sehayek, M. Ananthanarayanan, B. L. Shneider, F. J. Suchy, S. Shefer, J. S. Bollileni, F. J. Gonzalez, J. L. Breslow, and M. Stoffel. 2001. Hepatocyte nuclear factor-1a is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27:375-382. [DOI] [PubMed] [Google Scholar]
- 39.Summers, J., P. M. Smith, and A. L. Horwich. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 41.Uprichard, S. L., S. F. Wieland, A. Althage, and F. V. Chisari. 2003. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc. Natl. Acad. Sci. USA 100:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weimer, T., J. Salfeld, and H. Will. 1987. Expression of the hepatitis B virus core gene in vitro and in vivo. J. Virol. 61:3109-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieland, S. F., H. C. Spangenberg, R. Thimme, R. H. Purcell, and F. V. Chisari. 2004. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc. Natl. Acad. Sci. USA 101:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Buscher, R. Sprengle, R. Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada, G., Y. Sakamoto, M. Mizuno, T. Nishihara, T. Kobayashi, T. Takahashi, and H. Nagashima. 1982. Electron and immunoelectron microscopic study of Dane particle formation in chronic hepatitis B virus infection. Gastroenterology 83:348-356. [PubMed] [Google Scholar]
- 47.Zhang, Y. Y., and J. Summers. 2000. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J. Virol. 74:5257-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y. Z., B. H. Zhang, D. Theele, S. Litwin, E. Toll, and J. Summers. 2003. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc. Natl. Acad. Sci. USA 100:12372-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]