Abstract
The 3′-terminal end of the respiratory syncytial virus genomic RNA contains a 44-nucleotide leader (Le) region adjoining the gene start signal of the first gene. Previous mapping studies demonstrated that there is a promoter located at the 3′ end of Le, which can signal initiation of antigenome synthesis. The aim of this study was to investigate the role of the 3′ terminus of the RNA template in (i) promoter recognition and (ii) determining the initiation site for antigenome synthesis. A panel of minigenomes containing additional sequence at the 3′ end of the Le were analyzed for their ability to direct antigenome and mRNA synthesis. Minigenomes containing heterologous extensions of 6 nucleotides or more were unable to support efficient RNA synthesis. However, the activity of a minigenome with a 56-nucleotide extension could be restored by insertion of Le nucleotides 1 to 11 or 1 to 13 at the 3′ end, indicating that these nucleotides, in conjunction with the 3′ terminus, are sufficient to recruit polymerase to the template. Northern blot and 5′ rapid amplification of cDNA ends analysis of antigenome RNA indicated that antigenome initiation occurred at the first position of Le, irrespective of the terminal extension. This finding demonstrates that the 3′ terminus of the RNA is not necessary for determining the antigenome initiation site. Data are presented which suggest that following recruitment to a promoter at the 3′ end of Le, the polymerase is able to scan and respond to a promoter signal embedded within the RNA template.
Respiratory syncytial virus (RSV) is a major cause of human respiratory disease (6). It is a member of the subfamily Pneumovirinae in the family Paramyxoviridae in the order Mononegavirales. A feature shared by all mononegaviruses is their monopartite negative-sense RNA genome, which is encapsidated with multiple copies of nucleoprotein N. This ribonucleoprotein complex acts as a template for the viral RNA-dependent RNA polymerase to perform two processes: transcription, to produce capped and polyadenylated mRNAs, and RNA replication, to produce antigenome RNA, which in turn acts as a template for genome RNA synthesis (reviewed in references 20 and 30). Both transcription and replication initiation are dependent on cis-acting sequences in the leader (Le) region which lies adjacent to the 3′ terminus of the genome, upstream of the first gene (2, 14, 17, 21, 23, 40).
During transcription the polymerase initiates RNA synthesis at or near the 3′ terminus of the genome (9) and then progresses along the nucleocapsid, stopping and restarting RNA synthesis at the gene junctions in response to cis-acting gene end and gene start signals, respectively (1, 19, 33). During replication the polymerase disregards the gene junctions to produce the full-length complementary antigenome RNA. Antigenome RNA is encapsidated as it is synthesized and there is evidence that this increases polymerase processivity and allows RNA synthesis to continue through the gene end signals (16, 23, 36). The mechanisms for initiation of transcription and replication have yet to be fully elucidated. While it is commonly accepted that replication is initiated opposite the 3′ nucleotide of the Le region, there are two possible sites for transcription initiation. One long-standing model postulates that transcription is initiated at the 3′ terminus of the Le, similar to replication. According to this model, the polymerase releases the nascent transcript within a short distance and reinitiates RNA synthesis at the first gene start signal (reviewed in reference 18). However, recent data support an alternative model, in which transcription is initiated directly at the first gene start signal (3, 29, 42).
Based on our understanding of RNA synthesis, cis-acting sequences at the 3′ end of the mononegavirus genome could be responsible for several events, including polymerase binding, direction of accurate transcription and replication initiation, and antigenome encapsidation. In the case of RSV, the cis-acting sequences necessary for RNA synthesis have been identified and their roles partially dissected. Saturation mutagenesis showed that the first 11 nucleotides of Le are important for both transcription and replication, with nucleotides 3, 4, 5, 8, 9, 10, and 11 being particularly important for both processes, indicating that these nucleotides represent a key promoter element (14). Nucleotides 1, 2, 6, and 7 are required in addition for efficient replication. Subsequently it was shown that the first 15 nucleotides of Le are the only Le-specific sequence required for replication initiation. Taken together, these data suggest that nucleotides 1 to 11 contain the antigenome promoter.
Le nucleotides 16 to 34 are also important for replication. These nucleotides are necessary for efficient encapsidation of the nascent antigenome RNA and are required to allow antigenome synthesis to proceed efficiently through the gene end signals (23). With regard to transcription, the first 15 nucleotides of Le and the gene start signal are sufficient to direct mRNA synthesis from the gene start signal, and it was found that the relative spacing of these sequences is important (13, 23). These data indicate that either the first 15 nucleotides of Le contain the promoter for transcription and the gene start signal acts as a reinitiation signal, or that residues within the first 15 nucleotides of Le combine with the gene start signal to act as the transcription promoter.
Several mononegavirus genomes have promoter sequences at their 3′ termini, raising the possibility that the 3′ terminus of the nucleocapsid might also be an important feature of the promoter. Studies performed on Sendai virus, a member of the subfamily Paramyxovirinae, and vesicular stomatitis virus, a member of the family Rhabdoviridae, to address this question have given somewhat different results. In the case of Sendai virus, the polymerase prefers to utilize a promoter at the terminus of the template, but is able to recognize and initiate replication accurately at an internal promoter (37, 38). In contrast, even very short extensions at the 3′ terminus of a vesicular stomatitis virus minigenome template rendered it inactive (26), and extensions at the template 5′ terminus were tolerated, but removed during RNA replication, indicating that there was selection pressure to maintain the promoter at the terminus (26, 41).
With regard to RSV, a previous study indicated that there is some flexibility in the positioning of the promoter, as an extension of three cytosine residues at the 3′ end of the Le was tolerated (31). However, a minigenome containing an 11-nucleotide extension was not capable of replication (5). In this experiment, the effect of the 11-nucleotide extension on transcription could not be assessed, as in the minigenome system used, mRNA detection would have been dependent on the minigenome undergoing multiple cycles of replication. The aim of the present study was to investigate if the RSV promoter must be at, or near, the 3′ terminus of a template to be recognized for transcription as well as replication activity, and to ascertain if the polymerase uses the 3′ terminus to determine the antigenome initiation site.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
The plasmids used in Fig. 1 to 4 and Fig. 6 were derived from plasmid MP28Δ4H. MP28Δ4H was constructed from plasmid MP28, which has been described previously (14). Briefly, plasmid MP28 carries a dicistronic minigenome, flanked with a T7 promoter that directs synthesis of negative-sense minigenome RNA, and a hepatitis delta virus ribozyme to generate the 3′ terminus of the minigenome. In order to create plasmid MP28Δ4H, the region between the BglII site, immediately downstream of the internal gene junction in the minigenome, and the HindIII site, immediately downstream of the T7 promoter, was modified by substituting it with a PCR product generated using mutagenic oligonucleotides to introduce a 4-nucleotide deletion at the 5′ end of the RSV trailer (Tr) region and insert a hammerhead ribozyme and a short purine-rich sequence between the Tr region and the T7 promoter. Thus, the MP28Δ4H plasmid encodes a minigenome with precise ends that is limited to the antigenome synthesis step of replication. Its Le region corresponds to the wild-type RSV A2 sequence, except that it contains a G-to-C substitution at position 4 (in the negative sense) to augment replication, as described previously (14), and therefore its encoded minigenome is referred to as the wild type throughout this study.
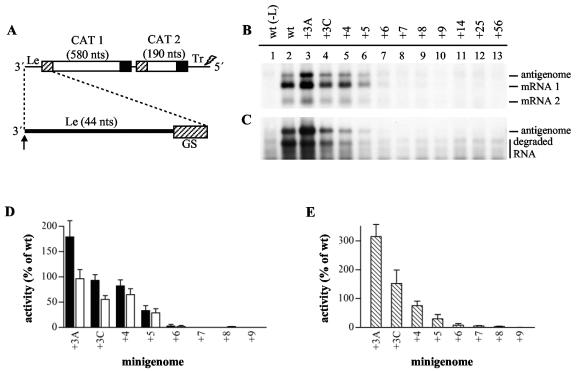
FIG. 1.
Effect of nucleotide additions at the 3′ terminus of a minigenome on RSV RNA synthesis. (A) Schematic diagram (not to scale) illustrating the dicistronic minigenome RNA used in this study and the site of nucleotide insertions. The gene start (GS) and gene end signals are indicated with small hatched and black boxes, respectively, and the genes (CAT 1 and CAT 2, encoding two different segments of the CAT gene) are indicated with white rectangles. The minigenome contains a 4-nucleotide deletion at the 5′ end of the Tr region, indicated with a lightning symbol. The 3′ end of the minigenome is enlarged and the site of insertions is indicated with an arrow. (B and C) Northern blot analysis of intracellular positive-sense RNA produced from minigenomes with additional nucleotides at the 3′ terminus of the Le region. Minigenomes containing extensions of from 3 to 56 nucleotides were analyzed. Lane 2 is a positive control showing RNA produced from a minigenome with a wild-type Le region, and lane 1 is a negative control of RNA from cells transfected with plasmid encoding a minigenome with a wild-type Le but no L polymerase. Panel B shows total intracellular RNA and panel C shows RNA from cell lysates that were treated with MCN prior to RNA purification. (D and E) Quantitation of antigenome and mRNA 1 generated from the mutant minigenomes shown in lanes 3 to 10 by phosphorimage analysis. Each RNA value was calculated as a percentage of the wild-type value (100%) and each error bar represents the standard error of the mean. Panel D shows analysis of antigenome RNA (black bars) and mRNA 1 (white bars) measured in the total RNA samples, and panel E shows analysis of encapsidated antigenome measured in the MCN-treated RNA samples.
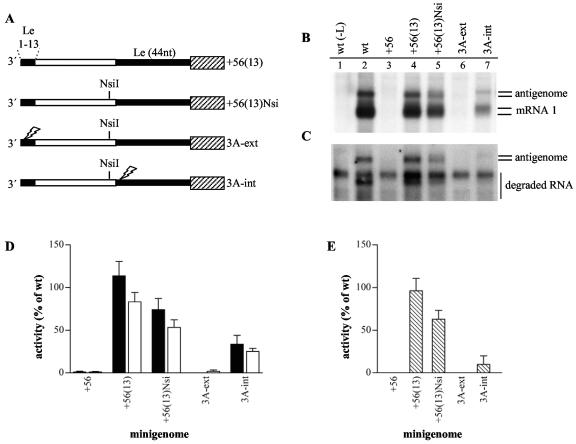
FIG. 4.
Both the external and internal promoter elements are necessary for efficient RNA synthesis. (A) Schematic diagram illustrating the sites of point mutations in the +56(13) minigenome. The first gene start sequence is illustrated with a hatched rectangle, Le-specific sequence is shown as a thin black rectangle, and nonspecific sequence is shown as a thin white rectangle. The position of the single nucleotide change to introduce an NsiI restriction site is indicated as NsiI, and the positions of the three C-to-A substitutions in the promoter elements are indicated with lightning symbols. (B and C) Northern blot analysis of intracellular positive-sense RNA produced from the wild-type (lane 2) and mutant (lanes 3 to 7) minigenomes, as indicated. Lane 1 is a negative control of RNA from cells transfected with plasmid encoding a minigenome with a wild-type Le but no L polymerase. Panel B shows total intracellular RNA and panel C shows RNA from cell lysates that were treated with MCN prior to RNA purification. (D and E) Quantitation of antigenome and mRNA 1 generated from the mutant minigenomes by phosphorimage analysis. Each RNA value was calculated as a percentage of the wild-type value (100%) and each error bar represents the standard error of the mean. It should be noted that the quantitation of antigenome and mRNA 1 includes quantitation of the more slowly migrating RNA species, in addition to the transcripts that migrated similarly to those from the wild-type minigenome. Panel D shows analysis of antigenome RNA (black bars) and mRNA 1 (white bars) measured in the total RNA samples, and panel E shows analysis of encapsidated antigenome measured in the MCN-treated RNA samples.
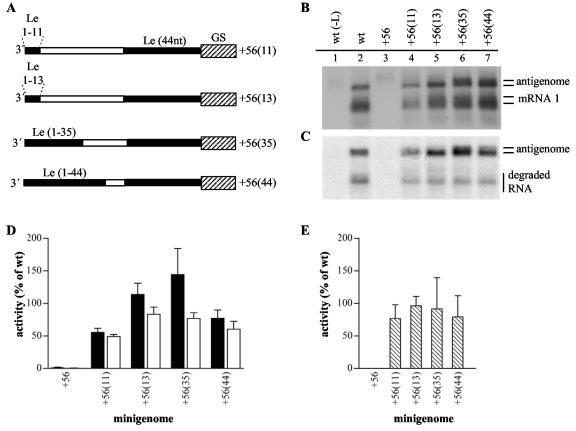
FIG. 6.
Insertion of 35 or 44 nucleotides of Le-specific sequence at the 3′ terminus of an extension allows efficient antigenome synthesis from the external promoter. (A) Schematic diagram illustrating the structures at the 3′ termini of minigenomes containing between 11 and 44 nucleotides of Le-specific sequence at the 3′ end within a 56-nucleotide extension. The gene start sequence is illustrated with a hatched rectangle, Le-specific sequence is shown as a thin black rectangle, and nonspecific sequence is shown as a thin white rectangle. (B and C) Northern blot analysis of intracellular positive-sense RNA produced from the wild-type (lane 2) and mutant (lanes 3 to 7) minigenomes, as indicated. Lane 1 is a negative control of RNA from cells transfected with plasmid encoding a minigenome with a wild-type Le but no L polymerase. Panel B shows total intracellular RNA and panel C shows RNA from cell lysates that were treated with MCN prior to RNA purification. (D and E) Quantitation of antigenome and mRNA 1 generated from the mutant minigenomes by phosphorimage analysis. Each RNA value was calculated as a percentage of the wild-type value (100%) and each error bar represents the standard error of the mean. Panel D shows analysis of antigenome RNA (black bars) and mRNA 1 (white bars) measured in the total RNA samples, and panel E shows analysis of encapsidated antigenome measured in the MCN-treated RNA samples.
Plasmid MP28Δ4H contains three unique restriction sites near the 3′ end of the minigenome sequence: an RsrII site in the hepatitis delta virus ribozyme region, a BtgI site at the junction between the hepatitis delta virus ribozyme and the Le region, and an XbaI site at the junction between the NS1 nontranslated region and the open reading frame of the chloramphenicol acetyltransferase (CAT) reporter sequence.
To construct plasmids encoding minigenomes with extensions of 3 to 25 nucleotides (Fig. 1 and 2), PCR products generated with mutagenic primers were inserted into either the RsrII-XbaI or BtgI-XbaI region of MP28Δ4H, as appropriate. The plasmid encoding minigenome +56 was created by amplifying nucleotides 5961 to 6009 of the RSV genome (RSV strain A2, GenBank accession number AF035006), consisting of the F gene sequence, into the BtgI site of plasmid MP28Δ4H. An additional nucleotide was also included to create a BamHI site. The +56 minigenome plasmid was used to generate plasmids encoding minigenomes +56(11), +56(13), +56(35), and +56(44) by inserting PCR products containing various amounts of Le, a BamHI site, and various amounts of F sequence into the RsrII and XbaI sites.
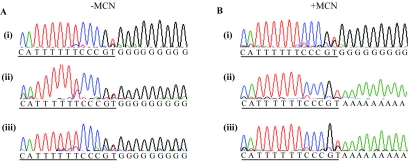
FIG. 2.
Sequence analysis of the 5′ end of antigenome RNA produced from minigenomes containing 3′-terminal extensions. Positive-sense total (panels A i to iii) or MCN-treated (panels B i to iii) RNA produced from minigenomes with nucleotide extensions consisting of AAA (panels i), CCC (panels ii), or UGCC (panels iii) was subjected to 5′ RACE and the cDNA was sequenced. The sequence of the negative-sense strand of DNA is shown, and the Le-specific sequence is underlined. The cDNA was tailed with either dGTP (panels A i to iii and panel B i) or dATP (panels B ii and iii).
Plasmid +56(13) was used to generate plasmid +56(13)Nsi, which was used in turn to generate plasmids 3A-ext and 3A-int by PCR mutagenesis. The minigenomes used in Fig. 5 each contained a wild-type Tr region and were encoded by different plasmid backbones than those used in the other figures. The wild-type minigenome in this experiment was encoded by plasmid C41, which has been described previously (14). Plasmid encoding minigenome +13only was constructed by PCR amplifying the Le-specific sequence and the nonspecific F sequence of minigenome +56(13) and inserting this PCR product into the BtgI and XbaI sites of C41. The negative control minigenome was encoded by a mutant version of plasmid MP28, containing a C-to-A mutation in position 3 of the Le (23). All sequences generated by PCR were confirmed by sequence analysis. The sequences inserted at the 3′ terminus of Le are shown in Table 1.
FIG. 5.
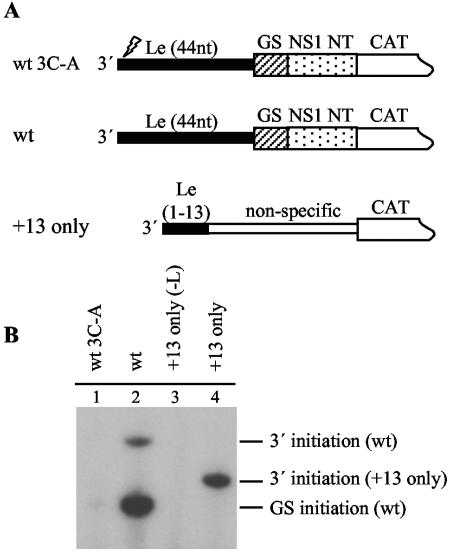
Primer extension analysis of RNA generated from a minigenome containing a minimal promoter at the 3′ terminus. (A) Schematic diagram illustrating the sequences at the 3′ termini of a wild-type minigenome and minigenome +13only, which contain 86 and 13 nucleotides, respectively, of wild-type RSV sequence at their 3′ termini. Le-specific sequence is shown as a thin black rectangle, the gene start sequence is illustrated with a hatched rectangle, the NS1 nontranslated region as a dotted rectangle, nonspecific sequence as a thin white rectangle, and CAT-specific sequence as a partial white rectangle. Minigenome wild-type 3C-A has the same 3′-terminal sequence as the wild-type minigenome except that it contains a C-to-A mutation at position 3 of the Le, indicated with a lightening symbol. (B) Primer extension analysis of total positive-sense RNA. Lane 1 represents RNA from cells transfected with a wild-type minigenome containing a C-to-A substitution at position 3 of the Le, lane 2 represents RNA produced from a wild-type minigenome, and lanes 3 and 4 represent RNA from cells transfected with the +13only minigenome, with lane 3 representing a negative control transfection in which the L plasmid was omitted.
TABLE 1.
Sequences inserted at the 3′ end of the minigenome Le region
| Minigenome | Sequence of 3′-terminal extensiona |
|---|---|
| +4 | UGCCb |
| +5 | UGCCA |
| +6 | UGCCAA |
| +7 | UGCCAAU |
| +8 | UGCCAAUA |
| +9 | UGCCAAUAG |
| +14 | UGCCAAUAGUGUUU |
| +25 | UGCCUUAGAUAACUCAAUAGUGUUU |
| +56 | UGCCUAGGUCGUUGUUUGUUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACUUAGG |
| +56(11) | UGCCCUUUUUUCCUAGGGUUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACUUAGG |
| +56(13) | UGCCCUUUUUUACCCUAGGUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACUUAGG |
| +56(13)Nsic | UGCCCUUUUUUACCCUAGGUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACgUAGG |
| 3Aextd | UGACCUUUUUUACCCUAGGUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACgUAGG |
| +13 only | UGCCCUUUUUUACCCUAGGUAGCUCGGUCUUCUCUUGAUGGUUCCAAAUACUUAGG |
| +56(35) | UGCCCUUUUUUACGCAUGUUGUUUGAACGUAUUUGCCUAGGUUCCAAAUACUUAGG |
| +56(44) | UGCCCUUUUUUACGCAUGUUGUUUGAACGUAUUUGGUUUUUUUACCUAGGCUUAGG |
| Wild typee | UGCCCUUUUUUACGCAUGUUGUUUGAACGUAUUUGGUUUUUUUACCCCGUUUAUUC |
The sequence is shown 3′ to 5′ as negative-sense RNA.
Nucleotides that share identity with the corresponding position in wild-type Le are shown in bold type.
The U-to-G substitution to introduce an NsiI site in the plasmid cDNA is in lowercase.
The C-to-A substitution at position 3 is underlined.
The sequence at the 3′ terminus of wild-type RSV is shown for comparison, with the Le sequence in bold type and the gene start sequence in italics.
Reconstituted minigenome transcription and replication.
HEp-2 cells in six-well dishes were transfected with (per well) 0.2 μg of the relevant minigenome plasmid, 0.4 μg of N, 0.2 μg of P, 0.1 μg of L, and 0.1 μg of M2-1-expressing plasmids, as described previously (11). The cells were coinfected with MVA-T7 to express the T7 RNA polymerase (43). After 42 to 48 h RNA was directly extracted to yield total intracellular RNA, or cells were lysed with nonionic detergent and treated with micrococcal nuclease (MCN) prior to RNA extraction, as described previously (11).
Northern blot hybridization.
The isolated RNA was electrophoresed and transferred to a nitrocellulose membrane as described previously (15). Negative- and positive-sense 32P-labeled CAT-specific riboprobes were synthesized by T7 RNA polymerase from templates prepared as described previously (23). Following phenol-chloroform extraction, the riboprobe was hybridized to the membrane in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 100 μg of sheared DNA per ml for a minimum of 12 h. The membranes were washed at 65°C in 2× SSC-0.1% SDS for 2 h and for 15 min in 0.1× SSC-0.1% SDS.
Rapid amplification of cDNA ends and sequence analysis.
Total intracellular or MCN-treated RNA representing one-tenth of a well of a six-well dish was annealed to a negative-sense CAT-specific primer (5′ CCAGCTCACCGTCTTT) and used as a template for Sensiscript reverse transcriptase (QIAGEN), according to the manufacturer's instructions. The cDNA was purified and tailed with dATP or dGTP using terminal transferase. The tailed product was then amplified by PCR using a CAT-specific primer (5′ GCGTCTGCAGCGGAATTCCGGATGA), which annealed upstream of the primer used for the reverse transcriptase reaction, and a primer which annealed to either the dATP or dGTP tail (5′ GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTT or 5′ GACCACGGTCGACCCCCCCCCCCCCCC, respectively). The PCR step was performed in triplicate and the products were pooled. The PCR products were purified by agarose gel electrophoresis and sequenced using a CAT-specific primer.
Primer extension analysis.
A negative-sense CAT specific primer (5′-GGGATATATCAACGGTGGTATATCCAGTG) was purified by polyacrylamide gel electrophoresis and 100 ng was end-labeled with 32P using polynucleotide kinase. The labeled primer was separated from unincorporated nucleotides using a Sephadex G25 spin column. Total RNA representing one fifth of a well of cells was annealed to one third of the 32P-labeled primer in 1× avian myeloblastosis virus cDNA synthesis buffer by heating the mixture to 65°C for 10 min and then cooling on ice for 5 min. The RNA-DNA hybrid was used as a template for reverse transcription using cloned avian myeloblastosis virus reverse transcriptase in 1× cDNA synthesis buffer, 1 mM dithiothreitol, deoxynucleoside triphosphates at 0.5 mM each and 40 U RNase inhibitor. Reverse transcription was carried out at 52°C for 2 h. The cDNA products were extracted with phenol and chloroform, precipitated with ethanol, and resuspended in 10 μl loading buffer (95% formamide, 20 mM EDTA, 0.05% xylene cyanol). Five microliters of the cDNA products were electrophoresed on a 6% polyacrylamide gel containing 7 M urea and analyzed by autoradiography. The cDNAs corresponding to RNA initiated at the 3′ terminus and the gene start signal of a wild-type minigenome generated using this primer were initially identified by comparison with a sequencing ladder run in an adjacent well (13).
RNA quantitation.
Phosphorimager analysis was carried out with a Molecular Imager FX and Quantity One quantitation software (Bio-Rad). Equivalent areas were selected for antigenome or mRNA 1 for the wild-type, a minigenome with a wild-type Le but no L polymerase [wt(−L)], and each of the mutant lanes, and the value for the a minigenome with a wild-type Le but no L polymerase was deducted from each measurement to account for background. Each RNA value was calculated as a percentage of the wild-type value of either mRNA or antigenome in the total RNA samples, or encapsidated antigenome in the MCN-treated RNA samples and the mean values were plotted. Each error bar represents the standard error of the mean from between three and seven independent experiments, depending on the mutation.
RESULTS
Polymerase can tolerate short but not long heterologous extensions at the 3′ end of the promoter.
To examine the importance of the 3′ terminus of the template for replication and transcription promoter activity, minigenomes were constructed which contained extensions at the 3′ end of Le and examined for their ability to direct antigenome and mRNA synthesis. The mutations were introduced into a dicistronic minigenome containing CAT-specific sequence in the first and second gene, as shown in Fig. 1A. The minigenome contained a 4-nucleotide deletion at the 5′ end of the Tr region, which inactivates the promoter at the 3′ end of the antigenome, limiting replication to the antigenome synthesis step (28) (data not shown). This modification conferred two advantages. First, it restricted analysis to RNA produced from the input mutant minigenome rather than from a minigenome generated under selective pressure during RSV-mediated RNA replication. This avoided the possibility that rare internal initiation events could result in “repaired” minigenomes, which could then act as efficient templates for RNA synthesis and create an impression of efficient internal initiation. Second, it allowed transcription and replication to be monitored as independent events, as has been described previously (11, 14, 23).
The minigenome plasmids were transfected into cells together with plasmids expressing the RSV N, P, L, and M2-1 proteins. Total intracellular RNA was isolated and analyzed by Northern blotting with a negative-sense CAT-specific riboprobe to examine antigenome and mRNA. Figure 1B, lane 2, shows total positive-sense RNA produced from a wild-type minigenome. The upper band is antigenome RNA and the lower band is mRNA transcribed from the first CAT 1 gene (mRNA 1), the smaller mRNA produced from the second CAT 2 gene is not readily detected and can only be seen as a faint band on this blot (mRNA 2).
To examine encapsidated antigenome, cell lysates from duplicate transfections were treated with micrococcal nuclease prior to RNA purification to digest unencapsidated RNA (Fig. 1C). In this and the following experiments, some mRNA appeared to be nuclease resistant. This is a common observation in experiments involving a nonreplicating minigenome and likely reflects nonspecific encapsidation of mRNA due to high levels of N protein relative to replicative product, as noted previously (23). In this and all subsequent experiments shown, duplicate blots of total and MCN-treated RNA samples were hybridized with a positive-sense CAT-specific riboprobe as a control to confirm that similar levels of minigenome template were produced by each of the minigenome cDNAs (data not shown). The Northern blots were analyzed by phosphorimager analysis to quantitate each of the RNAs, as described in the Materials and Methods. The bar charts in Fig. 1D and E show the levels of antigenome and mRNA in the total RNA samples (Fig. 1D) and encapsidated antigenome in the MCN-treated RNA samples (Fig. 1E).
To test if the polymerase could tolerate a small number of additional nucleotides at the 3′ terminus of the promoter, minigenomes containing either three adenine or cytosine nucleotides added to the 3′ end of the Le region were examined (minigenomes +3A or +3C). The +3A minigenome produced a higher level of antigenome and a similar level of mRNA compared to the wild-type control (Fig. 1B, compare lanes 2 and 3, and panel D). The +3C minigenome produced a similar amount of antigenome and slightly less mRNA than the wild-type control (Fig. 1B, compare lanes 2 and 4, and panels D and E). Examination of nuclease-resistant RNA showed that the antigenome RNA produced from these mutant minigenomes remained intact following MCN treatment, indicating that the mutations did not inhibit encapsidation (Fig. 1C, lanes 3 and 4, and panel E). Thus, these data confirm the finding of a previous study which showed that a tricytidylate extension permits a minigenome to function and suggest that antigenome synthesis is even enhanced by a triadenylate extension.
To determine if the promoter was functional if it was displaced further from the 3′ terminus, minigenomes containing 3′ extensions ranging from 4 to 56 nucleotides were analyzed. In each of these minigenomes, the 4 nucleotides at the 3′ end of the inserted sequence consisted of UGCC and the remainder was either random sequence (minigenomes +5, +6, +7, +8, +9, +14, and +25), or derived from the RSV F gene (minigenome +56; the sequences that were inserted are shown in Table 1). UGCC comprises the first four nucleotides of the optimal replication promoter (14, 28) and thus it was known that the polymerase is able to initiate with ACGG and that none of the extensions should inhibit RNA synthesis initiation per se.
Northern blot analysis of total and MCN-treated positive-sense RNA generated from these minigenomes demonstrated that the +4 minigenome generated similar levels of mRNA and antigenome as the wild-type Le minigenome (Fig. 1B and C, lane 5, panels D and E). However, as the length of the extension was increased from 5 to 56 nucleotides, there was a decrease in the levels of antigenome and mRNA that were generated, such that there was barely detectable RNA synthesis from minigenomes containing extensions of 6 nucleotides or more (Fig. 1B and C, lanes 6 to 13, panels D and E). Quantitative analysis of antigenome and mRNA produced from mutants containing an extension of up to 9 nucleotides showed that replication and transcription were affected similarly by each extension, with the exception of the +3A and +3C minigenomes (Fig. 1D).
To confirm that the decrease in RNA synthesis was not a sequence-specific effect, minigenomes containing extensions of 5 to 8 nucleotides with alternative random sequence were also tested. These minigenomes gave a similar pattern of RNA synthesis as the minigenomes shown, with the exception that the +6 minigenome generated marginally more antigenome and mRNA (data not shown). Therefore, these data show that extensions of 6 nucleotides or more at the 3′ terminus significantly inhibit all detectable RNA synthesis, indicating that although the promoter does not need to be directly adjacent to the end of the template, the 3′ terminus of the nucleocapsid does play an important role in promoter recognition, during both replication and transcription.
Polymerase can initiate RNA synthesis at an internal site.
It is assumed that under natural circumstances the polymerase initiates replication opposite the first nucleotide of the genome. One possibility is that the polymerase binds its promoter, locates the genome terminus, and initiates opposite the first nucleotide, regardless of the sequence at this site. Alternatively, it is possible that the promoter sequence in the template RNA determines the initiation site. To distinguish between these possibilities, the 5′ end of the antigenome RNA produced from the mutant minigenomes shown in Fig. 1 was examined. Unfortunately, it was not possible to use primer extension analysis to determine the antigenome initiation site because this approach is not sufficiently sensitive to detect the low levels of RNA produced from minigenomes limited to the antigenome step of replication, as described previously (23). Therefore, as an alternative approach, total and MCN-treated RNAs from cells transfected with the mutant minigenomes were used as templates for 5′ rapid amplification of cDNA ends (RACE) using a negative-sense CAT-specific primer for the reverse-transcriptase step. The 5′ RACE products were then sequenced to yield the consensus sequence at the 5′ end of the antigenome RNA.
Figure 2Ai shows sequence derived from total RNA produced from the +3A minigenome. The sequence is shown as negative-sense DNA and the Le sequence is underlined. In this case the cDNA was tailed with dGTP and the G residues can be seen as a homopolymeric tract. It can be seen from this analysis that the 5′ terminus of the antigenome sequence corresponded with wild-type Le sequence rather than with the AAA extension. A similar result was obtained using the MCN-treated RNA as a template (Fig. 2Bi). Analysis of total and MCN-treated RNA produced from the minigenomes containing cytosine and the 4-nucleotide UGCC extension demonstrated that the majority of these antigenome transcripts were also initiated opposite the first nucleotide of Le (Fig. 2A and B, panels ii and iii). The fact that these results were obtained with total as well as MCN-treated RNA confirms that the extension was not removed during nuclease treatment.
There was evidence that some antigenome RNAs contained the extension, for example in Fig. 2Ai and Bi three A residues could be detected beneath the homopolymeric G tail. However, assuming that 5′ RACE amplifies RNA sequences in proportion to their prevalence in the population, antigenomes containing the extension were only a minor fraction compared to those initiated correctly at nucleotide 1 of the Le region. Therefore, these data suggest that the antigenome initiation site is determined primarily by the promoter sequence, rather than by the 3′ terminus of the template.
Addition of 11 nucleotides of Le sequence to the 3′ end of a heterologous extension creates an efficient template for RNA synthesis.
As described in the introduction, mutation analysis of the RSV Le region suggested that there is a promoter element within the first 11 nucleotides of Le which is necessary for transcription and replication (14). One possible function of this element is to recruit polymerase to the template. We hypothesized that if this is the case, placing it at the 3′ end of the extension might restore template activity to an inactive minigenome.
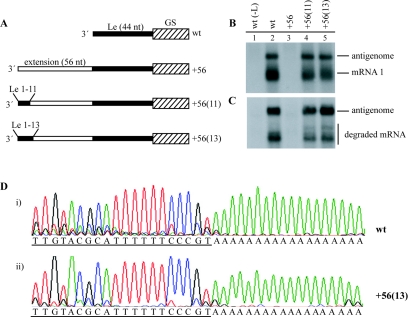
To test this hypothesis, the first 11 nucleotides of the +56 minigenome (used in Fig. 1B and C, lane 13) were replaced with nucleotides 1 to 11 of the RSV Le to create minigenome +56(11) (Table 1, Fig. 3A). This minigenome was analyzed in parallel with the wild-type and +56 minigenomes. Northern blot analysis of the RNAs produced showed that whereas the +56 minigenome produced no detectable antigenome or mRNA, the +56(11) minigenome produced significant amounts of both RNAs (Fig. 3B and C, compare lanes 3 and 4). In some experiments the levels of RNA produced from this minigenome were slightly lower than that produced from the wild-type control. Therefore, an additional minigenome was constructed containing Le nucleotides 1 to 13 at the 3′ terminus of the extension, minigenome +56(13) (Table 1, Fig. 3A). This minigenome consistently produced more RNA than the +56(11) minigenome and generated similar levels of mRNA and antigenome as the wild-type control (Fig. 3B and C, compare lanes 2 and 5; see Fig. 6 for quantitation of the RNAs produced from these minigenomes). These results suggest that residues within the first 13 nucleotides of Le function in concert with the 3′ terminus of the nucleocapsid to recruit polymerase to the template, and that most of this activity can be conferred by just nucleotides 1 to 11.
FIG. 3.
Insertion of Le-specific sequence at the 3′ terminus of an extension restores minigenome activity, but encapsidated antigenome is initiated at an internal site. (A) Schematic diagram illustrating the structure at the 3′ terminus of the wild-type, +56, +56(11), and +56(13) minigenomes. The first gene start (GS) sequence is illustrated with a hatched rectangle, Le-specific sequence is shown as a thin black rectangle, and nonspecific sequence is shown as a thin white rectangle. Note that the +56 minigenome has Le nucleotides 1 to 4 at the 3′ terminus. (B and C) Northern blot analysis of intracellular positive-sense RNA produced from the wild-type (lane 2) and mutant (lanes 3 to 5) minigenomes, as indicated. Lane 1 is a negative control of RNA from cells transfected with plasmid encoding a minigenome with a wild-type Le but no L polymerase. Panel B shows total intracellular RNA and panel C shows RNA from cell lysates that were treated with MCN prior to RNA purification. (D) Sequence analysis of the 5′ terminus of antigenome RNA produced from wild-type and +56(13) minigenomes (panels i and ii, respectively). The sequence of the negative-sense strand of DNA is shown and the Le-specific sequence is underlined. The 5′ RACE procedure was performed using MCN-treated RNA as a template and the cDNAs were tailed with dATP.
To formally exclude the possibility that the +56(11) and +56(13) minigenomes were functional due to restoration of some complementarity with the 5′ end of the minigenome, the wild-type, +56 and +56(13) minigenomes were modified to introduce a 22 nucleotide deletion at the 5′ end of the Tr region. These minigenomes were found to function similarly to their parental counterparts, confirming that addition of Le-specific sequence at the 3′ terminus of the minigenome restored template function independently of terminal complementarity (data not shown).
As described in the introduction, there is evidence that the promoter for antigenome initiation is contained within Le nucleotides 1 to 11. Thus, there were two possible initiation sites for antigenome synthesis from minigenome +56(11) and +56(13): the promoter element at the 3′ terminus of the extension, and the promoter in the internal Le sequence. To determine which promoter the encapsidated antigenome RNA was initiated from, MCN-treated RNA generated from minigenome +56(13) was analyzed by 5′ RACE and sequencing, as described above. Figure 3D shows the sequence trace that was derived from the +56(13)-encoded RNA (panel ii) below the trace derived from RNA produced by a wild-type minigenome (panel i). This analysis clearly shows that the 5′ terminus of the encapsidated antigenome produced from minigenome +56(13) corresponds with that produced from the wild-type minigenome. This result indicates that the embedded internal promoter of minigenome +56(13) was used to generate antigenome RNA.
Both the external and internal promoters are important for antigenome and mRNA synthesis.
The data presented above suggest that in minigenome +56(13), the 3′-proximal promoter sequence was necessary for template activity, but the promoter within the internal Le sequence was utilized to direct antigenome initiation. To confirm these findings and to examine if the internal promoter element was involved in transcription also, mutations were introduced into the internal or external promoter elements of minigenome +56(13). To facilitate mutagenesis, minigenome +56(13)Nsi was created by substituting a single nucleotide 5 nucleotides upstream of the internal Le region (Table 1, Fig. 4A), thus introducing an NsiI restriction site in the cDNA. Minigenome +56(13)Nsi was then modified to introduce a C-to-A substitution at position 3 of either the external or internal promoter element (3A-ext and 3A-int, respectively; Fig. 4A). Previous studies have shown that the C-to-A substitution at position 3 significantly inhibits both replication and transcription promoter activity (14, 23). The RNA produced from these minigenomes was analyzed by Northern blotting, as described above, except that it was migrated over a greater distance to improve separation of the different RNA species.
Surprisingly, the +56(13)Nsi minigenome generated somewhat lower levels of antigenome and mRNA than the parental +56(13) minigenome (Fig. 4B, lanes 4 and 5, panels D and E). In addition, whereas the +56(13) minigenome produced a single discrete band of antigenome, which comigrated with antigenome produced from the wild-type Le, the +56(13)Nsi minigenome generated a small amount of an additional slightly higher molecular weight band (Fig. 4B and C, compare lanes 2, 4, and 5). The most likely explanation for this difference is that all detectable antigenome generated from the +56(13) minigenome was initiated at the internal promoter, whereas a small amount of antigenome synthesized from the +56(13)Nsi minigenome was initiated from the external promoter, a hypothesis that is supported by the data described below.
As expected, the 3A-ext minigenome produced barely detectable levels of mRNA and antigenome (Fig. 4B and C, lane 6, panels D and E), confirming that there must be an intact promoter element near the 3′ terminus of the template for efficient transcription and replication to occur. Analysis of the 3A-int minigenome indicated that it produced less antigenome RNA than either the +56(13) or +56(13)Nsi minigenome (Fig. 4B and C, lane 7, and panels D and E). The antigenome that could be detected migrated more slowly than antigenome generated from the +56(13) or wild-type minigenome (Fig. 4B and C, compare lanes 2, 4 and 7) and correlated in size with the higher molecular weight antigenome band generated from the +56(13)Nsi minigenome (Fig. 4B, lanes 5 and 7). This indicated that this antigenome RNA was initiated from the intact external promoter element. Some of this RNA disappeared following MCN treatment, indicating that not all of it was encapsidated (Fig. 4C, lane 7, panel E). There was no evidence of a lower-molecular-weight antigenome band that correlated in size with that produced from the +56(13) or wild-type minigenome.
With regard to transcription, the 3A-int minigenome consistently generated at least twofold less mRNA 1 than the +56(13)Nsi minigenome, demonstrating that a significant proportion of transcription was also dependent on the internal promoter (Fig. 4B, lane 7, and panel D). The mRNA that was produced migrated more slowly than the major mRNA 1 band generated from the other minigenomes, indicating that it was initiated upstream of the gene start signal (Fig. 4B, compare lanes 2, 4, and 5 with lane 7). The size of the mRNA 1 transcript produced from the 3A-int minigenome is consistent with a transcript initiated at the 3′ terminus and terminated at the first gene end signal. Previous studies have identified unencapsidated, polyadenylated RNA initiated at the 3′ end of the Le and terminated at the first gene end signal, both in RSV-infected cells and in the minigenome system (4, 13, 19, 23). Thus, if the promoter element at the 3′ terminus of the minigenome was sufficient to direct RNA synthesis initiation, synthesis of such a transcript from the 3′ terminus of the minigenome would be expected.
Taken together these data confirm that the 3′-terminal promoter element was necessary for RNA synthesis to occur and suggest that this promoter element could direct RNA synthesis initiation, yielding a small amount of antigenome and a more significant amount of RNA terminated at the gene end signal. Importantly, the results indicate that an active internal promoter was also required to generate a significant proportion of the antigenome and mRNA that could be detected. These findings indicate that some polymerase recruited to the 13 nucleotides at the 3′ terminus was able to move to the internal promoter and use this to initiate antigenome and mRNA synthesis.
Le nucleotides 1 to 13 alone are sufficient to direct high levels of RNA synthesis initiation.
The data described above suggest that the polymerase is recruited by Le nucleotides 1 to 13 and then translocates to a downstream promoter to initiate RNA synthesis. If Le nucleotides 1 to 13 are sufficient to direct RNA synthesis initiation, this could provide the polymerase with the energy that would be required to dissociate and move away from its binding site at the 3′ terminus. The results from the +56(13)Nsi and 3A-int mutants shown in Fig. 4 suggest that some RNA is indeed initiated at the 13-nucleotide 3′-terminal promoter element. Therefore, to confirm that the 13-nucleotide promoter element alone is sufficient to direct RNA synthesis initiation, a minigenome containing Le nucleotides 1 to 13 but no other RSV-specific sequence at its 3′ terminus was examined (minigenome +13only; Table 1 and Fig. 5A).
To assess RNA synthesis initiation activity, primer extension analysis was employed. Because nonreplicating minigenomes do not generate sufficient RNA for primer extension analysis, this experiment was carried out using minigenomes with wild-type Tr regions. A wild-type minigenome, which contains 86 nucleotides of RSV-specific sequence at the 3′ terminus, and a minigenome containing wild-type 3′-terminal sequence except for a C-to-A mutation at position 3 of the Le region, were also analyzed for comparison. Total intracellular RNA from cells transfected with these minigenomes was hybridized with an excess of labeled negative-sense, CAT-specific primer. This primer generates products of 133 and 89 nucleotides from the wild-type minigenome, corresponding to antigenome RNA initiated at the 3′ terminus of the Le and mRNA initiated at the gene start signal, as described previously (13, 23), and would be expected to generate a product of 105 nucleotides from RNA initiated at the 3′ terminus of minigenome +13only. As this minigenome does not contain a gene start signal, no gene start initiation product would be expected.
As anticipated, primer extension on RNA from the wild-type minigenome yielded two bands, indicated as 3′ initiation (wt) and gene start initiation (wt) (Fig. 5B, lane 2). In contrast, there was no detectable RNA produced from a minigenome containing a C-to-A substitution at position 3 of the Le region, confirming that this substitution ablates RNA synthesis (Fig. 5B, lane 1). RNA from cells transfected with minigenome +13 only resulted in a single band that migrated appropriately to be derived from RNA initiated at the 3′ terminus of this minigenome (Fig. 5B, lane 4). This band was not detectable if cells were transfected with minigenome +13only but the L plasmid was omitted from the transfection, confirming that the RNA being detected was a product of the RSV polymerase (Fig. 5B, lane 3).
It should be noted that the stabilities of the RNA species detected in this experiment have not been determined and it might be expected that the RNA produced from the +13only minigenome would be relatively unstable due to the lack of an encapsidation signal (see below), and possibly also a 5′ cap. Therefore, these data do not provide information on the relative promoter activities of the wild-type and +13only minigenomes. However, they do clearly demonstrate that Le nucleotides 1 to 13 alone are sufficient to recruit the RSV polymerase and direct a high level of RNA synthesis initiation.
Increasing the amount of Le-specific sequence at the 3′ terminus increased the amount of antigenome RNA produced from the 3′ terminus of the template but did not eliminate antigenome initiation from the internal promoter.
As described in the introduction, a previous study showed that efficient encapsidation of antigenome RNA is dependent on Le nucleotides 16 to 34 (23). Thus, although the extension at the 3′ terminus of minigenome +56(13) contained sufficient sequence to direct RNA synthesis initiation, it did not contain all the cis-acting sequences necessary for efficient encapsidation and so it was not surprising that the external promoter of this minigenome did not yield significant amounts of full-length antigenome.
To determine if introducing all sequences required for encapsidation would allow full-length antigenome RNA to be generated efficiently from the 3′-terminal promoter, the +56(13) minigenome was modified to increase the length of RSV Le sequence in the extension from 13 to 35, or 44 nucleotides in minigenomes +56(35) and +56(44), respectively (Table 1 and Fig. 6A). These minigenomes were analyzed in parallel with the +56(11) minigenome for comparison of antigenome size.
Each of these minigenomes was able to produce significant levels of antigenome and mRNA, as expected (Fig. 6B, C, D, and E). The antigenome generated from the +56(11) and +56(13) minigenomes migrated similarly to that produced by the wild-type minigenome, indicating that it was initiated from the internal Le (Fig. 6B and C, compare lanes 2, 4 and 5). In contrast, minigenomes +56(35) and +56(44) produced antigenome RNAs that migrated more slowly, consistent with initiation from the 3′-terminal promoter (Fig. 6B and C, compare lanes 2, 4, and 5 with lanes 6 and 7). These RNAs were resistant to MCN treatment, indicating that they were encapsidated (Fig. 6C, lanes 6 and 7). These results confirm that full-length antigenome was efficiently produced from the 3′-terminal promoter provided the sequences required for efficient encapsidation were directly adjacent.
Interestingly, minigenomes +56(35) and +56(44) also generated antigenome RNA that migrated similarly to antigenome RNA produced from the wild-type minigenome (Fig. 6B and C, compare lanes 2, 6, and 7), indicating that it was initiated at the internal Le sequence. These data demonstrate that even in a situation in which the template contained the complete Le sequence at its 3′ terminus, a significant proportion of polymerase remained available to initiate RNA synthesis at an internal site.
DISCUSSION
A common feature of mononegavirus genomes is the location of an essential cis-acting element at the 3′ terminus of the Le region (14, 17, 21, 32, 40). The aim of this study was to determine if the 3′ end of the RSV genome RNA is important for promoter activity, either by contributing to the polymerase-binding site or by defining the antigenome initiation site. The data presented suggest that nucleotides 1 to 11 must be at or near the 3′ terminus to recruit polymerase to the template. However, provided the minigenome contained Le nucleotides 1 to 11 at the 3′ terminus, an internal promoter could be used to direct efficient and accurate RNA synthesis initiation, suggesting that following recruitment at the 3′ terminus, the polymerase is able to access embedded cis-acting sequences.
The importance of the genome 3′ terminus for Le function is clearly demonstrated by Fig. 1, which shows that although short 3′ extensions were tolerated, extensions of 6 nucleotides or more significantly inhibited RNA synthesis. Importantly, transcription was inhibited in addition to replication, indicating that the promoter for transcription must be at or near the 3′ end of the nucleocapsid, regardless of whether the polymerase initiates transcription at the 3′ terminus of the genome or internally at the gene start signal on the wild-type RSV genome. It is unclear why short extensions at the 3′ terminus were tolerated, whereas longer ones were not. One possibility is that the polymerase contacts the 3′ terminus and the promoter element in nucleotides 1 to 11 simultaneously and that it has sufficient flexibility to contact both these features provided they are not 6 nucleotides or more apart.
The positioning of the promoter region relative to the end of the genome has been shown to be important for both vesicular stomatitis virus and Sendai virus, indicating that the 3′ terminus plays a key role in mononegavirus RNA synthesis (26, 37, 38). However, it appears that the 3′ terminus plays a less important role in Sendai virus than in vesicular stomatitis virus and RSV. A possible explanation for this difference may be due to N phasing and the extent of the promoter sequence. In the case of Sendai virus (and other members of the Paramyxovirinae), recognition of cis-acting sequences is dependent not only on the nucleotide sequence of the RNA, but also on the relative positioning of the associated N protein (37, 38). Furthermore, these viruses have a bipartite replication promoter, in which two cis-acting elements lie on the same face of the helical nucleocapsid (17, 24, 25, 35). In rhabdo- and pneumoviruses, there is no evidence that relative phasing of the N subunits is significant, or that the replication promoter extends beyond the Le region (5, 7, 22, 27, 31). Therefore it is possible that the combination of RNA and protein signatures in Sendai virus results in a high-affinity binding site for the polymerase, which can be recognized as an internal element, whereas in RSV and vesicular stomatitis virus, the polymerase relies more heavily on the 3′ terminus for promoter recognition.
Whereas the previous studies on Sendai virus and vesicular stomatitis virus examined the juxtaposition of the complete Le region with the 3′ terminus, in this study we were able to demonstrate that only the first 11 nucleotides of RSV Le must be adjacent to (or near) the 3′ terminus to allow a minigenome to act as a template. As described in the introduction, residues in the first 11 nucleotides of Le were previously shown to be important for mRNA and antigenome synthesis (14), and the finding that insertion of nucleotides 1 to 11 restored both transcription and replication confirms that this element functions in both processes. These data indicate that residues within nucleotides 1 to 11 of Le combined with the 3′ terminus of the nucleocapsid function as a binding site to recruit both replication and transcription competent polymerases to the nucleocapsid.
Under natural circumstances, antigenome synthesis is initiated opposite the first nucleotide of the genome. There were two possible mechanisms by which this nucleotide is determined to be the initiation site. One possibility was that the polymerase binds its promoter, locates the genome terminus, and initiates opposite the first nucleotide, regardless of the sequence at this site. Alternatively, it was possible that the promoter sequence directs the polymerase to initiate opposite the first U residue. In this study, minigenomes containing short extensions at the 3′ terminus were found to direct antigenome synthesis from the first U residue of the internal Le sequence (Fig. 2), indicating that the promoter sequence is the primary determinant of the antigenome initiation site. It should be noted that there was some evidence of initiation at the terminal nucleotide of the minigenome (Fig. 2Ai and Bi), and a previous study showed that a trinucleotide extension at the 5′ end of an RSV miniantigenome was retained over two virus passages (31). Taken together, these data suggest that the promoter sequence is the dominant factor in antigenome start site selection but that the 3′ terminus might also contribute, albeit to a lesser extent.
The results presented here differ from those described by Samal and Collins, which indicated that the majority of minigenomes retained a trinucleotide extension (31). A possible explanation for the discrepancy is the methodology used for determining the initiation site of the encoded product. In the previous study, the sequences at the minigenome termini were determined using an intramolecular ligation technique. RNA ligation efficiency is highly dependent on the sequence at RNA termini, with a uridine residue at the 3′ terminus being particularly unfavorable (10). Therefore, it is possible that this method was biased towards sequencing a small proportion of RNA molecules initiated opposite the 3′ terminus of the extension (3′C), rather than those initiated opposite the first nucleotide of the Le (3′U). The possibility exists that the 5′ RACE procedure used in the present study is also prone to artifact, however, it should be noted that similar RACE results were obtained whether the cDNA was tailed with G or A residues, and the +4 minigenome directed internal initiation even though the first 4 nucleotides at the 5′ end of the antigenome would have been the same whether it was initiated at the 3′-terminal nucleotide or at the first nucleotide of the authentic Le sequence (Fig. 1Aiii and Biii). Furthermore, the finding that the polymerase could accurately initiate RNA synthesis at an internal promoter flanked with a 56-nucleotide extension, e.g., minigenome +56(13), supports the conclusion that the promoter sequence alone can dictate the antigenome initiation site.
The experiment shown in Fig. 4 demonstrates that the internal promoter of minigenome +56(13) generated most of the antigenome and mRNA that was produced. This result was substantiated with the data shown in Fig. 6, which showed that even if the minigenomes contain 35 or 44 nucleotides of Le sequence at the 3′ terminus, an internal promoter was utilized for antigenome synthesis. These data show that although an internal promoter could not be accessed independently, it could be recognized provided that there was a promoter at the 3′ terminus to recruit polymerase to the template.
It is of interest to speculate how the polymerase accessed the internal promoter in the minigenomes shown in this study. The findings that (i) Le nucleotides 1 to 13 are necessary for RNA synthesis and (ii) these nucleotides are sufficient to direct a high level of initiation suggest that the polymerase accessed the internal promoter by initiating RNA synthesis at the promoter at the 3′ terminus and then translocating to the promoter in the internal Le region and reinitiating RNA synthesis. This could occur if the polymerase released the transcript that it had initiated at the 3′-terminal promoter and then scanned for another initiation site. According to this model, all polymerase molecules (whether destined for replication or transcription) initiated RNA synthesis at the 3′ terminus. Some polymerase was able to continue RNA synthesis to the end of the genome to produce full-length antigenome or terminated at the first gene end signal, resulting in the higher-molecular-weight readthrough mRNA1 that could be detected from the 3A-int minigenome (Fig. 4B, lane 7). However, a significant proportion of polymerase released the nascent RNA shortly after initiation but remained attached to the template and scanned backwards or forwards until it encountered another cis-acting sequence. If it scanned backwards it encountered the 3′-proximal promoter and had another opportunity to initiate RNA synthesis at this site. If it scanned forwards, it reached the promoter contained in the internal Le sequence and could reinitiate RNA synthesis to produce either mRNA or antigenome.
Support for this model is derived from previous studies on RSV and vesicular stomatitis virus which demonstrated that short abortive transcripts could be generated during mononegavirus RNA synthesis initiation (8, 32), indicating that the mononegavirus polymerase sometimes releases the nascent RNA shortly after initiation. It is widely accepted that mononegavirus polymerases can remain attached to the template and scan for initiation sites following release of an RNA, for example, at intergenic regions (12, 34). In addition, there is evidence that the Sendai virus polymerase scans from the 3′ terminus of a genome template to locate the first internal gene start signal (39). Furthermore, in the current study, a single nucleotide change upstream of the internal Le region inhibited transcription and replication (minigenome +56[13]Nsi), which is consistent with initiation at the internal promoter being dependent on polymerase scanning from the 3′ end.
If this model is correct, it suggests that the polymerase was not committed to either transcription or replication before interacting with the template and initiating RNA synthesis. While it is possible that the polymerase employed an alternative mechanism to dissociate from the promoter at the 3′ terminus of the template and scan to the internal promoter, it is difficult to understand how it would have derived the energy and undergone the conformational changes that would be necessary to do this, while remaining attached to the template.
Regardless of the mechanism used to access the internal promoter, the data presented in this paper indicate that the 3′ terminus of the RSV nucleocapsid functions in conjunction with residues in the first 11 nucleotides of Le to recruit replication- and transcription-competent polymerases to the genome template and that, following recruitment, some polymerase is able to scan for internal cis-acting sequences.
Acknowledgments
We thank Peter Collins for providing plasmid MP28 and plasmids encoding the RSV L, P, N, and M2-1 proteins, Bernard Moss for providing MVA-T7, Andrew Cassidy for assistance in assembling Fig. 2 and 3, David McGivern and Frances Fuller-Pace for discussion, and Daniel Kolakofsky for helpful comments on the manuscript.
This work was funded by grants from the BBSRC (reference number 94/P17132) and the Wellcome Trust (reference number 065568).
REFERENCES
- 1.Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 1997. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 for polyadenylation. J. Virol. 71:8718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calain, P., and L. Roux. 1995. Functional characterisation of the genomic and antigenomic promoters of Sendai virus. Virology 211:163-173. [DOI] [PubMed] [Google Scholar]
- 3.Chuang, J. L., and J. Perrault. 1997. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J. Virol. 71:1466-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, P. L., and G. W. Wertz. 1985. Nucleotide sequences of the 1B and 1C non-structural protein mRNAs of human respiratory syncytial virus. Virology 143:442-451. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., M. A. Mink, and D. S. Stec. 1991. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc. Natl. Acad. Sci. USA 88:9663-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1433-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 7.Conzelmann, K. K., and M. Schnell. 1994. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J. Virol. 68:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase; phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 73:8384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson, S. U. 1982. Reconstitution studies detect a single RNA polymerase entry site on the vesicular stomatitis virus genome. Cell 31:635-642. [DOI] [PubMed] [Google Scholar]
- 10.Englund, T. E., and O. C. Uhlenbeck. 1978. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry 17:2069-2076. [DOI] [PubMed] [Google Scholar]
- 11.Fearns, R., M. E. Peeples, and P. L. Collins. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188-201. [DOI] [PubMed] [Google Scholar]
- 12.Fearns, R., and P. L. Collins. 1999. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J. Virol. 73:388-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearns, R., P. L. Collins, and M. E. Peeples. 2000. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J. Virol. 74:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearns, R., M. E. Peeples, and P. L. Collins. 2002. Mapping the transcription and replication promoters of respiratory syncytial virus. J. Virol. 76:1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins: transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubbay, O., J. Curran, and D. Kolakofsky. 2001. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J. Gen. Virol. 82:2895-2903. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, M. A., and A. K. Banerjee. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201-211. [DOI] [PubMed] [Google Scholar]
- 18.Kolakofsky, D., P. Le Mercier, F. Iseni, and D. Garcin. 2004. Viral RNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463-473. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effect of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.Li, T., and A. K. Pattnaik. 1999. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J. Virol. 73:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marriott, A. C., J. M. Smith, and A. J. Easton. 2001. Fidelity of leader and trailer sequences usage by the respiratory syncytial virus and avian pneumovirus replication complexes. J. Virol. 75:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGivern, D. R., P. L. Collins, and R. Fearns. 2005. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J. Virol. 79:2449-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, S. K., Y. Ito, and G. D. Parks. 1998. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J. Virol. 72:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, S. K., and G. D. Parks. 1999. RNA replication for the paramyxovirus simian virus 5 requires an internal repeated (CGNNNN) sequence motif. J. Virol. 73:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011-1020. [DOI] [PubMed] [Google Scholar]
- 27.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication and budding to generate infectious particles. Virology 206:760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qanungo, K. R., D. Shaji, M. Mathur, and A. K. Banerjee. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 101:5952-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1244. In D. M. Knipe, and P. M. Howley (ed.), Fields Virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Samal, S. K., and P. L. Collins. 1996. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J. Virol. 70:5075-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smallwood, S., and S. A. Moyer. 1993. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology 192:254-263. [DOI] [PubMed] [Google Scholar]
- 33.Stillman, E. A., and M. A. Whitt. 1997. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 71:2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stillman, E. A., and M. A. Whitt. 1998. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J. Virol. 72:5565-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tapparel, C., D. Maurice, and L. Roux. 1998. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J. Virol. 72:3117-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vulliemoz, D., and L. Roux. 2001. “Rule of six”: how does the Sendai virus RNA polymerase keep count? J. Virol. 75:4506-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vulliemoz, D., and L. Roux. 2002. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J. Virol. 76:7987-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vulliemoz, D., S. Corday, G. Mottet-Osman, and L. Roux. 2005. Nature of a paramyxovirus replication promoter influences a nearby transcription signal. J. Gen. Virol. 86:171-180. [DOI] [PubMed] [Google Scholar]
- 40.Whelan, S. P. J., and G. W. Wertz. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription, but not replication. J. Virol. 73:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan, S. P. J., and G. W. Wertz. 1999. The 5′-terminal trailer region of vesicular stomatitis virus contains a position dependent cis-acting signal for assembly of RNA into infectious particles. J. Virol. 73:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan, S. P. J., and G. W. Wertz. 2002. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc. Natl. Acad. Sci. USA 99:9178-9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]