Abstract
High-risk human papillomaviruses encode two oncogenes, E6 and E7, expressed in nearly all cervical cancers. Although E7 protein is best known for its ability to inactivate the retinoblastoma tumor suppressor protein, pRb, many other activities for E7 have been proposed in in vitro studies. Herein, we describe studies that allowed us to define unambiguously the pRb-dependent and -independent activities of E7 for the first time in vivo. In these studies, we crossed mice transgenic for human papillomavirus 16 E7 to knock-in mice genetically engineered to express a mutant form of pRb (pRbΔLXCXE) that is selectively defective for binding E7. pRb inactivation was necessary for E7 to induce DNA synthesis and to overcome differentiation-dependent cell cycle withdrawal and DNA damage-induced cell cycle arrest. While most of E7's effects on epidermal differentiation were found to require pRb inactivation, a modest delay in terminal differentiation with resulting hyperplasia was observed in E7 mice on the RbΔLXCXE mutant background. E7-induced p21 upregulation was also pRb dependent, and genetic Rb inactivation was sufficient to reproduce this effect. While E7-mediated p21 induction was partially p53 dependent, neither p53 nor p21 induction by E7 required p19ARF. These data show that E7 upregulates the expression of p53 and p21 via pRb-dependent mechanisms distinct from the proposed p19-Mdm2 pathway. These results extend our appreciation of the importance of pRb as a relevant target for high-risk E7 oncoproteins.
Human papillomaviruses (HPVs) are small DNA viruses and the causative agents of epithelial warts. The so-called “high-risk” HPVs, including HPV-16, infect the anogenital tract epithelium and are associated with almost all cases of cervical cancer, a leading cause of cancer mortality in women worldwide (64, 69). Although the HPV genome usually exists extrachromosomally, many HPV-associated cancers contain HPV genomes integrated into the host DNA (20). These integrated genomes invariably contain intact viral E6 and E7 genes, and integration causes their increased expression (35). These data suggest that E6 and E7 contribute to the development of cervical cancers.
HPV-16 E7 is a small nuclear phosphoprotein with potent transforming and tumorigenic properties. Coexpression of E6 and E7 is necessary and sufficient to transform primary human keratinocytes (48), and E7 acts robustly in a number of other in vitro transformation assays (6, 44, 52, 62, 63, 65). E7-expressing cells exhibit genomic instability in culture (17, 18, 56, 66, 67) and have impaired cell cycle arrest responses to DNA damage (12, 60). Additionally, expression of E7 in primary human keratinocytes or in transgenic mice results in abnormal centrosome synthesis, with associated multipolar mitoses and aneuploidy (3, 15-17). Previously, our laboratory generated mice transgenic for HPV-16 E7 under the control of the keratin 14 promoter, targeting E7 expression to the basal layer of stratified squamous epithelia such as the skin and cervical epithelium (30). These mice express E7 at levels similar to the levels seen in human cervical carcinoma cell lines and have a broad spectrum of phenotypes, including epithelial hyperplasia, increased cell cycle progression in all epithelial cell layers, disrupted epithelial differentiation, loss of DNA damage-induced cell cycle arrest, centrosome abnormalities, spontaneous skin tumors, and cervical cancers in estrogen-treated mice (3, 24, 30, 57, 61).
E7 has been reported to bind to over 20 cellular proteins (2, 8, 11, 47, 55). These putative targets of E7 include multiple cell cycle regulators, such as the pocket proteins pRb, p107, and p130 and the cyclin-dependent kinase inhibitors p21 and p27 (47). Additional binding partners of E7 include transcription factors (c-Jun, IRF-1, and MPP2), transcriptional cofactors and chromatin-remodeling enzymes (TBP, TAF-110, Skip, p300, pCAF, and Mi2β/histone deacetylase complexes), metabolic enzymes (M2 pyruvate kinase and acid α-glucosidase), F-actin, and the proteasome (2, 8, 47, 55). Thus, in vitro studies have suggested many possible mechanisms for E7 function.
Among the known E7 binding proteins, the retinoblastoma tumor suppressor protein, pRb, is perhaps the best characterized as a target of E7. Interaction between E7 and pRb disrupts the ability of pRb to bind cellular E2F transcription factors, and this inhibits pRb-mediated repression of E2F-responsive genes (10, 51), and results in proteasomal degradation of pRb in cultured cells (9, 26, 38). pRb has been connected to all of the processes disrupted by E7 in vivo, including cell cycle regulation, differentiation, DNA damage responses, centrosome synthesis, and tumorigenesis (3, 33, 46, 60). Furthermore, recombinational inactivation of the Rb gene in vivo recapitulates nearly all known effects of E7 on murine epidermis (3).
Despite the apparent importance of the E7-pRb interaction, several studies have indicated that E7's effects on targets other than pRb may contribute to its phenotypes. Two E7 mutants deficient in binding to pRb can cooperate with E6 in the immortalization of primary human keratinocytes (36), and E7's ability to transactivate certain E2F-responsive promoters may depend on binding to p107 rather than binding to pRb, as shown with the B-myb promoter (41). Furthermore, the E7Δ79-83 mutant, which binds and degrades pRb but is deficient in binding to TBP, acid α-glucosidase, and M2-PK, exhibits a decreased ability to transform baby rat kidney cells (29, 43, 70, 71). Another E7 mutant, E7CVQ68-70AAA, which is also able to bind and destabilize pRb but is deficient in p21 inactivation, fails to overcome differentiation-dependent cell cycle withdrawal or DNA damage-induced cell cycle arrest in human keratinocytes (28, 29). Additionally, E7 may contribute to transformation of rat embryo fibroblasts via activation of c-Jun independently of pRb inactivation (1), and E7 induces centrosome abnormalities in pRb-deficient cells (19). Finally, E7 expression in pRb-deficient epidermis in vivo results in phenotypes not observed with pRb loss alone (3). Thus, multiple studies indicate E7's effects on targets other than pRb make important contributions to E7 function.
In the current study, we sought to disrupt the E7-pRb interaction selectively. This separates E7's phenotypes into two groups: phenotypes that require pRb inactivation by E7 (pRb-dependent phenotypes), and phenotypes that do not require pRb inactivation by E7 (pRb-independent phenotypes). Interactions between E7 and pRb, p107, and p130 are dependent on an LxCxE sequence in E7, which binds to the conserved pocket domain for which the pocket proteins are named (42). Thus, mutations of E7's LxCxE sequence reduces or abolishes binding to pRb (4, 53). The use of such mutants to study the E7-pRb interaction is problematic, however, as mutations of this motif can also interfere with E7's ability to target p107, p130, p21, IRF-1, p300, TBP, acid α-glucosidase, and perhaps other targets and/or renders E7 unstable in cells (8, 21, 37, 41, 50, 53, 54, 70). Thus, more refined studies are necessary to separate the pRb-dependent and-independent consequences of E7 expression clearly.
In order to distinguish pRb-dependent and pRb-independent functions of E7 unambiguously, we used a knock-in mouse strain genetically engineered to carry a mutant Rb allele, RbΔLXCXE, which is mutated in the E7-binding site. Using a crystal structure of the human pRb pocket domain bound to the LxCxE peptide of HPV-16 E7, Dick et al. constructed the RbΔLXCXE allele, which produces a protein that does not bind E7 and yet maintains much of its normal cellular function (14). This allele contains three alanine mutations in the region of the pocket domain that was seen to interact closely with the LxCxE peptide in the crystal structure, and so fails to bind detectably to E7. In contrast, pRbΔLXCXE maintains much of the normal cellular function of wild-type pRb. pRbΔLXCXE binds E2Fs, induces G1 arrest in pRb-negative SAOS2 cells, and is phosphorylated and inactivated by cyclinD/cdk4 complexes similarly to wild-type pRb (14). pRbΔLXCXE also represses gene expression from E2F-responsive promoter constructs, though incrementally less effectively than wild-type pRb. Unlike wild-type pRb, however, pRbΔLXCXE-induced G1 arrest cannot be reversed by expression of HPV-16 or -18 E7 (14). Though pRbΔLXCXE is mutated in the highly conserved “pocket” domain, we obtained mice heterozygous or homozygous for the mutant allele with few overt phenotypes. Thus, expression of wild-type HPV-16 E7 in the stratified squamous epithelia of RbΔLXCXE mice allowed us to disrupt the E7-pRb interaction selectively without using problematic E7 mutants.
The presence of the RbΔLXCXE allele in young K14E7 mice prevented E7 from having any acute effect on the epidermis except for a minor, proliferation-independent thickening of the epidermis. Furthermore, we demonstrate that E7's ability to induce accumulation of the p21 tumor suppressor, previously hypothesized to result from p19ARF-induced p53 accumulation (5, 59), is pRb-dependent but p19 independent and occurs through both p53-dependent and -independent pathways. These data demonstrate that inactivation of pRb is required for nearly all acute in vivo effects of E7 and that pRb-independent mechanisms of E7 action have little acute in vivo effects when pRb function is maintained.
MATERIALS AND METHODS
Transgenic and knock-in mice.
All mice in this study have been previously described, including K14E7 (30), RbΔLXCXE (32), K14CreRbf/f (3), αAcryE7 mice (49), p19-null mice (40), and p53-null mice (34). Analysis of RbΔLXCXE mice was performed on a mixed 129/FVB/B6 background. Other experiments were performed on a mixed 129-FVB-C57/BL6 or FVB backgrounds, with all genotypes bred to contain the same levels of genetic heterogeneity within each experiment. Analyses were performed on mice between 8 and 28 days of age; each phenotype was analyzed at a time point for which the phenotype was clearly detected in K14E7RbWT/WT mice, or the time point at which the largest numbers of mice were generated. All mice were bred and maintained in the American Association for Accreditation of Laboratory Animal Care-approved McArdle Laboratory Cancer Center Animal Care Facility.
All mice were genotyped by PCR for E7, Rb, and p21 using the following primers: for RbΔLXCXE, oligonucleotides FD134 (5′-AGCTTCATACAGATAGTTGGG-3′) and FD135 (5′-CACACAAATCCCCATACCTATG-3′); for p19, oligonucleotides ARF1 (5′-AGTACAGCAGCGGGAGCATGG-3′), ARF2 (5′-TTGAGGAGGACCGTGAAGCCG-3′), and Neo2 (5′-ACCACACTGCTCGACATTGGG-3′); for p53, oligonucleotides p53-1 (5′-TATACTCAGAGCCGGCCT-3′), p53-2 (5′-ACAGCGTGGTGGTACCTTAT-3′), and p53-3 (5′-TCCTCGTGCTTTACGGTATC-3′), and for K14E7, K14Cre, and Rbflox as published previously (3). One hour prior to sacrifice, all mice were intraperitoneally injected with bromodeoxyuridine (BrdU, 10 μl per g body weight of 12.5 mg/ml solution). For the irradiation study, mice were exposed to 0 or 5 Gy ionizing radiation from a 137Cs source 24 h prior to BrdU administration.
Immunohistochemical and immunofluorescence analysis of epidermis.
Skin and ear epidermal samples were fixed in 10% phosphate-buffered formalin, embedded in paraffin, and cut into 5-μm sections. Serial sections were used for immunohistochemical staining for BrdU, p53, and p21, and immunofluorescent staining for keratin 14, keratin 10, and filaggrin.
For immunohistochemical stains, sections were deparaffinized in xylene and rehydrated through a graded series of ethanol/water solutions. Endogenous peroxidase activity was quenched by treatment in 3% H2O2 in methanol for 10 to 20 min. Slides were washed in phosphate-buffered saline and heated in boiling 10 mM sodium citrate pH 6.0 for 20 min. For BrdU staining, further unmasking was achieved with 20 min of immersion in 2 N HCl. Samples were blocked for 30 min at ambient temperature in 5% horse serum in phosphate-buffered saline (for BrdU) or 5% horse serum/5% milk/phosphate-buffered saline (for p21 and p53). Primary antibody was diluted in blocking buffer and applied: 1:40 anti-BrdU (Oncogene catalog number NA20-100UG) for 2.5 h at room temperature, 1:25 anti-p21 (Pharmingen catalog number 556430) overnight at 4 °C, or 1:500 anti-p53 (Novocastra CM5) 3 h at room temperature. After washes in phosphate-buffered saline, biotinylated secondary antibody and streptavidin-peroxidase conjugate were applied according to the Vectastain ABC kit instructions (Vector Labs catalog number PK-6200). Staining was developed in 3,3′-diaminobenzidine (DAB) solution (Vector Labs catalog number SK-4100) for 1 to 4 min, then quenched in H2O. Slides were counterstained with hematoxylin, dehydrated through a series of ethanols and xylenes, and coverslipped.
For immunofluorescent stains, sections were deparaffinized and rehydrated as above. Slides were blocked with 10% horse serum/3% bovine serum albumin for 30 min; 1:1,000 anti-mouse keratin 14 (Covance catalog number PRB-155P) or 1:100 anti-mouse filaggrin (Covance catalog number PRB-145P) in blocking solution was applied overnight at 4°C. Slides were then washed in phosphate-buffered saline and 1:250 biotinylated universal secondary antibody (Vector Labs catalog number PK-6200) was applied for 30 min at room temperature. After phosphate-buffered saline washes, 1:150 streptavidin-Texas Red (Vector Labs catalog number SA-5006) was applied for 30 min at room temperature, followed by additional washes. Slides were then incubated overnight with 1:200 fluorescein isothiocyanate-conjugated anti-keratin 10 (Covance catalog number FITC-159L) in blocking buffer. Slides were rinsed in phosphate-buffered saline, mounted in Vectashield mounting medium with DAPI (Vector Labs catalog number H-1200), and visualized using a Zeiss Axiophot fluorescent microscope.
Quantitation of cell proliferation and epidermal thickness.
BrdU incorporation into newly synthesized DNA was used as a measure of keratinocyte proliferation by counting BrdU-stained skin and ear sections. All keratinocyte nuclei in 10 visual fields were scored as either positive (brown) or negative (blue) for BrdU incorporation in both basal and suprabasal layers of the epidermis. For counting purposes, even slightly brown cells were counted as positive. Three to six mice were counted per genotype, and results are presented as the mean ± standard deviation. Epidermal hyperplasia was calculated as the ratio of the number of suprabasal cells to basal cells in each visual field and averaged over 8 to 10 visual fields per mouse. Statistical analyses of results were performed using the two-sided Wilcoxon rank sum test.
RESULTS
To determine which in vivo phenotypes of E7 require the E7-pRb interaction, we used knock-in mice carrying the RbΔLXCXE allele in place of the wild-type Rb allele (14, 32). RbΔLXCXE/WT mice are overtly normal and show no apparent phenotypes. In contrast to the lethal phenotypes seen in germ line Rb knockout mice (33), RbΔLXCXE/ΔLXCXE mice are also viable, fertile, and overtly normal on the mixed genetic background used in these studies.
Overt E7-induced phenotypes require inactivation of pRb.
To assess the effects of E7 expression when pRb function cannot be overridden by E7, K14E7 transgenic mice were crossed to mice carrying the RbΔLXCXE allele. The K14E7 transgene on an RbWT/WT background confers overt phenotypes, including stunted growth and a wrinkling of the skin caused by epithelial hyperplasia (30). These phenotypes were clearly evident in the K14E7RbWT/WT mice generated in this cross, yet K14E7RbΔLXCXE/WT and K14E7RbΔLXCXE/ΔLXCXE mice both had no overt phenotypes and could not be visually distinguished from mice lacking the K14E7 transgene.
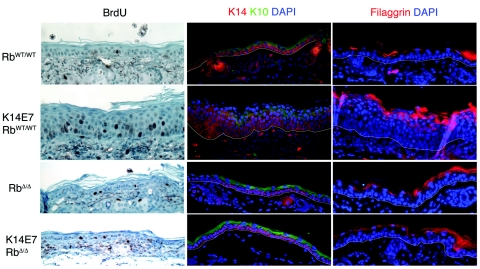
To evaluate further the effects of E7 expression in epidermis from mice carrying RbΔLXCXE and wild-type Rb, we collected ear and dorsal skin from 21-day-old mice and formalin fixed, embedded, and sectioned these epithelia. Histological analysis of the tissues from these mice revealed pronounced epithelial hyperplasia in the ear, but not the dorsal skin, of K14E7RbWT/WT mice (Fig. 1), as seen previously (3, 30). Due to the increased severity of E7 phenotypes in the ear epidermis over that seen in dorsal skin, we focused our study on E7's effects on ear epidermis. In contrast to the hyperplasia seen in K14E7RbWT/WT ear epidermis, a thin epithelium typical of normal mice was present in RbWT/WT, RbΔLXCXE/WT, and RbΔLXCXE/ΔLXCXE mice, indicating that the RbΔLXCXE allele has no effect on epithelial thickness in either the heterozygous or homozygous state (Fig. 1).
FIG. 1.
Proliferation and differentiation in RbWT/WT and RbΔ/Δ epidermis. Shown are immunohistochemistry and immunofluorescence images of RbWT/WT (top row), K14E7RbWT/WT (second row), RbΔ/Δ (third row), and K14E7RbΔ/Δ (bottom row) ear epidermis from 21-day-old mice. Left column: BrdU immunohistochemistry is brown with hematoxylin counterstain. Middle column: immunofluorescence stain for keratin 14 (K14, red) and keratin 10 (K10, green) with DAPI nuclear counterstain. Right column: immunofluorescence stain for filaggrin (red) with DAPI nuclear counterstain.
Inactivation of pRb is necessary for E7-induced DNA synthesis.
In order to search for any subtle pRb-independent effect of expression of E7 on epithelial cell proliferation, RbΔLXCXE mice with and without E7 were injected with the nucleotide analog BrdU 1 h prior to sacrifice and ear and dorsal skin sections were stained immunohistochemically for BrdU to stain all cells that synthesized DNA in the hour prior to sacrifice. As observed previously (30), control RbWT/WT ear epidermis incorporated BrdU at a low rate selectively in the basal layer of cells, whereas K14E7RbWT/WT epidermis exhibited dramatic increases in BrdU incorporation in both the basal and suprabasal layers of the epidermis (Fig. 1). Interestingly, no effect of E7 was seen on BrdU incorporation in RbΔLXCXE/WT or RbΔLXCXE/ΔLXCXE epidermis (Fig. 2, data not shown). This finding indicates that inactivation of pRb is necessary for E7-induced DNA synthesis in both the basal and suprabasal epidermal cell layers.
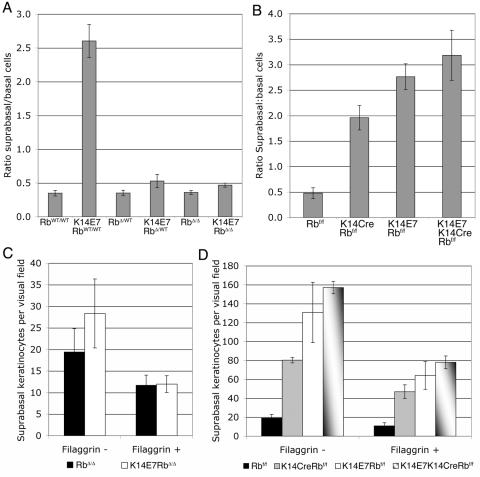
FIG. 2.
pRb inactivation is required for E7-induced cell cycle progression. Basal and suprabasal keratinocytes in ear epidermal sections from 21-day-old mice were quantified as BrdU positive or BrdU negative for four to six mice per genotype.
E7 subtly delays differentiation and induces hyperplasia in RbΔLXCXE epidermis.
Although E7 did not induce any proliferation in RbΔLXCXE epidermis, quantification of the number of basal and suprabasal cells per visual field revealed a 30 to 50% increase in epidermal thickness in K14E7RbΔLXCXE/WT and K14E7RbΔLXCXE/ΔLXCXE mice compared to that seen in mice lacking E7 (Fig. 3a, P < 0.01). Importantly, a similar pRb-independent hyperplastic effect of E7 is also evident in another mouse model system. Comparisons of ear epidermal thickness in 21-, 28-, and 56-day old mice either expressing E7 (K14E7 mice) or depleted of pRb by recombinational inactivation of the Rb gene (K14CreRbf/f mice) (3) show that inactivation of pRb results in significantly less hyperplasia that that induced by expression of E7 (Fig. 3B and data not shown, P < 0.005). Furthermore, expression of E7 in pRb-depleted epidermis (K14E7K14CreRbf/f mice) results in greater hyperplasia than that seen with inactivation of pRb alone (Fig. 3B, P < 0.003). In both model systems, this pRb-independent hyperplasia does not result from E7-induced proliferation (Fig. 2) (3); we therefore hypothesized that E7 may induce epidermal thickening by disrupting the process of keratinocyte differentiation. Thus, we examined the ability of E7 to disrupt epithelial differentiation in both wild-type Rb and RbΔLXCXE epidermis.
FIG. 3.
E7 induces a pRb-independent delay in keratinocyte terminal differentiation. Epidermal hyperplasia was measured as the ratio of suprabasal to basal cells in ear epidermis. (A) E7 induces a major increase in epithelial thickness in RbWT/WT mice, and a minor thickening of both RbΔ/Δ and RbΔ/WT epidermis (P = 0.025 for RbΔ/Δ versus E7RbΔ/Δ and P = 0.004 for RbΔ/WT versus E7RbΔ/WT). (B) E7 expression induces more severe hyperplasia than that resulting from pRb loss alone. P < 0.005 for K14CreRbf/f vs K14E7Rbf/f. Data are shown from 28-day-old mice; similar results were seen at 21 and 56 days. (C and D) pRb-independent hyperplasia in K14E7 mice results primarily from expansion of the filaggrin-negative suprabasal cell layer. (C) P = 0.028 for RbΔ/Δ versus E7RbΔ/Δ filaggrin-negative layer. A significant expansion of the filaggrin-negative layer was also seen in RbΔ/WT versus E7RbΔ/WT (not shown, P = 0.034). (D) P < 0.04 for K14E7Rbf/f versus K14CreRbf/f in both filaggrin-negative and filaggrin-positive cell layers.
RbWT/WT, RbΔLXCXE/WT, and RbΔLXCXE/ΔLXCXE ear epidermis samples all show normal expression of the differentiation markers keratin 14, keratin 10, and filaggrin (Fig. 1). In these mice, keratin 14 is confined to the poorly differentiated, proliferating basal layer of cells with only a few occasional cells above the basal layer staining positive for keratin 14. Staining for keratin 10 exhibits a complementary pattern, where essentially all suprabasal cells are strongly positive for keratin 10. Staining for filaggrin is absent in the basal and lower suprabasal “spinous” cells, but is strongly positive in the more differentiated upper suprabasal “granular” cells (Fig. 1).
As demonstrated previously (27, 30), E7 expression in RbWT/WT epidermis disrupts this pattern of differentiation. This disruption is evidenced by an expansion of the basal-like, keratin 14-positive cell compartment to three to five cell layers, altered expression of keratin 10 resulting in several keratin 10-negative cell layers, and expansion of the filaggrin-positive cell layer (Fig. 1). In contrast, expression of E7 in epidermis carrying the RbΔLXCXE allele did not induce any expansion of the keratin 14-positive cell compartment or alter the expression of keratin 10 (Fig. 1). This result demonstrates that E7's ability to disrupt the onset of keratinocyte differentiation requires inactivation of pRb. It also indicates that the observed pRb-independent hyperplasia is not due to an expansion of the keratin 14-positive basal-like cell layer.
Expression of filaggrin was also overtly normal in K14E7RbΔLXCXE mice. However, quantification of suprabasal keratinocytes as either filaggrin-negative spinous cells or filaggrin-positive granular cells revealed a 30 to 50% increase in the thickness of the spinous suprabasal cell layer in both K14E7RbΔLXCXE/WT and K14E7RbΔLXCXE/ΔLXCXE mice compared to RbΔLXCXE/WT and RbΔLXCXE/ΔLXCXE mice, respectively (Fig. 3C and data not shown, P < 0.03). E7 did not induce any change in the number of filaggrin-positive granular cells in RbΔLXCXE/WT or RbΔLXCXE/ΔLXCXE mice (Fig. 3C and data not shown). These data indicates that E7 has a pRb-independent activity capable of inducing epidermal hyperplasia by expanding the thickness of the spinous suprabasal cell layer. This finding was confirmed by quantification of filaggrin-positive cells in K14E7Rbf/f, K14CreRbf/f and K14E7K14CreRbf/f epidermis (Fig. 3D). Here E7 induced a prominent expansion of the filaggrin-negative suprabasal cell layer beyond that seen with inactivation of Rb alone (P = 0.034). In this model, we also saw a significantly thicker filaggrin-positive granular epidermal layer in K14E7Rbf/f and K14E7K14CreRbf/f mice than in K14CreRbf/f mice, suggesting E7's pRb-independent activities can delay multiple stages of epidermal differentiation (Fig. 3D, P = 0.034).
Inactivation of pRb is necessary for E7 to disrupt DNA damage-induced cell cycle arrest.
One mechanism by which E7 may contribute to tumorigenesis is by blocking the ability of DNA-damaged cells to undergo G1 arrest. To determine if E7 can disrupt DNA damage-induced cell cycle arrest by pRb-independent mechanisms, RbΔLXCXE/ΔLXCXE and RbWT/WT mice with or without E7 were irradiated with 5 Gy ionizing radiation at 21 days of age, injected with BrdU 24 h later, and sacrificed 1 hour after BrdU administration. Ear epidermal sections were stained for BrdU and the incorporation of BrdU with and without irradiation was quantified (Fig. 4).
FIG. 4.

E7 overcomes DNA damage-induced cell cycle arrest in a pRb-dependent manner. We exposed 21-day-old mice to 0 or 5 Gy ionizing radiation, BrdU was injected 24 h later, and mice were sacrificed 1 hour later. BrdU incorporation in ear epidermis was quantified.
In K14E7RbWT/WT mice, E7 had the expected effect of maintaining DNA synthesis in irradiated cells, whereas control RbWT/WT epidermis exhibited little to no incorporation of BrdU. In contrast, both RbΔLXCXE/ΔLXCXE and K14E7RbΔLXCXE/ΔLXCXE epidermis exhibited nearly complete loss of BrdU-positive nuclei after irradiation (Fig. 4). This demonstrates both that pRbΔLxCxE is capable of mediating proper cell-cycle arrest in response to DNA damage in vivo and that the pRb-independent functions of E7 are not sufficient to disrupt DNA damage-induced cell cycle arrest.
E7 induces p21 by multiple pRb-dependent mechanisms.
One nonpocket protein target of E7 that has attracted much attention is the cyclin-dependent kinase inhibitor p21, both because p21 is an important tumor suppressor and because of the paradoxical observations that E7 both upregulates the expression of p21 and inhibits the function of p21 (25, 37). It has been proposed that E7 upregulates expression of p21 by increasing E2F-dependent expression of p19ARF. p19ARF could then inhibit Mdm2-mediated degradation of p53, resulting in accumulation of p53 and transactivation of the p21 promoter (5). Indeed, multiple studies have observed accumulation of p53 in E7-expressing cells (13, 38, 58). However, in vitro studies have shown that both E7 and E2F1 can increase levels of p53 in a p19-independent manner (58, 68). Furthermore, regardless of how p53 is upregulated, it is unclear whether the accumulated p53 is transcriptionally active (22, 58). Finally, accumulation of p21 in E7-expressing fibroblasts occurs with only small increases in p21 mRNA (39, 58), but the stability of p21 protein is greatly increased (39). These observations suggest that E7 upregulates p21 by a novel p19- and perhaps p53-independent mechanism, but have not been confirmed in the relevant cell types in vivo. In order to clarify the mechanism by which E7 upregulates p21, we tested the pRb, p53, and p19 dependence of p21 upregulation in vivo by immunohistochemistry.
As reported previously, little p21 was detectable in RbWT/WT epidermis, whereas a striking upregulation of p21 was evident in the suprabasal layers of K14E7RbWT/WT ear epidermis (Fig. 5A). In contrast, expression of E7 had no effect on staining for p21 in RbΔLXCXE/ΔLXCXE epidermis; both RbΔLXCXE/ΔLXCXE and K14E7RbΔLXCXE/ΔLXCXE epidermis showed little to no staining for p21 (Fig. 5A). These data indicate that induction of p21 by E7 is pRb dependent. This finding was confirmed in immunohistochemical analyses of K14CreRbf/f and K14E7Rbf/f mice (Fig. 5B). While control Rbf/f epidermis exhibits little to no staining for p21, K14CreRbf/f and K14E7Rbf/f epidermis both show similar up-regulation of p21 in the suprabasal epidermal layers. Thus, inactivation of pRb by E7 is necessary and deficiency in pRb function is sufficient for induction of p21.
FIG. 5.
E7 induces p21 in a pRb-dependent manner. Ear epidermal sections from 21-day-old mice were stained for p21 (brown) and counterstained with hematoxylin. (A) E7 induces p21 in RbWT/WT but not RbΔ/Δ epidermis. (B) Inactivation of the Rb gene and expression of E7 similarly induce p21 in epidermis.
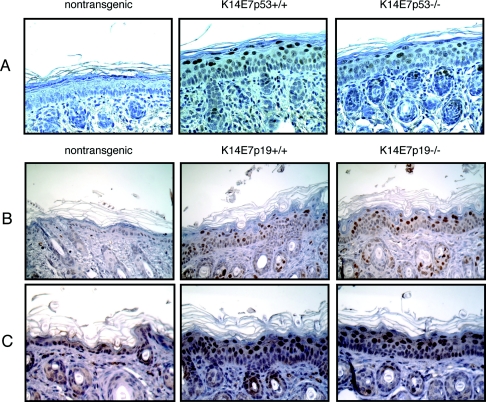
As mentioned above, it has been hypothesized that E7 induces p21 by upregulating expression of p19ARF and inhibiting destabilization of p53 by Mdm2. To test this hypothesis, we monitored E7's capacity to induce p21 in p53-null mice. E7 retained the ability to induce expression of p21 in p53−/− ear epidermis to a level clearly higher than that seen in mice lacking E7 (Fig. 6A, left and right panels). However, this p53-independent induction of p21 was at a reduced level compared to the p21 levels seen in K14E7p53+/+ epidermis (Fig. 6A, right and middle panels). p21 positive staining in suprabasal cells was quantified in the two mice per genotype available in archival samples: two samples of K14E7p53+/+ epidermis exhibited 86% and 85% positive staining, while two samples of K14E7p53−/− epidermis exhibited 58% and 63% positive staining. Thus, p21 induction by E7 occurs through both p53-dependent and-independent pathways. The relative importance of these two pathways may vary depending upon the epidermal site examined; E7's ability to induce p21 was not visibly diminished in p53−/− dorsal skin epidermis (data not shown).
FIG. 6.
p19- and p53-independent induction of p21. Ear epidermal sections from 8-day-old mice of the indicated genotypes were stained for p21 (A and B) or p53 (C) in mice of the indicated genotypes. Brown precipitate indicates positive staining, with blue hematoxylin counterstain.
To determine if the p53-dependent induction of p21 resulted from the proposed p19-mediated inhibition of Mdm2, E7's effect on p19-null epidermis was examined. Interestingly, robust induction of p21 was seen in K14E7p19−/− mice (Fig. 6B). Furthermore, p19 was not necessary for E7-mediated upregulation of p53 protein (Fig. 6C). Thus, E7 induces accumulation of p53 by a mechanism independent of p19 function.
DISCUSSION
We have found that pRb inactivation is necessary for nearly all of the phenotypes resulting from expression of HPV-16 E7 in young mice. The use of the RbΔLXCXE mutant mouse allowed the E7-pRb interaction to be specifically disrupted in vivo without compromising E7's effects on other targets or E7 protein stability.
pRb-independent activities of E7 are insufficient to induce DNA synthesis or prevent cell cycle arrest.
Two consequences of the expression of E7 that are likely to be important both in the life cycle of HPVs and in cervical carcinogenesis are accelerated cell cycle progression and resistance to cell cycle arrest. Previous analyses have suggested that non-pRb targets of E7 may be important for these functions, as some E2F-responsive promoters are activated specifically by inactivation of p107 (41), and because an E7 mutant which binds and degrades pRb, p107, and p130, but is impaired in inhibition of p21 (E7CVQ68-70AAA) and is unable to overcome differentiation- or DNA damage-induced cell cycle arrest (28, 29). While these studies indicated that non-pRb targets of E7 are necessary for these phenotypes, no previous analysis had determined if they are sufficient.
In this study, E7's effects on cell cycle progression were studied in the context of the RbΔLxCxE/ΔLxCxE epidermis. Importantly, RbΔLxCxE/ΔLxCxE epidermis halted proliferation normally in response to differentiation or irradiation. Interestingly, we observed a small increase in proliferation in the basal layer of RbΔLxCxE/WT compared to RbWT/WT epidermis (P < 0.04, data not shown). While this indicates some defect in cell cycle control by the RbΔLxCxE allele exists, the effect was subtle enough that it did not interfere with our analyses of E7 function.
In contrast to the effects of E7 on RbWT/WT epidermis, E7 expression in RbΔLxCxE/ΔLxCxE epidermis failed to induce increased or aberrant cell cycle progression. In RbΔLXCXE epidermis, E7 failed to increase cell cycle progression of the normally cycling, basal layer of keratinocytes (Fig. 1 and 2), failed to overcome differentiation-induced cell cycle arrest in suprabasal keratinocytes (Fig. 1 and 2), and failed to overcome DNA-damage induced cell cycle arrest in epidermis exposed to ionizing radiation (Fig. 4). By contrast, E7 potently induces cell growth under all these conditions in wild-type Rb epidermis. Thus, the combined effects of all pRb-independent activities of E7 are not sufficient to induce any detectable increase in cell cycle progression or to overcome cell cycle arrest in vivo. As noted above, however, in vitro data also suggest that binding and degradation of pRb are not sufficient to reproduce E7's full effect on these phenotypes. Combined, these data indicate that E7 has evolved to target multiple regulators of cell cycle progression and arrest because E7 cannot efficiently disrupt normal cell cycle checkpoint controls via its effects on any one of these targets alone.
pRb-independent effect on epidermal differentiation and hyperplasia.
In spite of E7's inability to upregulate DNA synthesis in RbΔLXCXE epidermis, a 30 to 50% increase in the number of suprabasal keratinocytes per visual field was observed in both K14E7RbΔLXCXE/WT and K14E7RbΔLXCXE/ΔLXCXE mice compared to RbΔLXCXE/WT and RbΔLXCXE/ΔLXCXE controls (Fig. 3A). This pRb-independent, DNA synthesis-independent hyperplastic activity of E7 was also observed when comparing the effects of E7 expression to the effects of pRb depletion in murine epidermis at 21, 28, or 56 days of age (Fig. 3B and data not shown). Examination of the differentiation markers keratin 14, keratin 10, and filaggrin in both the RbΔLXCXE and Rbflox mouse models indicated that E7 possesses a pRb-independent, proliferation-independent ability to expand the lower suprabasal (spinous) epidermal layer (Fig. 3C and D), and possibly also the filaggrin-positive granular layer (Fig. 3D). This expansion is the only in vivo activity of E7 identified thus far that does not require inactivation of pRb. The molecular mechanism of this effect and its significance for the HPV life cycle and HPV-associated carcinogenesis are currently unclear and under investigation.
p21 induction by E7.
It has long been known that expression of E7 induces the accumulation of two important tumor suppressors, p53 and p21. While the induction of p53 by E7 may be compensated for by degradation of p53 by HPV E6, the upregulation of p21 poses an interesting paradox in that E7 both induces the expression of and inhibits the function of p21 (25, 37). Thus, it is important to understand the mechanism by which E7 induces p21. p53 regulates p21 expression at the transcriptional level, so an obvious possibility was that p21 accumulates as a direct consequence of E7-induced upregulation of p53. Accumulation of p53, in turn, has been hypothesized to result from E2F-dependent expression of p19ARF, which inhibits the p53-degrading protein Mdm2 (5). In vitro studies have challenged this model, though, by demonstrating both p19- and p53-independent induction of p21 by E7 in fibroblasts (39, 58).
Our analysis of p21 expression in vivo in K14E7RbΔLXCXE and K14CreRbf/f epidermis confirmed that inactivation of pRb is necessary and sufficient for induction of p21 (Fig. 5). Furthermore, we found that induction of p21 was partially p53 dependent, although also partially p53 independent in ear epidermis (Fig. 6A). This observation implies that E7 can upregulate p21 through at least two pathways downstream of inactivation of pRb. This is consistent with previous reports which have shown a moderate increase in p21 mRNA upon E7 expression (58) as well as posttranscriptional stabilization of p21 protein by E7 (39). Moreover, in the latter study Jones et al. observed that E7 increases p21 protein levels in both p53-functional and p53-inhibited cells. However, both the basal and E7-induced levels of p21 are lower in p53-inhibited cells than in p53-functional cells (39). These data mirror our observation that E7 induces p21, but to a diminished extent, in p53-null epidermis. Importantly, the relative importance of p53-dependent versus p53-independent induction of p21 appears to vary depending on the epidermal site examined, as induction of p21 in dorsal skin epidermis was unaffected by p53 status (data not shown).
Given that the inactivation of pRb correlates with E7's induction of p21 (Fig. 5), an obvious downstream target that might mediate E7's induction of p21 are the E2F family of transcription factors, which are normally regulated by pRb and its related proteins p107 and p130. We therefore compared the level of p21 induction in E2F1-sufficient versus E2F1-null mice expressing E7 in the mouse lens from the αAcry promoter. In the lens of αAcryE7 mice, as in the epidermis of K14E7 mice, E7 induces cell hyperproliferation that is associated with the induction of DNA synthesis in the terminally differentiating cell compartment (49). Thus, the lens provides a similar readout of biological activities for E7 on epithelial tissues.
In a prior study, the ability of E7 to induce hyperproliferation in the lens was reduced by 50% on an E2F1-null background (45). This is consistent with a model in which E7's phenotypes are partially dependent upon E2F1 activation following inactivation of pRb. As in the epidermis, the induction of hyperproliferation by E7 in the lens of αAcryE7 mice is associated with an induction of p21 protein levels as detected by immunohistochemistry (data not shown). Given the availability of E7 transgenic lens tissues on an E2F1-null versus E2F1-sufficient background, we quantified the frequency of p21-positive cells to learn whether E2F1 status influences E7's ability to induce p21. While the frequency of p21-positive cells in the differentiated lens fiber cell compartment of control E2F1+/+ mice was less than 0.5% (i.e., below the level of detection), αAcryE7E2F1+/+ exhibited p21 staining in 6.4 ± 0.22% of lens fiber cells. However, this induction of p21 was not dependent upon E2F1, as the frequency of p21-positive lens fiber cells in αAcryE7E2F1−/− mice was 8.7 ± 2.13% (P = 0.323 for αAcryE7E2F1+/+ versus αAcryE7E2F1−/−, analysis of variance, single factor). Thus, E2F1 is not a critical mediator of E7's induction of p21 in the lens.
Although this study clearly shows that induction of p21 by E7 results from inactivation of pRb, the pathways connecting inactivation of pRb to induction of p21 remain unclear. Our data agree with others who have shown that induction of p21 and p53 do not occur via the proposed p19-Mdm2 mechanism (58), because E7 was found to be capable of inducing p21 and p53 in p19−/− mice (Fig. 6). As an alternative, p53-independent induction of p21 may be partly explained by the direct transactivation of the p21 promoter by E2F (31). However, as mentioned above, induction of p21 may also occur via stabilization of p21 protein posttranscriptionally (39). p53-dependent induction of p21 may occur via E2F-mediated ATM upregulation (7). Indeed, E7 expression has been demonstrated to elevate ATM protein levels and induce phosphorylation of p53 (7). However, these possibilities are complicated by the observations that induction of p53 and p21 by E7 are differentiation dependent (Fig. 6) and that E7's induction of p21 in the mouse lens is not dependent on E2F1 function. Thus, the mechanisms linking inactivation of pRb by E7 to induction of p21 are likely to be complex and may be indirect. Indeed, upregulation of p21 in suprabasal keratinocytes is also observed when proliferation of these cells is driven by expression of the HPV-16 E5 or E6 gene, suggesting induction of p21 may be a general response to proliferation in this normally quiescent portion of the epithelium (Anny Shai, Sybil M. G. Williams, and Paul Lambert, unpublished data).
It is useful to combine the current observations with published data (3) comparing E7 expression to pRb inactivation in 21-day-old murine ear epidermis (Table 1). The combined data show that inactivation of pRb by E7 contributes to every phenotype resulting from E7 expression. Furthermore, several of E7's phenotypes are fully reproduced by pRb inactivation alone and are completely absent when E7 cannot bind pRb. These phenotypes are (i) increased proliferation in basal cells, (ii) failure of differentiation-induced cell cycle arrest in suprabasal cells, (iii) expansion of the keratin 14-positive epidermal cell layer, (iv) failure of DNA damage-induced cell cycle arrest, and (v) p21 protein accumulation. In contrast, pRb-independent functions of E7 are necessary for the full extent of E7-induced hyperplasia and disruption of terminal differentiation. Furthermore, the pRb-independent functions of E7 are sufficient to induce a mild hyperplasia and disruption of terminal differentiation. Finally, the fact that both the K14CreRbf/f and K14E7RbΔLXCXE models yielded the same conclusions indicates that the mutations made in the RbΔLXCXE allele did not result in unexpected effects that interfered with our analysis.
TABLE 1.
pRb-dependent and pRb-independent phenotypes of E7 detected in 21-day-old murine ear epidermisa
| Biological property | Mouse genotype
|
|||||
|---|---|---|---|---|---|---|
| Control | K14E7 | Rb−/− | K14E7 Rb−/− | RbΔ/Δ | K14E7 RbΔ/Δ | |
| Basal proliferation | + | +++ | +++ | +++ | + | + |
| Suprabasal proliferation | − | +++ | +++ | +++ | − | − |
| Hyperplasia | − | +++ | ++ | +++ | − | + |
| Expanded keratin 14-positive layer | − | +++ | +++ | +++ | − | − |
| Expanded keratin 10-positive layer | − | +++ | ++ | +++ | − | + |
| Expanded filaggrin-positive layer | − | +++ | ++ | +++ | − | − |
| DNA synthesis after irradiation | − | +++ | +++ | +++ | − | − |
| p21 induction | − | +++ | +++ | +++ | − | − |
Control, nontransgenic littermates; Rb−/−, K14CreRbf/f mice; −, absence of a phenotype; +++, phenotype severity equal to that seen in K14E7 mice; +, a phenotype is present, but with less than 35% of the severity observed in K14E7 mice; ++, a phenotype is present, but with 35 to 70% of the severity observed in K14E7 mice.
Future studies are now needed to identify the pathways by which E7 mediates its pRb-independent activities, to determine if the conclusions reported here in ear epidermis also apply to cervical epidermis, and to determine whether E7's ability to promote cervical carcinogenesis is pRb dependent.
Acknowledgments
We thank the UWCCC histology facility for expert histological support and Bill Sugden for critical analysis of the manuscript.
This work was supported by NIH grants CA64402, CA098428, CA22443, and EY09091 and by UW Comprehensive Cancer Center grant P30 CA014520. F.D. is a research scientist of the Canadian Cancer Society through an award from the National Cancer Institute of Canada. L.F. and S.B. were supported by NIH grants T32 CA009135 and T32 GM00721, respectively.
REFERENCES
- 1.Antinore, M., M. Birrer, D. Patel, L. Nader, and D. McCance. 1996. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 15:1950-1960. [PMC free article] [PubMed] [Google Scholar]
- 2.Avvakumov, N., J. Torchia, and J. Mymryk. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833-3841. [DOI] [PubMed] [Google Scholar]
- 3.Balsitis, S. J., J. Sage, S. Duensing, K. Munger, T. Jacks, and P. F. Lambert. 2003. Recapitulation of the effects of the HPV-16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol. Cell. Biol. 23:9094-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, L., C. Edmonds, and K. H. Vousden. 1990. Ability of the HPV16 E7 protein to bind Rb and induce DNA synthesis is not sufficient for efficient transforming activity in NIH3T3 cells. Oncogene 5:1383-1389. [PubMed] [Google Scholar]
- 5.Bates, S., A. C. Phillips, P. A. Clarke, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 6.Bedell, M. A., K. H. Jones, S. R. Grossman, and L. A. Laimins. 1989. Identification of human papillomavirus type 18 transforming genes in immortalized and primary cells. J. Virol. 63:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkovich, E., and D. Ginsberg. 2003. ATM is a target for positive regulation by E2F-1. Oncogene 22:161-167. [DOI] [PubMed] [Google Scholar]
- 8.Bernat, A., N. Avvakumov, J. Mymryk, and L. Banks. 2003. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 22:7871-7881. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 10.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Luca, A., R. Mangiacasale, A. Severino, L. Malquori, A. Baldi, A. Palena, A. M. Mileo, P. Lavia, and M. G. Paggi. 2003. E1A deregulates the centrosome cycle in a Ran GTPase-dependent manner. Cancer Res. 63:1430-1437. [PubMed] [Google Scholar]
- 12.Demers, G. W., S. A. Foster, C. L. Halbert, and D. A. Galloway. 1994. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc. Natl. Acad. Sci. USA 91:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demers, G. W., C. L. Halbert, and D. A. Galloway. 1994. Elevated wild-type p53 protein levels in human epithelial cell lines immortalized by the human papillomavirus type 16 E7 gene. Virology 198:169-174. [DOI] [PubMed] [Google Scholar]
- 14.Dick, F., E. Sailhamer, and N. Dyson. 2000. Mutagenesis of the pRb pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duensing, S., A. Duensing, C. P. Crum, and K. Munger. 2001a. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 16.Duensing, S., A. Duensing, E. R. Flores, A. Do, P. F. Lambert, and K. Munger. 2001b. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinoctyes. J. Virol. 75:7712-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Pinoonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duensing, S., and K. Munger. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 62:7075-7082. [PubMed] [Google Scholar]
- 19.Duensing, S., and K. Munger. 2003. Human Papillomavirus Type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J. Virol. 77:12331-12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durst, M., A. Kleinheinz, M. Hotz, and L. Gissman. 1985. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J. Gen. Virol. 66:1515-1522. [DOI] [PubMed] [Google Scholar]
- 21.Edmonds, C., and K. H. Vousden. 1989. A point mutational analysis of human papillomavirus type 16 E7 protein. J. Virol. 63:2650-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichten, A., M. Westfall, J. A. Pietenpol, and K. Munger. 2002. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology 295:74-85. [DOI] [PubMed] [Google Scholar]
- 23.Flores, E. R., L. Allen-Hoffman, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazier, I. Personal communication.
- 25.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulliver, G. A., R. L. Herber, A. Liem, and P. F. Lambert. 1997. Both conserved region 1 (CR1) and CR2 of the human papillomavirus type 16 E7 oncogene are required for induction of epidermal hyperplasia and tumor formation in transgenic mice. J. Virol. 71:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helt, A., J. O. Funk, and D. A. Galloway. 2002. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J. Virol. 76:10559-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helt, A., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herber, R., A. Liem, H. Pitot, and P. F. Lambert. 1996. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 70:1873-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiyama, H., A. Iavarone, and S. Reeves. 1998. Regulation of the cdk inhibitor p21 during cell cycle progression is under the control of the transcription factor E2F. Oncogene 16:1513-1523. [DOI] [PubMed] [Google Scholar]
- 32.Isaac, C. E., A. Martens, L. M. Julian, L. A. Seifried, U. K. Binne, A. Naar, P. Sicinski, N. Dyson, and F. Dick, in press.
- 33.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 34.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jewers, R. J., P. Hildebrandt, W. Ludlow, B. Kell, and D. McCance. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, D. L., and K. Munger. 1997. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 71:2905-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, D. L., D. A. Thompson, E. Suh-Burgmann, M. Grace, and K. Munger. 1999. Expression of the HPV E7 oncoprotein mimics but does not evoke a p53-dependent cellular DNA damage response pathway. Virology 258:406-414. [DOI] [PubMed] [Google Scholar]
- 40.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 41.Lam, E., J. Morris, R. Davies, T. Crook, R. Watson, and K. H. Vousden. 1994. HPV-16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J. 13:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, J., A. Russo, and N. Pavletich. 1998. Structure of the retinoblastoma tumor-suppressor pocket domain bound to a peptide from HPV E7. Nature 391:859-865. [DOI] [PubMed] [Google Scholar]
- 43.Massimi, P., D. Pim, and L. Banks. 1997. Human papillomavirus type 16 E7 binds to the conserved carboxy-terminal region of the TATA box binding protein and this contributes to E7 transforming activity. J. Gen. Virol. 78:2607-2613. [DOI] [PubMed] [Google Scholar]
- 44.Matlashewski, G., J. Schneider, L. Banks, N. Jones, A. Murray, and L. Crawford. 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J. 6:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaffrey, J., L. Yamasaki, N. Dyson, E. Harlow, and A. Griep. 1999. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell. Biol. 19:6458-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulligan, G., and T. Jacks. 1998. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 14:223-229. [DOI] [PubMed] [Google Scholar]
- 47.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 48.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan, H., and A. Griep. 1994. Altered cell cycle regultion in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 8:1285-1299. [DOI] [PubMed] [Google Scholar]
- 50.Park, J., E. Kim, H. Kwon, E. Hwang, S. Namkoong, and S. Um. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. J. Biol. Chem. 275:6764-6769. [DOI] [PubMed] [Google Scholar]
- 51.Phelps, W. C., S. Bagchi, J. A. Barnes, P. Raychaudhuri, V. Kraus, K. Munger, P. M. Howley, and J. R. Nevins. 1991. Analysis of trans activation by human papillomavirus type 16 E7 and adenovirus 12S E1A suggests a common mechanism. J. Virol. 65:6922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phelps, W. C., K. Munger, C. L. Lee, J. A. Barnes, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 53.Phelps, W. C., K. Munger, C. Yee, J. A. Barnes, and P. M. Howley. 1992. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 66:2418-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips, A., and K. H. Vousden. 1997. Analysis of the interaction between human papillomavirus type 16 E7 and the TATA-binding protein, TBP. J. Gen. Virol. 78:905-909. [DOI] [PubMed] [Google Scholar]
- 55.Prathapam, T., C. Kuhne, and L. Banks. 2001. The HPV-16 E7 oncoprotein binds Skip and suppresses its transcriptional activity. Oncogene 20:7677-7685. [DOI] [PubMed] [Google Scholar]
- 56.Reznikoff, C. A., C. Belair, E. Savelieva, Y. Zhai, K. Pfeifer, T. Yeager, K. J. Thompson, S. DeVries, C. Bindley, and M. A. Newton. 1994. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 8:2227-2240. [DOI] [PubMed] [Google Scholar]
- 57.Riley, R., S. Duensing, T. Brake, K. Munger, P. F. Lambert, and J. Arbeit. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 63:4862-4871. [PubMed] [Google Scholar]
- 58.Seavey, S. E., M. Holubar, L. J. Saucedo, and M. E. Perry. 1999. The E7 oncoprotein of human papillomavirus type 16 stabilizes p53 through a mechanism independent of p19ARF. J. Virol. 73:7590-7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 60.Slebos, R., M. H. Lee, B. S. Plunkett, T. D. Kessis, B. O. Williams, T. Jacks, L. Hedrick, M. B. Kastan, and K. R. Cho. 1994. p53-dependent G1 arrest involves pRb-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc. Natl. Acad. Sci. USA 91:5320-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song, S., G. A. Gulliver, and P. F. Lambert. 1998. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways. Proc. Natl. Acad. Sci. USA 95:2290-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka, A., T. Noda, H. Yajima, M. Hatanaka, and Y. Ito. 1989. Identification of a transforming gene of human papillomavirus type 16. J. Virol. 63:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vousden, K. H., J. Doniger, J. A. DiPaolo, and D. R. Lowy. 1988. The E7 open reading frame of human papillomavirus type 16 encodes a transforming gene. Oncogene Res. 3:167-175. [PubMed] [Google Scholar]
- 64.Walboomers, J., M. Jacobs, M. Manos, F. Bosch, J. Kummer, K. Shah, P. Snijders, J. Peto, C. Meijer, and N. Munoz. 1999. Human Papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe, S., and K. Yoshiike. 1988. Transformation of rat 3Y1 cells by human papillomavirus type-18 DNA. Int. J. Cancer 41:896-900. [DOI] [PubMed]
- 66.White, A. E., E. M. Livanos, and T. D. Tlsty. 1994. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 8:666-677. [DOI] [PubMed] [Google Scholar]
- 67.Xiong, Y., D. Kuppuswamy, Y. Li., E. M. Livanos, M. Hixon, A. White, D. Beach, and T. D. Tlsty. 1996. Alteration of cell cycle kinase complexes in human papillomavirus E6- and E7-expressing fibroblasts precedes neoplastic transformation. J. Virol. 70:999-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signalling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.zur Hausen, H. 1996. Papillomavirus infections-a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]
- 70.Zwerchke, W., B. Mannhardt, P. Massimi, S. Nauenburg, D. Pim, W. Nickel, L. Banks, A. Reuser, and P. Jansen-Durr. 2000. Allosteric activation of acid alpha-glucosidase by the human papillomavirus E7 protein. J. Biol. Chem. 275:9534-9541. [DOI] [PubMed] [Google Scholar]
- 71.Zwerchke, W., S. Mazurek, P. Massimi, L. Banks, E. Eigenbrodt, and P. Jansen-Durr. 1999. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc. Natl. Acad. Sci. USA 96:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]