Abstract
Several complex enveloped viruses assemble in the membranes of the secretory pathway, such as the Golgi apparatus. Among them, bunyaviruses form immature viral particles that change their structure in a trans-Golgi-dependent manner. To identify key Golgi factors for viral structural maturation, we have purified and characterized the three viral forms assembled in infected cells, two intracellular intermediates and the extracellular mature virion. The first viral form is a pleomorphic structure with fully endo-β-N-acetylglucosaminidase H (Endo-H)-sensitive, nonsialylated glycoproteins. The second viral intermediate is a structure with hexagonal and pentagonal contours and partially Endo-H-resistant glycoproteins. Sialic acid is incorporated into the small glycoprotein of this second viral form. Growing the virus in glycosylation-deficient cells confirmed that acquisition of Endo-H resistance but not sialylation is critical for the trans-Golgi-dependent structural maturation and release of mature viruses. Conformational changes in viral glycoproteins triggered by changes in sugar composition would then induce the assembly of a compact viral particle of angular contours. These structures would be competent for the second maturation step, taking place during exit from cells, that originates fully infectious virions.
Viruses are molecular machines built up by viral and occasionally cellular macromolecules. Their functions depend on very stable intermolecular interactions among their components, which must be ready to revert during infection of a new cell. These apparently contradictory requirements are satisfied by molecular mechanisms that are understood in only a few cases. Many enveloped viruses bud through the plasma membrane, a process that serves for virus assembly, acquisition of envelope, and exit from the cell. However, there are a number of enveloped viruses, some of them important pathogens for animals and humans, that replicate and bud into intracellular membranes (16, 27, 38). These viruses frequently build a complex structure known as the viral factory, where the virus anchors its replication complexes and assembly sites. The mechanisms of formation of these factories and how replication, transport, and assembly of viral elements are coordinated inside are poorly understood, but they usually involve the recruitment and association of membranes, mitochondria, and cytoskeletal elements (34).
Some of these complex viruses use the Golgi apparatus as a key element of the viral factory (41, 43, 44). The reasons for this preference are not clear since it adds a potential difficulty in terms of efficient transport of large structures, such as viral particles, along the Golgi stack. Moreover, the Golgi complex is a highly dynamic organelle, subjected to constant changes associated with anterograde and retrograde transport of proteins (2, 31).
During transport of enveloped viruses along the Golgi stacks, posttranslational modifications, such as glycosylation, proteolysis, and phosphorylation, are performed on accessible viral proteins by the enzymes located at the lumen of the different Golgi subcompartments. Since these enzymes must act on large structures, modifications take place with a variable efficiency. In the case of flaviviruses, posttranslational modifications in Golgi are crucial for the infectivity of the final products of assembly. For coronaviruses and bunyaviruses, major structural changes of viral particles inside the Golgi stack take place in a trans-Golgi-dependent manner (43, 44).
We have been studying the structure and assembly pathways of several complex enveloped viruses to understand how viral factors interact with cellular elements to generate infectious virions (11, 34, 40, 41, 43, 44). Bunyaviruses in particular are an adequate structural model to analyze how the viral factory is built and how Golgi factors participate in viral assembly and maturation.
Bunyaviruses form a large family of viruses with worldwide distribution, the Bunyaviridae, that contains more than 350 members, most of them transmitted by arthropods. Some of these viruses cause severe disease in humans, including hemorrhagic fevers and encephalitis (14). The virions are spherical, 80 to 120 nm in diameter, display surface glycoprotein projections, and contain three single-stranded RNA genome segments of negative polarity designated large (L), medium (M), and small (S). Four structural proteins are inserted into mature virions: nucleocapsid (N), polymerase (L), and two glycoproteins (Gc and Gn) (45). Bunyamwera virus, the prototype of the family, encodes two additional nonstructural proteins named NSs and NSm. NSs acts as a virulence factor that inhibits cellular protein translation, acting also on the activity of the viral polymerase (6). The role of NSm, a protein that accumulates in the Golgi complex (32), is unknown.
Most bunyaviruses assemble in the Golgi complex due to a Golgi retention signal in the Gn glycoprotein (26, 42, 48). In the case of Uukuniemi virus, assembly has been also observed in membranes of the pre-Golgi intermediate compartment (19). In a previous study from our laboratory, that included processing of cells by cryomethods with a high preservation of structures (44), we detected two unknown maturation steps in the morphogenesis of Bunymawera virus in BHK-21 and Vero cells. The first maturation takes place inside the Golgi stack, where annular particles transform into dense, compact structures. The second structural change, which consists of detectable changes in the surface coat of spikes, most probably takes place during the egress of viral particles from cells. A functional trans-Golgi is needed for the first maturation, as demonstrated when infection is done in the presence of drugs that reversibly disrupt the trans-side of the Golgi complex (44). These results, together with data obtained from some other unrelated viruses, show that the Golgi complex contains key factors for the structural transformation of complex viruses.
To understand how bunyaviruses assemble and mature, we have purified the three types of viral particles previously identified in infected cells by transmission electron microscopy (TEM). Structural and biochemical characterization of the three viral forms, together with the assembly of the virus in glycosylation-deficient cells, indicates that conformational changes of viral glycoproteins induced by modifications in sugar composition after acquisition of endo-β-N-acetylglucosaminidase H (Endo-H) resistance, must trigger the formation of a proteinaceous geometric envelope. These particles are then prepared to undergo the second structural change during exit from the cell, which makes them fully infectious.
MATERIALS AND METHODS
Cell lines, viruses, antibodies, and conjugates.
BHK-21 (C-13) and Vero cells, supplied by the American Type Culture Collection, were grown in Dulbecco's modified Eagle's medium supplemented with 10% (BHK-21) or 5% (Vero) fetal calf serum from Reactiva S.A. (Barcelona, Spain). The CHO pro5 cell line and the derived Lec1 and Lec2 cell lines, deficient in N-acetylglucosaminyltransferase I activity and CMP-sialic acid transport, respectively (8, 13, 51), were also purchased from the American Type Culture Collection and propagated in minimal essential medium supplemented with 10% fetal calf serum. The BHK Ricr 14-deficient cell line, originally generated in R. C. Hughes' laboratory (28), was further characterized later (35) and kindly supplied by P. Gleeson (Monash University Medical School, Victoria, Australia). This cell line was grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum.
Bunyamwera virus (originally V-565-001-522), obtained from the American Type Culture Collection, was propagated and subjected to titer determination in BHK, Vero, and CHO cells by a lysis-plaque-forming assay, as described previously (55).
The antiserum against Bunyamwera virus and the monoclonal antibody against Bunyamwera Gc protein were described previously (21, 55, 56). The 7H12.1 monoclonal antibody, specific for Bunyavirus Gn glycoprotein (22), obtained with sonicated La Crosse bunyavirus as the immunogen, was purchased from Chemicon International (Temecula, CA). Secondary antibodies conjugated with rhodamine or fluorescein were supplied by Southern Biotechnology Associates, Inc. (Birmingham, AL). Mitochondria were localized with the probe for live cells Mitotracker Green FM (Molecular Probes, Eugene, Oregon). Secondary antibodies conjugated with colloidal gold particles of 10 nm were provided by Biocell Research Laboratory (Cardiff, United Kingdom). The conjugate of the Triticum vulgaris agglutinin and 10-nm colloidal gold was purchased from EY Laboratories (San Diego, CA).
Infections and treatments with drugs.
For production of Bunyamwera virus infectious stocks, confluent monolayers were infected at 32°C and a multiplicity of infection of 0.001 PFU per cell. Supernatants were processed as described (44). Virus yields (PFU per milliliter) were determined by titer determination on cell monolayers using a lysis-plaque-forming assay, as described (55). For the studies of viral assembly, cell monolayers were infected at 37°C and 1 PFU/ml. At different times postinfection cells were processed for immunofluorescence or fixed for electron microscopy analysis. Some cultures were subjected to treatments with the drug megalomycin, kindly supplied by B. Alarcón (Centro de Biología Molecular Severo Ochoa, Madrid, Spain). Treatments with megalomycin were done as described (44).
Immunofluorescence and confocal microscopy.
Cell monolayers grown on coverslips were washed with phosphate-buffered saline (PBS) and incubated at −20°C in a mixture of methanol and acetone (1:1) before treating with PBS containing 2% bovine serum albumin and adding the anti-Gc monoclonal antibody diluted 1:200 in PBS containing 0.1% bovine serum albumin. Coverslips were then incubated with the corresponding secondary antibody diluted 1:600 in PBS containing 0.1% bovine serum albumin, washed with the same buffer, and mounted on glass slides with Mowiol (Aldrich Chemical Co., Milwaukee, WI). Samples were studied under a Zeiss Axiophot fluorescence microscope, and images were collected with a MicroMax digital camera system.
To localize the viral protein Gc and mitochondria simultaneously, the probe Mitotracker green (Molecular Probes/Invitrogen Corp.) was added to live cell cultures at a concentration of 200 nM in Dulbecco's modified Eagle's medium containing 2% fetal calf serum. After 45 min at 37°C, the medium was removed, and the cells were fixed 20 min at −20°C with methanol-acetone (1:1) and incubated with the corresponding primary and secondary antibodies, washed, and mounted as described above. Images were obtained in a Bio-Rad Radiance 2100 confocal laser microscope.
Transmission electron microscopy of cell cultures.
Ultrastructural analysis of infected cell monolayers was done by conventional embedding in epoxy-resin EML-812 (Taab Laboratories, Adermaston, Berkshire, United Kingdom) using procedures previously described in detail (39, 43). Ultrathin sections were collected in Formvar-coated electron microscopy grids, stained with uranyl acetate and lead citrate, and studied in a JEOL 1200-EX II electron microscope operating at 100 kV.
Isolation of the three viral forms by centrifugation in Optiprep gradients.
Cell monolayers (BHK-21 or Vero cells) were infected at a multiplicity of infection of 0.001 PFU/cell and maintained 55 h at 32°C. Culture medium was removed and clarified by centrifugation at 3,700 × g, 20 min at 4°C. Supernatant was centrifuged 2.5 h at 67,000 × g through a 30% (wt/vol) sucrose cushion made in TEN buffer (0.01 M Tris-HCl, pH 7.4 containing 0.1 M NaCl and 1 mM EDTA) with the Roche Complete Mini protease inhibitor cocktail (1 tablet per 20 ml of buffer). The pellet was resuspended in 200 μl of TEN with protease inhibitors and applied to a 13 to 22% (vol/vol) Optiprep iodixanol density gradient (Sigma). The gradient was made by placing 10 layers of 0.9 ml, which differed in 1% iodixanol, in a centrifuge tube, starting with the bottom layer (22%). Each layer was frozen in dry ice before adding a new one. These tubes were kept frozen and placed at room temperature overnight before use. During melting, a continuous gradient is formed. Centrifugation of virus preparation was performed for 1.5 h at 250,000 × g. Fractions of 250 μl were collected from the top of the gradient and processed for structural and biochemical characterization.
To purify the intracellular viral forms, cell monolayers were washed twice with TEN buffer containing protease inhibitors after removing the cell culture medium at 55 h postinfection. Cells were collected in the buffer, transferred to Falcon tubes, and frozen at −80°C. After three consecutive cycles of freezing and thawing to break the cells and release the viral particles, the rest of the procedure was done as described for extracellular viral particles.
Electron microscopy of isolated viral forms.
Purified viruses were processed by negative staining, particle counting, freeze-etching, thin sectioning, immunogold labeling, and lectin-gold detection before TEM. Negative staining with uranyl acetate was performed after adsorbing viral particles on EM grids made hydrophilic by glow discharge by standard procedures. The number of particles in cell supernatants was calculated after making a series of dilutions of the virus preparations and a standard solution of latex spheres, as described (44). The standard provided by Ted Pella, Inc. (Redding, CA), contained latex spheres 93 nm in diameter and known concentration (2,415 × 1011 particles/ml). After negative staining, samples were studied by TEM and counting was done in the samples containing similar numbers of viral particles and latex spheres. Correlation with values of PFU/ml from lysis-plaque-forming assays allowed calculation of the number of infectious particles.
The surface of viral particles was studied by freeze-etching. Particles were adsorbed to mica sheets and fast frozen in liquid ethane. Samples were then transferred to a BAF 060 freeze fracture unit (BAL-TEC, Liechtenstein), where the temperature was switched from −150°C to −80°C. Etching was prolonged for 2 h. Metal replicas of the exposed surfaces were obtained by evaporating 2 nm of tantalum at an angle of 45° and 20 nM of carbon at 90°. Replicas were floated in commercial bleach, extensively washed in distilled water, picked up in Formvar-coated EM grids, and studied by EM.
For studying the internal structure of isolated viruses, they were processed by conventional embedding and thin sectioning as described above after mixing the viral particles with fixed noninfected cells, to increase the total mass of the sample. Thin sections were stained with uranyl acetate and lead citrate and studied by TEM. For immunogold labeling of isolated viruses, particles were adsorbed on EM grids coated with Formvar and carbon made hydrophilic by glow discharge. Viruses were then incubated with saturation buffer, primary antibodies (anti-Gc or anti-Gn) and secondary antibodies conjugated with 10-nm colloidal gold particles, as described (39).
For detection of sialic acid on the surface of viral particles, they were labeled with a 10-nm gold conjugate of Triticum vulgaris agglutinin. Viruses adsorbed to EM grids were washed with TEN buffer and incubated 10 min with the lectin-gold conjugate diluted 1:40 in TEN. After washing in the same buffer and distilled water, samples were processed by negative staining with uranyl acetate. As a control some samples were treated with the lectin conjugate diluted 1:40 in 1 mg/ml sialic acid in TEN.
Biochemical analysis of isolated viruses.
To study the protein composition of purified viruses, samples were processed by electrophoresis in polyacrylamide gels in the presence of sodium dodecyl sulfate (SDS) under standard conditions. Aliquots of purified viruses were boiled for 5 min in protein dissociation buffer containing 62.5 mM Tris (pH 6.8), 2% (wt/vol) SDS, 0.25% (wt/vol) bromophenol blue, 5% (vol/vol) glycerol, and 5% (vol/vol) 2-mercaptoethanol. Proteins were resolved by polyacrylamide gel electrophoresis (PAGE) in the presence of SDS using 8 or 11% acrylamide gels and visualized by Coomassie blue staining. Unidentified bands in gels were processed by matrix-assisted laser desorption ionization (MALDI) peptide mass fingerprinting and database searching as described (33).
For Western blot analysis, proteins were transferred from gels to nitrocellulose membranes by standard blotting procedures. Membranes were saturated (overnight at 4°C) with PBS containing 5% nonfat dry milk and 0.05% Tween 20, and incubated 1 h at room temperature with an anti-Bunyamwera polyclonal antiserum diluted 1:2,000 in saturation buffer. After washing with this buffer, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody diluted 1:2,000. Immunoreactive bands were detected by chemiluminescence (ECL kit, Amersham Biosciences, United Kingdom).
Treatment with endoglycosidase H (Roche Diagnostics GmbH, Mannheim, Germany) was done after incubating the samples in denaturation solution (0.15% SDS with 0.01% β-mercaptoethanol in water). Samples were incubated 10 min at room temperature in this buffer and then sonicated three times for 5 seconds each. The same volume of reaction buffer (0.5 M sodium citrate containing 0.6% EDTA, pH 5.5) was then added to the samples, as well as 2.5 mU of Endo-H per sample. Control samples received the same volume of reaction buffer instead. Incubation was then maintained 2 h at 37°C. Samples were then sonicated and 0.5 mU of enzyme was added (or an equivalent volume of reaction buffer to control samples). Incubation was done 1 h at 37°C and sample buffer was added to all samples before processing by SDS-PAGE and Western blot. Transferrin and RNase B, provided by Roche in the Endo-H kit, were included as negative and positive controls for Endo-H sensitivity (not shown).
Detection of sialic acid in viral proteins was done with a mixture of lectins conjugated with digoxigenin from the DIG glycan differentiation kit (Roche Diagnostics GmbH, Mannheim, Germany). After SDS-PAGE, proteins were transferred from gels to nitrocellulose membranes that were incubated overnight with saturation buffer as described above. After washing in TBS (0.05 M Tris-HCl, 0.15 M NaCl, pH 7.5) membranes were incubated 2 h with a mixture of Sambucus nigra agglutinin and Maackia amurensis agglutinin diluted 1:500 and 1:100, respectively, in TBS containing 1 mM MgCl2, 1 mM MnCl2 and 1 mM CaCl2. After washing with TBS three times for 10 min each, membranes were incubated 1 h with an antidigoxigenin antibody conjugated with alkaline phosphatase, diluted 1:1,000 in TBS. After washing in TBS, the staining reaction was performed by adding the staining solution, prepared with 10 ml Tris buffer (0.1 M Tris-HCl, 0.05 M MgCl2, 0.1 M NaCl, pH 9.5) plus 200 μl nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolylphosphate (X-phosphate) solution. The color reaction was stopped with distilled water.
RESULTS
Isolation of three viral structures from Bunyamwera virus-infected cells.
Bunyamwera virus induces the formation of a large perinuclear structure, the viral factory, in BHK-21 cells. This structure is defined by the accumulation of viral proteins and particular cellular organelles, such as mitochondria (Fig. 1A) around the Golgi complex, where the virus initiates its morphogenesis (26, 34). In the viral factory (Fig. 1B) the virus replicates and initiates assembly, building two intermediates and the extracellular virion as the final product. The structural components of mature virions are represented in Fig. 1C and include an envelope derived from Golgi membranes, where the spikes are formed by heterodimers of the viral glycoproteins Gc and Gn, and the internal component with three ribonucleoproteins (negative-sense viral RNA segments) bound to the nucleocapsid N protein and the RNA polymerase (L protein). These elements get together to assemble a first structure that we have named type I intracellular virus, formerly named intracellular annular virus (44).
FIG. 1.
Isolation of three viral forms from Bunyamwera virus-infected BHK-21 cells. (A) At 8 h postinfection, confocal microscopy shows the large perinuclear structure (arrow), known as the viral factory (vf), where viral proteins (here Gc is labeled in red) and mitochondria (green) are recruited. Labeling was performed as described in Materials and Methods. Small red dots seen on the cell periphery correspond to secretory vesicles filled with viruses. (B) Electron microscopy shows the viral factory around the nucleus (N) as associations of mitochondria (mi) and membranes (arrows). In these factories viral particles are assembled by recruitment of the structural components represented in C: an envelope with spikes (made of Gc and Gn glycoproteins) and an internal core containing three ribonucleoproteins (RNPs) of RNA, nucleocapsid protein (N), and RNA polymerase (L). (D) Viral particles with three different morphologies are seen in thin sections of infected cells: intracellular viruses on the left correspond either to type I (annular structures that correspond to immature precursors) or type II (an intermediate dense form thatassembles in a trans-Golgi-dependent manner) morphology. Extracellular virions (E) on the right are also dense particles but with a more defined coat of spikes (arrowheads). Higher-magnification fields on the bottom show the structural characteristics of the viral particles in more detail. (E) Isolation of these three viral forms gave homogeneous populations of type I (left), type II (middle), and extracellular (right) viruses. Viral structural proteins were separated by SDS-PAGE and analyzed by Coomassie blue staining (CB) and Western blotting (WB). The asterisk on the gel for extracellular viruses marks a band of seralbumin identified by MALDI peptide mass fingerprinting. Thin sections of purified viruses (high-magnification EM fields on the bottom) show the internal structure of the three purified viral forms. Bars: 1 μm in B, 100 nm in D and E.
In thin sections these structures have an annular morphology and a light core (Fig. 1D, arrow in left panel). Type I viruses transform into a more compact, denser structure that we have named type II intracellular virus (Fig. 1D, arrowhead in left panel). Type II virus (formerly intracellular dense virus) seems to change the organization of the envelope at the moment of secretion from the cell, since extracellular E viruses (formerly extracellular dense virus) have a neater coat of spikes (Fig. 1D, arrowheads in right panel). Higher-magnification fields in Fig. 1D show the structural characteristics of the three viral morphologies.
The isolation of these three viral forms from infected cells is shown in Fig. 1E. Intracellular particles were isolated from infected cell monolayers after removing the culture supernatant that was used to purify the extracellular virions. Three rounds of freezing and thawing were applied to the cells, followed by several steps of clarification that ended up with centrifugation in Optiprep gradients as described in Materials and Methods. These gradients allow the purification of very intact viruses in terms of structural and functional integrity and have been used successfully to separate viral intermediates that are very similar in size and density (53).
Negative staining of fractions from the gradients showed complete separation of both intracellular viral forms, I and II. Type I viruses look pleomorphic and heterogeneous in size (Fig. 1E, left). Type II viruses are more compact, homogeneous in size (Fig. 1E, center), and very similar to purified extracellular virions (Fig. 1E, right). The presence of the viral structural proteins was confirmed by SDS-PAGE (Coomassie blue) and Western blotting using a polyclonal antiserum against whole extracellular virions. Further purification steps were not systematically applied to preparations of intracellular viruses after observing that they damaged their structural integrity (not shown). Extracellular virions were purified in much larger amounts with almost no contaminants, as shown by Coomassie blue staining after SDS-PAGE, with the exception of seralbumin (marked with an asterisk) identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) peptide mass fingerprinting (Fig. 1E, right). Thin sections of purified viruses show the exclusive internal features of the three viral forms (Fig. 1E, high-magnification fields) that correlate to the structures previously described in situ (Fig. 1D, high-magnification fields).
Structural and biochemical characterization of isolated viruses.
Structural characterization included negative staining, freeze-etching, and immunolabeling of viral glycoproteins(Fig. 2). Negative staining shows that type I virus has a round or pleomorphic contour, while type II virus has a more structured envelope with angular contours. The extracellular virions also exhibit an envelope with angular contours, and in addition, spikes are clearly seen on the surface of the particles (Fig. 2A, first column on the left). These features are confirmed by freeze-etching, which also shows a close-packed pattern for the spikes of the extracellular virions, instead of a random surface distribution (Fig. 2A, second column).
FIG. 2.
Structural and biochemical characterization of isolated viral forms. (A) EM shows the characteristics of type I (upper line), type II (middle), and extracellular (bottom) viruses. Contour and surface analyses by negative staining and freeze-etching, respectively (two columns on the left), show that type I virus is pleomorphic with a rounded contour and a smooth surface, while type II virus has acquired a more compact, angular contour. In addition, E virions have neat surface spikes. Immunogold labeling with anti-Gc and anti-Gn antibodies (two columns on the right) shows full recognition of Gc on E viruses, while Gn was detected on type I viruses exclusively. Values for infectivity (expressed as number of infectious particles/total number of particles) are indicated on the right. The large EM field shows extracellular virions treated with 1 M sucrose. Particles are opened in one spot and are releasing the RNPs (arrows), leaving a thick, geometrical envelope (arrowheads). (B) Posttranslational modifications in bunyavirus structural proteins were analyzed in isolated viral forms. Sensitivity to endoglycosidase H (on the left) distinguishes immature from mature glycoproteins. Total sensitivity is seen as a decrease in molecular weight for the corresponding band after Endo-H digestion, SDS-PAGE, and Western blot, while partial sensitivity is shown as an increase in band width by Western blot due to the generation of two or more bands after digestion (see Coomassie blue for Gc of E viruses on the right). Both Gc and Gn glycoproteins were totally sensitive to Endo-H in type I virus while sensitivity was partial in type II and E viruses. The presence of sialic acid is shown on the right. Lectin binding to proteins transferred to nitrocellulose showed reactivity in Gn of type II and extracellular viruses, but no reaction in the position corresponding to Gc. Binding of a conjugate of Triticum vulgaris agglutinin and 10-nm gold particles showed strong exposure of sialic acid on the surface of type II and E viruses, while type I virus was totally devoid of labeling. Bars: 100 nm.
The accessibility of the Gc and Gn epitopes on the viral surface was studied with specific antibodies against mature glycoproteins (obtained with extracellular mature virions as the immunogens). Immunogold detection showed that the “mature” Gc epitope starts to be recognized on the surface of type II viruses, while extracellular virions exhibit full recognition. On the other hand, the small Gn glycoprotein was successfully recognized with a monoclonal antibody that labeled the surface of approximately 25% of type I viral particles. However, type II and extracellular viruses were not labeled. This indicates that the Gn epitope is hidden to antibodies in mature viral forms, probably due to interactions with Gc in the mature heterodimer or to changes in the folding of Gn during maturation.
According to the number of infectious particles (calculated as described in Materials and Methods), isolated extracellular viruses are fully infectious (1 infectious particle out of 40), while type I viruses are noninfectious (1 out of 50,000). Type II viruses represent an intermediate situation, with low infectivity rates (1 out of 1,600). Treatment of purified extracellular viruses with 1 M sucrose in TEN buffer for 3 min at room temperature opened the viral particles, apparently through one single spot, which provoked the release of the internal ribonucleoproteins, while a thick, compact envelope of angular contours is still distinguishable (Fig. 2A, large field on the right).
Biochemical analysis of the three viral forms revealed a clear difference between the pleomorphic type I precursor and the two denser viral forms. Both Gc and Gn glycoproteins are glycosylated in the three viral forms since they were fully sensitive to Endo-F digestion (not shown). However, full sensitivity to Endo-H was found in both Gc and Gn glycoproteins from the precursor type I virus, while partial sensitivity in both glycoproteins was found in type II and extracellular viruses (Fig. 2B, left). Full sensitivity is seen as a displacement of the whole band, which shows higher mobility. This is clearly seen in both Gc and Gn of the type I precursor (Fig. 2B, left). Partial sensitivity means that some molecules remain sensitive to the enzyme, while the rest become resistant. This is visualized as a double band by Coomassie staining (Fig. 2B, right) and as a wider band by Western blot (Fig. 2B, central panels).
The presence of sialic acid in viral glycoproteins was analyzed in the three purified viral forms using specific lectins bound to digoxigenin as described in Materials and Methods (Fig. 2B, right). No reaction was detected for Gc protein from the three viral forms. Gn from type II and extracellular viruses exhibited a weak reaction, while no signal was detected for Gn from type I viruses. Bands in other positions must correspond to sialylated cellular proteins that copurify with intracellular viruses (see Coomassie-stained gels of purified intracellular viruses in Fig. 1E).
The accessibility of sialic acid on the virus surface was analyzed with a gold conjugate of the lectin Triticum vulgaris agglutinin and negative staining (Fig. 2B, micrographs on the right). Both type II and extracellular viruses exhibited a strong reaction that was completely eliminated when the lectin-gold conjugate was preincubated with sialic acid (not shown). Type I viruses did not show any binding of Triticum vulgaris agglutinin-gold conjugate.
In summary, glycoproteins from pleomorphic type I precursors contain the typical modifications of proteins that have not reached the trans-side of the Golgi stack, while compact, angular type II intermediate viruses already contain the modifications typical of trans-Golgi, also present in mature extracellular virions.
Growth of Bunyamwera virus in cells with impaired glycosylation steps.
Bunyavirus Gn and Gc glycoproteins exhibit various degrees of glycosylation depending on the specific virus and the cell line used (45). Biochemical analysis of the isolated viral forms pointed to trans-Golgi-associated posttranslational modifications as potential key factors for the structural transformation of the type I viral precursor to the second intermediate structure. This is also suggested by the results obtained when the virus was grown in the presence of megalomycin, a trans-Golgi-disrupting drug (5) (Fig. 3). Immunofluorescence detection of viral Gc shows that infection progresses in the presence of the drug (Fig. 3A and B). EM analysis showed that while the three typical viral forms are detected in normally infected BHK-21 cells and numerous type II intracellular viruses and extracellular virions are seen at 14 h postinfection (Fig. 3A), an accumulation of type I-like particles is observed in Golgi-derived membranes of cells treated with megalomycin (Fig. 3B). A careful EM examination confirmed that the internal structure of these accumulated particles, which are not able to exit the cells, is very similar to that described for isolated type I particles from untreated infected cells (compare Fig. 3B with Fig. 1D).
FIG. 3.
Viral forms produced at 14 h postinfection in BHK-21 cells, BHK-21 cells treated with the drug megalomycin (MGM), and BHK Ricr 14 cells. (A) Characteristic viral factory induced by Bunyamwera virus in BHK-21 cells, as seen by immunofluorescence (top), is seen as accumulation of Gc protein close to the nucleus (arrow). EM fields of these areas show numerous type II viruses at 14 h postinfection, as well as typical extracellular (E) virions, with dense internal structure. Geometrical contours, as seen by negative staining (NS) and freeze-etching (FE),are typical of a mature, functional morphology. (B) In the presence of the drug megalomycin, infection progresses, as indicated by immunofluorescence of Gc (top). As seen by EM, type I virus-like particles are the only morphology assembled and accumulated in these cells. No extracellular forms are detected. (C) The GlcNAc-TI-deficient BHK Ricr 14 cell line also supports virus growth, as seen by immunofluorescence of Gc (top), but only type I-like viruses accumulate intracellularly and exit the cells (central EM fields). Their morphology is undistinguishable from that corresponding to immature type I viruses assembled in normally infected BHK-21 cells (Fig. 1) and from the viral particles accumulated in megalomycin-treated BHK-21 cells shown in B. Extracellular “immature” viruses are designated E*. As indicated at the bottom, infectivity in supernatants of Ricr 14 cells is significantly lower compared to that of supernatants from infected BHK-21 cells at the same times postinfection. (D) Endo-H sensitivity analyzed in cell monolayers showed partial sensitivity in glycoprotein Gc of infected BHK-21 cells while fully sensitive Gc is detected in infected Ricr14 cells. (E) Quantification of viral forms (expressed as the percentage of viruses with a particular morphology after counting more than 500 viruses for each cell line in thin sections and TEM) showed the presence of numerous mature virions in BHK-21 cells, while immature viruses accumulate and are secreted from BHK Ricr 14 cells. Bars: 150 nm.
To test if the trans-Golgi-associated factor involved in the transformation of type I to type II virus is the processing of sugar chains, viruses were grown in BHK Ricr 14 cells, which are deficient in N-acetylglucosaminyltransferase I (GlcNAc-TI), the enzyme that catalyzes the first step in the conversion of high-mannose N-glycans into complex and hybrid N-glycans (8, 35). In these cells, infection progresses as suggested by immunofluorescence of Gc (Fig. 3C, top) and viral particles accumulate in the Golgi complex, where they exhibit an immature morphology typical of type I viruses (Fig. 3C, middle). These particles are able to exit the cells and are seen in the extracellular environment, where they maintain a pleomorphic shape with a low-density core (Fig. 3C, bottom).
Endo-H sensitivity confirmed that the viruses secreted by these cells have a fully sensitive Gc glycoprotein compared with the partially sensitive Gc from virions secreted by BHK-21 cells (Fig. 3D). Quantification of viral forms in thin sections of infected cells confirmed the blockade in structural maturation at the type I virus stage in BHK Ricr 14 cells (Fig. 3E). While in infected BHK-21 cells more than 80% of viral particles correspond to type II and mature extracellular virions, in BHK Ricr 14 cells more than 70% are intracellular type I viruses and more than 20% are immature extracellular viruses. Values for infectivity indicate that the viruses with immature morphology released by these cells have low infectivity rates.
Addition of sialic acid is the final step of glycosylation and takes place in the trans-Golgi (18). We have detected sialic acid in Gn from type II and extracellular viruses (Fig. 2B). To check if sialic acid is important for transformation of type I viruses, we have grown the virus in cell lines derived from Chinese hamster ovary cells (CHO) (Fig. 4). CHO pro5 cells support the growth of Bunyamwera virus and formation of the three viral morphologies (Fig. 4A). In CHO Lec 1 cells, a cell line derived from CHO pro5 with the same deficiency as BHK Ricr 14 cells (no GlcNAc-TI activity), infection progressed, as suggested by immunofluorescence of viral Gc (Fig. 4B, top). Type I-like viruses accumulated intracellularly and exited the cells (Fig. 4B, top EM fields).
FIG.4.
Viral forms assembled in CHO pro5 cells and two derived glycosylation-deficient cell lines, Lec1 and Lec2. (A) Perinuclear accumulation of viral proteins is seen in infected CHO pro5 cells as seen by immunofluorescence of Gc (top). EM fields show the two typical intracellular morphologies (I and II) as well as extracellular virions (E) with a dense interior. Negative staining (NS) and freeze-etching (FE) show compact particles with rough surfaces, but the layer of spikes is not clearly seen. (B) The N-acetylglucosaminyltransferase I-deficient CHO Lec1 cell line also accumulates viral proteins in the perinuclear area, rendering a Golgi-like pattern by immunofluorescence of Gc (top). A few dots representing secretory vesicles with viruses are observed. At the EM level, type I-like annular intracellular viruses accumulated inside infected cells (middle panel). These immature forms are able to exit the cells and they exhibit a pleomorphic, round morphology (E*). These particles are labile and easily disrupted, as observed by negative staining (arrows). Freeze-etching shows a smooth surface for these particles. (C) The sialylation-deficient CHO Lec2 cell line exhibits patterns similar to the parental cell line by both immunofluorescence and TEM (top). Intracellular viruses correspond to both type I and type II morphologies, while only normal extracellular virions (E), with compact and dense internal structure, are detected. (D) Quantification of viral forms assembled in CHO pro5, Lec1, and Lec2 cells shows similar patterns for CHO pro5 and Lec2 cells, while Lec1 accumulates immature forms at both intracellular and extracellular locations. Quantification was done on thin sections of infected cells and is represented as the percentage of each morphology after counting more than 500 viral particles for each cell line. Bars: 150 nm.
As shown by negative staining, extracellular viral particles are pleomorphic and rather labile since they get easily disrupted during the staining procedure (arrows). Both negative staining and freeze-etching showed smooth, round viral particles (Fig. 4B, bottom EM panels). When the virus was grown in the CHO Lec2 cell line, derived from the parental CHO pro5 cell line and deficient in the translocase of CMP N-acetylneuraminic acid (CMP-sialic acid) (13), immunofluorescence showed perinuclear accumulation of viral antigens (Fig. 4B, top). In these sialylation-deficient cells, the three viral forms were detected ultrastructurally (Fig. 4C, middle EM panels). Extracellular viruses looked very similar to the particles released by CHO pro5 cells: compact angular contours and poorly defined spikes after freeze-etching. Infectivity was low in CHO pro5, Lec 1, and Lec 2 supernatants: 1 out of 4,200, 1 out of 10,200, and 1 out of 18,000, respectively.
Quantification of viral forms also confirmed the block in structural maturation of the virus in the Lec1 but not in the Lec 2 cell line (Fig. 4D).
DISCUSSION
Viruses are genetically simple associations of macromolecules that exhibit a variable complexity from a structural point of view. Complex enveloped viruses frequently use intracellular membranes for replication and assembly. How they connect these processes inside the cells is largely unknown for most of them.
Bunyaviruses have an interesting assembly pathway that starts in the Golgi complex, a key element of the viral factory. Inside the Golgi stack the immature precursors transform into a different structure in a process that depends on a functional trans-Golgi. Posttranslational modifications identified in bunyavirus structural proteins include palmitoylation and processing of sugar chains in both glycoproteins (7, 45). No proteolytic processing, phosphorylation, or sulfation has been described for any of the bunyavirus structural proteins. Being a process associated with transport to the trans-side of the Golgi stack, processing of sugar chains into complex, Endo-H-resistant forms and sialylation were the first candidates to be tested as potential key factors for viral structural transformation.
N-linked glycosylation is important not only for correct protein folding but also for glycoprotein function (18, 36, 49). The analysis of isolated viral intermediates and the growth of the virus in glycosylation-deficient cell lines indicated that correct folding of bunyavirus glycoproteins, which most probably create particular lateral interactions in the envelope, is obtained through acquisition of Endo-H resistance in a percentage of glycoprotein molecules. This biochemical modification is a key Golgi factor for envelope structural transformation and release of infectious virions. Interestingly, the related hantaviruses have fully Endo-H-sensitive glycoproteins (47). A detailed structural analysis of hantaviruses is needed, although preliminary observations suggest that they can be pleomorphic structures (15).
Although a trans-Golgi-associated factor, sialylation does not seem to participate in the first structural maturation of these viruses. This was confirmed when the virus was grown in the sialylation-deficient cell line CHO Lec2. However, due to the semipermissive character of parental CHO cells to Bunyamwera infection, we do not know if sialylation is important for infectivity.
The surface structure of viruses released by CHO pro5 cells does not match the well-organized coat of spikes described for viruses released by BHK-21 and Vero cells. Additional factors must be participating in the construction of the fully functional particle, probably during the second maturation step. This takes place when the virus exits the cells. Differences in composition between the content of secretory vesicles and the extracellular environment must be related to the observed changes in the organization of the spikes, which make the virus fully infectious.
Palmitoylation is the other posttranslational modification detected in bunyavirus glycoproteins. Intracellular location of palmitoylation is not completely understood, but it has not been described as a trans-Golgi-associated process (4). Thus, this modification was not considered a good candidate for participating in the first structural maturation of Bunyamwera virus. In fact, when the virus was grown in the presence of high concentrations of palmitoylation inhibitors, a reduction in virus production was observed but viral morphogenesis proceeded normally and the three viral morphologies were observed (Novoa et al., unpublished results).
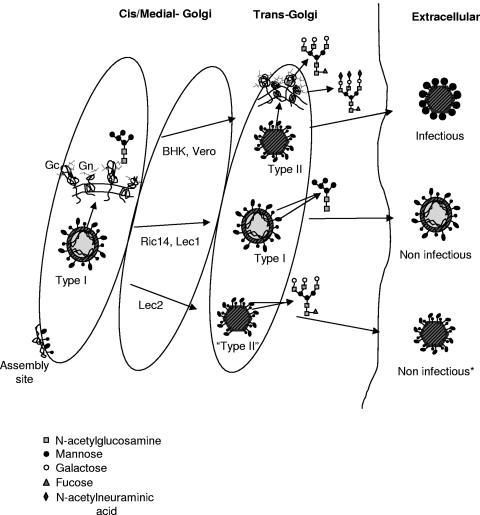
Our data are summarized in Fig. 5 as a proposal for the potential connection between the trans-Golgi-associated molecular changes in viral glycoproteins and the generation of a mature viral structure. Molecular modifications associated with transport of viral precursors to the trans-Golgi, in particular acquisition of Endo-H resistance in viral glycoproteins, trigger a major structural transformation that generates prefunctional particles, ready to exit the cells and become fully infectious. This is unique compared with the processes operating for most viruses, which usually mature by an irreversible proteolytic processing of polypeptide precursors (10, 12, 50, 57, 58).
FIG. 5.
Proposal for the structural changes taking place during the generation of the three viral forms, and the potential molecular changes involved, associated with modifications of viral glycoproteins in the trans-Golgi subcompartment, according to the results obtained in this study. The shape and final state of sugar chains are indicated for the three viral forms. The asterisk marks the viral form released by Lec2 cells that exhibits a normal shape but an abnormal surface of spikes and low infectivity values.
The observed changes in the bunyaviral envelope suggest that maturation must be driven by lateral interactions of the glycoproteins within the lipid bilayer. Lateral interactions between envelope proteins play a dominant role in viral assembly for some other enveloped viruses, such as coronaviruses and flaviviruses (1, 54). However, envelopes with internal symmetry have not been demonstrated for large viruses, such as 100-nm bunyaviruses. Among geometrical shapes in viruses, icosahedral symmetry is the most frequent since it allows the formation of a core shell with the smallest number of identical units. Icosahedral symmetry has not been demonstrated for bunyaviruses, although in the present work the hexagonal and pentagonal profiles of isolated bunyavirus particles point to a potentially icosahedral symmetry for intracellular type II and extracellular viral particles. Some other viruses, smaller than bunyaviruses, have been reported to contain an icosahedral envelope tightly attached to other icosahedral components, as described for togaviruses and flaviviruses (9, 20, 23).
In the case of bunyaviruses, there are no data supporting the existence of an internal capsid. In fact, when the isolated virions are disrupted, they open in a single spot and release the internal RNPs, while the envelope maintains the hexagonal and pentagonal profiles (Fig. 2). This strongly suggests that the internal RNPs do not participate in the angular shape of the viral envelope.
It is unclear why these viruses use the Golgi complex, since they have to anchor the assembly sites in a structure subjected to constant changes. New insights into Golgi architecture might help us to understand this apparent contradiction. It has been reported that Golgi structure is maintained by a matrix that would nucleate the Golgi assembly (24, 46). We can speculate that macromolecular complexes associated with viral assembly could preferentially associate with the most stable framework of this organelle. On the other hand, over the past few years, considerable evidence has accumulated for a nonvesicular mechanisms of transport through the Golgi (30, 37). This is relevant to understanding how large viral particles (average diameter, ≥100 nm) can be efficiently transported through the stack, since vesicles in the vicinity of the Golgi area have an average diameter of ∼50 nm (31). In fact, a considerable plasticity of the Golgi structure with respect to cargo load can be predicted from three-dimensional maps of Golgi stacks. Direct connections can form transiently between Golgi cisternae that normally remain distinct from one another (25).
Whatever the molecular interactions that allow the maintenance of assembly sites, the formation and maturation of viral particles inside the stack have a favorable consequence for the viruses, since they are kept hidden from the immune system until they complete maturation. Although advantageous for the virus, this would not explain why the assembly locates in the Golgi membranes. We believe that the location of the replication complexes must be determining where the assembly sites are built. Our preliminary data strongly suggest that replication complexes are peculiar tubular structures assembled inside the Golgi stack (44; Novoa et al., in preparation). It would be of considerable help to understand why replication is located in the Golgi, how replication complexes spatially connect with assembly sites, and where maturation occurs.
To this purpose we are working to obtain a three-dimensional map of the structures that participate in replication and assembly inside the Golgi. Electron tomography is becoming the method of choice for this type of study at the cellular level (3, 52). New aspects about the architecture of cells (29) and the structure of complex viruses, such as herpesvirus virus (17) and vaccinia virus (11), have recently been obtained using this technology. Together with high-resolution analysis of purified viral intermediates that participate in assembly, it will help us to define how changes in the sugar composition of viral glycoproteins generate a functional viral structure and how the Golgi apparatus participates in this complex process.
Acknowledgments
We express our gratitude to Silvia Gutierrez Erlandsson for expert assistance with confocal microscopy, to P. Gleeson for kindly providing the BHK Ricr 14 cell line, and to J. R. Castón, A. Fraile-Ramos, X. Shi, and J. L. Carrascosa for critically reading the manuscript and useful suggestions. Our gratitude also to Juan Fontana for many useful discussions.
R.R.N. is the recipient of a fellowship for postgraduate students (FPI Program) from the Ministerio de Ciencia y Tecnología of Spain. This work was supported by grants BMC2000-0555 and BMC2003-01630 from the Ministerio de Ciencia y Tecnología of Spain and 07B/0039/2002 from the Comunidad de Madrid (to C.R.).
REFERENCES
- 1.Allison, S. L., Y. J. Tao, G. O'Riordain, C. W. Mandl, S. C. Harrison, and F. X. Heinz. 2003. Two disinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 77:11357-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altan-Bonnet, N., R. Sougrat, and J. Lippincott-Schwartz. 2004. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16:364-372. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister, W. 2002. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 12:679-684. [DOI] [PubMed] [Google Scholar]
- 4.Bijlmakers, M. J., and M. Marsh. 2003. The on-off story of protein palmitoylation. Trends Cell Biol. 13:32-42. [DOI] [PubMed] [Google Scholar]
- 5.Bonay, P., S. Munro, M. Fresno, and B. Alarcon. 1996. Intra-Golgi transport inhibition by megalomycin. J. Biol. Chem. 271:3719-3726. [DOI] [PubMed] [Google Scholar]
- 6.Bridgen, A., W. Friedemann, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cash, P., L. Hendershot, and D. H. L. Bishop. 1980. The effects of glycosylation inhibitors on the maturation and intracellular polypeptide synthesis induced by snowshoe hare bunyavirus. Virology 103:235-240. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W., and P. Stanley. 2003. Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology 13:43-50. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H. K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway, J. F., R. L. Duda, N., Cheng, R. W. Hendrix, and A. C. Steven. 1995. Proteolytic and conformational control of virus capsid maturation: the bateriophage HK97 system. J. Mol. Biol. 253:86-99. [DOI] [PubMed] [Google Scholar]
- 11.Cyrklaff, M., C. Risco, J. J. Fernández, M. V. Jiménez, M. Esteban, W. Baumeister, and J. L. Carrascosa. 2005. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. USA 102:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Han, C. A. M., K. Staedler, G. -J. Godeke, B. Jan Bosch, and P. J. M. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78:6048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deustcher, S. L., N. Nuwayhid, P. Stanley, E. I. Briles, and C. B. Hirschberg. 1984. Translocation across Golgi vesicle membranes: a CHO glycosilation mutant deficient in CMP-sialic acid transport. Cell 39:295-299. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, R. M. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3:572-577. [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith, C. S., L. H. Elliott, C. J. Peters, and S. R. Zaki. 1995. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch. Virol. 140:2107-2122. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, G., and P. Rottier. 1992. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grünewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 18.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 19.Jantti, J., P. Hilden, H. Ronka, V. Makiranta, S. Keranen, and E. Kuismanen. 1997. Immunocytochemical analysis of Uukuniemi virus budding compartments: role of the intermediate compartment and the Golgi stack in virus maturation. J. Virol. 71:1162-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn, R. J., W. Zhang, M. G. Rossman, S. V. Pletner, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus. Implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lappin, D. F., G. W. Nakitare, J. W. Palfreyman, and R. M. Elliott. 1994. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J. Gen. Virol. 75:3441-3451. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, G. V., B. A. Israe, B. M. Christensen, T. M. Yuill, and K. T. Schultz. 1991. Monoclonal antibodies directed against the envelope glycoproteins of La Crosse virus. Microb. Pathog. 11:411-421. [DOI] [PubMed] [Google Scholar]
- 23.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryoelectron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 24.Marsh, B. J., and K. E. Howell. 2002. The mammalian Golgi -complex debates. Nat. Rev. Mol. Cell. Biol. 3:789-795. [DOI] [PubMed] [Google Scholar]
- 25.Marsh, B. J., N. Volkmann, J. R. McIntosh, and K. E. Howell. 2004. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc. Natl. Acad. Sci. USA 101:5565-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka. Y., S. Y. Chen, and R. W. Compans. 1994. A signal for Golgi retention in the bunyavirus G1 glycoprotein. J. Biol. Chem. 268:22565-22573. [PubMed] [Google Scholar]
- 27.Mckenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meager, A., A. Ungkitchanukit, and R. C. Hughes. 1976. Variants of hamster fibroblasts resistant to Ricinus communis toxin (ricin). Biochem. J. 154:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medalia, O., I. Weber, A. S. Frangakis, D. Nicastro, G. Gerish, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298:1209-1213. [DOI] [PubMed] [Google Scholar]
- 30.Mironov, A. A., P. Weidman, and A. Luini. 1997. Variations on the intracellular transport theme: maturing cisternae and trafficking tubules. J. Cell Biol. 138:481-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogelsvang, S., B. J. Marsh, M. S. Ladinsky, and K. E. Howell. 2004. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic 5:338-345. [DOI] [PubMed] [Google Scholar]
- 32.Nakitare, G. W., and R. M. Elliott. 1993. Expression of the Bunyamwera virus M genome segment and intracellular localization of NSm. Virology 195:511-520. [DOI] [PubMed] [Google Scholar]
- 33.Navarro-Lérida, I., M. Martínez-Moreno, F. Roncal, F. Gavilanes, J. P. Albar, and I. Rodríguez-Crespo. 2004. Proteomic identification of brain proteins that interact with dynein light chain LC8. Proteomics 4:339-346. [DOI] [PubMed] [Google Scholar]
- 34.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cells organelles for viral replication and morphogenesis. Biol. Cell. 97:147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opat, A. S., H. Puthalakath, J. Burke, and P. A. Gleeson. 1998. Genetic defect in N-acetylglucosaminyltransferase I gene of a ricin-resistant baby hamster kidney mutant. Biochem. J. 336:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parodi, A. J. 2000. Protein glycosylaton and its role in protein folding. Annu. Rev. Biochem. 69:69-93. [DOI] [PubMed] [Google Scholar]
- 37.Pelham, H. R., and J. E. Rothman. 2000. The debate about transport in the Golgi -two sides of the same coin? Cell 102:713-739. [DOI] [PubMed] [Google Scholar]
- 38.Prentice, E., W. G. Jerome, T. Yoshimori, M. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risco, C., L. Menéndez-Arias, T. D. Copeland, P. Pinto da Silva, and S. Oroszlan. 1995. Intracellular transport of murine leukemia virus during acute infection of NIH-3T3 cells: nuclear import of nucleocapsid protein and integrase. J. Cell Sci. 108:3039-3050. [DOI] [PubMed] [Google Scholar]
- 40.Risco, C., J. R. Rodríguez, C. López-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Risco, C., J. L. Carrascosa, and T. K. Frey. 2003. Structural maturation of rubella virus in the Golgi complex. Virology 312:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronnholm, R. 1992. Localization to the Golgi complex of Uukuniemi virus glycoproteins G1 and G2 expressed from cloned cDNAs. J. Virol. 66:4525-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 73:7952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salanueva, I. J., R. R. Novoa, P. Cabezas, C. López-Iglesias, J. L. Carrascosa, R. M. Elliott, and C. Risco. 2003. Polymorphism and structural maturation of Bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 77:1368-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmaljohn, S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In D. M. Knipe and P. M. Holey (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 46.Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407:1022-1026. [DOI] [PubMed] [Google Scholar]
- 47.Shi, X., and R. M. Elliott. 2004. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J. Virol. 78:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi, X., D. F. Lappin, and R. M. Elliott. 2004. Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J. Virol. 78:10793-10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiro, R. G. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43-56. [DOI] [PubMed] [Google Scholar]
- 50.Staedler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley, P. 1989. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol. Cell. Biol. 9:377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramaniam, S., and J. L. S. Milne. 2004. Three-dimensional electron microscopy at molecular resolution. Annu. Rev. Biophys. Biomol. Struct. 33:141-155. [DOI] [PubMed] [Google Scholar]
- 53.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vennema, H., G.-J. Godeke, W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope proteins. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watret, G. E., G. R. Pringle, and R. M. Elliott. 1985. Synthesis of bunyavirus-specific proteins in a continuous cell line (XTC-2) derived from Xenopus laevis. J. Gen. Virol. 66:473-482. [DOI] [PubMed] [Google Scholar]
- 56.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA polymerase in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 57.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: Implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., J. Corver, P. R. Chapman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]