Abstract
The detection and identification of retroviral transcripts in brain samples, cerebrospinal fluid, and plasma of individuals with recent-onset schizophrenia and schizoaffective disorders suggest that activation or upregulation of distinct human endogenous retroviruses (HERVs) may play a role in the etiopathogenesis of neuropsychiatric diseases. To test this hypothesis, we performed a comprehensive microarray-based analysis of HERV transcriptional activity in human brains. We investigated 50 representative members of 20 HERV families in a total of 215 brain samples derived from individuals with schizophrenia or bipolar disorders and matched controls. A characteristic brain-specific retroviral activity profile was found that consists of members of the class I families HERV-E, HERV-F, and ERV9 and members of HERV-K taxa. In addition to these constitutively expressed HERVs, a number of differentially active HERV elements were identified in all brain samples independent of the disease pattern that may reflect differences in the genetic background of the tested individuals. Only a subgroup of the HML-2 family (HERV-K10) was significantly overrepresented in both bipolar-disorder- and schizophrenia-associated samples compared to healthy brains, suggesting a potential association with disease. Real-time PCR analysis of HERV env transcripts with coding capacity potentially involved in neuroinflammatory conditions revealed that env expression of HERV-W, HERV-FRD, and HML-2 remains unaffected regardless of the clinical picture. Our data suggest that HERV transcription in brains is weakly correlated with schizophrenia and related diseases but may be influenced by the individual genetic background, brain-infiltrating immune cells, or medical treatment.
Schizophrenia is a highly complex and pervasive neuropsychiatric disorder of uncertain etiology (23). Current data from family, twin, and adoption studies, as well as from epidemiological surveys, suggest that the etiopathogenesis involves the interplay of complex polygenic influences and environmental risk factors operating on brain maturational processes. Among the environmental factors, winter-spring births, perinatal infections, household crowding, upbringing in urban areas, and pet ownership support the concept that schizophrenia could be triggered by infectious agents affecting the brains of genetically susceptible individuals (28, 53, 54, 57, 61).
Viruses are possible infectious agents in chronic nervous system diseases of unknown etiology because of their potential for neurotropism and latency (50). Recently, an involvement of retroviruses in the pathogenesis of schizophrenia has been hypothesized (17, 34, 60, 61). It is well established that human retroviruses, such as human immunodeficiency virus (HIV) and human T-cell leukemia virus, can replicate in cells of the central nervous system (CNS), thereby causing neurological and psychiatric symptoms in some infected individuals (for a review, see reference 1).
Infections could also arise from polytropic retroviruses derived from domesticated animals (pets) or pest animals that are in close contact with humans (24, 56). HIV types 1 and 2 are the best known examples of zoonotic retroviruses that have crossed the species barrier into humans during the last century (9). Neurotropic retroviruses that cause spongiform degeneration and neuron loss in the CNSs of susceptible animals are known from rodents, the main pest animals in human habitats (13, 30, 36, 38, 48). A prenatal or early postnatal silent infection of human brain cells by such retroviruses would be consistent with the hypothesis that schizophrenia represents the interaction of both genetic susceptibility and an infectious viral agent (55).
On the other hand, the human genome itself harbors an immense reservoir of endogenous retroviral sequences that might have pathogenic effects on the host cell under certain conditions. Human endogenous retroviruses (HERVs) are normal components of the human genome and are considered to be remnants of ancient germ line infections with exogenous retroviruses that have been genetically fixed and transmitted in a Mendelian fashion (for a review, see references 25 and 52). During evolution, these elements have been amplified and spread throughout the genome by repeated events of retrotransposition and/or reinfection. Completion of the human genome sequencing project revealed that 8 to 9% of the human genome is of retroviral origin (20). Around 8,100 elements contain pol-related sequences, 3,661 with full or partial open reading frames (11).
The majority of HERVs are assumed to be noninfectious replication-defective retroviral fossils that were primarily active in the early evolutionary history of primates. Recent studies, however, have revealed that at least some members of each HERV family are still transcriptionally active and display tissue-specific expression profiles (47, 51). Most active members of HERV families were found in skin, placenta, and tissues of reproductive organs. Of 19 human tissues investigated, none could be found that lacks HERV transcription, confirming that human endogenous retroviruses are permanent components of the human transcriptome (47).
A few HERV elements have been reported that possess intact open reading frames and the capability to encode functional proteins with pathogenic and nonpathogenic activities. Screening human sequence databases for HERV elements with complete envelope genes identified 16 candidate genes that have the potential to encode functional Env proteins (7). In at least two cases, HERV Env proteins have been shown to be involved in physiological and/or pathological processes (2, 3, 19, 26). HERV envelope proteins have been associated with several chronic human diseases, including various autoimmune disorders and neurological diseases (for a review, see references 6, 31, and 37). However, the causative or disease-promoting role of HERV proteins in these disorders has yet to be conclusively demonstrated.
Recently, indirect evidence for a possible role of retroviral elements in neurological diseases, such as schizophrenia and bipolar disorders, has been provided by several studies (for a review, see reference 60). (i) Retroviral nucleic acids have been detected in the brains and cerebrospinal fluid (CSF) of affected individuals, and increased levels of virally encoded reverse transcriptase were observed (60, 61). HERV-W, ERV9, and HERV-FRD pol transcripts have been identified by PCR in 10 of 35 (29%) cell-free CSF samples of individuals with recent-onset schizophrenia but not in neurologically unaffected individuals. This suggests that the transcriptional activation of certain retroviral elements within the nervous system may be associated with the development of schizophrenia (17). (ii) In a further study of patients with schizophrenia, schizoaffective psychosis, or schizophreniform disorder, conducted by the same group, HERV-W-related RNA was detected in the plasma of 9/54 individuals with recent-onset schizophrenia (16). (iii) Primate retrovirus-directed antibodies have been found at greater frequency in the sera of affected individuals than in control groups (24, 60). (iv) Syncytin, the envelope protein of the ERVWE1 locus, is capable of causing cell fusion and generation of syncytia (26, 39) and mediates neuroinflammation and death of oligodendrocytes in a mouse model of multiple sclerosis (2, 10). The proinflammatory properties of syncytin support the hypothesis that HERV elements can act as auto-, super-, or neoantigens with the potential to enhance inflammatory responses or induce autoimmune reactions (6, 12).
Here, we report a comprehensive study of HERV transcription in human brain samples (The Stanley Brain Collection) by means of retrovirus-specific microarray analysis (46, 47). HERV pol expression profiles of prefrontal-cortex samples derived from individuals with schizophrenia or bipolar disorders and from unaffected controls were identified and compared. HERV elements found to be differentially regulated, as determined with the DNA chip, were further investigated by an env-specific quantitative real-time PCR (QRT-PCR). To test the hypothesis of zoonosis, we established an animal retrovirus-specific microarray.
MATERIALS AND METHODS
Human brain RNA samples.
Postmortem human brain tissue specimens were obtained from the Stanley Foundation Brain Collection, Bethesda, MD. For comparison of retroviral activities in different brain regions, total RNA was prepared from 110 frozen tissue blocks cut from the prefrontal cortex, orbitofrontal cortex, temporal cortex, parietal cortex, occipital cortex, corpus callosum, thalamus, cerebellum, caudate nucleus, and putamen, each derived from well-matched groups of healthy controls (n = 4) and patients with bipolar disorders (n = 3) and with schizophrenia (n = 4). In brief, blocks of approximately 0.5 g were ground in liquid nitrogen and subsequently treated according to a guanidinium isothiocyanate-cesium chloride ultracentrifugation protocol (41). In addition, 105 samples of total RNA (The Stanley Array Collection, provided by the Stanley Medical Research Institute, Bethesda, MD) derived from the prefrontal cortex (Brodmann's area 46) in well-matched groups of 35 healthy controls, 35 patients with bipolar disorders, and 35 patients with schizophrenia were included in the study (27, 58). To exclude genomic-DNA contamination, all samples (n = 215) were treated with 100 units/μg RNase-free DNase (Roche Molecular Biochemicals, Mannheim, Germany) in 100 mM sodium acetate, pH 5.0, 5 mM MgSO4. Subsequently, 100 ng of each total-RNA preparation was tested by PCR with mixed oligonucleotide primers, omitting the reverse transcription step to ensure the use of DNA-negative RNA preparations in the subsequent chip hybridization experiments.
Animal DNA samples.
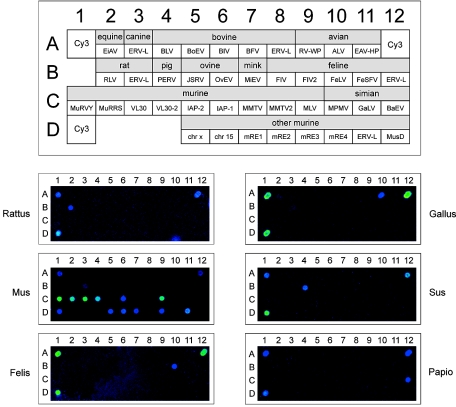
For establishment and validation of an animal retrovirus-specific microarray (pet chip), DNAs of animals served as targets in PCRs. If not otherwise stated, genomic DNA was prepared from fresh EDTA peripheral blood obtained from local veterinarians. DNAs from cattle (Bos primigenius taurus), pig (Sus scrofa domestica), and chicken (Gallus gallus domesticus) were extracted from fresh liver tissue derived from local butchers according to the proteinase K-based standard protocol described by Sambrook and colleagues (41). Ovine fetal liver cells were obtained from Bernd Aigner, Institut für Tierzucht und Genetik, Veterinärmedizinische Universität Wien, Vienna, Austria. Feline T cells (FL4) were a kind gift of Rüdiger Dörries, Institut für Virologie, Universitätsklinikum Mannheim der Universität Heidelberg. Rodent genomic DNA prepared from rat (208F) and murine (RAW264) cell lines was donated by Alex D. Greenwood (Technical University of Munich, Munich, Germany). Primate genomic DNA represented by DNA samples of gibbon (Hylobates concolor), rhesus macaque (Macaca mulatta), and hamadryas baboon (Papio hamadryas) was kindly provided by Jürgen Blusch (Novartis, Basel, Switzerland).
Chip design and hybridization.
Compared to the HERV chip described previously (47), some variations in number and arrangement of capture probes were introduced to expand the spectrum of HERV targets and to minimize chip redundancy (Table 1). Capture probes corresponding to Seq65 (dot code E11), HS49C23 (dot code F4), HERV-Z (dot code F5), HERV-F2 (dot code G9), HERV-F (dot code G10), NMWV7 (dot code C12), NMWV3 (dot code C11), and NMWV9 (dot code C10) were included, and RPL19 (dot code A4) replaced the housekeeping gene alpha-tubulin.
TABLE 1.
Origins and classification of human retrovirus-specific capture probes
| Retrovirus class | Family or groupa | RepBaseb name | Sequence source (accession no.)a | Dot code |
|---|---|---|---|---|
| Class I retroviruses | HERV-I | HERVI | HERV-I (M92067) | E9 |
| (gammaretrovirus-like) | HERV-IP-T47D (U27241) | E10 | ||
| HERVIP10F | Seq65 (AP000842) | E11 | ||
| HERV-T | HERVS71 | S71pCRTK6 (U12969) | F1 | |
| S71pCRTK1 (U12970) | F2 | |||
| HERV-FRD | MER50I | HERV-FRD (U27240) | F3 | |
| HS49C23 (Z93019) | F4 | |||
| HERV-Z (Z69907) | F5 | |||
| HERV-E | HERVE | E4-1 (M10976) | F7 | |
| Seq32 (AC010636) | F8 | |||
| HERV-H | HERVH | RGH2 (D11078) | F9 | |
| HERV-H (AF026252) | F10 | |||
| Seq66 (AL359740) | F11 | |||
| HERV-F | HERVH48I | HERV-F2 (AC002416) | G9 | |
| HERVFH19I | HERV-F (Z94277) | G10 | ||
| HERVFH21 | HERV-Fb (AC000378) | G11 | ||
| HERV-W | HERV17 | HERV-W (AF009668) | G3 | |
| HERV-R | HERVR | ERV-3 (AC004609) | F6 | |
| ERV9 | HERV9 | Seq64 (AC005253) | G4 | |
| HERV17 | Seq63 (AC018926) | G5 | ||
| ERV9 (X57147) | G6 | |||
| HERV9 | Seq59 (AC006397) | G7 | ||
| HERVFH19I | Seq60 (AL135749) | G8 | ||
| Class II retroviruses | HML-1 | HERVK14I | HML-1 (U35102) | A8 |
| (betaretrovirus-like) | Seq29 (S77579) | A9 | ||
| HML-2 | HERVK | HERV-K10 (M14123) | B1 | |
| HERV-K2.HOM (U87592) | B2 | |||
| HERV-KHP1 (U87588) | B3 | |||
| HERV-KD1.2 (U87595) | B4 | |||
| HML-3 | HERVK9I | Seq26 (AC073115) | B5 | |
| Seq34 (AL592449) | B6 | |||
| HML-3 (U35236) | B7 | |||
| HERV1 (S66676) | B8 | |||
| Seq43 (AF047595) | B9 | |||
| HML-4 | HERVK13I | Seq10 (AF047591) | B10 | |
| HERV-K-T47D (AF020092) | B11 | |||
| HML-5 | HERVK22I | HML-5 (U35161) | B12 | |
| HML-6 | HERVK3I | HML-6 (U60269) | A10 | |
| Seq38 (AC010328) | A11 | |||
| Seq56 (AC018558) | A12 | |||
| HML-7 | HERVK11DI | NMWV7 (AP003171) | C12 | |
| HML-8 | HERVK11I | NMWV3 (AL513321) | C11 | |
| HML-9 | NMWV9 (AC025569) | C10 | ||
| HML-10 | HERVKC4 | HERV-KC4 (U07856) | C8 | |
| Seq31 (AL162734) | C9 | |||
| Class III retroviruses | HERV-L | HERVL | HERV-L (G895836) | E2 |
| (spumavirus-like) | Seq39 (AC091914) | E3 | ||
| Seq45 (AC006971) | E4 | |||
| Seq51 (AL353741) | E5 | |||
| Seq58 (AL590730) | E6 | |||
| Human exogenous retroviruses | HIV-1 (K02013) | H2 | ||
| HIV-2 (J04542) | H3 | |||
| HTLV-1 (M81248) | H4 | |||
| HTLV-2 (M10060) | H5 | |||
| HFV (Y07725) | H6 | |||
| Human housekeeping genes | Ubiquitin (U49869) | A2 | ||
| GAPDH (NM_002046.1) | A3 | |||
| RPL19 (NM_000981) | A4 | |||
| Beta-actin (E01094) | A5 | |||
| HPRT (NM_000194) | A6 |
HML, human mouse mammary tumor virus-like; HFV, human foamy virus; RPL19, ribosomal protein L19.
RepBase, Genetic Information Research Institute, Sunnyvale, CA (http://www.girinst.org); Jurka (15).
For the design of an animal retrovirus-specific microarray (pet chip), reverse transcriptase (RT)-related DNA sequences were selected from databases (http://www.ncbi.nlm.nih.gov/) and the mouse genome database (http://www.ensembl.org/mus_musculus/) according to a strategy described in detail previously (45). DNA sequences of preferably complete, functional retroviral genomes were selected to design capture probes (n = 41) specific for exogenous and endogenous animal retroviruses of the avian genera Columba and Gallus; the mammalian genera Equus, Canis, Bovis, Rattus, Sus, Ovis, Mustela, Felis, and Mus; and the primate genera Macaca, Hylobates, and Papio (Table 2). For chip validation and assay standardization, several sets of experiments were performed with various species-specific primer cocktails using DNA from the respective animal species as a template. For example, rat leukemia virus, feline leukemia virus, and avian leukosis virus were detected with high specificity in DNAs from the corresponding animals. In DNA from the murine cell line RAW 264.7 (BALB/c mice), the presence of 11 families of endogenous retroviruses and retrovirus-like elements could be verified (Fig. 1).
TABLE 2.
Origins and classification of animal retrovirus-specific capture probes
| Animal genus | Retrovirus or retroelement | Sequence source (accession no.) | Dot code (grid locus) |
|---|---|---|---|
| Equus | EiAV, equine infectious anemia virus | M87581 | A2 |
| Canis | ERV-L, canine endogenous retrovirus type L | AJ233669 | A3 |
| Bovis | BLV, bovine leukemia virus | K02120 | A4 |
| BoEV, bovine endogenous retrovirus | X99924 | A5 | |
| BIV, bovine immunodeficiency virus | M32690 | A6 | |
| BFV, bovine foamy virus | AY134750 | A7 | |
| ERV-L, bovine endogenous retrovirus type L | AJ233662 | A8 | |
| Columba | RVwp, wood pigeon retrovirus | AJ236133 | A9 |
| Gallus | ALV, avian leukosis virus | Z46390 | A10 |
| EAV-HP, endogenous avian retrovirus type HP | AJ292966 | A11 | |
| Rattus | RLV, rat leukemia virus | M77194 | B2 |
| ERV-L, rat endogenous retrovirus type L | AJ233604 | B3 | |
| Sus | PERV, porcine endogenous retrovirus, types A,B,C | AF038600 | B4 |
| Ovis | JSRV, Jaagsiekte retrovirus | A27950 | B5 |
| OvEV-2, ovine endogenous retrovirus | X99932 | B6 | |
| Mustela | MiEV-1, mink endogenous retrovirus | X99931 | B7 |
| Felis | FIV, feline immunodeficiency virus | M59418 | B8 |
| FIV2, feline immunodeficiency virus type 2 | U56928 | B9 | |
| FeLV, feline leukemia virus | L06140 | B10 | |
| FeSFV, feline syncytium-forming virus | U78765 | B11 | |
| ERV-L, feline endogenous retrovirus type L | AJ233664 | B12 | |
| Mus | MuRVY, murine endogenous retrovirus from chr Y | X87639 | C1 |
| MuRRS, murine retroviral-related sequence | X02487 | C2 | |
| VL30, murine virus-like element encoding 30S RNA | AF053745 | C3 | |
| VL30-like, murine virus-like element for 30S RNA-like | AL844168 | C4 | |
| IAP-2, intracistemal particle type 2 | U58494 | C5 | |
| IAP-1, intracistemal particle type 1 | X87638 | C6 | |
| MMTV, mouse mammary tumor virus | M15122 | C7 | |
| MMTV-like murine retroelement | AL606472 | C8 | |
| MLV, murine leukemia virus | J02255 | C9 | |
| chr x, unclassified murine retroelement on chr X | AL672245 | D5 | |
| chr 15, unclassified murine retroelement on chr 15 | AL513352 | D6 | |
| mRE1, unclassified murine retroelement | XM_143828 | D7 | |
| mRE2, unclassified murine retroelement | XM_136338 | D8 | |
| mRE3, unclassified murine retroelement | XM_146956 | D9 | |
| mRE4, unclassified murine retroelement | XM_141084 | D10 | |
| ERV-L, murine endogenous retrovirus type L | Y12713 | D11 | |
| MusD (1/2), type D-like murine endogenous retrovirus | AF246632/3 | D12 | |
| Macaca | MPMV, Mason-Pfizer monkey virus | M12349 | C10 |
| Hylobates | GaLV, gibbon ape leukemia virus | M26927 | C11 |
| Papio | BaEV, baboon endogenous retrovirus | D10032 | C12 |
FIG. 1.
Validation of the animal retrovirus-specific microarray (pet chip). Genomic DNAs (100 ng) derived from avian (Gallus gallus domesticus) and mammalian (Rattus rattus, Mus musculus, Felis catus, Sus scrofa domestica, and Papio hamadryas) genera were amplified with primers using the standardized amplification protocol. The names and corresponding grid locations of animal retrovirus-specific oligonucleotides (capture probes) are shown in the top panel. Grid locators represented by a spotted Cy3-labeled arbitrary oligonucleotide are designated Cy3. For the origins and identities of all capture probes, see Table 2.
Robotic preparation of microarrays was carried out under standardized conditions as described previously (46). DNA chips of the same lot number, checked for quality consistency, were used for all experiments. Primer sequences were selected to match highly conserved regions present in the RT genes of HERVs and animal retroviruses. Reverse primers were 5′ modified with Cy3 fluorochrome. For amplification of HERV RT sequences, two separate multiplex PCRs were performed using primer mixtures for either class I or class II/III retroviruses (49). The fluorochrome-labeled PCR products were combined and used as probes for chip hybridization. Primers designed for the animal retrovirus-specific microarray are listed in Table 3. Pilot experiments were performed with a mixture of all primers and with distinct subsets of oligonucleotides corresponding to the intended target of amplification (avian, rodent, or nonrodent mammalian genera). For amplification of murine retroviral RT sequences, a mixture of 16 different forward and 10 reverse primers was used (mix A, forward primers 2, 4, 9 to 15, 17, 18, 20, and 26 to 29; reverse primers 30, 33 to 35, 41, 42, and 45 to 56). For nonmurine targets, a primer cocktail combining 25 forward and 25 reverse primers (mix B, forward primers 1, 3, 5 to 8, 16, 19, and 21 to 25; reverse primers 31, 32, 36 to 40, 43, 44, and 57 to 59) was employed. Amplification of the hybridization probes and chip hybridization were performed according to standardized procedures described previously (46, 47).
TABLE 3.
Retrovirus-specific primers used for amplification of pet chip hybridization probes
| No. | Nucleotide sequencea | Target of amplification (accession no.)b |
|---|---|---|
| Forward primers | ||
| 1 | GGAGAATAGGTTCTTCCTCAGAAG | Canine ERV-L (AJ233669) |
| 2 | GGAGAATAGAGACTACCACAGGGG | VL30 (AF053745) |
| 3 | GGAGAATAGGTTTTGCCTCGAGGA | Rat ERV-L (AJ233604) |
| 4 | GGAGAATAGGTTTTGCCCCAGGGT | MMTV (M15122) |
| 5 | GGAGAATAGGTTTTACCACAAGG | EAV-HP (AJ292966); MPMV (M12349); FeSFV (U78765) |
| 6 | GGAGAATAGGTTTACCACAAGGA | EiAV (M87581); FIV2 (U56928) |
| 7 | GGAGAATAGGTCTTACCACAGGGA | IAP-2 (U58494) |
| 8 | GGAGAATAGGTTCTACCACAAGGA | BIV (M32690) |
| 9 | GGAGAATAGGTTCTCCCBCAGGG | MusD1 (AF246632); JSRV (A27950); BFV (AY134750) |
| 10 | GGAGAATAGTATCTCCCACAGGGC | MusD2 (AF246633) |
| 11 | GGAGAATAGGTTCTTTCTTAAGAC | MMTV-like mRE (AL606472) |
| 12 | GGAGAATAGGTTCTACCCCAACTC | mRE (XM_141084) |
| 13 | GGAGAATAGGTTCTGCCTCGGGGA | mRE (XM_146956) |
| 14 | GGAGAATAGTTTCTACCTCAAGGA | mRE (XM_143828) |
| 15 | GGAGAATAGGTTTTGCCTCAAGTA | MuERVL (Y12713) |
| 16 | GGAGAATAGAGCTTACCACAGGG | FIV (M59418); RLV (M77194); MiEV (X99931) |
| 17 | GGAGAATAGAGACTCCCACAGGG | MLV (J02255); VL30-like mRE (AL844168) |
| 18 | GGAGAATAGGTCCTACCTCAGGG | IAP-1 (X87638); feline ERV-L (AJ233664); BLV (K02120); bovine ERV-L (AJ233662) |
| 19 | GGAGAATAGCGGCTACCACAAGGG | GaLV (M26927) |
| 20 | GGAGAATAGAGACTTCCACAAGGA | mRE (XM_136338) |
| 21 | GGAGAATAGCAACTTCCACAAGGA | OvEV (X99932) |
| 22 | GGAGAATAGCGCTTCCCACAAGGA | BoEV (X99924) |
| 23 | GGAGAATAGCGCCTTCCHCAAGGG | RVwp (AJ236133); FeLV (L06140); BaEV (D10032) |
| 24 | GGAGAATAGGTCTTGCCCCAAGGG | ALV (Z46390) |
| 25 | GGAGAATAGCGACTGCCCCAAGGG | PERV (AF038600) |
| 26 | GGAGAATAGAGGTCGCGACAGAGG | MuRVY (X87639) |
| 27 | GGAGAATAGTGGTAACCCCAGGAG | MuRRS (X02487) |
| 28 | GGAGAATAGGTTCTGCTGGAGGTT | mRE (AL513352) |
| 29 | GGAGAATAGGTCTGTAGTCAGATG | mRE (AL672245) |
| Reverse primers | ||
| 30 | GGAGAACATCAAGACATCATCTGTGAA | MusD1/2 (AF246632/3) |
| 31 | GGAGAAGAAAAGGATATCGTCCATATA | BLV (K02120) |
| 32 | GGAGAAATATATATATCATCCACATA | JSRV (A27950); FIV (M59418); FIV2 (U56928); FeSFV (U78765) |
| 33 | GGAGAAAAGAGGATGTCATCCATGTA | MMTV (M15122/AL606472); IAP-2 (U58494); can. ERV-L (AJ233669); MPMV (M12349) |
| 34 | GGAGAAAATCAAAATGTCATCCATATA | IAP-1 (X87638) |
| 35 | GGAGAACAGCATAATGTCATCAATATA | MuERV-L (Y12713) |
| 36 | GGAGAATAGCAAAAGATCATCCATATA | EAV-HP (AJ292966); ALV (Z46390) |
| 37 | GGAGAAAATCAACAAATCATCCATATA | BIV (M32690) |
| 38 | GGAGAACACGAACAAATCATCCATATA | EiAV (M87581) |
| 39 | GGAGAACAGAAGCAGGTCATCCACGTA | PERV (AF038600) |
| 40 | GGAGAATAGAAGAAGGTCATCAACAAA | RLV (M77194) |
| 41 | GGAGAACAGCAGTAAGTCATCYACGTA | MLV (J02255) |
| 42 | GGAGAACAGCAGTARGTCATCTACATA | FeLV (M18247) |
| 43 | GGAGAACAAGAGGAGGTCATCTACATA | BaEV (D10032) |
| 44 | GGAGAACACATAGACGTCATCCACATA | BFV (AY134750) |
| 45 | GGAGAACAGTAGTATGTCATCCACGTA | OvEV (X99932); MiEV (X99931) |
| 46 | GGAGAAACCAAGAGGTCGTCCACATA | BoEV (X99924); GaLV (M26927); VL30-like mRE (AL844168) |
| 47 | GGAGAAGATTAAAAGATCATCTACGTA | VL30 (AF053745); bovine ERV-L (AJ233662) |
| 48 | GGAGAAAAGGATTAAATCATCTCCATA | MuRVY (X87639) |
| 49 | GGAGAAAAGGAGCAAGTCATCAACATA | MuRRS (X02487) |
| 50 | GGAGAACAAAAGAAGGTCATCTATATA | mRE (XM_136338) |
| 51 | GGAGAACAATAGGAGATCATCTACATA | mRE (AC127359.4) |
| 52 | GGAGAAACAGAAGACATCATCATCATA | mRE (XM_146956) |
| 53 | GGAGAAGAGAAGGACATCATCATCATA | mRE (XM_143828) |
| 54 | GGAGAAGAGAAGAATATCATCTATATA | mRE (XM_141084) |
| 55 | GGAGAAAATCAAAATATCATCCATACT | mRE (AL513352) |
| 56 | GGAGAAATAAAAAATATCACCCATACT | mRE (AL672245) |
| 57 | GGAGAACATAATGTCATCCAGGTA | Bovine ERV-L (AJ233662) |
| 58 | GGAGAAGTCATCCAGGTAATCCAG | Bovine ERV-L (AJ233662) |
| 59 | GGAGAATAGCAACAAATACCATTG | RVwp (AJ236133) |
Analysis of microarray data.
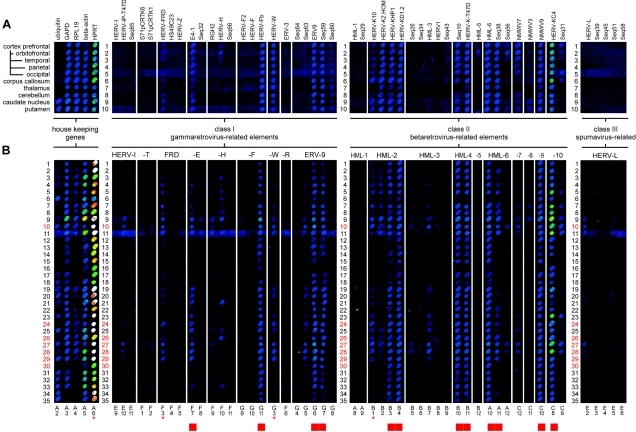
The hybridized chips were scanned using an Affymetrix Scanner GMS 418 (laser power settings, 100%; gain, 50%), and the resulting images (16-bit TIFF) were subjected to densitometric analysis using ImaGene 4.0 software (Biodiscovery Inc., Los Angeles, CA). Due to the systematic limitation of the assay (discussed in detail in references 45 and 47), the densitometric data were not used for signal quantification but for defining a cutoff value that allowed discrimination of positive signals from the cross-hybridization levels observed in the pilot experiments (47). Relative signal intensities (signalmean) were corrected by subtracting the corresponding signal background values (backgroundmean). Consecutively, signals were further corrected by subtraction of the corresponding relative signal intensities obtained from the negative control chip (using water instead of cDNA as a template). An arbitrary cutoff value of 1,200 relative signal intensity units was set up for discrimination of negative (<1,200) from positive (≥1,200) signals. This cutoff value was defined by weak cross-hybridization observed among related HERV subgroups. The influence of such signal blurring has been validated for various class II retrovirus capture probes by single Cy3-oligonucleotide hybridization in pilot experiments (data not shown). This cutoff proved to be in good agreement with the optical appearance of the raw images when observed on color-calibrated monitors in a darkened room. False-color printing (Fig. 2) may be inadequate to display all signal gradations, especially those of weak signals.
FIG. 2.
Alignment of false-color chip data sets corresponding to HERV class I, II, and III transcriptional activities observed in 10 different brain regions of a healthy human brain (A) and in prefrontal-cortex brain samples from 35 healthy individuals (B) by HERV microarray hybridization. A housekeeping-gene panel served as an internal control. For the origins and identities of the dots, see Table 1. QRT-PCR was performed for a subset of seven samples indicated by red numbers and a subset of three HERV elements (HERV-W, HERV-FRD, and HML-2) and the housekeeping gene HPRT, marked by red asterisks. HERV elements representing the brain-specific HERV activity profile are indicated by red boxes (bottom line).
QRT-PCR of HERV env transcripts.
For amplification of coding envelope sequences, the specific primers designed by de Parseval and coworkers (7) were used. In addition to the three HERV taxa HERV-W, HERV-FRD, and HML-2, two housekeeping genes (G6PD and hypoxanthine phosphoribosyl transferase [HPRT]) were included in the analysis as described previously (47). QRT-PCR was performed based on the protocol of de Parseval and coworkers (7) using 5 μl of 1:100 cDNA sample dilutions and primers at 1 μM in LightCycler FastStart DNA MasterPlus SYBRGreen I ready-to-use hot-start PCR mix containing Taq DNA polymerase, reaction buffer, dUTP, and the deoxyribonucleoside triphosphates dATP, dCTP, and dGTP; the dye SYBR Green I; and MgCl2 (Roche Diagnostics GmbH, Mannheim, Germany). Amplification was performed using a 2-min step at 50°C and then a 10-min denaturation step at 95°C, followed by 29 cycles of 15 s of denaturation at 95°C, 1 min of primer annealing, and a polymerization step at 60°C. Relative quantification of HERV env transcription was performed as described previously (47).
RESULTS
The human brain displays a distinct HERV activity signature.
The study was initiated to establish an overall HERV expression profile for the human brain and to evaluate the possible implications of different HERV elements in neuropsychiatric diseases suggested by previous studies. The rationale was to identify the constitutive HERV activity and among-individual variations in different brain areas of healthy persons and to compare the incidence of differentially expressed HERVs with those in patients affected with schizophrenia and bipolar disorders.
All experiments were performed with RNA samples from the Stanley Array Collection (58). This specimen collection was specifically assembled for high-throughput array technologies and combines a total of 105 high-quality RNA samples extracted from the dorsolateral prefrontal cortex (Brodmann's area 46), a region of the human brain associated with schizophrenia. The collection contains samples from 35 individuals in each of the three diagnostic groups, schizophrenia, bipolar disorder, and unaffected controls, which are matched by age, sex, race, postmortem interval, pH, side of brain, and RNA quality. A summary of demographic details of the Stanley Array Collection can be found on the website of the Stanley Medical Research Institute (http://www.stanleyresearch.org/programs/brain_collection.asp).
To examine the transcriptome of human endogenous retroviral pol sequences, we employed a recently established retrovirus-specific microarray (HERV chip) that allows simultaneous detection and identification of a wide variety of HERV elements and human exogenous retroviruses (46, 47). The microarray consists of 50 representative HERV RT-derived sequences from 20 major HERV families and five RT-derived sequences from human exogenous retroviruses. According to the hybridization conditions, the microarray discriminates between HERV family members with more than 20% sequence divergence within the selected pol region. Depending on the copy number and degree of divergence of a given HERV family, transcripts from multiple loci may hybridize to a single sequence on the chip.
After RNA quality control, all 105 encoded brain samples were tested twice in a blind study with the HERV chips according to our standardized protocol (46). A digitally processed alignment of a representative image data set for 35 healthy-brain RNA specimens is shown in Fig. 2B. A panel of five human housekeeping genes served as an internal control for RNA quality. Interestingly, two of these genes, ubiquitin (dot code A2) and RPL19 (dot code A4), seem to be differentially regulated in human brain, whereas glycerol aldehyde 3-phosphate dehydrogenase (GAPDH; dot code A3), β-actin (dot code A5), and HPRT (dot code A6) showed reliable and more uniform transcriptional activity. Qualitative evaluation of hybridization signals by signal processing and cutoff calculation revealed retroviral activity profiles that are summarized in Fig. 3 in taxonomic order. For assignment of hybridization signals to HERV taxa, subgroup levels, and capture probes, see Table 1.
FIG. 3.
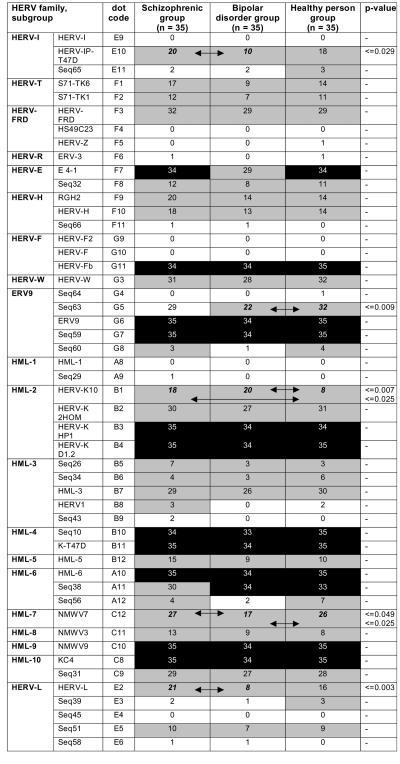
Incidences of HERV transcripts in 105 brain RNA samples. Ubiquitously active HERVs are marked with black boxes (incidence, 33 to 35/35), and differentially active (incidence, 3 to 32/35) and inactive (incidence, 0 to 2/35) HERV elements are depicted by gray and white boxes, respectively. “-” means that the P value was not significant in Fisher's exact test.
Analysis of 35 RNAs derived from healthy human prefrontal cortex revealed a brain-specific HERV transcription signature that is clearly distinct from that found in other human tissues (47). As expected from previous studies (46), not all HERV taxa appear to be equally active in the healthy human brain. Differential transcriptional activity was observed, ranging from HERV elements with ubiquitous or frequent activity to HERV family members with rare or no detectable expression in all samples under investigation. According to the arbitrarily set cutoff limit, we were able to cluster HERV taxa into three groups corresponding to elements with (i) ubiquitous (incidence, 33 to 35 of 35 samples), (ii) differential (frequent to rare incidence, 3 to 32 of 35 samples), and (iii) very rare or no (incidence, 0 to 2 of 35 samples) transcriptional activity. HERV taxa, transcripts of which were detected in more than 33 of the 35 healthy specimens, were considered ubiquitously active and are stated core components of the brain transcriptome. These HERV sequences are marked in Fig. 2B (bottom line) and in Fig. 3.
The retroviral core activity signature is composed of 12 proviruses from eight families of both class I and II HERV elements. Ubiquitous elements are HERV-E (E4-1), HERV-F (HERV-Fb), and ERV9 (ERV9 and Seq59) and members of the class II betaretrovirus-related elements, including HML-2 (HERV-KHP1 and HERV-KD1.2) HML-4 (Seq10 and HERV-K-T47D), HML-6 (HML-6 and Seq38), HML-9 (NMWV9), and HML-10 elements (HERVKC4). Class II HERVs appear to be active with higher incidence than class I elements. Spumavirus-related class III elements are not constitutively active in the human brain.
In addition, some differentially active HERVs were identified that are expressed in brain samples with varying frequencies (incidence, 3 to 32 of 35 samples). These transcripts originate from the class I HERV families HERV-I (HERV-IP-T47D; Seq65), HERV-T (S71pCRTK6; S71pCRTK1), HERV-FRD (HERV-FRD), HERV-E (Seq32), HERV-H (RGH2; HERV-H AF026252), HERV-W, and ERV9 (Seq60); the class II families HML-2 (HERV-K10; HERV-K2.HOM), HML-3 (Seq26; Seq34; HML-3; HERV1), HML-5, HML-6 (Seq38; Seq56), HML-7, HML-8, and HML-10 (Seq31); and class III elements (HERV-L G895836; Seq39; Seq51). The remaining HERVs represented by our DNA chip (class I Seq77, HERV-I, Seq65, HS49C23, HERV-Z, ERV-3, Seq66, HERV-F2, HERV-F, Seq64, Seq63, and Seq60; class II HML-1, Seq29, Seq43, and Seq56; and class III Seq39, Seq45, and Seq58) are considered transcriptionally inactive. Human exogenous retroviruses were not detected (image data not shown).
Different brain areas of each individual show the same HERV expression signature.
HERV elements are not necessarily transcribed at the same levels in different areas of the human brain, and thus, a pathological transcription profile could manifest in other brain areas besides the dorsolateral prefrontal cortex. We therefore examined the brain area-specific HERV activities of a series of 110 RNA samples isolated from 10 different brain regions, each from 11 individuals. Thus, one sample set consisted of RNAs from 10 brain areas of one individual (prefrontal cortex, orbitofrontal cortex, temporal cortex, parietal cortex, occipital cortex, corpus callosum, thalamus, cerebellum, caudate nucleus, and putamen). The series included RNA material from unaffected individuals (n = 4) and patients with schizophrenia (n = 4) and bipolar disorders (n = 3).
A comparative alignment of area-specific HERV transcription levels within the brain of one healthy individual is shown in Fig. 2A. Hybridization signals are consistent, pointing to a uniform transcriptional activity of HERV elements among all 10 brain areas. Conspicuous differences between distinct brain regions were not observed. The brain-specific basal HERV activity described (Fig. 2B) was confirmed for each of the 11 sample sets independent of the clinical picture. Comparing Fig. 2A and B, it becomes apparent that the interindividual differences in HERV expression (Fig. 2B) are more evident than the slight among-area variation observed in each single individual (Fig. 2A). This variation—for example, the differential activities of HERV-H and HML-10 in samples 20 and 21, respectively—may reflect the individual genetic background (47).
Differences in HERV transcription profiles are detectable between diagnostic groups.
In order to search for disease-related HERV elements, we compared the expression patterns of prefrontal-cortex samples from 35 patients with schizophrenia, 35 patients with bipolar disorders, and 35 healthy controls. HERV expression profiles of patient groups were generally similar to those of the control group. As shown in Fig. 3, the ubiquitously active HERV elements (HERV-E, HERV-F, ERV9, HML-2, HML-4, HML-6, HML-9, and HML-10) that make up the characteristic signature are equally represented in all three diagnostic groups. The only exceptions are E4-1, which is underrepresented in the bipolar-disorder group, and Seq38, which displays a lower incidence in schizophrenia compared with both the bipolar-disorder and healthy-control groups. However, both differences are not statistically significant.
Within the group of differentially active HERV elements (Fig. 3), the expression of five HERV elements (HERV-IP-T47D, Seq63, HERV-K10, NMWV7, and HERV-L) varied significantly (P ≤ 0.049). Of these, HERV-IP, a subgroup of HERV-I elements; NMWV7, representing the HML-7 family; and HERV-L, a subfamily of class III HERVs, are significantly overrepresented in schizophrenia compared to bipolar-disorder specimens. Furthermore, Seq63, representing a subgroup of ERV9 elements, and NMWV7 are underrepresented in samples obtained from patients with bipolar disorders compared to healthy-brain samples. Finally, HERV-K10 is significantly overrepresented in both bipolar-disorder- (P ≤ 0.007) and schizophrenia-associated (P ≤ 0.025) samples compared to healthy brains.
Establishment of an animal retrovirus-specific microarray (pet chip).
Prenatal exposure to viruses has been considered a potential risk factor for schizophrenia (56). Since domestic animals represent a potential source of transmissible viruses, we extended our efforts to screen 105 brain samples from the Stanley Array Collection for the occurrence of animal-related retroviral sequences. According to the strategy described earlier (45, 46), we established an animal retrovirus chip for detection of 41 exogenous and endogenous retroviruses from various domestic animals (Table 2). Using the standardized protocol, all 105 brain samples were tested for the occurrence of animal-derived pol transcripts (data not shown). No significant signals could be detected in repeated experiments, indicating the complete lack of animal retroviral sequences in the brain cDNA samples under investigation.
We obtained signals related to murine leukemia virus (MLV; pet chip dot code C9) with an incidence of about 27% in all tested samples irrespective of the type of diagnostic group. MLV was also occasionally detectable in our water controls. Cloning by ligation-mediated PCR (8, 42) and sequencing of the MLV-related amplicons revealed that MLV RT expression vector molecules occurred in our assay. These have been introduced by the use of a commercial MLV RT (Invitrogen Inc., Carlsbad, CA) for cDNA synthesis that was produced by recombinant bacteria. The enzyme preparations sometimes contained traces of bacterial expression plasmid, causing false-positive signals with our microarray.
Identification and quantification of coding HERV env transcripts.
Env proteins of retroviruses in particular have been associated with neurological and neuropsychiatric diseases because of their potential to induce cell-cell fusion and elicit immunosuppressive and neuroinflammatory effects (2, 17). Therefore, we extended our investigation to HERV env transcripts that possess open reading frames by QRT-PCR (7). We selected HERV-K10, a member of the HML-2 family, for our experiments. The HML-2 family contains six members with coding env genes (7). Since pol transcripts of HERV-K10 were found to be overrepresented in both patient groups compared to unaffected individuals, it was of special interest to assess HML-2 env transcription in a subset of brain samples. We further included HERV-W and HERV-FRD in our study. Both families have one member encoding a functional Env protein, also known as syncytin 1 (HERV-W) and syncytin 2 (HERV-FRD). Both proteins are able to induce cell-cell fusion in vitro (3, 5) and confer infectivity on pseudotypes generated with lentivirus virions (4, 22).
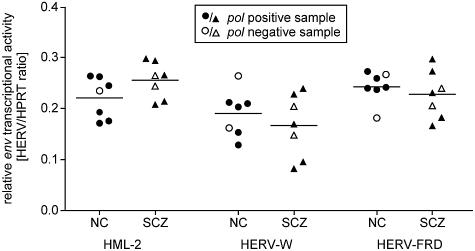
QRT-PCR experiments were performed at least in triplicate on brain RNA sample sets from seven healthy and seven schizophrenic individuals (Fig. 4). Specific primer pairs were used that ensure discrimination between Env coding-competent and -incompetent proviral copies (7). QRT-PCR revealed that (i) the env genes of HERV-W, HERV-FRD, and HML-2 are transcriptionally active but (ii) no significant differences in env transcription levels between healthy controls and schizophrenic-patient samples can be observed. Since invariant env transcript levels were found in pol-positive and -negative samples, pol and env gene transcriptions are likely independently regulated.
FIG. 4.
Relative quantification of HERV env transcriptional activity by QRT-PCR. Transcriptional activities of HML-2, HERV-W, and HERV-FRD were analyzed in a subset of seven healthy (NC) and seven schizophrenia-derived (SCZ) brain samples. The relative abundance of HERV transcripts in each sample was normalized by HPRT levels and represents the mean value of at least triplicate experiments.
DISCUSSION
The present study is the first to comprehensively investigate the transcriptional activities of endogenous and exogenous retroviral elements in the CNSs of healthy individuals and patients affected with schizophrenia. Two microarray-based assays were employed in this study. The HERV chip carries capture probes for 50 representative members of 20 HERV families and five exogenous human retroviruses. This microarray has been a powerful tool for establishing the specific HERV expression profiles of various human tissues (46, 47). Since prenatal exposure to viruses has been postulated as a risk factor for schizophrenia, a second chip was designed for this study to detect zoonotic infections. Based on the same principle and technology as the HERV microarray, the pet chip identifies putative polytropic retroviruses from domesticated or pest animals that are in close contact with humans. The microarray contains 41 mammalian and avian retroviral pol sequences from 14 genera as capture probes. No transcripts of any animal retroviruses were detected with the pet chip in all 105 human brain samples tested, arguing against a zoonotic infection. However, due to the limitations of our assay (target, pol gene only) and the given sample type (RNA only) we cannot exclude the possibility that, subsequent to prenatal or early postnatal infection, inactivated proviruses may persist in the genomes of infected cells.
Surveying the brain transcriptome with the HERV chip revealed, irrespective of the brain area and the disease pattern, a core transcription signature consisting of members of the class I families HERV-E, HERV-F, and ERV9 and the class II families HML-2, -4, -6, -9, and -10. These constitutively expressed HERVs were also seen in a brain-specific HERV expression profile described previously (47). A further important outcome of this study is the definition of differentially active HERVs, which include elements of all three HERV classes and may reflect the individual genetic background. Variation in transcriptional regulation may also be due to individual differences in the HERV methylation status (18, 21) and in the availability or levels of cellular transcription factors (33, 43). A correlation with individual parameters, such as sex, age, race, or alcohol and drug abuse, however, was not found (data not shown). Among the variably active HERVs, transcripts of the HERV-IP, ERV9, HML-7, and HERV-L families occur with significantly different incidences between the diagnostic groups; however, they cannot be clearly associated with disease (Fig. 3). Only some elements of the HML-2 family (subtype HERV-K10) are significantly overrepresented in both patient groups in comparison to unaffected individuals.
Functional HERV Env proteins have been postulated to cause neuropathogenic effects (2, 35, 40). Among the HML-2 family members, six proviruses with a coding env gene were identified (7). Therefore, we tested pol-positive and -negative RNA samples from patients with schizophrenia and healthy individuals by QRT-PCR for coding HML-2 (HERV-K10) env transcripts. We further included HERV-W env (syncytin 1) and HERV-FRD env (syncytin 2), which have been shown to induce cell fusion (3, 5) and to confer infectivity on pseudotyped retroviral particles (4, 22) and which may be involved in demyelination (2). Assuming that one of these envelope proteins could be implicated in the particle formation observed in patients with schizophrenia (16, 17), we expected it to be detectable with QRT-PCR. However, no significant differences in env transcription were found. The expression level of HML-2 env transcripts did not correlate with the overrepresentation of HML-2 pol sequences in patients with schizophrenia. However, it should be taken into account that only about 10% of the approximately 60 members of the HML-2 family are detected by the specific primers used for real-time PCR (7). The higher incidence of HML-2 pol transcription in patients could reflect the transcriptional activities of other family members. Furthermore, full-length pol-containing transcripts and spliced env transcripts may be differentially regulated.
Differential expression patterns of HERV elements in association with schizophrenia have been reported in several previous studies. Particle-associated viral RNA was predominantly detected in CSFs and plasma of patients with recent-onset schizophrenia or schizoaffective disorders (16, 17). The identified sequences are related to the HERV-W, ERV9, and HERV-FRD family transcripts, which were detected with high, although not disease-relevant, incidence in 215 brain samples from 116 individuals investigated in our study. In addition, the authors reported differential HERV pol expression in frontal-cortex tissue from five patients with schizophrenia and six healthy individuals using RT-PCR with degenerate primers and cloning and sequencing of the amplification products. A direct comparison with our microarray results, however, is not possible, since the reported sequence data were pooled within the two diagnostic groups and data on the incidence of HERV transcripts for each individual are not available. In total, the analyzed transcripts (ERV9, HERV-W, HERV-FRD, HERV-IP, HERV-E, HML-2, and HML-4) correlate with our microarray data and comprise constitutively and differentially expressed HERVs in human brain. The prevalence of HERV-W transcripts in patients with schizophrenia and of ERV9 transcripts in unaffected individuals, however, was not confirmed by our investigation. One explanation is that low sample numbers and the pooling of sequence data may lead to an inadvertent overrepresentation of some individual transcripts. Furthermore, different mixtures of degenerate primers and reaction conditions were used in both studies, which may have preferentially amplified different subspecies of HERVs. The differential occurrences of HERV transcripts in brain tissue and particle-associated HERV RNAs in the CSFs observed by Karlsson et al. (17) could be explained by selective packaging of viral RNAs, as has been previously demonstrated for particle-releasing human cell lines (44).
Further complicating the comparison of the data in our study and previous investigations is the influence of antipsychotic medications on HERV activity. Inhibitory effects of typical and atypical antipsychotics, such as haloperidol and clozapine, on retroviruses have been observed in vitro (14, 59). Therefore, the type and period of medication could also influence the individual HERV expression profiles.
A number of studies have suggested that HERV activity in the human brain is associated with inflammatory diseases, such as multiple sclerosis (for a review, see reference 6). HERVs that have been linked to multiple sclerosis are similar to those that have been associated with schizophrenia (17) and comprise the HERV families HERV-W, ERV9, HERV-H, ERV-3, and HML-2. With the exception of ERV-3, these HERVs correspond to the brain-specific HERV expression signature defined with our microarray. ERV9 and some HML-2 members belong to the constitutive HERVs. HERV-W, HERV-H, and HERV-K10 belong to the variably expressed elements. So far, a causal mechanism leading to neuroinflammation and death of oligodendrocytes has been demonstrated only for HERV-W (syncytin 1) (2). In a further study, an increase of HERV expression activity was reported for brain tissues from patients with multiple sclerosis, Alzheimer's disease, and AIDS (12). The transcripts examined consisted of the HERV families HML-2 (subtype HERV-K10), HERV-E, HERV-W, and HERV-H. HERV-W, HERV-K10, and HERV-H levels were found to be increased in HIV-infected and multiple sclerosis patients and to a lesser extent in Alzheimer patients. The authors suggested that increased macrophage activity might contribute to elevated HERV expression in inflammatory brain diseases, because they observed the same effects on HERV transcription in monocytic cell lines stimulated with phorbol-12-myristate-13-acetate or lipopolysaccharide.
These observations could also apply to schizophrenia. During acute psychotic episodes, an accumulation of monocytes/macrophages is observed in the CSF of schizophrenic patients (29, 32). Furthermore, schizophrenia and multiple sclerosis share a number of epidemiological features, such as similarities in ages of onset and geographic distributions (17). Analysis of the monocytic cell lines M8166 and SKNSH with the HERV-specific microarray revealed that members of the HML-2 family, including HERV-K10, are expressed in both cell lines even in a nonactivated state (46). Thus, the higher incidence of HERV-K10 transcripts in the brains of patients with schizophrenia and bipolar disorders could also be a consequence of increased immune activity (12).
In conclusion, we established a HERV expression profile specific for the human brain that consists of constitutively and differentially active HERVs. The expression profile may be useful for the investigation of many diseases of the CNS that are thought to be associated with retroviral activity. We could not confirm an essential role for specific HERV elements, such as HERV-W (syncytin 1), which is potentially involved in multiple sclerosis and has also been associated with schizophrenia in previous studies. In contrast to neuroinflammatory diseases, the molecular background, localization, and type of cells involved in neuropsychiatric diseases are not well known. Therefore, further studies of well-defined clinical states are required to define the role, if any, of HERVs in schizophrenia.
Acknowledgments
Postmortem brain samples were donated by the Stanley Medical Research Institute's Brain Collection courtesy of Michael B. Knable, E. Fuller Torrey, Maree J. Webster, Serge Weis, and Robert H. Yolken. We are very grateful for statistical analysis kindly performed by Christel Weiss (Department of Biostatistics, Medical School Mannheim, University of Heidelberg, Mannheim, Germany). We thank Alex D. Greenwood (Technical University of Munich, Munich, Germany) for critically reading the manuscript and for helpful discussions.
This work was supported by grant 02R-164 to W.S. from the Stanley Medical Research Institute (The Theodore and Vada Stanley Foundation, Bethesda, MD) and the Bayerische Forschungsverbund Prionen (FORPRION), grant TUM7.
REFERENCES
- 1.Albright, A. V., S. S. Soldan, and F. Gonzalez-Scarano. 2003. Pathogenesis of human immunodeficiency virus-induced neurological disease. J. Neurovirol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 2.Antony, J. M., G. Van Marle, W. Opii, D. A. Butterfield, F. Mallet, V. W. Yong, J. L. Wallace, R. M. Deacon, K. Warren, and C. Power. 2004. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 7:1088-1095. [DOI] [PubMed] [Google Scholar]
- 3.Blaise, S., N. de Parseval, L. Benit, and T. Heidmann. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 100:13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaise, S., A. Ruggieri, M. Dewannieux, F. L. Cosset, and T. Heidmann. 2004. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J. Virol. 78:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clausen, J. 2003. Endogenous retroviruses and MS: using ERVs as disease markers. Int. MS J. 10:22-28. [PubMed] [Google Scholar]
- 7.de Parseval, N., V. Lazar, J. F. Casella, L. Benit, and T. Heidmann. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77:10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank, O., C. Rudolph, C. Heberlein, N. von Neuhoff, E. Schröck, A. Schambach, B. Schlegelberger, B. Fehse, W. Ostertag, C. Stocking, and C. Baum. 2004. Tumor cells escape suicide gene therapy by genetic and epigenetic instability. Blood 104:3543-3549. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 10.Hayward, P. 2004. Active HERV protein implicated in demyelination. Lancet Neurol. 3:637. [DOI] [PubMed] [Google Scholar]
- 11.Jern, P., G. O. Sperber, and J. Blomberg. 2004. Definition and variation of human endogenous retrovirus H. Virology 327:93-110. [DOI] [PubMed] [Google Scholar]
- 12.Johnston, J. B., C. Silva, J. Holden, K. G. Warren, A. W. Clark, and C. Power. 2001. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann. Neurol. 50:434-442. [DOI] [PubMed] [Google Scholar]
- 13.Jolicoeur, P., C. Hu, T. W. Mak, J. C. Martinou, and D. G. Kay. 2003. Protection against murine leukemia virus-induced spongiform myeloencephalopathy in mice overexpressing Bcl-2 but not in mice deficient for interleukin-6, inducible nitric oxide synthetase, ICE, Fas, Fas ligand, or TNF-R1 genes. J. Virol. 77:13161-13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones-Brando, L. V., J. L. Buthod, L. E. Holland, R. H. Yolken, and E. F. Torrey. 1997. Metabolites of the antipsychotic agent clozapine inhibit the replication of human immunodeficiency virus type 1. Schizophr. Res. 25:63-70.9176928 [Google Scholar]
- 15.Jurka, J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- 16.Karlsson, H., J. Schroder, S. Bachmann, C. Bottmer, and R. H. Yolken. 2004. HERV-W-related RNA detected in plasma from individuals with recent-onset schizophrenia or schizoaffective disorder. Mol. Psychiatry 9:12-13. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson, H., S. Bachmann, J. Schroder, J. McArthur, E. F. Torrey, and R. H. Yolken. 2001. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. USA 98:4634-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khodosevich, K., Y. Lebedev, and E. D. Sverdlov. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knerr, I., E. Beinder, and W. Rascher. 2002. Syncytin, a novel human endogenous retroviral gene in human placenta: evidence for its dysregulation in preeclampsia and HELLP syndrome. Am. J. Obstet. Gynecol. 186:210-213. [DOI] [PubMed] [Google Scholar]
- 20.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 21.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette, D., M. Marin, A. Ruggieri, F. Mallet, F. L. Cosset, and D. Kabat. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76:6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis, D. A., and J. A. Lieberman. 2000. Catching up on schizophrenia: natural history and neurobiology. Neuron 28:325-334. [DOI] [PubMed] [Google Scholar]
- 24.Lillehoj, E. P., G. M. Ford, S. Bachmann, J. Schroder, E. F. Torrey, and R. H. Yolken. 2000. Serum antibodies reactive with non-human primate retroviruses identified in acute onset schizophrenia. J. Neurovirol. 6:492-497. [DOI] [PubMed] [Google Scholar]
- 25.Mager, D. L., and P. Medstrand. 2003. Retroviral repeat sequences, p. 57-63. In D. Cooper (ed.), Nature encyclopedia of the human genome. Nature Publishing Group, London, United Kingdom.
- 26.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 27.Miller, C. L., S. Diglisic, F. Leister, M. Webster, and R. H. Yolken. 2004. Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. BioTechniques 36:628-633. [DOI] [PubMed] [Google Scholar]
- 28.Mortensen, P. B., C. B. Pedersen, T. Westergaard, J. Wohlfahrt, H. Ewald, O. Mors, P. K. Andersen, and M. Melbye. 1999. Effects of family history and place and season of birth on the risk of schizophrenia. N. Engl. J. Med. 340:603-608. [DOI] [PubMed] [Google Scholar]
- 29.Müller, N., M. Riedel, R. Gruber, M. Ackenheil, and M. J. Schwarz. 2000. The immune system and schizophrenia. An integrative view. Ann. N. Y. Acad. Sci. 917:456-467. [DOI] [PubMed] [Google Scholar]
- 30.Münk, C., V. Prassolov, M. Rodenburg, V. Kalinin, J. Löhler, and C. Stocking. 2003. 10A1-MuLV but not the related amphotropic 4070A MuLV is highly neurovirulent: importance of sequences upstream of the structural Gag coding region. Virology 313:44-55. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa, K., and L. C. Harrison. 1996. The potential roles of endogenous retroviruses in autoimmunity. Immunol. Rev. 152:193-236. [DOI] [PubMed] [Google Scholar]
- 32.Nikkila, H. V., K. Muller, A. Ahokas, K. Miettinen, R. Rimon, and L. C. Andersson. 1999. Accumulation of macrophages in the CSF of schizophrenic patients during acute psychotic episodes. Am. J. Psychiatry 156:1725-1729. [DOI] [PubMed] [Google Scholar]
- 33.Okahara, G., S. Matsubara, T. Oda, J. Sugimoto, Y. Jinno, and F. Kanaya. 2004. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84:982-990. [DOI] [PubMed] [Google Scholar]
- 34.O'Reilly, R. L., and S. M. Singh. 1996. Retroviruses and schizophrenia revisited. Am. J. Med. Genet. 67:19-24. [DOI] [PubMed] [Google Scholar]
- 35.Perron, H., E. Jouvin-Marche, M. Michel, A. Ounanian-Paraz, S. Camelo, A. Dumon, C. Jolivet-Reynaud, F. Marcel, Y. Souillet, E. Borel, L. Gebuhrer, L. Santoro, S. Marcel, J. M. Seigneurin, P. N. Marche, and M. Lafon. 2001. Multiple sclerosis retrovirus particles and recombinant envelope trigger an abnormal immune response in vitro, by inducing polyclonal Vβ16 T-lymphocyte activation. Virology 287:321-332. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, K. E., J. S. Errett, T. Wei, D. E. Dimcheff, R. Ransohoff, W. A. Kuziel, L. Evans, and B. Chesebro. 2004. MCP-1 and CCR2 contribute to non-lymphocyte-mediated brain disease induced by Fr98 polytropic retrovirus infection in mice: role for astrocytes in retroviral neuropathogenesis. J. Virol. 78:6449-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portis, J. L. 2002. Perspectives on the role of endogenous human retroviruses in autoimmune diseases. Virology 296:1-5. [DOI] [PubMed] [Google Scholar]
- 38.Portis, J. L., and W. P. Lynch. 1998. Dissecting the determinants of neuropathogenesis of the murine oncornaviruses. Virology 247:127-136. [DOI] [PubMed] [Google Scholar]
- 39.Prudhomme, S., G. Oriol, and F. Mallet. 2004. A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression. J. Virol. 78:12157-12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolland, A., E. Jouvin-Marche, M. Saresella, P. Ferrante, R. Cavaretta, A. Creange, P. Marche, and H. Perron. 2005. Correlation between disease severity and in vitro cytokine production mediated by MSRV (Multiple Sclerosis associated RetroViral element) envelope protein in patients with multiple sclerosis. J. Neuroimmunol. 160:195-203. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schmidt, M., G. Hoffmann, M. Wissler, N. Lemke, A. Mussig, H. Glimm, D. A. Williams, S. Ragg, C. U. Hesemann, and C. von Kalle. 2001. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 12:743-749. [DOI] [PubMed] [Google Scholar]
- 43.Schön, U., W. Seifarth, C. Baust, C. Hohenadl, V. Erfle, and C. Leib-Mösch. 2001. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology 279:280-291. [DOI] [PubMed] [Google Scholar]
- 44.Seifarth, W., H. Skladny, F. Krieg-Schneider, A. Reichert, R. Hehlmann, and C. Leib-Mösch. 1995. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J. Virol. 69:6408-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifarth, W., U. Krause, C. Hohenadl, C. Baust, R. Hehlmann, and C. Leib-Mösch. 2000. Rapid identification of all known retroviral reverse transcriptase sequences with a novel versatile detection assay. AIDS Res. Hum. Retrovir. 16:721-729. [DOI] [PubMed] [Google Scholar]
- 46.Seifarth, W., B. Spiess, U. Zeilfelder, C. Speth, R. Hehlmann, and C. Leib-Mösch. 2003. Assessment of retroviral activity using a universal retrovirus chip. J. Virol. Methods 112:79-91. [DOI] [PubMed] [Google Scholar]
- 47.Seifarth, W., O. Frank, U. Zeilfelder, B. Spiess, A. D. Greenwood, R. Hehlmann, and C. Leib-Mösch. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharpe, A. H., and R. Jaenisch. 1993. Retroviral spongiform degenerative disease produced by the murine neurotropic retrovirus Cas-Br-E. Dev. Biol. Stand. 80:45-52. [PubMed] [Google Scholar]
- 49.Shih, A., R. Misra, and M. G. Rush. 1989. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J. Virol. 63:64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector, D. H. 1992. Potential host range expansion of the retroviruses. Dev. Biol. Stand. 76:153-164. [PubMed] [Google Scholar]
- 51.Stauffer, Y., G. Theiler, P. Sperisen, Y. Lebedev, and C. V. Jongeneel. 2004. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 4:2. [PubMed] [Google Scholar]
- 52.Sverdlov, E. D. (ed.). 2004. Retroviruses and primate genome evolution. Landes Bioscience, Georgetown, Tex.
- 53.Torrey, E. F., and R. H. Yolken. 1995. Could schizophrenia be a viral zoonosis transmitted from house cats? Schizophr. Bull. 21:167-171. [DOI] [PubMed] [Google Scholar]
- 54.Torrey, E. F., and R. H. Yolken. 1998. At issue: is household crowding a risk factor for schizophrenia and bipolar disorder? Schizophr. Bull. 24:321-324. [DOI] [PubMed] [Google Scholar]
- 55.Torrey, E. F., and R. H. Yolken. 2001. The schizophrenia-rheumatoid arthritis connection: infectious, immune, or both? Brain Behav. Immun. 15:401-410. [DOI] [PubMed] [Google Scholar]
- 56.Torrey, E. F., R. Rawlings, and R. H. Yolken. 2000. The antecedents of psychoses: a case-control study of selected risk factors. Schizophr. Res. 46:17-23. [DOI] [PubMed] [Google Scholar]
- 57.Torrey, E. F., J. Miller, R. Rawlings, and R. H. Yolken. 1997. Seasonality of births in schizophrenia and bipolar disorder: a review of the literature. Schizophr. Res. 28:1-38. [DOI] [PubMed] [Google Scholar]
- 58.Torrey, E. F., M. Webster, M. Knable, N. Johnston, and R. H. Yolken. 2000. The Stanley foundation brain collection and neuropathology consortium. Schizophr. Res. 44:151-155. [DOI] [PubMed] [Google Scholar]
- 59.Wunderlich, V., F. Fey, and G. Sydow. 1980. Antiviral effect of haloperidol on Rauscher murine leukemia virus. Arch. Geschwulstforsch. 50:758-762. [PubMed] [Google Scholar]
- 60.Yolken, R. 2004. Viruses and schizophrenia: a focus on herpes simplex virus. Herpes 11(Suppl. 2):83A-88A. [PubMed] [Google Scholar]
- 61.Yolken, R. H., H. Karlsson, F. Yee, N. L. Johnston-Wilson, and E. F. Torrey. 2000. Endogenous retroviruses and schizophrenia. Brain Res. Brain Res. Rev. 31:193-199. [DOI] [PubMed] [Google Scholar]