Abstract
Previous studies have shown that herpes simplex virus type 1 (HSV-1) infection alters the phosphorylation of the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAP II), creating a new form of the enzyme known as RNAP III. However, the specific phosphorylation changes induced by HSV-1 have not been characterized. In this study, we used phospho-specific anti-CTD antibodies to probe the structure of the postinfection RNAP II. We find that RNAP III is phosphorylated on serine-5 (Ser-5) of the CTD consensus repeat but generally lacks phosphorylation on serine-2 (Ser-2). Since Ser-2 phosphorylation is normally associated with efficient transcriptional elongation and the recruitment of pre-mRNA processing factors, our results suggest that RNAP III may have altered elongation properties and decreased interactions with the mRNA processing machinery. The viral factors responsible for the reduction in Ser-2 CTD phosphorylation were studied. We found that viral immediate-early (IE) gene expression is required and sufficient, in the context of infection, for loss of Ser-2 phosphorylation. However, studies with viral mutants failed to implicate a single IE protein (among ICP0, ICP4, ICP22, and ICP27) in this process. Although most Ser-2-phosphorylated RNAP II is lost after infection, our immunofluorescence analyses identified a small subfraction that escapes loss and relocalizes to splicing antigen-rich nuclear speckles. A similar phenomenon is seen in uninfected cells after various treatments that inhibit RNAP II transcription. We hypothesize that the HSV-1-induced relocalization of residual Ser-2-phosphorylated RNAP II to nuclear speckles reflects a host response to the inhibition of cellular gene transcription.
Herpes simplex virus type 1 (HSV-1) is a common human pathogen that is associated with mild to severe disease. Its robust growth in cell culture has made it a valuable model for understanding how alphaherpesviruses replicate in their host cells. During productive infection, HSV-1 utilizes the host RNA polymerase II (RNAP II) and associated machinery to transcribe its approximately 80 genes in a coordinately regulated cascade that consists of three phases: immediate-early (IE), delayed-early (DE), and late (L) (reviewed in references 69 and 70). The five IE genes are expressed first. The promoters of these genes resemble cellular promoters, containing TATA boxes and upstream binding sites for cellular transcription factors such as Sp1. Although these promoters can be utilized to some extent by RNAP II when they are experimentally introduced into uninfected cells, their high-level transcription during infection is dependent upon a virus-encoded virion protein, VP16, which binds in a complex with host cell proteins to TAATGARAT motifs in the upstream regions of IE promoters. DE and L promoters are simpler than IE promoters. DE promoters resemble IE promoters but lack TAATGARAT motifs. L gene promoters are the least complex, consisting only of TATA boxes and downstream elements. In general, DE and L promoters are weak or inactive when introduced into uninfected cells. During infection, their utilization requires the action of IE proteins, in particular ICP4. ICP4 is a DNA-binding protein that interacts with RNAP II general transcription factors (for recent discussion of ICP4, see reference 72). Through these interactions, ICP4 promotes the formation of RNAP II preinitiation complexes on DE and L gene promoters. The IE proteins ICP0, ICP22, and ICP27 also have been implicated in the transcription of DE and L genes (19, 21, 53), but their mechanisms of action are less well characterized. At the same time that HSV-1 efficiently recruits RNAP II to viral promoters, it strongly inhibits RNAP II transcription on most host cell genes (22, 33, 45, 62-64). The mechanisms by which viral infection results in a global shift of RNAP II from host to viral genes are poorly understood.
RNAP II is a multiprotein complex composed of 12 subunits (reviewed in reference 14). The C-terminal domain (CTD) of RNAP II is a component of the largest subunit (known as the large subunit or LS) and contains multiple repeats (52 in mammalian cells) of the heptapeptide consensus sequence YSPTSPS. A minimum of 28 heptapeptide repeats is required for mammalian cell survival. The CTD is extensively phosphorylated, and this alteration regulates transcription, in part by helping to recruit pre-mRNA processing factors to nascent transcripts (5, 24, 40, 46, 47, 75). In normal cells, RNAP II exists in two predominant forms: a hypophosphorylated form, RNAP IIA, and a hyperphosphorylated form, RNAP IIO. The corresponding LS species present in these forms are referred to as IIa and IIo, respectively.
The phosphorylation status of the CTD correlates with the stage of transcription (24, 40). RNAP IIA is responsible for promoter-binding, whereas RNAP IIO is involved in promoter clearance and elongation. A number of cellular kinases and phosphatases have been implicated in mediating the phosphorylation and dephosphorylation of the CTD during normal cell growth. The CTD kinases predominantly target serine-2 (Ser-2) or serine-5 (Ser-5) of the CTD consensus repeat. Two of the most physiologically important CTD kinases are the cyclin-dependent kinase 7 (cdk7) subunit of the general transcription factor TFIIH, and the cdk9 subunit of positive transcription elongation factor b (P-TEFb). cdk7 preferentially phosphorylates Ser-5 (50, 57, 67), causing the release of the initiation complex and promoter clearance (reviewed in reference 11) and promoting the efficient recruitment of mRNA capping factors (17). In contrast, cdk9 preferentially phosphorylates Ser-2 during elongation, allowing RNAP II to overcome transcriptional blocks imposed by negative elongation factors and increasing the efficiency of 3′-end processing by recruiting polyadenylation factors (1, 52, 68). Consistent with this model, it has been found that RNAP II at the promoter/5′ end of a gene has high levels of phosphorylated Ser-5 (Ser-5P), whereas RNAP II in coding regions is enriched for phosphorylated Ser-2 (Ser-2P) (25).
We have previously shown that HSV-1 infection dramatically alters both the nuclear localization and phosphorylation of RNAP II (56). By 4 to 6 h after infection, the majority of RNAP II is recruited into virus-induced globular regions known as replication compartments. These structures form early in infection (48), exclude cellular chromatin (36, 51), and are sites of viral DNA synthesis and accumulation (9, 44). They also appear to be sites of viral genome transcription at late times (23, 44, 51). HSV-1 also alters the phosphorylation of the CTD, causing the loss of RNAP IIO and the appearance of a novel hyperphosphorylated form known as RNAP III (56). RNAP III contains LS species (collectively designated IIi), which migrate on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in between the positions of IIo and IIa, probably because they have intermediate levels of phosphorylation. The HSV-1-induced changes to RNAP II phosphorylation appear to be mediated by two distinct pathways (30, 53). One pathway is dependent on as-yet-unidentified IE proteins and results in the loss of RNAP IIO. A second pathway is dependent on ICP22 and the virion protein kinase UL13 and is responsible for inducing RNAP III. The loss of RNAP IIO correlates with host cell genome transcriptional shutoff (63), whereas the induction of RNAP III correlates with efficient viral L gene transcription in certain cell lines (30, 53).
As discussed above, Ser-2 and Ser-5 CTD phosphorylation are mediated by different cellular CTD kinases and have distinct roles in transcription. In the present study, we used phospho-specific CTD antibodies to determine whether the HSV-1-induced changes to RNAP II involve specific effects on Ser-2 or Ser-5 phosphorylation. We find that Ser-2 CTD phosphorylation is rapidly lost from RNAP II after infection, whereas Ser-5 phosphorylation is maintained. Heretofore, the absence of Ser-2 CTD phosphorylation in cells has been associated with transcriptionally inactive RNAP II (3, 32, 39, 66). However, our results suggest that the transcription of HSV-1 genes can proceed quite efficiently without this modification. This in turn suggests that the RNAP II transcription cycle in HSV-1-infected cells is significantly different from that in uninfected cells.
MATERIALS AND METHODS
Cells, viruses, and infections.
Vero cells (African green monkey kidney cells) and HEL (human embryonic lung) cells were obtained from the American Type Culture Collection. Vero cells were grown in Dulbecco modified Eagle medium supplemented to contain 5% heat-inactivated fetal bovine serum, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. The medium for HEL cells was the same as for Vero cells except that it contained 10% heat-inactivated fetal bovine serum. All tissue culture reagents except bovine serum were purchased from Life Technologies/Invitrogen (Carlsbad, CA). Bovine serum was purchased from HyClone (Logan, UT).
Strain KOS1.1 (18) was the HSV-1 wild-type (WT) strain used in these studies. Virus mutants in the ICP0 (n212) (6), ICP4 (d120) (10), ICP22 (d22lacZ) (30), ICP27 (d27-1), (54), and UL13 (d13lacZ) (30) genes have all been previously described. The derivation of the HSV-1 ICP22-UL13 double mutant (d22/13) is described below. All infections were carried out in phosphate-buffered saline (PBS) containing 0.1% glucose and 0.1% heat-inactivated newborn calf serum. Viral adsorption was for 1 h at 37°C, after which time the viral inoculum was replaced with 199 medium containing 2% heat-inactivated newborn calf serum, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. Infections were incubated at 37°C.
The ICP22/UL13 double-mutant d22/13 was constructed by using a marker transfer protocol (54), as follows. First, an ICP22 deletion virus, designated d22, was constructed. This mutant contains the same ICP22 gene deletion as d22lacZ (30) but lacks its E. coli lacZ gene marker. To generate d22, infectious d22lacZ DNA was cotransfected into Vero cells with plasmid DNA from pUCNSΔ, an HSV-1 BamN-derived plasmid carrying a deleted version of the ICP22 gene (30). The cultures were harvested after 6 days, and viral stocks were made. Recombinants that did not express β-galactosidase were identified by plaque assay in Vero cells in the presence of 300 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml. The resulting isolates were plaque purified three times in Vero cells and characterized by diagnostic Southern blotting. An isolate which had the expected genomic structure was designated d22. To construct d22/13, viral DNA from d22 was cotransfected with plasmid DNA from pBglOZ, which contains a lacZ gene-disrupted UL13 gene (30). The transfected cultures were harvested after several days, and β-galactosidase-expressing recombinants were identified by X-Gal plaque assay. Two independent isolates (obtained from different transfections) were plaque purified three times and designated d22/13 and d22/13b. Their genomic structures were confirmed by Southern blotting. Both isolates were found to replicate efficiently in Vero cells but to be compromised for growth in HEL cells, similar to the phenotype of ICP22 mutants (data not shown).
Analysis of viral mutants.
Analysis of protein expression by immunoblotting was carried out as previously described (30, 42). Briefly, mock- or virus-infected cells were scraped in PBS containing protease (50 μg of TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone2] per ml and 25 μg of phenylmethylsulfonyl fluoride per ml) and phosphatase inhibitors (1 mM sodium orthovanadate, 25 mM sodium fluoride, 50 mM tetrasodium phosphate, 50 mM sodium pyrophosphate). The cells were pelleted by low-speed centrifugation and lysed immediately in SDS-polyacrylamide gel sample buffer. Proteins were separated by SDS-6% PAGE, electrophoretically transferred to TransBlot nitrocellulose membranes (Bio-Rad; Hercules, CA), and probed with anti-RNAP II LS specific antibodies. The antibodies used were ARNA3 (26), purchased from Research Diagnostics (Flanders, NJ), and H5 and H14 (4), purchased from Covance (Denver, PA). All antibodies were diluted 1:500 in PBS. As a loading control, the levels of the cellular endosomal antigen EEA1 (37) were also determined. An antibody specific for EEA1 was purchased from BD Biosciences (San Jose, CA) and used at a 1:2,500 dilution. The secondary antibody used for immunoblot detection was horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G, purchased from Jackson Immunoresearch (West Grove, PA) and diluted 1:7,500. It was detected by using enhanced chemiluminescence (ECL) Western blotting detection reagents (catalog no. RPN2106; Amersham; Piscataway, NJ). For the quantitative immunoblot analysis shown in Fig. 3, immunoblot detection was carried out with ECL-Plus reagents (catalog no. RPN2132; Amersham). Direct chemifluorescence detection of the immunoblot signals was done by using a Molecular Dynamics Storm 840 Imager. Signal intensities for LS and EEA1 were quantitated by using ImageQuant image analysis software (Amersham). The LS signal in each sample was normalized to the corresponding EEA1 signal for that sample.
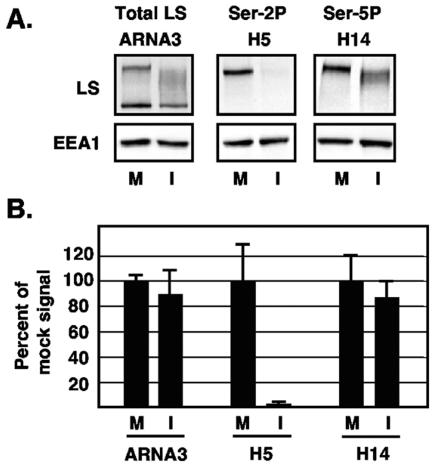
FIG. 3.
Quantitative immunoblot analysis of RNAP II LS forms after HSV-1 infection. Triplicate cultures of Vero cells were either mock infected (M) or infected with WT HSV-1 (I), and protein extracts were prepared at 8 hpi. Equal fractions of each extract were analyzed by immunoblotting with MAb ARNA3 to detect all forms of LS or MAbs H5 or H14 to detect Ser-2- or Ser-5-phosphorylated LS, respectively. As a loading control, cellular EEA1 was also analyzed. Signal intensities were determined by direct chemifluorescence imaging of the immunoblots using a Molecular Dynamics Storm 840 Imager. (A) Representative digital images of the data show the approximate areas of the blots that were quantitated. (B) Quantification of LS forms after HSV-1 infection. The chemifluorescence imaging data were quantitated by using ImageQuant analysis software. The RNAP II LS signal for each sample was normalized to the corresponding EEA1 signal to correct for protein loading variations. The mean values for mock-infected cells were defined as 100%, and the error bars represent standard deviations.
Indirect immunofluorescence and confocal microscopy.
Vero cells, grown to ∼80% confluence on coverslips, were mock infected or infected with HSV-1. In most experiments (including those shown in Fig. 6 and 7), samples were processed for immunofluorescence as described by Zeng et al. (73). Briefly, the cells were permeabilized for 2 min with 0.5% Triton X-100 in CSK buffer (10 mM PIPES [pH 7], 1 mM EGTA, 3 mM MgCl2, 20% sucrose) and then fixed in 3.7% formaldehyde in the same buffer for 20 min. In other experiments (not shown), the cells were processed for immunofluorescence by immediate fixation in 3.7% formaldehyde, followed by acetone permeabilization (48). For fluorescence staining, coverslips were incubated at 37°C for 1 h with ARNA3 (diluted 1:100), 8WG16 (diluted 1:200), H5 (diluted 1:100), or H14 (diluted 1:300). 8WG16 was a gift from Nancy Thompson. In some experiments, cells were costained with rabbit antisera specific for ICP8 (a gift from David Knipe; diluted 1:800) or an antibody specific for SC35 (Sigma; diluted 1:1,000). After primary antibody incubation, cells underwent secondary staining with a 1:2,000 dilution of Cy3-conjugated goat anti-mouse immunoglobulin G. For double-labeling experiments analyzing ICP8, 1:1,000 Cy2-conjugated goat anti-rabbit immunoglobulin G was also included. In experiments in which RNAP II and SC35 were analyzed, staining was done with a 1:1,000 dilution of Cy3-conjugated goat anti-mouse immunoglobulin M μ chain antibody, and a 1:200 dilution of Cy2-conjugated anti-mouse immunoglobulin G Fc fragment antibody. All secondary antibodies were purchased from Jackson Immunoresearch. In all double-labeling studies, control experiments were performed to ensure that staining was dependent on the relevant primary and secondary antibodies. Dual immunofluorescence detection was performed with a MRC-1024 confocal microscope (Bio-Rad, Hercules, CA) equipped with a krypton-argon laser. Series of 0.5-μm optical sections were acquired by using LaserSharp (version 2.1) acquisition software and analyzed with Confocal Assistant software (T. C. Belije, University of Minnesota). Images were then pseudocolored and merged by using Adobe Photoshop software (Adobe Systems, Inc., Mountain View, CA).
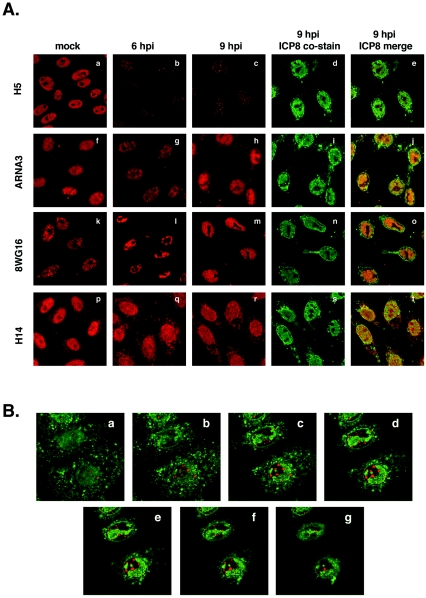
FIG. 6.
Ser-2-phosphorylated LS levels decline in infected cells but residual species relocalize to nuclear foci. (A) Localization of various RNAP II forms after viral infection. Vero cells were mock infected or infected with HSV-1. At 6 or 9 hpi, cells were fixed and processed for immunofluorescence with anti-RNAP II LS antibodies H5 (a to e), ARNA3 (f to j), 8WG16 (k to o), or H14 (p to t). Cells were costained for ICP8, a marker for viral replication compartments. (B) Residual Ser-2-phosphorylated RNAP II is associated with replication compartments. Cells were infected with WT HSV-1 and processed for immunofluorescence at 6 hpi with H5 (red signal) and a rabbit antiserum specific for ICP8 (green signal), a marker for HSV-1 replication compartments. Confocal microscopy was used to capture a series of seven continuous 0.5-μm optical sections.
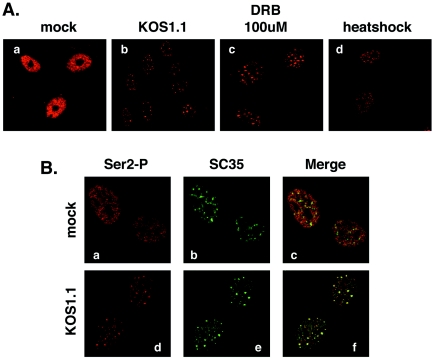
FIG. 7.
Both HSV-1 infection and RNAP II transcription inhibitors cause Ser-2-phosphorylated RNAP II to relocalize to splicing antigen-rich nuclear speckles. (A) Localization of Ser-2-phosphorylated RNAP II after HSV-1 infection or treatments which lead to transcriptional inhibition. Vero cells were mock-infected (a) or infected with HSV-1 KOS1.1 (b) for 6 h. Replicate uninfected cultures were treated with 100 μM DRB for 2 h (c) or subjected to heat shock for 1 h at 45° (d). All samples were processed for immunofluorescence and stained with MAb H5. (B) Foci containing Ser-2-phosphorylated RNAP II after HSV-1 infection correspond to splicing antigen-rich nuclear speckles. Vero cells were mock infected (a to c) or infected for with HSV-1 KOS1.1 for 6 h (d to f). Cells were processed for immunofluorescence with H5 and an antibody specific for splicing factor SC35.
RESULTS
Loss of CTD Ser-2 phosphorylation after HSV-1 infection.
Our past results indicate that HSV-1 infection alters RNAP II phosphorylation, causing a loss of the normal RNAP IIO form and the induction of a novel form, RNAP III (55). Although RNAP IIO is primarily phosphorylated on Ser-2 and Ser-5 of the CTD heptapeptide consensus repeat (YSPTSPS), the status of the infected cell RNAP III in regard to Ser-2 and Ser-5 phosphorylation is unknown. We decided to address this question through the use of phospho-specific anti-CTD monoclonal antibodies (MAbs) (reviewed in reference 40). Our previous studies on the effects of HSV-1 on RNAP II utilized either ARNA3, a MAb specific for the body of the LS, or 8WG16, an anti-CTD MAb that reacts strongly to the HSV-1-induced IIi form of LS. Although the reactivity of 8WG16 is influenced by phosphorylation, the epitope recognized is not well defined (40). More helpful in determining the phosphorylation status of RNAP II are MAbs H5 and H14 (4), which are specific for the Ser-2- and Ser-5-phosphorylated CTD, respectively. Therefore, we used these two antibodies to characterize the postinfection RNAP II.
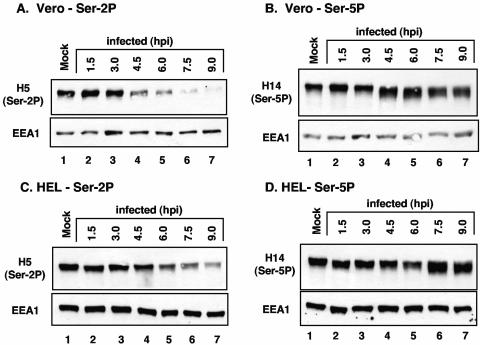
To carry out the analysis, Vero or HEL cells were mock-infected or infected with WT HSV-1 (strain KOS1.1) at a multiplicity of infection of 10. Total protein extracts were prepared every 1.5 h for 9 h and subjected to immunoblotting with either H5 or H14. In both Vero and HEL cells, HSV-1 infection caused a dramatic decline in CTD phosphorylation on Ser-2, as judged by the decrease in H5 reactivity of the LS (Fig. 1A and C, respectively, lanes 2 to 7). This was evident by 4.5 h postinfection (hpi) in Vero cells and by 6 hpi in HEL cells. In other experiments, loss of Ser-2 phosphorylation in Vero cells was observed as early as 3 hpi (data not shown). In both cell lines, phosphorylation on Ser-2 had dramatically declined by 9 hpi. In contrast, analysis with the Ser-5P-specific MAb H14 suggested that RNAP II remains phosphorylated on Ser-5 after HSV-1 infection (Fig. 1B and D, respectively, lanes 2 to 7). However, it was noted that the electrophoretic mobility of the LS species containing Ser-5P increases as infection proceeds; subsequent experiments demonstrated that this faster-migrating species is the HSV-1-induced IIi form of the LS (see below). In this and other experiments, we found that the RNAP II modifications induced by HSV-1 were qualitatively similar in Vero and HEL cells. However, the kinetics of RNAP II modification were consistently more rapid in Vero than in HEL cells.
FIG. 1.
Loss of RNAP II Ser-2 phosphorylation during HSV-1 infection. Vero (A and B) or HEL (C and D) cells were mock infected or infected with WT HSV-1 (strain KOS1.1) at a multiplicity of infection of 10, and total cell proteins were harvested at the times indicated. Equal fractions of each protein extract were analyzed by immunoblotting with H5 (A and C) or H14 (B and D), which recognize Ser-2- and Ser-5-phosphorylated RNAP II LS, respectively. As a loading control, a separate portion of each filter was probed for the cellular endosomal protein EEA1, the abundance of which does not change significantly during HSV-1 infection.
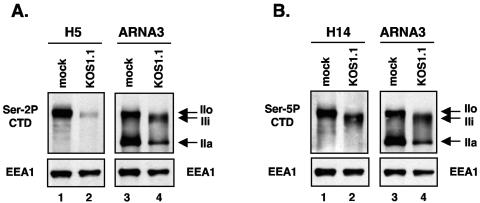
To confirm the identity of the H5- and H14-reactive LS forms seen in infected cells, we compared their electrophoretic mobilities to those of known LS forms. To do this, we probed replicate immunoblots of mock- and HSV-1-infected samples with H5 (or H14) and the anti-LS MAb ARNA3. Since ARNA3 recognizes the body of the LS, its binding is independent of phosphorylation. The H5/ARNA3 analysis (Fig. 2A) revealed that the Ser-2P-containing species seen in mock-infected cells (lane 1) corresponds to IIo (lane 3). This species was dramatically reduced in abundance after HSV-1 infection (lane 2), as observed in the previous experiment. Notably, little if any of the Ser-2P-containing material comigrated with IIi (lane 4), indicating that RNAP III generally lacks Ser-2 phosphorylation. Analysis of the protein samples using H14 and ARNA3 (Fig. 2B) indicated that the Ser-5P-containing species seen in mock-infected cells (lane 1) also corresponds to IIo (lane 3). In addition, the H14/ARNA3 analysis showed that the Ser-5P-containing species comigrates with IIi (compare lanes 2 and 4). This indicates that RNAP III is phosphorylated on Ser-5.
FIG. 2.
Identification of phospho-specific RNAP II forms seen in HSV-1-infected cells. Proteins were harvested from mock- or KOS1.1-infected Vero cells at 6 hpi. (A) Duplicate immunoblots were prepared from the same gel were probed with H5 (left) to detect Ser-2-phosphorylated LS forms and ARNA3 (right) to detect all forms. (B) Analysis was as in panel A, except that the H14 antibody was used to identify Ser-5-phosphorylated LS forms. As a loading control, EEA1 levels were also determined.
These results suggest that HSV-1 infection causes a dramatic and specific reduction in the level of Ser-2-phosphorylated RNAP II. To verify this and to measure the degree of the effect, we carried out a quantitative immunoblotting analysis. Triplicate cultures of Vero cells were mock infected or infected with HSV-1, and protein extracts were prepared at 8 hpi. The proteins were subjected to immunoblotting with ARNA3 to detect total LS and with H5 and H14 to detect Ser-2- and Ser-5-phosphorylated LS, respectively. To avoid quantitation problems associated with X-ray film exposures, signal intensities were obtained directly from the immunoblots by chemifluorescence imaging. To correct for variations in protein loading, each LS signal was normalized to the corresponding EEA1 signal for that sample. The results of this analysis, shown in Fig. 3, indicate that by 8 hpi, HSV-1 infection causes a more than 40-fold reduction in Ser-2 phosphorylated LS (2.1% of the mock-infected level). In contrast, both total LS levels and Ser-5-phosphorylated LS levels are only slightly affected by HSV-1 infection (89.6 and 86.9%, respectively, of mock-infected values).
Together, the experiments described above indicate that HSV-1 infection causes a specific loss of Ser-2 phosphorylation on RNAP II, such that the major hyperphosphorylated form in infected cells, RNAP III, bears Ser-5 but not Ser-2 CTD phosphorylation. This conclusion is consistent with our past analyses of RNAP II in infected cells using the 8WG16 MAb (56), which has been reported to not recognize the CTD when Ser-2 is phosphorylated (8, 41). We found that 8WG16 fails to recognize the IIo form of LS from uninfected Vero cells, a finding consistent with that form bearing Ser-2P. In contrast, 8WG16 binds strongly to IIi, a finding consistent with that phosphorylated form lacking modification on Ser-2.
Viral IE protein expression induces loss of CTD Ser-2 phosphorylation.
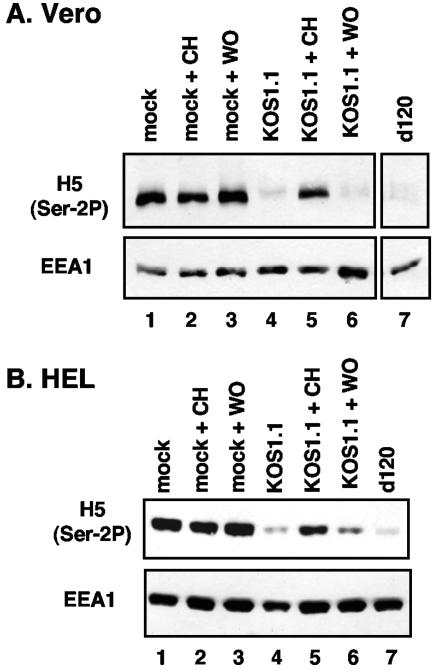
We next sought to determine whether viral gene expression is required for the loss of Ser-2-phosphorylated RNAP II. Vero cells were mock infected or infected with WT HSV-1 or d120, an ICP4 deletion mutant that is unable to express DE or L genes (10). Some of the infections were carried out in the presence of cycloheximide (CH) to prevent protein synthesis, and in some cases the CH was washed out at 8 hpi. Total protein extracts were prepared at 10 hpi and analyzed by immunoblotting with H5 (Fig. 4A). As expected, Ser-2P was lost in the WT (lane 4) but not the mock infections (lanes 1 to 3). Viral protein synthesis is required for loss of Ser-2P, since near-normal levels were maintained when WT infections were treated continuously with CH (lane 5). Consistent with this conclusion, CTD Ser-2 phosphorylation was lost within 2 h when cycloheximide was washed out, allowing translation of accumulated mRNAs (lane 6). Interestingly, Ser-2 CTD phosphorylation was lost in cells infected with d120 (lane 7). This result indicates that infection need not progress past the IE stage to induce the loss of Ser-2P and that ICP4 is not required. An identical experiment was carried out in HEL cells, with very similar results (Fig. 4B). Together, the results from the cycloheximide reversal and d120 infections indicate that IE proteins are required and sufficient in infected cells to induce the loss of Ser-2 phosphorylation from the RNAP II CTD.
FIG. 4.
Loss of phosphorylation on Ser-2 of the RNAP II CTD is dependent on expression of HSV-1 IE proteins. Vero (A) or HEL (B) cells were mock infected (lanes 1 to 3), infected with KOS1.1 (lanes 4 to 6), or infected with the ICP4 mutant d120 (lane 7). Some infections (lanes 2, 3, 5, and 6) were carried out in the presence of 50 μg of cycloheximide (CH)/ml to inhibit protein synthesis. In some cases (lanes 3 and 6), the cycloheximide was washed out at 8 hpi (WO). At 10 hpi, all protein samples were harvested. Immunoblotting was carried out with H5 to detect Ser-2-phosphorylated LS forms. As a control, EEA1 levels were also determined.
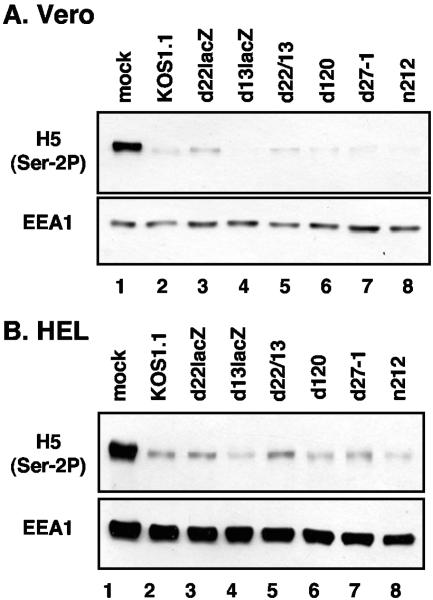
To see whether a specific IE protein is required, we utilized various HSV-1 IE gene mutants: d120, d22lacZ (an ICP22 mutant), d27-1 (an ICP27 mutant), and n212 (an ICP0 mutant). We also analyzed the role of UL13, since this virion-localized protein kinase is present at IE times and has been implicated in altering RNAP II (30). To study UL13, we used the UL13 mutant d13lacZ (30), as well as d22/13, a recently isolated ICP22/UL13 double mutant (described in Materials and Methods). Vero cells were mock infected or infected with KOS1.1 or the various mutants. Total proteins were collected at 9 hpi and immunoblotted with H5 (Fig. 5A). As expected, WT HSV-1 infection led to a striking reduction in Ser-2-phosphorylated LS (lane 2) compared to the mock infection (lane 1). All mutant infections had the same effect (lanes 3 to 8). This experiment was repeated in HEL cells, with very similar results (Fig. 5B). We conclude that ICP4, ICP22, ICP27, ICP0, and UL13 are not individually required for the loss of CTD Ser-2 phosphorylation after HSV-1 infection. In addition, the combined absence of ICP22 and UL13 does not prevent the loss of Ser-2-phosphorylated RNAP II.
FIG. 5.
Role of specific IE proteins and UL13 in causing the loss of Ser-2 phosphorylation on the CTD. Vero (A) or HEL (B) cells were mock infected (lanes 1), infected with WT HSV-1 (lane 2), or infected with the virus mutants indicated (lanes 3 to 8). Protein extracts were prepared at 9 hpi and analyzed by immunoblotting with MAb H5 to detect Ser-2-phosphorylated LS. Cellular protein EEA1 was also analyzed.
Relocalization of residual Ser-2-phosphorylated RNAP II to nuclear speckles.
The immunoblotting experiments indicate that the levels of Ser-2-phosphorylated RNAP II decline dramatically after HSV-1 infection. To confirm this, we carried out an immunofluorescence analysis of infected cells. Vero cells were mock infected or infected with HSV-1 and then processed for immunofluorescence at 6 and 9 hpi. Cells were stained with H5 to assess Ser-2-phosphorylated RNAP II or with other LS-specific MAbs (ARNA3, 8WG16, or H14) for comparison. All samples were costained for the viral ICP8 single-stranded DNA-binding protein, which serves as a marker for HSV-1 replication compartments (49). The result of the H5 staining (Fig. 6Aa to e) confirmed that the overall level of Ser-2-phosphorylated RNAP II declines significantly after infection. However, we noted that there was residual staining of this form of RNAP II in small nuclear foci which appeared to localize outside of replication compartments (Fig.6Ae). In contrast, and consistent with previously published results (56), both ARNA3 and 8WG16 staining showed recruitment of much of RNAP II into viral replication compartments after infection (Fig. 6Af to j and k to o, respectively). The H14 staining (Fig. 6Ap to t), specific for Ser-5-phosphorylated RNAP AII, gave a somewhat different pattern in infected cells than did ARNA3 or 8WG16. Staining was distributed throughout most of the nucleus, including but not limited to viral replication compartments (Fig.6At). Several conclusions can be made from this experiment and several repeat analyses. First, consistent with the immunoblotting experiments, the abundance of the Ser-2-phosphorylated form of RNAP II declines significantly after HSV-1 infection. Second, a subfraction of this form of RNAP II escapes loss and localizes to small nuclear foci. Third, Ser-5-phosphorylated RNAP II is present in viral replication compartments, as one might expect if RNAP III mediates viral genome transcription. However, Ser-5 phosphorylated RNAP II is also found in the other areas of the nucleus.
We used confocal microscopy to further analyze the spatial relationship between the nuclear foci containing Ser-2-phosphorylated RNAP II and viral replication compartments. Optical sectioning of multiple infected cells indicated that nearly all of the foci localized outside of replication compartments but in close apposition to them. This is shown for one field in Fig. 6Ba to g.
The Ser-2-phosphorylated RNAP II-bearing foci seen in HSV-1-infected cells are reminiscent of similar foci that have been described in uninfected cells after treatments that inhibit RNAP II transcription (4, 73). Specifically, when host transcription is blocked by various means, including heat shock or the addition of metabolic inhibitors, Ser-2P-bearing RNAP II moves from a mostly diffuse nuclear localization into cellular structures known as nuclear speckles. Nuclear speckles are thought to be storage, assembly, and/or modification sites for pre-mRNA splicing factors and some other proteins involved in RNAP II-directed gene expression (reviewed in reference 27). We carried out an experiment to see if the H5 antigen-positive nuclear foci observed in HSV-1-infected cells are similar in shape, size, and distribution to those induced in uninfected cells upon RNAP II transcriptional inhibition. To do this, Vero cells were mock infected or infected with WT HSV-1 for 6 h. Replicate uninfected cultures were incubated with 5,6-dichloro-1-β-d-ribobenzimidazole (DRB), a chemical inhibitor of transcription, or subjected to heat shock for 1 h at 45°C. All cells were fixed in parallel and stained for Ser-2P-bearing RNAP II using H5 (Fig. 7A). As expected, Ser-2P-containing RNAP II in uninfected/untreated Vero cells had a diffuse nuclear, non-nucleolar localization (Fig.7Aa), whereas HSV-1 infection caused a diminished signal that was concentrated in nuclear foci (Fig.7Ab). Treatment of uninfected cells with either DRB or heat shock had very similar effects on Ser-2 CTD phosphorylated RNAP II, i.e., a diminished signal and relocalization of the antigen into nuclear foci (Fig. 7Ac and d, respectively). These foci closely resembled those formed in HSV-1-infected cells.
To see whether the H5-positive foci seen in HSV-1-infected cells correspond to splicing antigen-rich nuclear speckles, mock- or HSV-infected Vero cells were costained with H5 and an antibody specific for splicing factor SC35, a marker for nuclear speckles (27) (Fig. 7B). As expected, nuclear speckles were seen in uninfected cells, but these did not obviously colocalize with Ser-2 phosphorylated RNAP II (Fig. 7Ba to c). However, after HSV-1 infection, the H5 and SC35 signals substantially overlapped (Fig. 7Bd to f). Because fixation conditions can influence the degree to which H5 stains nuclear speckles (13), we tested whether the HSV-1-induced relocalization of the H5 antigen to speckles was fixation dependent. The results were independent of fixation, in that similar H5-positive speckles were observed in infected cells when the cells were immediately fixed in formaldehyde, as well as when they were first permeabilized with Triton X-100 prior to fixation, the latter being conditions that give optimal H5 staining of speckles (4) (data not shown). We conclude from these experiments that HSV-1 infection causes a subfraction of Ser-2P-bearing RNAP II to migrate into splicing antigen-rich nuclear speckles, which is similar to what happens in uninfected cells when transcription is inhibited.
DISCUSSION
RNAP II in HSV-1-infected cells lacks Ser-2 phosphorylation.
Our studies demonstrate that the major hyperphosphorylated form of RNAP II in HSV-1-infected cells, RNAP III, lacks Ser-2 phosphorylation on its CTD. This contrasts with the usual situation in uninfected cells, wherein active RNAP II acquires Ser-2 phosphorylation as part of the normal transcription cycle. Although rare, global deficiencies in CTD Ser-2 phosphorylation are not unique to HSV-1-infected cells. This RNAP II modification has been reported to be missing during early stages of embryogenesis in Xenopus laevis (39) and Drosophila melanogaster (32), in mammalian cells after ERK-type mitogen-activated protein kinase signaling (3), and in bunyameravirus-infected cells (66). In all of these cases, however, the lack of Ser-2 phosphorylation was associated with a transcriptionally inactive RNAP II. In contrast, photoaffinity cross-linking experiments demonstrate that RNAP III can be cross-linked to nascent mRNA and therefore is transcriptionally active (63). Thus, our results reveal a novel situation in which RNAP II transcription occurs efficiently in the absence of Ser-2 CTD phosphorylation. This suggests that the transcription cycle in HSV-1-infected cells is significantly different from that in uninfected cells.
The HSV-1-induced loss of Ser-2 CTD phosphorylation happens under IE conditions prior to the global shift in transcription from host to viral genes. We therefore suspect that this phenomenon reflects an active viral strategy to modify RNAP II. We previously proposed a model (30, 53) in which HSV-1 infection modifies RNAP II via two distinct pathways: (i) an IE protein-dependent pathway that results in RNAP IIO loss and (ii) an ICP22/UL13-dependent pathway that induces RNAP III. Our present results can be placed in the context of this model by proposing that the HSV-1-induced loss of Ser-2 CTD phosphorylation is involved in the pathway leading to loss of RNAP IIO. The evidence for this is based on our finding that IE genes are required and sufficient, in the context of infection, for loss of Ser-2 phosphorylation (Fig. 4). In addition, we find that ICP0, ICP4, ICP22, and ICP27 are not singly responsible for the effect (Fig. 5). The very same findings apply to the physical loss of RNAP IIO. That is, IE protein expression is required and sufficient for loss of RNAP IIO, but ICP0, ICP4, ICP22, and ICP27 are not individually required (53). Based on this correlation, we suggest that the loss of Ser-2P from the CTD and the physical loss of RNAP IIO are mechanistically related events.
It is important to emphasize that HSV-1-mediated loss of Ser-2 CTD phosphorylation on RNAP II is insufficient to explain the induction of RNAP III. Indeed, in the case of ICP22 or UL13 mutants, Ser-2 phosphorylation is efficiently lost (Fig. 5), but little Ser-5 phosphorylated intermediately migrating LS is produced, as determined by H14 immunoblotting (K. Fraser and S. Rice, unpublished data). Thus, ICP22 and UL13 are required for enhanced levels of Ser-5-phosphorylated RNAP II. The mechanisms and CTD kinases involved in this process are as yet unknown.
Possible mechanisms and viral factors involved in loss of Ser-2-phosphorylated RNAP II.
There are several possible mechanisms by which HSV-1 infection could cause a decrease in the levels of Ser-2-phosphorylated RNAP II. First, infection could inhibit the activity or levels of P-TEFb, the major Ser-2 CTD kinase in mammalian cells. The inhibition of cdk9 subunit of P-TEFb by various pharmacological inhibitors has been shown to lead to the loss of the IIo LS form (28), similar to what happens in infected cells. Second, HSV-1 could activate a CTD phosphatase that removes Ser-2P from the CTD. Third, HSV-1 could induce the proteolysis of Ser-2-bearing RNAP II. In such a scenario, the loss of Ser-2P would be due to the targeted degradation of RNAP II, rather than to an effect on its phosphorylation. Consistent with this, preliminary evidence indicates that proteasome inhibitors prevent the loss of Ser-2P from the CTD following infection (Fraser and Rice, unpublished). It should be noted that a proteolysis mechanism is not mutually exclusive with other mechanisms, such as inhibition of a CTD kinase. There is evidence that the proteasome tracks with RNAP II during transcription, and it has been proposed that the proteasome functions generally to remove RNAP II complexes that have become arrested during transcription (12, 65). Thus, if HSV-1 IE proteins cause transcribing RNAP II to stall (e.g., by inhibiting P-TEFb), this could lead to proteolysis of the arrested transcription complexes.
Although we have implicated IE proteins in the loss of Ser-2-phosphorylated RNAP II, it is not clear which IE protein (or proteins) carries out this function. Our mutant data show that the IE proteins ICP0, ICP4, ICP22, and ICP27 are not singly responsible. It is possible that the fifth known IE protein, ICP47, is the relevant factor. However, this protein is not known to affect gene expression but rather works in the cytoplasm to prevent the transport of class I major histocompatibility complex antigens to the cell surface (16, 71). Another possibility is that the IE gene regulatory proteins are redundant in their ability to cause Ser-2P loss, thus explaining why mutants singly disrupted for ICP0, ICP4, ICP22, or ICP27 are still able to induce the loss of Ser-2-phosphorylated RNAP II.
Relocalization of residual Ser-2P-containing RNAP II to nuclear speckles.
Although most Ser-2-phosphorylated RNAP II disappears after infection, we identified a subfraction that relocalizes to splicing antigen-rich nuclear speckles. These structures are storage, assembly, and/or modification sites for splicing proteins and some other factors involved in mRNA production, including certain transcription and polyadenylation factors (27). These are not believed to be active sites of transcription or splicing. A possible clue to the biological significance of our observations comes from studies in uninfected cells which show that Ser-2-phosphorylated RNAP II migrates into nuclear speckles when RNAP II activity is inhibited (4, 73). Although RNAP II is not globally inhibited in infected cells, it is redirected from cellular to viral chromatin. Thus, the HSV-1-induced relocalization of Ser-2-phosphorylated could reflect a normal cellular response to the inhibition of RNAP II activity on cellular chromatin.
We found that nuclear speckles in HSV-1-infected cells are distinct from replication compartments. This is partially consistent with the results of Phelan et al. (44), who found that most (70 to 80%) of the sites of HSV-1 DNA synthesis, as defined by in situ incorporation of biotinylated-dUTP, are distinct from the localization of the B" component of the U2 snRNP localization (44). These investigators, however, identified a minority of replication foci (20 to 30%) that were coincident with snRNP staining. In our experiments, we did not observe any overlap between replication compartments and speckles. The reason for this difference is unknown but could reflect the different techniques and antibodies used in the two studies.
Although RNAP II-containing nuclear speckles are localized outside of viral replication compartments, we found that they are usually closely apposed to them. Previous studies have shown that nuclear speckles increase in size and move to the nuclear periphery in HSV-1-infected cells in an ICP27-dependent fashion (31, 43, 58). To our knowledge, their proximity to viral replication compartments has not been previously reported. Although the significance of this localization is unknown, it has been hypothesized that in uninfected cells the speckles supply splicing factors to active sites of transcription (27). With this in mind, it is conceivable that the infected cell speckles supply replication compartments with needed mRNA processing and transcription factors, possibly including RNAP II itself.
Model for the nuclear relocalization of RNAP II forms after HSV-1 infection.
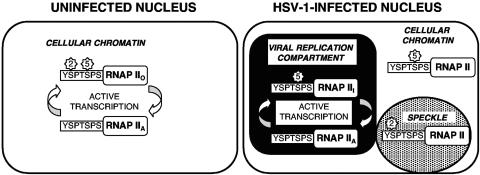
In Fig. 8 we present a model for how HSV-1 infection alters the localization of various forms of RNAP II. In uninfected cells, active transcription of genes resident on cellular chromatin involves a cycling between RNAP IIA and RNAP IIO and involves phosphorylation on both Ser-2 and Ser-5 of the CTD (Fig. 8, left). After HSV-1 infection (Fig. 8, right), however, the bulk of RNAP II moves into viral replication compartments. This is demonstrated by our present (Fig. 6) and past (55) immunofluorescence analyses using ARNA3 MAb, which detects all forms of RNAP II. We hypothesize that the phosphorylation of RNAP II is altered in replication compartments, such that the hyperphosphorylated form, RNAP III, acquires Ser-5 but not Ser-2 phosphorylation. Our immunofluorescence studies indicate that there are at least two distinct forms of RNAP II outside of replication compartments. First, there is a subfraction of Ser-2-phosphorylated RNAP that relocalizes to transcriptionally inactive nuclear speckles. Second, analysis with the H14 antibody showed that there is a significant amount of Ser-5-phosphorylated RNAP II that localizes outside of replication compartments, possibly associated with cellular chromatin. This finding was somewhat unexpected, given that both ARNA3 and 8WG16 predominantly stain replication compartments. However, these latter antibodies differ significantly from H14 in that they also recognize the hypophosphorylated RNAP IIA form, which may be very abundant in replication compartments. The significance of the localization of Ser-5-phosphorylated RNAP II outside of replication compartments is unknown. Since many if not most cellular genes are transcriptionally inhibited after HSV-1 infection, we suspect that this form is not transcriptionally active. However, it is possible that there is some residual transcription of cellular genes in virus-infected cells and that this is mediated by Ser-5-phosphorylated RNAP II.
FIG. 8.
Model for localization of various forms of RNAP II in HSV-1-infected cells. See the text for details.
Implications for the switch from host to viral transcription.
We speculate that the HSV-1-induced loss of Ser-2P from the CTD represents a specific viral mechanism that helps promote the global switch from host to viral gene transcription. This phenomenon could be involved in this switch in two ways. First, it could help induce shutoff of host cell gene transcription. In fact, there is some evidence linking loss of Ser-2P to host transcription shutoff, since both phenomena (along with loss of RNAP IIO) require IE protein expression but still occur in the individual absences of ICP0, ICP4, ICP22, or ICP27 (Fig. 5) (63). The loss of Ser-2-phosphorylated RNAP II in HSV-1-infected cells suggests that P-TEFb is inhibited or that its effects are rapidly reversed. Since P-TEFb activity is essential for most RNAP II transcription in eukaryotic cells (7, 60), our work provides a possible mechanism by which host transcription might be inhibited. However, it is also possible that loss of Ser-2 phosphorylation is an effect, rather than a cause, of the inhibition of host cell mRNA synthesis. Determining the mechanism by which Ser-2P loss occurs should help elucidate the connection between this event and the shutoff of host transcription.
Second, loss of Ser-2 phosphorylation could favor viral transcription if RNAP II transcription of the viral genome does not require this modification, as our results imply. In uninfected cells, Ser-2 CTD phosphorylation by P-TEFb promotes transcriptional elongation by relieving the pausing caused by inherent signals in the gene and/or the action of negative transcription elongation factors (61). It is possible that HSV-1 genes, which are short and not configured in standard nucleosomes (29, 38), may not be subject to transcriptional pausing and thus may not need P-TEFb. Consistent with this idea, nuclear run-on transcription studies show that HSV-1 transcription is relatively resistant to DRB, an inhibitor of P-TEFb (C. Spencer, unpublished data). Another role of CTD Ser-2 phosphorylation is to recruit polyadenylation and possibly splicing factors to elongating transcription complexes (1, 2). How could HSV-1 transcription proceed in the absence of this recruitment? Since nearly all HSV-1 DE and L genes are intronless, viral transcription could conceivably dispense with the recruitment of splicing factors. However, viral genes have canonical polyadenylation sites and therefore need to access the polyadenylation machinery (34). In this regard, it is intriguing that some (34, 35, 59) but not all (15) studies suggest that ICP27 increases the efficiency of polyadenylation. Given that ICP27 also physically interacts with RNAP II complexes (20, 74), it is conceivable that ICP27 compensates for the loss of Ser-2P by itself recruiting polyadenylation factors to transcribing RNAP II.
In summary, our results show that HSV-1 infection leads to a striking loss of Ser-2 phosphorylation on the RNAP II CTD. We speculate that this helps to create a form of RNAP II that is able to transcribe viral but not host genes. In addition, we show that a fraction of Ser-2-phosphorylated RNAP II escapes loss and moves into nuclear speckle structures. This latter effect may represent a host response to the viral inhibition of host cell genome transcription.
Acknowledgments
We thank Charlotte Spencer for stimulating discussions, a review of the manuscript, and sharing unpublished data; Leslie Schiff for helpful comments on the manuscript; members of the Rice and Schiff labs for interest and encouragement; Joy Lengyel, Julia Meyer, and Linse Lahti for expert technical help; Neal DeLuca and Priscilla Schaffer for viral mutants; and David Knipe and Nancy Thompson for antibodies.
This research was supported by a grant from the NIH (RO1-AI50127) to S.A.R.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 24:8963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, F., M. Vigneron, O. Bensaude, and M. F. Dubois. 1999. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2). Nucleic Acids Res. 27:4399-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10:679-680. [DOI] [PubMed] [Google Scholar]
- 6.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 8.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpesvirus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 10.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dvir, A. 2002. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta 1577:208-223. [DOI] [PubMed] [Google Scholar]
- 12.Gillette, T. G., F. Gonzalez, A. Delahodde, S. A. Johnston, and T. Kodadek. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. USA 101:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillot, P. V., S. Q. Xie, M. Hollinshead, and A. Pombo. 2004. Fixation-induced redistribution of hyperphosphorylated RNA polymerase II in the nucleus of human cells. Exp. Cell Res. 295:460-468. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11:394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann, L. E., W. J. Cook, S. L. Uprichard, D. M. Knipe, and D. M. Coen. 1998. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 17.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 icp27 is required for transcription of two viral late (γ2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins, H. L., and C. A. Spencer. 2001. RNA polymerase II holoenzyme modifications accompany transcription reprogramming in herpes simplex virus type 1-infected cells. J. Virol. 75:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, R., and P. A. Schaffer. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemp, L. M., and D. S. Latchman. 1988. Induction and repression of cellular gene transcription during herpes simplex virus infection are mediated by different viral immediate-early gene products. Eur. J. Biochem. 174:443-449. [DOI] [PubMed] [Google Scholar]
- 23.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 25.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer, A., R. Haars, R. Kabisch, H. Will, F. A. Bautz, and E. K. Bautz. 1980. Monoclonal antibody directed against RNA polymerase II of Drosophila melanogaster. Mol. Gen. Genet. 180:193-199. [DOI] [PubMed] [Google Scholar]
- 27.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 28.Lavoie, S. B., A. L. Albert, H. Handa, M. Vincent, and O. Bensaude. 2001. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J. Mol. Biol. 312:675-685. [DOI] [PubMed] [Google Scholar]
- 29.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 51:45-59. [DOI] [PubMed] [Google Scholar]
- 30.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, T. E., S. C. Barghusen, G. P. Leser, and P. G. Spear. 1987. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J. Cell Biol. 105:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinho, R. G., P. S. Kunwar, J. Casanova, and R. Lehmann. 2004. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol. 14:159-165. [DOI] [PubMed] [Google Scholar]
- 33.Mayman, B. A., and Y. Nishioka. 1985. Differential stability of host mRNAs in Friend erythroleukemia cells infected with herpes simplex virus type 1. J. Virol. 53:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 70:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 66:6939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell. Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 37.Mu, F. T., J. M. Callaghan, O. Steele-Mortimer, H. Stenmark, R. G. Parton, P. L. Campbell, J. McCluskey, J. P. Yeo, E. P. Tock, and B. H. Toh. 1995. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 270:13503-13511. [DOI] [PubMed] [Google Scholar]
- 38.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palancade, B., S. Bellier, G. Almouzni, and O. Bensaude. 2001. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J. Cell Sci. 114:2483-2489. [DOI] [PubMed] [Google Scholar]
- 40.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270:3859-3870. [DOI] [PubMed] [Google Scholar]
- 41.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 42.Perkins, K. D., J. Gregonis, S. Borge, and S. A. Rice. 2003. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 77:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phelan, A., M. Carmo-Fonseca, J. McLaughlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 90:9056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pizer, L. I., and P. Beard. 1976. The effect of herpesvirus infection on mRNA in polyoma virus transformed cells. Virology 75:477-480. [DOI] [PubMed] [Google Scholar]
- 46.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 48.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 49.Quinlan, M. P., and D. M. Knipe. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol. 5:957-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 51.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 52.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 53.Rice, S. A., M. C. Long, V. Lam. P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice, S. A., and V. Lam. 1994. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rickert, P., J. L. Corden, and E. Lees. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093-1102. [DOI] [PubMed] [Google Scholar]
- 58.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 69:6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandri-Goldin, R. M., and G. E. Mendoza. 1992. A herpesvirus regulatory protein appears to act posttranscriptionally by affecting mRNA processing. Genes Dev. 6:848-863. [DOI] [PubMed] [Google Scholar]
- 60.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the Caenorhabditis elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 62.Smibert, C. A., and J. R. Smiley. 1990. Differential regulation of endogenous and transduced beta-globin genes during infection of erythroid cells with a herpes simplex virus type 1 recombinant. J. Virol. 64:3882-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spencer, C. A., M. E. Dahmus, and S. A. Rice. 1997. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 71:2031-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stenberg, R. M., and L. I. Pizer. 1982. Herpes simplex virus-induced changes in cellular and adenovirus RNA metabolism in an adenovirus type 5-transformed human cell line. J. Virol. 42:474-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svejstrup, J. Q. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116:447-451. [DOI] [PubMed] [Google Scholar]
- 66.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471-31477. [DOI] [PubMed] [Google Scholar]
- 67.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 273:6769-6775. [DOI] [PubMed] [Google Scholar]
- 68.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acids Res. Mol. Biol. 51:123-165. [DOI] [PubMed] [Google Scholar]
- 70.Weir, J. P. 2001. Regulation of herpes simplex virus gene expression. Gene 271:117-130. [DOI] [PubMed] [Google Scholar]
- 71.York, I. A., C. Roop, D. W. Andrews, S. R. Riddell, F. L. Graham, and D. C. Johnson. 1994. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77:525-535. [DOI] [PubMed] [Google Scholar]
- 72.Zabierowski, S., and N. A. DeLuca. 2004. Differential cellular requirements for activation of herpes simplex virus type 1 early (tk) and late (gC) promoters by ICP4. J. Virol. 78:6162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng, C., E. Kim, S. L. Warren, and S. M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16:1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 76:5893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296:91-97. [DOI] [PubMed] [Google Scholar]