Abstract
Virus infections have dramatic effects on structural and morphological characteristics of the host cell. The gene product of open reading frame 3 in the early region 4 (E4orf3) of adenovirus serotype 5 (Ad5) is involved in efficient replication and late protein synthesis. During infection with adenovirus mutants lacking the E4 region, the viral genomic DNA is joined into concatemers by cellular DNA repair factors, and this requires the Mre11/Rad50/Nbs1 complex. Concatemer formation can be prevented by the E4orf3 protein, which causes the cellular redistribution of the Mre11 complex. Here we show that E4orf3 colocalizes with components of the Mre11 complex in nuclear tracks and also in large cytoplasmic accumulations. Rearrangement of Mre11 and Rad50 by Ad5 E4orf3 is not dependent on interactions with Nbs1 or promyelocytic leukemia protein nuclear bodies. Late in infection the cytoplasmic inclusions appear as a distinct juxtanuclear accumulation at the centrosome and this requires an intact microtubule cytoskeleton. The large cytoplasmic accumulations meet the criteria defined for aggresomes, including γ-tubulin colocalization and formation of a surrounding vimentin cage. E4orf3 also appears to alter the solubility of the cellular Mre11 complex. These data suggest that E4orf3 can target the Mre11 complex to an aggresome and may explain how the cellular repair complex is inactivated during adenovirus infection.
There are numerous approaches that viruses utilize to modify the host cell environment and promote efficient viral replication. Viral infections show remarkable spatial regulation and are often accompanied by dynamic rearrangement of cellular structures. Adenovirus replicates within the nucleus of the host cell and induces distinct sites called replication centers, where viral transcription and replication occur (36). The earliest detected ultrastructural changes in adenovirus-infected cells are small masses of thin fibrils (37, 38). As infection progresses these structures rapidly increase in size and are observed as pleiomorphic shapes of crescents and rings (36, 38) that can be localized by immunostaining with an antibody to the viral single-stranded DNA binding protein. Cellular proteins involved in viral replication are also recruited to these replication foci (2). In addition to the formation of viral replication centers, infection is also accompanied by disruption of certain cellular structures. During adenovirus infection discrete nuclear structures containing the promyelocytic leukemia protein PML and known as oncogenic domains (PODs/ND10) are disrupted (8, 12), and the PML protein is redistributed into track-like structures.
Early region 4 (E4) of adenovirus serotype 5 (Ad5) encodes at least six gene products (reviewed in references 28 and 46). Deletions of the E4 region result in a number of severe phenotypes, including defects in viral mRNA accumulation, transcription, splicing, late protein synthesis, host cell shutoff, and viral DNA replication (20, 22). During infection with an E4-deleted adenovirus the viral genome becomes joined together into large concatemers (48). Concatemerization requires cellular proteins involved in the nonhomologous end-joining pathway (3, 43), including a cellular repair complex containing the Mre11, Rad50, and Nbs1 proteins that is referred to as the Mre11 complex (43). Concatemerization can be prevented by E4 gene products, which lead to the mislocalization and degradation of the Mre11 complex (43). The product of open reading frame 3 of the E4 region in Ad5 (Ad5 E4orf3) can redistribute Mre11, Rad50, and Nbs1 from their normal diffuse nuclear localization into large nuclear and cytoplasmic accumulations (43). The E4orf3 protein is also the viral factor responsible for reorganization of the PODs/ND10 (8, 12). Redistribution of cellular proteins by E4orf3 may play an important role in aiding viral replication and inactivating cellular antiviral defenses (12, 15, 43).
The E4orf3 protein is tightly associated with the nuclear matrix and localizes mainly to the nucleus, but is also found in cytoplasmic accumulations (8, 26, 40, 41). E4orf3 physically interacts with the adenoviral E1b55K protein (29). A complex distribution pattern has been reported for the E1b55K protein during adenovirus infections (18, 35). The protein is found in the cytoplasm, in a perinuclear body, in nuclear tracks and spicules, and at viral replication centers (12, 35). At late times of infection E1b55K becomes associated with viral replication centers and this is dependent upon the E4orf6 protein (19, 35). In cells infected with an E4-deleted adenovirus, the E1b55K protein is found in a diffusely nuclear pattern (29, 35). When expressed alone by transfection or in stable cell lines (7, 11, 51), the E1b55K protein is predominantly found in cytoplasmic bodies but can be transported into the nucleus by expression of either E4orf3 (26) or E4orf6 (9, 11, 19, 34). The E1b55K protein has also been suggested to associate with the nuclear matrix independently of the E4orf3 protein and this is inhibited by E4orf6 interaction (30).
Cellular proteins that become misfolded and aggregated are normally targeted for proteasomal degradation (25, 50). Aggregated proteins can also become sequestered into specialized cytoplasmic structures termed aggresomes (reviewed in references 17 and 27). Particles of aggregated proteins form and are transported by the motor protein dynein along microtubules towards the microtubule-organizing center or centrosome, where they assemble into a single spherical aggresome or in some cases an extended ribbon structure. Aggresome formation can be inhibited by drugs such as nocodazole, which depolymerize microtubules (23). Chaperones and proteasomes accumulate at aggresome structures, where they may aid clearance of the aggregated protein. In cells with aggresomes the cytoskeleton is rearranged, and the intermediate filament protein vimentin forms a cage around the aggresome (23). It has been observed that the phase-dense cytoplasmic accumulations of E1b55K are in close proximity to the centrosome and costain with cellular proteins, including p53 and hsp70 (1, 5, 51). These cytoplasmic structures are highly insoluble and may represent aggregates of misfolded protein.
The connections between E4orf3, E1b55K, and aggresomes have not been investigated. In this study we have further analyzed the cytoplasmic accumulation of E4orf3 and the mechanism by which it targets the Mre11 complex. We find that the Mre11/Rad50/Nbs1 proteins colocalize with E4orf3 in nuclear tracks and at the cytoplasmic sites. The large cytoplasmic aggregates that are juxtanuclear also costain with the centrosomal marker γ-tubulin. Accumulation at a single cytoplasmic aggregate is blocked by the microtubule-depolymerizing drug nocodazole. Although the adenoviral E1b55K protein is also found at the centrosome/microtubule-organizing center (5) accumulation of E4orf3 and the Mre11 complex can occur independently of E1b55K. Analysis of the requirements for redistribution of Mre11 and Rad50 by Ad5 E4orf3 demonstrated that it is not dependent on interactions with Nbs1 or PML nuclear bodies. Together these results suggest that E4orf3 is sufficient to form an aggresome and that it targets members of the Mre11 complex to these sites during infection and transfection.
MATERIALS AND METHODS
Cell lines.
HeLa, 293, A549, and U2OS cells were obtained from the American Type Culture Collection. The cell line W162 was used for growth of E4-deleted viruses and was obtained from G. Ketner (49). Parental Nijmegen breakage syndrome (NBS) cells (GM7166) were obtained from the Coriell Institute and ataxia-telangiectasia-like disorder (A-TLD) cell lines were obtained from J. Petrini. Mutant cells were transduced with retroviruses carrying wild-type cDNAs for Nbs1 and Mre11 as previously described (7, 43). All cells were maintained as monolayers in Dulbecco's modified Eagle's medium supplemented with 10 or 20% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids.
Plasmids containing the cDNAs for E4orf3 (12) and E1b55K (44) of Ad5 expressed from the cytomegalovirus promoter were used for transfections. Cells in monolayers were transfected with calcium phosphate or Effectene (QIAGEN) or Lipofectamine (Invitrogen) according to standard protocols and the manufacturers' protocols.
Viruses and infections.
Wild-type adenovirus serotype 5 was obtained from the American Type Culture Collection and propagated in 293 cells. The E4 mutant viruses dl1004 and dl1017 were gifts of G. Ketner and were propagated on W162 and 293 cells, respectively. Recombinant adenovirus expressing Ad5 E4orf3 was generated using the Ad-Easy kit (Quantum) and was a gift from Sarah Malpel. All viruses were purified by two sequential rounds of ultracentrifugation in CsCl gradients and stored in 40% glycerol at −20°C. HeLa cells were infected at a multiplicity of infection of 25 and NBS and A-TLD cells were infected at a multiplicity of infection of 100. Cells were harvested for immunofluorescence 24 h following infection unless otherwise indicated.
Antibodies.
The following antibodies were obtained from commercial sources and used at the indicated dilutions: Rad50 (Novus Biologicals, 1:300), Mre11 (Novus Biologicals, 1:300), Nbs1 (Novus Biologicals, 1:500), PML (Santa-Cruz, 1:200), γ-tubulin (Sigma, 1:2000), and vimentin (Amersham, 1:200). Other antibodies to cellular proteins were polyclonal rabbit antiserum to RPA32 (a gift from T. Melendy; 1:1,000) and giantin and the coat protein β-COP (both gifts from V. Malhotra; 1:1,000). Antibodies to viral proteins included a monoclonal mouse antibody to E1b55K (a gift from A. Levine, 1:200 of supernatant fluid), and rabbit polyclonal antiserum to E4orf3 (a gift from G. Ketner, 1:2000) and DNA-binding protein (a gift from P. van der Vliet, 1:1,000). Secondary antibodies for immunofluorescence and immunoblotting were obtained from Sigma or Jackson ImmunoResearch.
Immunofluorescence.
Cells were grown on glass coverslips in 24-well dishes and infected with virus or transfected. Where indicated the drug nocodazole (Sigma) was included at a concentration of 10 mg/ml. Cells were harvested for immunofluorescence at 24 h after infection/transfection unless indicated otherwise. Cells were washed in phosphate-buffered saline (PBS) and in most cases cells were fixed for 10 min at −20°C in ice-cold methanol-acetone (1:1). Cells were rehydrated in PBS and blocked with PBS-B (5% bovine serum albumin in PBS) for 30 min.
In the experiments to look at the solubility of the Mre11 complex, HeLa cells were washed and permeabilized prior to fixation by a 7-min incubation in 0.5% Triton X-100 in 20 mM HEPES-KOH (pH 7.9), 50 mM NaCl, 3 mM MgCl2 and 300 mM sucrose. Fixation with 4% paraformaldehyde was done at room temperature for 20 min followed by three washes in PBS. Cells were extracted for a second time by a 10-min incubation in 0.5% Triton X-100. Primary antibodies were incubated for 1 h at room temperature in PBS-B and then washed three times in PBS. Secondary antibodies were incubated with the cells for 1 h at room temperature in PBS-B. Cells were washed four times in PBS and nuclear DNA was stained with 4′,6′-diamidino-2-phenylindol (DAPI). Coverslips were mounted using Fluoromount-G (Southern Biotechnology Associates). In all cases, control staining experiments showed no cross-reaction between the fluorophores, and images obtained by staining with individual antibodies were the same as those shown for double labeling. Immunoreactivity was visualized using a Nikon microscope in conjunction with a charge-coupled device camera (Cooke Sensicam). Images were obtained in double or triple excitation mode and processed using SlideBook and Adobe Photoshop. Where quantitation was required, 200 cells were counted under the microscope and scored for the presence of multiple cytoplasmic aggregates or a single large aggresome.
Protein solubility.
HeLa cells were transfected with the E4orf3 expression vector and harvested for analysis of protein solubility after 24 h. Cells were lysed in PBS containing 1% NP-40 and 0.1% sodium dodecyl sulfate. Supernatant and pellet fractions were obtained by centrifugation (13 krpm, 20 min, 4°C) and protein concentrations were determined by the standard Lowry assay (Bio-Rad). Equal amounts of protein were loaded (40 μg) for analysis of supernatant fractions on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Pellets were resuspended in 50 μl NuPAGE LDS sample buffer (Invitrogen) with 50 mM dithiothreitol and normalized to their respective supernatant fractions for total protein. Proteins were separated by electrophoresis and transferred to Hybond ECL membranes. Membranes were blocked overnight in PBS with 5% dry milk. Primary antibodies were incubated with the membrane for 1 h at room temperature in PBS with 3% bovine serum albumin. Proteins were visualized by enhanced chemiluminescence (Perkin Elmer) after incubation with secondary antibodies coupled to horseradish peroxidase (1:200 dilution) for 1 h at room temperature.
RESULTS
Ad5 E4orf3 protein is located in intranuclear tracks and cytoplasmic aggresomes.
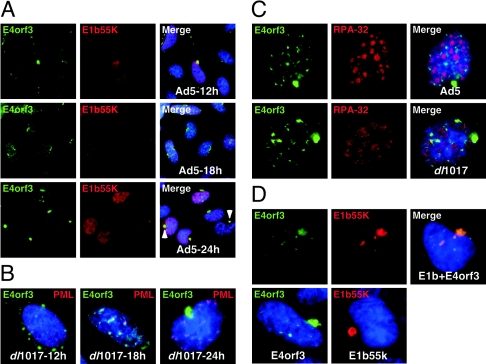
We used immunofluorescence to analyze the localization of E4orf3 protein throughout infection relative to other viral and cellular proteins. As previously reported (8, 12, 14, 29, 41), E4orf3 was located in nuclear tracks and cytoplasmic accumulations during Ad5 infection (Fig. 1A). As infection progressed the majority of the protein was found at later times in a single cytoplasmic accumulation in close proximity to the nucleus. A physical interaction between E1b55K and E4orf3 has been previously reported (29), but E1b55K is not required for the formation of nuclear track-like structures during infection (12). Costaining with an antibody to Ad5-E1b55K revealed this protein to be in multiple patterns as previously reported, and at late times colocalization with E4orf3 was observed at the large cytoplasmic perinuclear bodies (Fig. 1A).
FIG. 1.
Localization of Ad5 E4orf3 in infected and transfected cells. (A) HeLa cells were infected with Ad5 and analyzed by immunofluorescence at the indicated times after infection. The E4orf3 protein is found in nuclear track structures and cytoplasmic accumulations. At later times the E4orf3 and E1b55K proteins are found colocalized in a large perinuclear cytoplasmic accumulation as indicated by arrows. (B) E1b55K is not required for cytoplasmic accumulation of E4orf3. Similar patterns of localization were observed for E4orf3 when cells were infected with the mutant virus dl1017, which does not express E1b55K and E4orf6. The PML protein partially colocalizes with E4orf3 in nuclear tracks but is not found at the cytoplasmic accumulations. (C) E4orf3 is excluded from viral replication centers (stained with an antibody to RPA32) during infections with wild-type Ad5 or dl1017. (D) Localization of E4orf3 and E1b55K proteins when expressed alone or together by transfection of expression plasmids.
We investigated whether E1b55K affected the localization of E4orf3 by infection of cells with the virus dl1017, which is mutated for E1b55K and E4orf6 (4). The patterns observed with this virus were similar to those seen for wild-type Ad5, with E4orf3 in nuclear tracks and at cytoplasmic sites (Fig. 1B). At late times the E4orf3 protein was found accumulated at a single juxtanuclear cytoplasmic accumulation. We also costained for E4orf3 together with an antibody to the cellular PML protein. The PML protein was redistributed from PODs/ND10 bodies into intranuclear track-like structures that partially overlapped E4orf3, but it did not appear in the cytoplasmic sites (Fig. 1B).
We also examined the relative localization of E4orf3 with respect to viral replication centers, which we detected using an antibody to the cellular RPA32 protein that is recruited to viral centers (45). With both wild-type and dl1017 viruses the E4orf3 protein was excluded from viral replication centers (Fig. 1C). Finally, E4orf3 localization was detected after transfection of expression vectors (Fig. 1D). The E4orf3 protein was located at both nuclear tracks and cytoplasmic accumulations, independently of the presence of E1b55K. When expressed alone the E1b55K protein could be found at cytoplasmic accumulations at a single site, as previously reported (5). When E1b55K and E4orf3 were coexpressed they colocalized in nuclear tracks and at a large juxtanuclear accumulation.
In these transient transfection experiments we observed some variability in staining patterns for E1b55K and E4orf3, with cytoplasmic accumulations appearing as either discrete punctate structures or as larger globular structures. This degree of structural heterogeneity correlated with the variability of protein expression levels inherent to transient transfections. Together these results demonstrate that localization of E4orf3 in nuclear tracks and cytoplasmic accumulations occurs independently of E1b55K expression and can be detected outside the context of viral infection.
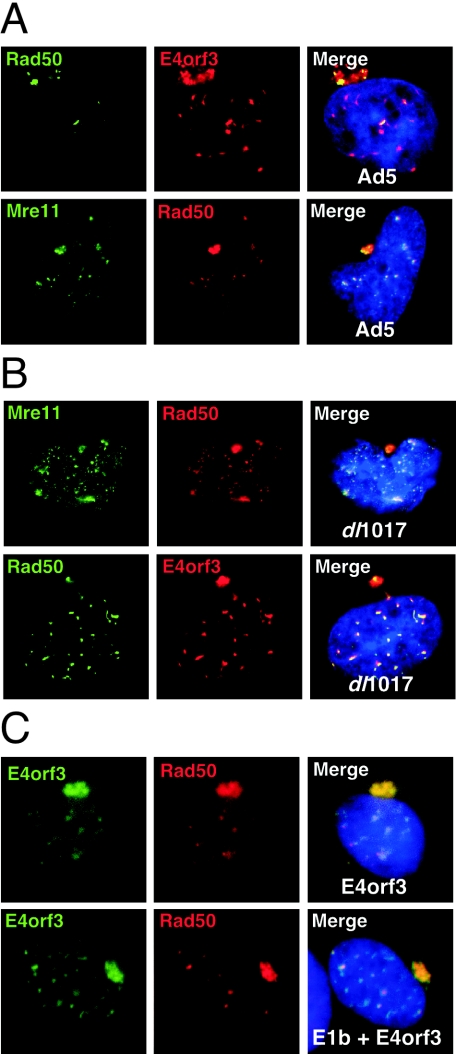
Ad5 E4orf3 protein colocalizes with Mre11/Rad50/Nbs1 at nuclear and cytoplasmic sites.
We have previously reported that upon either transfection or infection, the Ad5 E4orf3 protein can reorganize the cellular distribution of the Mre11/Rad50/Nbs1 complex factors (43). We therefore examined the relationship between the structures formed by E4orf3 expression and the nuclear and cytoplasmic speckles observed for Mre11 complex members. During infection with wild-type Ad5 we observed partial colocalization of Mre11, Rad50, and Nbs1 with both the nuclear tracks and cytoplasmic accumulations of E4orf3 (Fig. 2A and 3A). Relocalization of Mre11 and Rad50 by E4orf3 was not dependent upon the viral E1b55K protein, as it was still seen in cells infected with the dl1017 virus lacking E1b55K and E4orf6 (Fig. 2B). Similar results were obtained by transfection of an E4orf3 expression construct (43) in the presence and absence of E1b55K (Fig. 2C).
FIG. 2.
E4orf3 protein of Ad5 partially colocalizes with the Mre11 complex. (A) Infection with wild-type Ad5 leads to redistribution of the Mre11 complex into nuclear tracks and cytoplasmic accumulations that partially colocalize with E4orf3. (B) Colocalization of E4orf3 with the Mre11 complex is not dependent upon E1b55K as it is still observed in cells infected with dl1017. (C) The Mre11 complex is redistributed by E4orf3 transfection into nuclear tracks and large cytoplasmic accumulations, in the absence and presence of E1b55K.
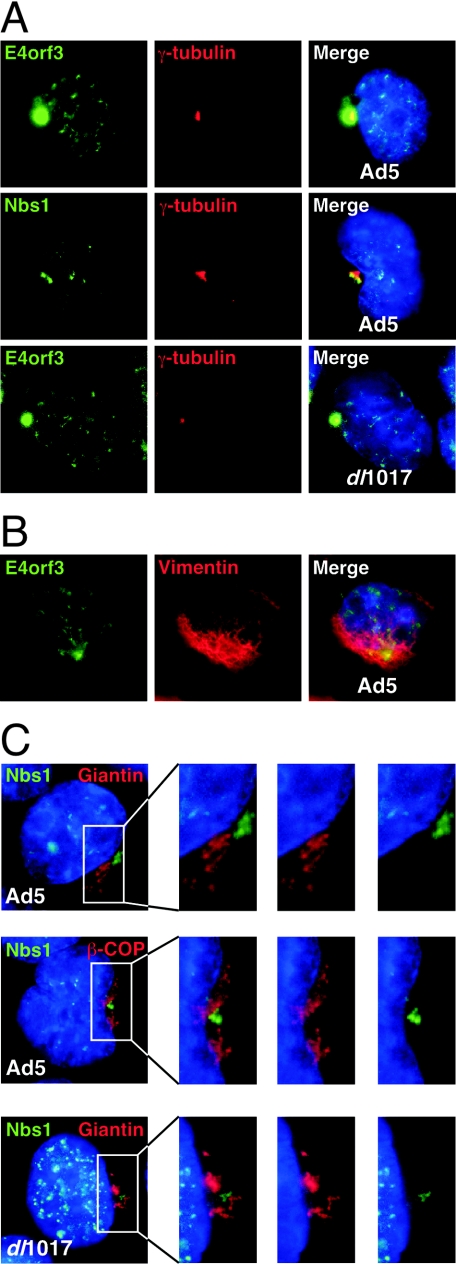
FIG. 3.
E4orf3 and the Mre11 complex accumulate at the centrosome. Cells were infected with wild-type Ad5 or the dl1017 mutant that lacks E1b55K. (A) The E4orf3 and Nbs1 proteins accumulate at the centrosome and costain with γ-tubulin independently of E1b55K. (B) The cytoplasmic accumulation of E4orf3 is surrounded by vimentin. (C) The cytoplasmic accumulation of Nbs1 induced by E4orf3 does not correspond to the Golgi apparatus as assessed by staining for giantin and β-COP.
Cytoplasmic accumulations of E4orf3 and the Mre11 complex colocalize with centrosome markers.
The formation of a single large juxtanuclear cytoplasmic accumulation of E4orf3 localized together with the Mre11 complex at later times of infection was observed in many different cell types, including HeLa, A549, U2OS, and primary IMR90 cells (data not shown). The regularity and proximity to a nuclear invagination suggested that this was not a random localization. As the E1b55K protein has been reported to localize to the centrosome in infected and transfected cells (5), we examined E4orf3 localization with respect to centrosome markers. We costained E4orf3 with an antibody to the centrosomal marker γ-tubulin. The perinuclear accumulation of the E4orf3 and Mre11/Rad50/Nbs1 proteins colocalized with the centrosome during infections with wild-type Ad5 and the dl1017 mutant virus (Fig. 3A and data not shown).
We explored the possibility that these accumulations might represent aggresomes. Aggresome formation at the centrosome/microtubule-organizing center correlates with changes in the distribution of the intermediate filament protein vimentin (23). In cells infected with Ad5, vimentin was found accumulated around the staining observed for E4orf3 (Fig. 3B), supporting the idea that this represents an aggresome. Similar results were obtained by transfection of an E4orf3 expression vector (data not shown).
The cellular Golgi apparatus is localized around the microtubule-organizing center (31). To differentiate between Golgi and aggresomes, we costained infected cells with Nbs1 and antibodies to the Golgi markers giantin and β-COP (Fig. 3C). The Nbs1 protein was localized in a discrete site, often surrounded by the Golgi apparatus, and no colocalization was observed. These results revealed that the accumulation of Nbs1 is distinct from the Golgi apparatus, consistent with previous reports of aggresome formation (23). This was observed for wild-type virus and also in the absence of E1b55K in infections with dl1017 (Fig. 3C). Together these results demonstrated that the perinuclear aggregates of E4orf3 with the Mre11 complex are clustered around the centrosome.
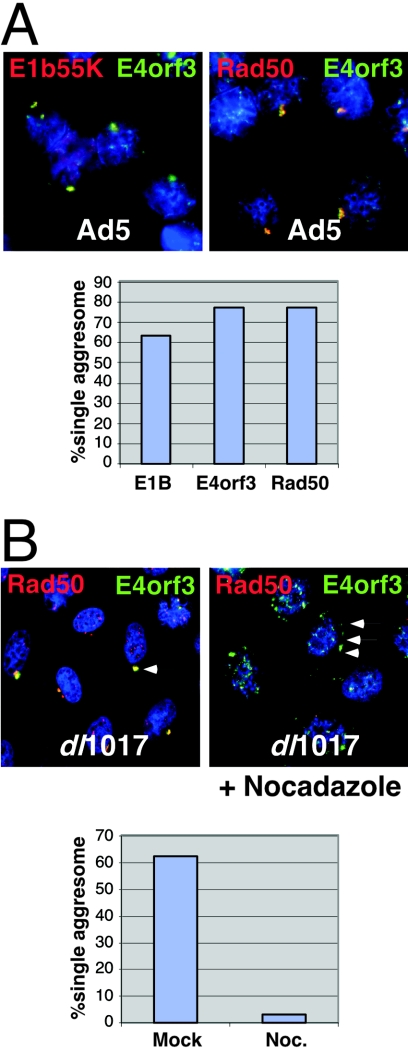
We quantitated the accumulation of viral and cellular proteins at a single large cytoplasmic aggregate (Fig. 4A). In wild-type virus infections, 78% of cells showed a single aggresome for E4orf3 and Rad50. The accumulation of E1b55K at a single cytoplasmic site was observed in 64% of infected cells. Formation of a single aggresome of E4orf3 and Rad50 was also observed in 62% of cells infected with the dl1017 virus (Fig. 4B), indicating that aggresome formation with these proteins is independent of E1b55K. Formation of aggresomes at the centrosome/microtubule-organizing center is a directed process that requires transport on microtubules (23). Therefore we examined the effect of microtubule depolymerization using the drug nocodazole (Fig. 4B). Infection in the presence of nocodazole resulted in E4orf3 localization in multiple sites dispersed throughout the cytoplasm, and there were very few cells with a single large aggresome of E4orf3 and Rad50. These results further demonstrate that accumulation of E4orf3 and the Mre11 complex at a single perinuclear accumulation requires an intact microtubule-based cytoskeleton, and is consistent with the notion that it represents an aggresome.
FIG. 4.
Quantitation of viral and cellular proteins at a single juxtanuclear cytoplasmic aggregate during virus infections. (A) HeLa cells were infected with Ad5 for 24 h and analyzed by immunofluorescence for E4orf3, E1b55K, and Rad50. The presence of a single juxtanuclear aggregesome compared to multiple cytoplasmic aggregates was determined for 200 cells. (B) Nocodazole prevents the formation of a single large aggresome. HeLa cells were infected with dl1017 for 24 h in the presence or absence of nocodazole. E4orf3 and Rad50 were localized by immunofluorescence, and 200 cells were counted and quantitated for the presence of a single cytoplasmic aggresome.
Determinants of Mre11/Rad50/Nbs1 relocalization.
To determine which components of the Mre11 complex were important for its relocalization upon E4orf3 expression, we analyzed infections in fibroblasts from Nijmegen breakage syndrome (NBS) and ataxia-telangiectasia-like disorder (A-TLD) patients, which harbor characterized mutations in the genes for Nbs1 and Mre11, respectively (6, 42). NBS cells contain wild-type Rad50 and Mre11 proteins that are detected by immunofluorescence predominantly in the cytoplasm (Fig. 5) due to a truncation mutation in NBS1 that impairs their nuclear localization (6, 10, 32).
FIG. 5.
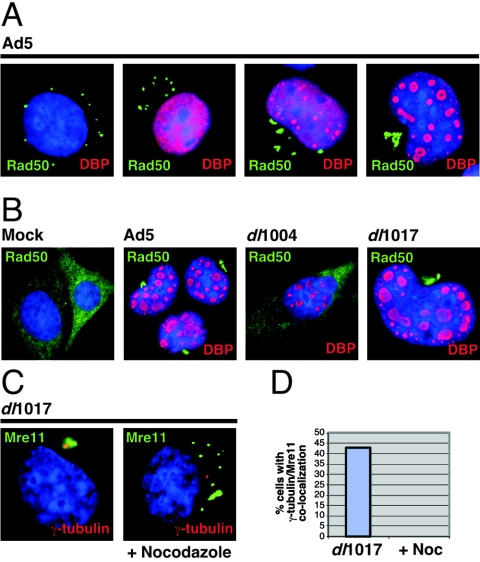
Reorganization of Rad50 and Mre11 in NBS cells. NBS fibroblasts were infected with wild-type Ad5 or mutants and analyzed by immunofluorescence at 48 and 72 h postinfection. (A) In adenovirus-infected cells, Rad50 accumulates in cytoplasmic clusters and migrates progressively to the centrosome. Representative images are shown for progressive stages of the life cycle by DNA-binding protein staining. (B) Rad50 reorganization is dependent on E4orf3 expression. In uninfected (mock) cells, Rad50 localizes diffusely to the cytoplasm due to the NBS1 mutation. After 48 h of infection, adenovirus mutants that lack E4orf3 (dl1004) fail to relocate Rad50, while E4orf3-containing mutants (dl1017) display the same pattern as with wild-type Ad5. (C) Infection of NBS cells with dl1017 results in accumulation of Mre11 at the centrosome and is inhibited by nocodazole. (D) Quantitation of the effect of nocodazole on accumulation of Mre11 at a single cytoplasmic aggregate in NBS cells infected with dl1017.
Infection of NBS cells with Ad5 results in reorganization of the cytoplasmic Rad50 and Mre11 proteins (Fig. 5A and data not shown). The Rad50 accumulations migrate progressively towards the centrosome as adenovirus replication centers (stained with an antibody to the viral replication protein DNA-binding protein) form in the nucleus (Fig. 5A). Conversely, infection with the E4-deleted virus dl1004 has no effect on Rad50 localization (Fig. 5B), demonstrating that relocalization of the Mre11 complex is dependent on expression of E4 proteins. The requirement for E4orf3 to rearrange cytoplasmic Rad50 localization was confirmed by infection with the E4orf3-containing dl1017 virus, in which Rad50 accumulation at the centrosome was similar to that with Ad5 (Fig. 5B). An E1-deleted recombinant adenovirus vector expressing only E4orf3 gave similar results (data not shown), demonstrating that E4orf3 expression alone is sufficient to cause redistribution of Rad50. Similar observations were made for Mre11 localization in NBS cells (Fig. 5C and data not shown). Staining for Mre11 and γ-tubulin showed colocalization, suggesting that the Mre11/Rad50 complex is accumulating at the centrosome in these cells (Fig. 5C). Addition of the drug nocodazole inhibited the translocation of Mre11 to a single centrosomal location, as revealed by quantitation of the immunofluorescence patterns (Fig. 5D). Together these results demonstrate that cytoplasmic Mre11/Rad50 are directed into aggresomes by E4orf3 expression.
There are two distinct mutations found in different family groups of A-TLD patients (42). The A-TLD1 allele contains a premature stop codon (633R→STOP) that results in a C-terminal truncation, and the A-TLD3 allele contains a missense mutation (117N→S) within the N-terminal nuclease domain (42). These cells express mutant Mre11 proteins with weakened interaction with Nbs1 and have reduced levels of Rad50 and Nbs1 proteins (42). In these cells, staining for the Rad50 protein shows a predominantly diffuse cytoplasmic pattern, whereas the Nbs1 protein remains nuclear (Fig. 6A). Expression of wild-type Mre11 in A-TLD1 and A-TLD3 (43) cells from a retrovirus vector restores the nuclear localization of Rad50 (Fig. 6A).
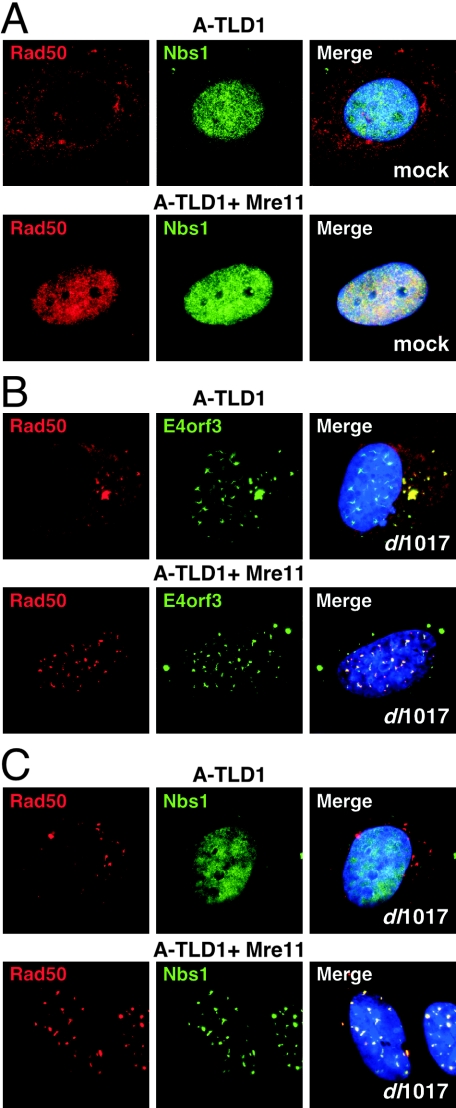
FIG. 6.
Rad50 localization is altered by Ad5 infection in both the nucleus and cytoplasm of A-TLD cells. (A) The Rad50 protein is localized in the cytoplasm of A-TLD1 cells that express a truncated form of Mre11 (top panels). In A-TLD1 cells complemented with wild-type Mre11 from a retrovirus the Rad50 protein is recruited back into the nucleus (lower panels). In both cases the Nbs1 protein is nuclear. (B) In cells infected with the dl1017 virus for 48 h the E4orf3 protein is found in nuclear and cytoplasmic speckles, and these colocalize with rearranged Rad50 whether it is in the cytoplasm (in A-TLD1) or in the nucleus (in Mre11-complemented A-TLD1 cells). (C) Nbs1 localization is unaffected by virus infection in the absence ofMre11 (A-TLD1) but is disrupted into speckles in cells expressing wild-type Mre11, where it colocalizes with Rad50.
When A-TLD1 cells were infected with the dl1017 adenovirus, we detected E4orf3 in the characteristic nuclear track structures and cytoplasmic speckles (Fig. 6B). In infected cells we observed reorganization and aggregation of the cytoplasmic Rad50 protein (Fig. 6B). Similar results were obtained in A-TLD3 cells and also with Ad5 infection (data not shown). When Rad50 was restored to the nucleus by Mre11 expression, infection also led to accumulation of Rad50 in nuclear tracks (Fig. 6B). However, in infected A-TLD1 cells the Nbs1 protein was not found in nuclear tracks unless the cells had been complemented with wild-type Mre11 (Fig. 6C). Together these data show that expression of E4orf3 can alter the distribution of Rad50 independently of its cellular compartment or association with the Nbs1 protein. Nbs1, on the other hand, appears to be affected by E4orf3 only by virtue of its interaction with Mre11 and/or Rad50.
E4orf3 expression reduces the solubility of the Mre11 complex.
We investigated whether the reorganization induced by E4orf3 correlated with changes in solubility of the targeted proteins in response to E4orf3 expression. HeLa cells were transfected with an E4orf3 expression vector and the relative abundance of individual proteins was determined for the soluble supernatant fraction and the insoluble pellet. In mock-transfected cells the Mre11 complex is present mostly in the soluble supernatant fraction (Fig. 7A). However, upon E4orf3 expression the solubility of the complex is altered, and the Mre11, Rad50, and Nbs1 proteins were each detected in the insoluble pellet fraction. As expected, E4orf3 itself was present only in the pellet fraction. Controls included Ku86, which was present only in the soluble fraction, and RPA32, which was present in both fractions. Neither of these two controls changed significantly in solubility upon E4orf3 expression.
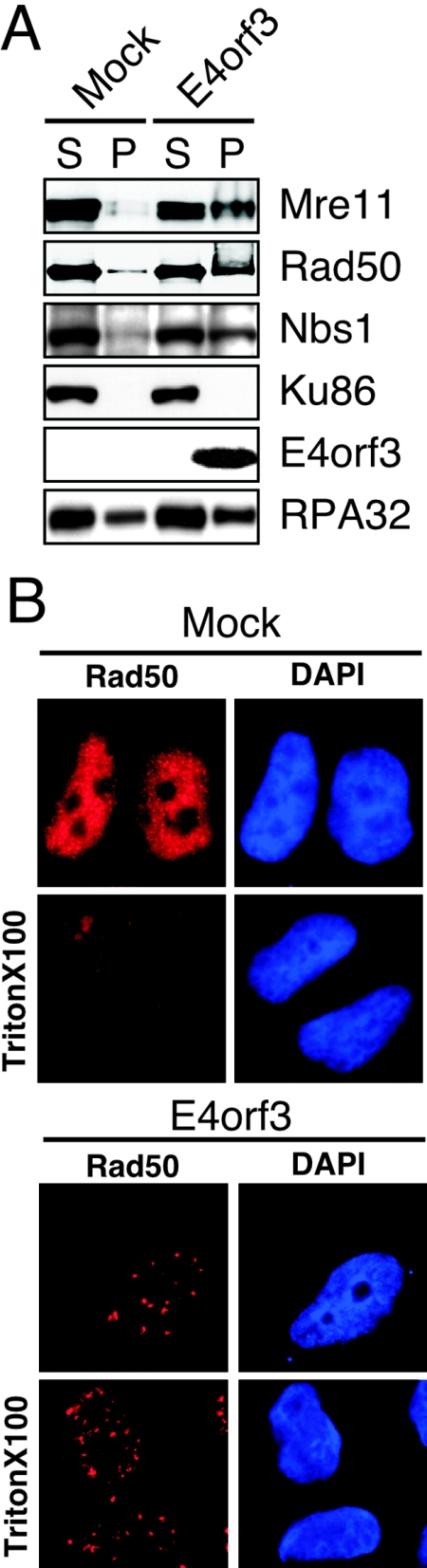
FIG. 7.
E4orf3 expression alters the solubility of the Mre11 complex. (A) The relative abundance of proteins in soluble (S) and insoluble pellet (P) fractions was assessed by immunoblotting of lysates from transfected cells. Mre11, Rad50, and Nbs1 proteins are found mostly in the soluble fraction in mock-transfected cells. Following E4orf3 expression Mre11, Rad50, and Nbs1 are found both in the soluble fraction and in the insoluble pellet. In contrast, the solubility of Ku86 and RPA32 is unchanged by E4orf3 expression. (B) HeLa cells that were either untreated (mock) or transfected with an E4orf3 expression vector were extracted with Triton X-100 prior to fixation. The detergent treatment extracts the soluble Mre11 complex, as indicatedby the diminished staining for Rad50 in the mock sample. In the presence of E4orf3 the Rad50 protein remains in nuclear tracks even after detergent treatment.
The effect of E4orf3 on solubility of the Mre11 complex was also examined using an immunofluorescence assay. HeLa cells were transfected with E4of3 and immunofluorescence for Rad50 was performed with or without detergent extraction with Triton X-100 prior to fixation with paraformaldehyde (Fig. 7B). Detergent treatment led to diminished antibody staining for Rad50 in mock-transfected cells. In contrast, after E4orf3 expression the Rad50 protein remained in nuclear tracks even after detergent extraction. Together these results indicate that E4orf3 alters the solubility of the Mre11 complex proteins, and this correlates with the formation of nuclear tracks and cytoplasmic aggregates.
DISCUSSION
The Ad5 E4orf3 protein induces dramatic changes in nuclear architecture and triggers redistribution of specific cellular proteins. It has been shown that Ad5 E4orf3 expression leads to disruption of PODs and reorganization of PML into nuclear tracks (12). We also reported that some, but not all, of the other cellular factors reported to be associated with PODs are also rearranged upon E4orf3 expression (43, 45). In this report we have shown that E4orf3 partially colocalizes with PML in the nuclear track structures but that it also forms cytoplasmic aggregates that coalesce to form an aggresome. Although PML is not found in the cytoplasmic aggregates, the cellular Mre11 complex is redistributed into both the nuclear tracks and the cytoplasmic accumulations in the presence of E4orf3. Reorganization of Mre11 and Rad50 by Ad5 E4orf3 does not appear to require Nbs1, as it was observed in cells harboring a mutant NBS1 allele.
The results from the experiments using A-TLD cells suggest that reorganization of Nbs1 protein may be mediated through its interaction with the Mre11/Rad50 complex. The redistribution of Mre11 and Rad50 in NBS cells, where they are predominantly cytoplasmic, suggests that nuclear localization of the complex is not required for E4orf3 to induce its relocalization. It also implies that association of Mre11/Rad50 with POD/ND10 structures is not required for E4orf3 to affect their subcellular localization. This suggests that redistribution of PML and the Mre11 complex by Ad5 E4orf3 may be independent events. This conclusion is supported by data showing that the E4orf3 proteins from some adenovirus serotypes cannot affect the localization of the Mre11 complex (45) and that specific mutations can separate redistribution of Mre11 from PML (15).
We observed large accumulations of Ad5 E4orf3 and the Mre11 complex proteins at the centrosome during infections of a variety of cell types. Protein aggregates do not normally accumulate in unstressed cells due to cellular degradation of misfolded proteins (50). Aggresomes form when the cell's capacity to degrade misfolded proteins is exceeded. It has been proposed that misfolded proteins become ubiquitinylated and then form aggregates on microtubules that are transported in a retrograde fashion to the proteasome-enriched centrosome. Accumulation of the Mre11 complex at aggresomes in the presence of adenoviral E4orf3 protein may result from a structural change in Mre11 that leads to its destabilization and aggregation. Alternatively, E4orf3 may target a cellular protein required to maintain the integrity of the Mre11 complex, and in its absence the other complex members are destabilized. Our data from nocodazole experiments suggest that small aggregates of E4orf3 and associated proteins join together to form larger single aggresomes via active transport on microtubules. We have also observed that the cytoplasmic structures induced by E4orf3 accumulate proteasomes, although proteasome inhibitors had no effect on aggresome formation by E4orf3 (F. D. Araujo and M. D. Weitzman, unpublished observations).
Cytoplasmic aggresomes have been described for a number of unstable or misfolded proteins (16, 27). Aggresomes also resemble intracellular inclusions observed in diseases such as Parkinson's and polyglutamine disease, as well as amyotrophic lateral sclerosis (39). Whether these aggregates are a cause or consequence of the disease state remains unknown. Although the formation of aggresomes may have evolved as a cellular response designed to handle misfolded proteins, viruses seem to have developed ways to exploit this pathway to aid their growth. There are examples of aggresome-like structures that are involved in virus assembly (21, 33). High levels of expression of Ad2 and Ad5 E1b55K in transformed cells also leads to the formation of aggresomes and sequestration of p53 (1, 51), although it is not clear that these structures are required for transformation (13, 47).
The induction of aggresomes by E4orf3 will provide a powerful system to investigate the mechanism of aggresome formation in a biologically relevant setting. A recent report suggests a role for histone deacetylase 6 in aggresome assembly (24). We have seen no effect of short interfering RNA to histone deacetylase 6 or histone deacetylase inhibitors, such as trichostatin A, on aggresome formation induced by E4orf3 (F. D. Araujo and M. D. Weitzman, unpublished observations). There may be multiple routes to aggresome formation, and therefore it will be interesting to investigate how they accumulate E4orf3 and what determines which associated cellular proteins become targeted to the aggresome.
The products of the E4 region are required for efficient replication and late protein synthesis during adenovirus infection (28). In the absence of E4 proteins, the adenoviral genomic DNA is joined together into concatemers in a process that requires the Mre11 complex (43). The E4orf6 and E4orf3 proteins display redundancy in preventing concatemer formation, and both can target the Mre11 complex. Redistribution of the Mre11 complex by E4orf3 may be required for both stimulating viral replication (14, 15) and preventing concatemers (43, 45).
The mechanisms by which the E4orf3 protein prevents the concatemerization mediated by the Mre11 complex is unclear. One possibility is that E4orf3 induces modifications to one or more members of the Mre11 complex and that this triggers the relocalization into nuclear tracks and cytoplasmic aggresomes. This is supported by our observation that E4orf3 alters the solubility of Mre11 complex members. Infection with an E4-deleted virus also leads to accumulation of the Mre11 complex at sites of viral replication (43) and induction of signal transduction cascades characteristic of the DNA damage response (7). In the presence of E4orf3 the Mre11 complex is excluded from viral replication centers (43, 45) and this correlates with inhibition of signaling (C. Carson and M. D. Weitzman, unpublished observations). The redistribution of the Mre11 complex may prevent it from responding to DNA damage and this may explain its inactivation by E4orf3.
Further analysis of E4orf3 function using proteins from different serotypes (45) together with directed mutagenesis (15) will shed light on how this protein achieves its many roles in aiding viral replication and countering cellular antiviral responses.
Acknowledgments
We are grateful to G. Ketner, A. Levine, V. Malhotra, T. Melendy, J. Petrini, and P. van der Vliet for generous gifts of reagents. We thank David Ornelles and members of the Weitzman lab for discussions and critical reading of the manuscript. We acknowledge the James B. Pendleton Charitable Trust for providing the Pendleton Microscopy Facility.
This work was supported in part by NIH grants AI43341 and CA97093 (M. D. Weitzman), and by gifts from the Joe W. and Dorothy Dorsett Brown Foundation and the Lebensfeld Foundation. T.H.S. was supported by an NIH Graduate Training Grant to UCSD and by fellowships from the H. A. and Mary K. Chapman Charitable Trust, the Legler Benbough Foundation, and the Salk Institute Association. C.T.C. was supported by the Timken-Sturgis Foundation and by an NIH Training Grant to the Salk Institute.
REFERENCES
- 1.Blair Zajdel, M. E., and G. E. Blair. 1988. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene 2:579-584. [PubMed] [Google Scholar]
- 2.Bosher, J., A. Dawson, and R. T. Hay. 1992. Nuclear factor I is specifically targeted to discrete subnuclear sites in adenovirus type 2-infected cells. J. Virol. 66:3140-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 4.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. R., S. J. Doxsey, E. White, and W. J. Welch. 1994. Both viral (adenovirus E1B) and cellular (hsp 70, p53) components interact with centrosomes. J. Cell Physiol. 160:47-60. [DOI] [PubMed] [Google Scholar]
- 6.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 7.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho, T., J.-S. Seeler, K. Öhman, P. Jordan, U. Pettersson, G. Akusjärvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of Adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai-Mehta, A., K. M. Cerosaletti, and P. Concannon. 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21:2184-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 13.Endter, C., B. Hartl, T. Spruss, J. Hauber, and T. Dobner. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 24:55-64. [DOI] [PubMed] [Google Scholar]
- 14.Evans, J. D., and P. Hearing. 2003. Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Mata, R., Y. S. Gao, and E. Sztul. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3:388-396. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenoviral early region 4 encodes functions required for efficient DNA replication, late expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, M.-H., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., J. J. Kovacs, A. McLaurin, J. M. Vance, A. Ito, and T. P. Yao. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115:727-738. [DOI] [PubMed] [Google Scholar]
- 25.Kirschner, M. 1999. Intracellular proteolysis. Trends Cell Biol. 9:M42-45. [PubMed] [Google Scholar]
- 26.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B 55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 28.Leppard, K. N. 1997. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen. Virol. 78:2131-2138. [DOI] [PubMed] [Google Scholar]
- 29.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 30.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J Gen. Virol. 84:259-268. [DOI] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz, J. 1998. Cytoskeletal proteins and Golgi dynamics. Curr. Opin. Cell Biol. 10:52-59. [DOI] [PubMed] [Google Scholar]
- 32.Maser, R. S., R. Zinkel, and J. H. Petrini. 2001. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 27:417-421. [DOI] [PubMed] [Google Scholar]
- 33.Nozawa, N., Y. Yamauchi, K. Ohtsuka, Y. Kawaguchi, and Y. Nishiyama. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299:486-497. [DOI] [PubMed] [Google Scholar]
- 34.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul, F., J. Pedron, and C. Cajean-Feroldi. 1984. Identification of intranuclear structures containing the 72K DNA-binding protein of human adenovirus type 5. Eur. J. Cell Biol. 34:313-322. [PubMed] [Google Scholar]
- 38.Puvion-Dutilleul, F., and E. Puvion. 1990. Analysis by in situ hybridization and autoradiography of sites of replication and storage of single- and double-stranded adenovirus type 5 DNA in lytically infected HeLa cells. J. Struct. Biol 103:280-289. [DOI] [PubMed] [Google Scholar]
- 39.Ross, C. A., and M. A. Poirier. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl.):S10-17. [DOI] [PubMed] [Google Scholar]
- 40.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 41.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 78:9924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577-587. [DOI] [PubMed] [Google Scholar]
- 43.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 44.Stracker, T. H., G. D. Cassell, P. Ward, Y. M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzman. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1-23. [DOI] [PubMed] [Google Scholar]
- 47.van den Heuvel, S. J., T. van Laar, W. M. Kast, C. J. Melief, A. Zantema, and A. J. van der Eb. 1990. Association between the cellular p53 and the adenovirus 5 E1B-55kd proteins reduces the oncogenicity of Ad-transformed cells. EMBO J. 9:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 51.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]