Abstract
Herpes simplex virus (HSV) infects both epithelial cells and neuronal cells of the human host. Although HSV assembly has been studied extensively for cultured epithelial and neuronal cells, cultured neurons are biochemically, physiologically, and anatomically significantly different than mature neurons in vivo. Therefore, it is imperative that viral maturation and assembly be studied in vivo. To study viral assembly in vivo, we inoculated wild-type and replication-defective viruses into the posterior chamber of mouse eyes and followed infection in retinal ganglion cell bodies and axons. We used PCR techniques to detect viral DNA and RNA and electron microscopy immunohistochemistry and Western blotting to detect viral proteins in specific portions of the optic tract. This approach has shown that viral DNA replication is necessary for viral DNA movement into axons. Movement of viral DNA along ganglion cell axons occurs within capsid-like structures at the speed of fast axonal transport. These studies show that the combined use of intravitreal injections of replication-defective viruses and molecular probes allows the genetic analysis of essential viral replication and maturation processes in neurons in vivo. The studies also provide novel direct evidence for the axonal transport of viral DNA and support for the subassembly hypothesis of viral maturation in situ.
Herpes simplex virus 1 (HSV-1) infects mucous membranes at sites of entry into the host. After infection of the epithelium, the virus spreads to other cells in the epithelium and to the endings of sensory neurons, where it is taken up. The nucleocapsid of the virus moves by retrograde transport within the peripheral axons to the nuclear compartment of the sensory cell (for reviews, see references 32 and 36). Upon replication in the neuronal cell body, new components of the virion move by anterograde transport from the neuron cell body to both peripheral and central branches of the neuron (12). Envelope components, as marked by envelope-specific proteins, are transported independently of capsid protein components, as marked by capsid-specific proteins (11, 27, 29). However, little is known about the exact mechanisms of viral DNA transport or release from the axon. It has been assumed that viral DNA, which is contained in reassembled envelope and capsid components, must be released along the axon shaft and at axon terminals, because the glial cells which envelop the infected axons also become infected with HSV (23, 39).
Previous models of HSV DNA transport are inferences based on indirect evidence. First, morphological identification of capsids has been based on the presence of a dense core within a small particle. However, uninfected sensory axons contain granular vesicles that have dense cores and that are approximately the same diameter (75 to 95 nm) and morphologically similar to viral nucleocapsids. Second, staining of small vesicles (∼100 nm wide) within axons with a monoclonal antibody to VP5, the major capsid protein, suggests that capsids are transported (19). However, not all capsids contain DNA. Some capsids, such as B capsids, may contain tegument proteins, as well as histones and other immunoantigens, without DNA (21, 37). Our understanding of viral-DNA transport kinetics and identification of the morphological compartment(s) in which it is transported have been hampered by the lack of an adequate in vivo system for study.
Although HSV assembly has been studied extensively in cultured cells and even more relevantly in cultured neurons, there are significant differences between cultured neurons and mature neurons in situ. For example, mature uninjured vertebrate axons in vivo have few ribosomes and few of the other factors necessary to produce proteins locally (5). In contrast, embryonic neurites in vitro contain the cell machinery necessary for synthesis of new proteins (13). Theoretically, viral protein synthesis might also occur in situ within the neurites. Therefore, to fully understand HSV infection in mature neurons, in vivo studies are necessary.
To understand viral pathogenesis in neurons in vivo, our strategy has been to use viral strains with known genetic mutations and defects in essential viral functions. However, the analysis of viral functions may be complicated because the same functional defects may also prevent the spread of the virus to the cell types of interest, e.g., neurons. For example, replication-defective mutant viruses show little spread between cells or to sensory neurons, although nonessential functions can be studied in vivo (16, 32, 39). Thus, while replication-defective mutant viruses are important tools for the study of viral gene function in culture, they have had fewer applications in vivo.
Recently, we developed a model of murine retinal ganglion cell infection by directly injecting virus into the posterior chamber of the eye, thereby delivering virus to the retinal ganglion cells and producing an infection (11). Current studies use this system to examine the infection of neurons in situ by replication-competent and replication-defective mutant HSV strains. We tested whether injected virus was taken up by ganglion cells and transported anterograde by a transcytotic mechanism. Second, we also described the viral structures that are involved in the axonal spread in viral DNA of an HSV infection in vivo. Last, we used electron microscopy (EM) immunohistochemistry, PCR, and EM in situ hybridization to document the following aspects of viral maturation and egress: (i) the rate of transport of viral DNA in the axon and (ii) the identity of the structure within which viral DNA is transported.
MATERIALS AND METHODS
Antibodies and culture supplies.
Monoclonal antibody (gD-1D3) specific for HSV-1 glycoprotein D (gD) was provided by Gary Cohen and Roselyn Eisenberg, University of Pennsylvania, Philadelphia. An additional monoclonal antibody specific for HSV-1 gD (MAB8684) was purchased from Chemicon International, Temecula, CA. Two monoclonal antibodies specific for the HSV major capsid protein ICP5 were also obtained (no. C050IM [Biodesign International, Saco, ME] and ICP5 [Virusys Corp., North Berwick, ME]). Polyclonal rabbit anti-HSV-horseradish peroxidase (HRP) (no. AXL298P) and rabbit anti-digoxigenin (DIG)-HRP (no. AXL5227P) were from Accurate Chemical and Scientific, Westbury, NY. Sheep anti-mouse immunoglobulin G-HRP (NA-931) was from purchased Amersham Biosciences, United Kingdom. For protein assays, we used the BCA kit and protocol from Pierce (Rockford, IL). All culture supplies were obtained from the Cell Culture Facility, UCSF.
Preparation of viral stocks.
African green monkey kidney (Vero) cells were grown and maintained as described previously (11). Vero cells were infected at a multiplicity of infection of 0.01 with the HSV wild-type (wt) KOS strain. After 48 h, 100% of the cells showed cytopathic effects. Progeny virus was harvested by pooling cells and medium and freeze-thawing three times. Cellular debris was removed by filtration through a 0.45-μm sterile filter (Millipore, Billerica, MA). The filtrate was centrifuged at 25,000 rpm in a Beckman SW28 rotor for 2 h at 4°C through a step gradient of 10%, 30%, and 60% sucrose. Virus was collected from the 30% to 60% interface, suspended in phosphate-buffered saline (PBS), and collected by centrifugation for 1 h at 4°C. The pellet was suspended in minimal essential medium without serum and stored at −80°C. The viral titer was determined by a standard plaque assay.
Two other HSV preparations were used in this study. We exposed aliquots of the wt strain to UV light at a 254-nm wavelength to inactivate viral infectivity (33). After 20 min of exposure, the titer dropped 1,000-fold. We used this UV-treated KOS (KOS-UV) as a control for virus that is replication competent. The HSV-1 KOS UL29 gene mutant, 8lacZ, was constructed by insertion of an ICP8-lacZ fusion cassette into the ICP8/UL29 gene, as described previously for HSV-1 KOS1.1 (4). This viral strain is capable of expressing immediate early, early, and some late viral gene products but is replication defective in normal cells.
Intraocular injection.
All procedures involving animals adhered to the Declaration of Helsinki and the Guiding Principles in the Care and Use of Animals and to the guidelines of the UCSF Committee on Animal Research. Eyes of male BALB/c mice (5 to 6 weeks of age) were infected with equivalent titers of virus in 4 μl of sterile minimal essential medium without serum at concentrations of approximately 9 × 104 PFU/μl, according to a standard procedure (11). We introduced valacyclovir hydrochloride (1 mg/ml) (Valtrex; Glaxo Wellcome, Inc., Greenville, NC) into the drinking water 24 h after ocular infection to block further replication (11). This infection protocol allows a restricted time period of viral replication in retinal ganglion cells and interferes with replication in astrocytes in the optic nerve (11).
Retinal mRNA analysis.
Total RNA was purified from infected mouse retinas by using the Trizol reagent and protocol (Invitrogen Corp., Carlsbad, CA). The integrity of the mRNA was verified using primers to mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The RNA was reverse transcribed into cDNA by using random hexamer primers according to the Invitrogen Superscript first-strand synthesis system for reverse transcription (RT). The resulting cDNA from the RT reaction was tested for the presence of Us8 (gE) by using gE gene-specific primers in a PCR (6). The gE forward primer was 5′-AGCTCCCAGAATGTCTGTCC-3′, and the reverse primer was 5′-GATACAGGCCGGAGTGTTGT-3′.
Tissue isolation.
At 2, 3, or 5 days postinfection (dpi), the animals were anesthetized, and each mouse was perfused intracardially with 20 ml of saline. The optic nerves, extending from the orbit to the optic chiasm (OC), were removed by following a standard protocol (11). In brief, the retinal ganglion cell axons were dissected into four segments. The portion extending from the orbit to the OC was bisected into two segments, optic nerve 1 (ON1, 3 mm) and optic nerve 2 (ON2, 3 mm). The OC (2 mm) was dissected from the hypothalamus, and the optic tract (OT, 5 mm) was removed to the point where the axons enter the dorsal thalamus. As a negative control, uninfected mice were prepared similarly. The same experiment was repeated three to four times for each time point. To control for possible vascular transfer of virus, we took samples of cerebellum from infected mice; we failed to find any viral DNA in these control samples. Aliquots of viral stocks were used as positive controls.
Polyacrylamide gel electrophoresis and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with antibodies to VP5, the major capsid protein, and to gD, one of the major viral envelope proteins, were performed as described previously (11). Equivalent amounts of protein were loaded for each lane. When we omitted the primary monoclonal antibody in Western blotting and incubated in the secondary antibody only, i.e., sheep anti-mouse antiserum, we consistently found a band at ∼55 kDa in all lanes. This represents the nonspecific binding of the second antibody to the heavy chain of the immunoglobulin G reacting to the mouse. When we omitted the second antibody, we saw no bands on the immunoblots.
DNA preparation and PCR procedures.
We pooled tissue from 10 eyes to normalize the amount of viral DNA for each treatment condition and time point. Genomic DNA from the nerve specimens was isolated as previously described (25). The resulting pellet was suspended in Tris-EDTA, and the DNA concentration was determined by A260 measurement. One hundred nanograms of DNA was used for the PCR analysis. To determine the limit of detection, we serially diluted 10-fold the PCR product made using the gD primers, starting at 100 ng. The lower limit of detection was ∼1 fg of DNA, which is the equivalent of 3,400 copies of DNA.
For PCR analysis, forward and reverse primers targeting the Us6 (gD) sequence of HSV-1 were used. Primer sequences are 5′-ATGGGAGGCAACTGTGCTAT-3′ for the gD forward primer and 5′-CTCGGTGCTCCAGGATAAAC-3′ for the gD reverse primer. These primers yield a gD-specific PCR product of 250 bp. Twenty-microliter PCR mixes contained the following: 100 ng DNA, 1× QIAGEN PCR buffer (final MgCl concentration, 1.5 mM), 0.15 μm gD forward primer, 0.15 μm gD reverse primer, 5% dimethyl sulfoxide, 0.2 mM deoxynucleoside triphosphates, and 0.5 U Hot Star polymerase (QIAGEN, Valencia, CA). PCR mixes were subjected to thermal cycling under the following conditions: 15 min at 94°C and 25 cycles of 94°C for 30 s/52°C for 30 s/72°C for 1 min. A final extension reaction was carried out at 72°C for 6 min. The PCRs were run on a 1% agarose gel and stained with ethidium bromide according to standard procedures (31). Aliquots of wt and ICP8− stocks of HSV were used as positive controls. Relative estimates of the amounts of viral DNA were obtained by using NIH ImageJ to quantify the density of bands in the films.
EM immunohistochemistry.
The mice were anesthetized, perfused, and dissected as previously described (11). The segments of the optic path were treated for EM immunohistochemistry by using an HRP-tagged polyclonal antiserum against human HSV according to standard procedures (22). In some experiments, the segments were treated with a monoclonal antibody to ICP5 (Virusys Corp.) at a dilution of 1:100 overnight, followed by use of a mouse-to-mouse detection kit (no. 2700; Chemicon), according to the instructions of the manufacturer. Thin sections for EM were not counterstained with uranyl acetate or lead citrate. When the primary antibody was omitted, we found no immunostained structures in the retinal ganglion cells or glial endfeet (data not shown).
Production of in situ probe.
Primers for Us6, as described above, were used to prepare the PCR product for DIG labeling. Approximately 4 × 105 PFU per reaction of wt virus was used as template DNA in 50-μl reaction volumes containing 1× Hot Star (QIAGEN) buffer, 5% dimethyl sulfoxide, 2 mM deoxynucleoside triphosphates, 0.15 μM of each primer, and 1.0 U Hot Star Taq DNA polymerase. Cycling conditions were as follows: 15 min at 94°C and 40 cycles of 94°C for 30 s/52°C for 30 s/72°C for 1 min. A final extension reaction was carried out at 72°C for 6 min. Ten of these PCRs were pooled and purified using a QIAquick PCR purification kit (QIAGEN). The final volume was 10% of the original pooled volume. Quantitation was estimated by running 1 μl of the purified PCR product on a 1% agarose gel with subsequent ethidium bromide staining. The intensity of the band was compared to that of the stained standard DNA. The integrity of the PCR product was verified by sequencing analysis (Biomolecular Resource Center, UCSF). An aliquot (300 to 600 ng) of each PCR product was labeled with DIG-dUTP by using a DIG DNA labeling kit from Roche and following the manufacturer's protocol. The labeled probe was quantitated using a DIG nucleic acid detection kit from Roche.
EM in situ hybridization.
Five days after inoculation, mice were anesthetized and perfused according to standard procedures (11). The tissues were fixed for 24 h to 3 weeks at 4°C, and the optic nerves and tracts were dissected and cut manually into thin slices. We modified the procedure described by Hayat (7) as follows. The tissues were rinsed three times for over 30 min in PBS, incubated in 10 mM sodium citrate (pH 6.0), and then heated to boiling for 1 min. The sections were incubated in 1.0 M sodium thiocyanate for 10 min at 80°C and then washed two times for 5 min each in distilled water. The sections were incubated at 78°C for 10 min to denature the DNA and then incubated overnight at 37°C in the hybridization buffer. This buffer was composed of the following: deionized formamide (65%), 2× SSC buffer (0.3 M sodium chloride and 0.03 M sodium citrate), 10% dextran, salmon sperm DNA (1 μg/μl), and DIG-labeled probe (40 μl at 20 ng/ml).
The tissues were rinsed two times for 10 min each in 2× SSC buffer with 50% formamide at 40°C and rinsed three times for 5 min each in PBS. To block endogenous peroxidases, the tissues were rinsed in 0.3% hydrogen peroxide for 15 min and rinsed in PBS. To block nonspecific binding, the samples were incubated in 4% normal rabbit serum in PBS for 1 h at room temperature. After a rinse in PBS, the tissue was incubated overnight at room temperature in HRP-tagged rabbit anti-DIG antiserum. The tissue was washed three times for 5 min each in PBS and processed for EM immunohistochemistry according to standard procedure (22). Thin sections were not counterstained. Positive controls included optic tracts from wt virus-infected animals that were not treated with valacyclovir. We found labeled viral DNA in capsids in astrocyte nuclei throughout the optic pathway (data not shown). Negative controls included tissue from uninfected animals, tissue from infected animals in which the primary antibody was omitted, and tissue from infected animals in which the DIG probe was omitted. We found no in situ staining in any of the negative controls.
RESULTS
We wished to determine the role of viral-DNA replication in viral assembly and axonal transport in murine visual system neurons in situ following injection of the HSV strains into the posterior chamber of the eye. Our first step was to characterize the infection model and the assays used to detect viral DNA, RNA, and proteins.
Characterization of retinal infections.
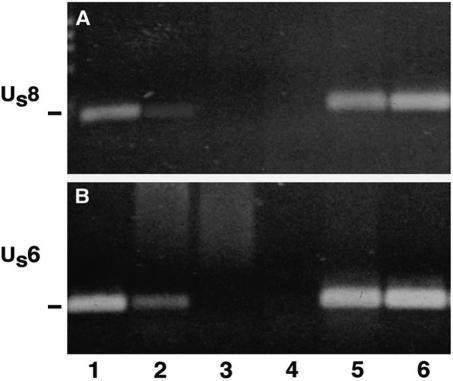
In these studies, we used PCR to measure viral-DNA movement in axons in vivo. We first determined that the assay was sensitive enough to detect input viral genomes. We infected mice with wt or ICP8− virus and, 24 h later, euthanized the animals. The posterior eye was dissected and DNA was isolated. To determine if we could detect viral genomes from the ICP8− mutant virus in the retina, we used primers for gE and gD gene sequences for PCR amplification. We detected both DNA sequences in wt virus-infected eyes and, to a lesser degree, in ICP8− virus-infected eyes (Fig. 1A and B, lanes 1 and 2). Based on densitometric scans of the gel, about sixfold more of the Us8 gene sequence and about two- to fivefold more of the Us6 sequence were detected after infection with wt virus than after infection with the ICP8−, as a result of the impaired DNA replication of this strain. No DNA was detected in uninfected eyes or in the cocktail-injected eyes. Thus, our PCR could detect input viral genomes.
FIG. 1.
(A and B) Viral DNA was detected in retinal extracts at 24 h postinfection. Bands migrating at approximately 219 and 250 bp indicate the presence of Us8 for gE (A) and Us6 for gD (B) DNA, respectively. We detected DNA fragments in wt-infected (lane 1) and ICP8−-infected (lane 2) retinas. There was no viral DNA detected in uninfected eyes (lane 3) or in eyes injected with cocktail alone (lane 4). Positive controls included DNA from the wt (lane 5) and ICP8− (lane 6) viral stocks.
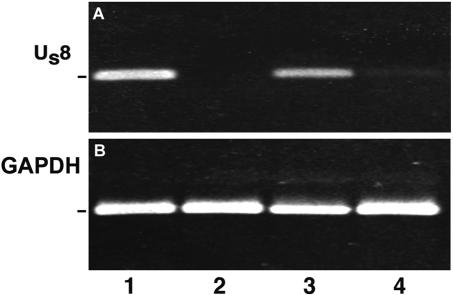
To further test that the viruses introduced into the eye were capable of infecting retinal cells, we infected mice with the wt, KOS-UV, or ICP8− mutant strain and euthanized the mice at 24 h postinfection. The posterior portions of the eyes were dissected, retinal RNA was purified and subjected to RT, and the resulting product was amplified by PCR with primers for Us8 (gE), a γ-1 or early/late viral gene (30). We measured the expression of a γ-1 or leaky late gene, because γ-1 gD was one of the proteins assayed for transport in axons. Both wt virus-infected and ICP8− virus-infected retinas expressed gE mRNA (Fig. 2, lanes 1 and 3). Neither KOS-UV nor control (uninfected) retinas expressed gE mRNA (Fig. 2, lanes 2 and 4). GAPDH mRNA was assayed as a positive control. These results indicated that wt and ICP8− strains were capable of infecting retinal tissue by 24 h after injection.
FIG. 2.
(A and B) Viral mRNA was detected in retinal extracts at 24 h postinfection. mRNA of Us8 (gE) was found in retinas in the wt (lane 1) or in the ICP8− (lane 3) strain of HSV. There was no expression in retinas infected with the KOS-UV-treated strain (lane 2) or in cerebellar tissue from mice infected with the wt viral strain (lane 4). (B) The GAPDH mRNA internal control was positive.
To define the cellular localizations of the wt and ICP8− viruses in retinal ganglion cells, we examined retinal tissues after EM immunohistochemical staining. Retinal ganglion cells are the sole source of axons to the optic nerve. Although potential amplification of virus might also occur in non-ganglion cells, these cells cannot contribute to viral transport into the optic pathway, because they do not have processes that extend beyond the retina. Thus, we focused on infection in retinal ganglion cell bodies.
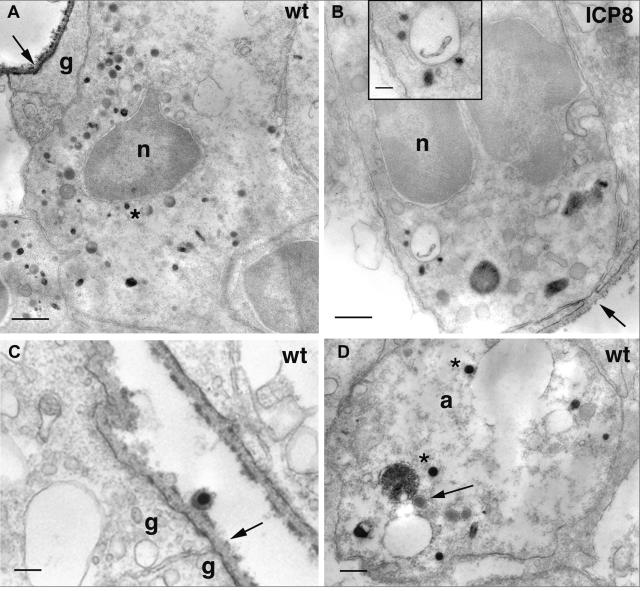
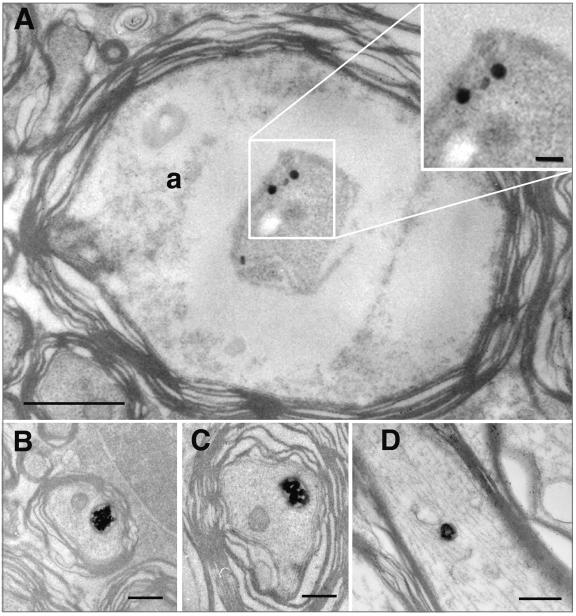
At 24 h postinfection with wt virus, we found evidence of HSV antigens at the vitreal surface and on the surface of Müller glial cell endfeet that are extensions of the cytoplasm of the Müller cells, the major glial cell type of the retina (Fig. 3A and C). In retinal ganglion cell bodies, we found a variety of immunopositive vesicles and nucleocapsids, including one that was located adjacent to the nucleus (Fig. 3A). The most proximal portion of ganglion cell axons in the ganglion cell layer of the retina also contained a variety of immunopositive organelles, including some that were about 110 nm in diameter (similar in size to capsids) and some that were significantly larger (Fig. 3D). Dense cored vesicles about 110 nm in diameter that were not immunopositive were also noted in the axon (Fig. 3D). At 24 h after infection with the ICP8− mutant virus, we found less intense immunoreactive product coating the inner limiting membrane of the retina and the surface of Müller glial cell endfeet (Fig. 3B). We were unable to identify any infected retinal ganglion cell bodies after injection with KOS-UV virus.
FIG. 3.
A variety of particles were immunopositive in retinal ganglion cell bodies 24 h after infection with the wt (A, C, and D) or the ICP8− (B) strain of HSV. (A) HSV immunostaining coats the inner limiting membrane of the retina (arrow) and the surface of Müller glial endfeet. Various immunopositive particles occupy the ganglion cell cytoplasm. One immunopositive particle (asterisk) is located in close proximity to the nucleus. Bar = 600 nm. (B) Labeled particles are also found in the cytoplasm of ganglion cells infected with the ICP8− virus. A magnified image is shown in the inset. The inner limiting membrane is interrupted in some sites (arrow). This could allow access of virus to the neuronal cell surface and eliminate the usual requirement that the virus pass through the glial cell before reaching the neuron. Bars = 300 nm (B) and 100 nm (inset). (C) A virion lies adjacent to the Müller glial endfeet of the inner limiting membrane. The endfeet are sealed at a junctional complex (arrow). Bar = 200 nm. (D) Capsids (asterisks) are clustered in a ganglion cell axon in the optic fiber layer of the retina. An unlabeled, dense core vesicle is also present (arrow). Bar = 200 nm. a, axon; g, glial endfoot; n, nucleus.
Replication is necessary for viral transport.
We tested whether injected virus was taken up by ganglion cells and transported anterograde by a transcytotic mechanism. This process is defined as delivery of a protein or complex from one pole of a cell to the other pole of the cell, via vesicle-mediated, fast axonal transport, and by this mechanism, no DNA replication would be necessary. Thus, if a replication-defective HSV is anterograde transported, then transcytosis could be a mechanism for viral targeting to the axon.
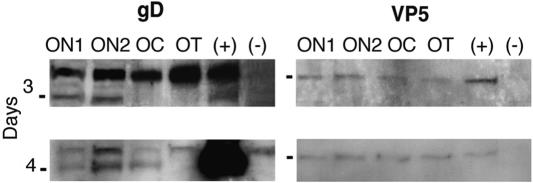
After retinal ganglion cell infection, we used Western blots and antibodies to VP5 and gD, markers of the capsid and envelope subassemblies, respectively, to determine that the viral proteins were synthesized and transported in the ganglion cell axons. By 3 dpi with wt virus, we found gD in the ON1 and ON2 segments (Fig. 4). We detected VP5 in ON1, ON2, and the OC in two of three experiments at 3 dpi; in a third experiment, we identified VP5 as far as the OT (Fig. 4). By 4 dpi, we detected gD in ON1, ON2, and the OC and VP5 throughout the pathway. Thus, we found that the transport of gD was somewhat slower than that of VP5. This finding differs from the result we obtained earlier, in which we found that gD was detected in the axon of retinal ganglion cells infected with the F strain of HSV at least a day earlier than the VP5 protein could be detected (11). In the current study, we used the KOS strain, the background strain of the ICP8− mutant strain. When we injected ICP8− or KOS-UV virus, we found no VP5 or gD proteins in Western blots of optic pathways from animals at 3, 4, or 5 dpi (data not shown). These results indicate that viral replication is a necessary step for the expression, targeting, or axonal transport of viral proteins.
FIG. 4.
The axonal transport of the wt proteins was assayed by Western blotting of segments of the optic pathway. The envelope protein, gD (left panel), was detected in the ON1 and ON2 segments of the optic pathway at 3 dpi. The capsid protein, VP5 (right panel), was detected throughout the pathway by 3 dpi. gD was detected in ON, ON2, and the OC by 4 dpi. VP5 persisted throughout the pathway. (+), viral stock aliquots; (−), optic pathway homogenates from uninfected animal. Markers = 50 kDa (left panels) and 155 kDa (right panels).
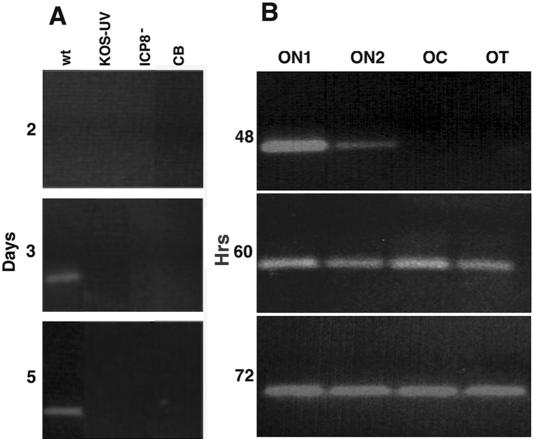
We used PCR to measure the viral-DNA content in each section of the optic pathway, to assay the rate of DNA transport, and to test whether viral replication was necessary for DNA delivery to the mouse optic tract. In contrast to the appearance of viral DNA in the ICP8−-infected retinas (Fig. 1), no viral DNA was detected in the optic tracts at any time point. No evidence of a PCR product for the Us6 gene (gD) was found in the optic tracts of either ICP8−-infected or KOS-UV-infected mice at 2, 3, or 5 dpi (Fig. 5A). However, a PCR product was found in the optic tracts at 3 and 5 dpi with wt HSV. There was no evidence of viral DNA in tissue from the cerebellum, which was assayed as a control for possible systemic or viremic delivery. These findings confirm that replication of new virus is essential for the transport of viral DNA to the axon and that transcytosis of DNA from the original injected viral stock does not play a role in anterograde transport.
FIG. 5.
The axonal transport of viral DNA was assayed for PCR product primed off of Us6. (A) Comparison of the transport of viral DNA from different viral strains to the OT segment 2, 3, or 5 dpi. Positive bands were found in the OT from animals infected with wt virus at 3 and 5 dpi, but no DNA was found in the tracts of mice 2, 3, or 5 days after ICP8− or KOS-UV infection. The cerebellum (CB) from the wt was uninfected. (B) Translocation over time of wt DNA in axons. Viral DNA was detected in the proximal (ON1 and ON2) optic pathway 48 h after infection. By 60 h, the DNA was detected in all four segments and persisted for an additional 12 h.
Temporal course of transport of wt viral DNA.
By 48 h after infection, viral DNA was detected in the ON1 and ON2 segments of the optic nerve that are closest to the retinal ganglion cell bodies (Fig. 5B). By 60 h, we detected viral DNA in these segments and in the remaining portions of the optic nerve. The viral DNA was concentrated about three- to fivefold greater in ON1 than in ON2 at 48 and 60 h after infection (Fig. 5B). By 72 h, it was more homogenously distributed throughout the pathway. Assuming that the front of DNA transport travels a distance of about 6 mm (from the OC to the length of the OT) in 12 h (from 48 to 60 h postinfection), this result suggested a transport rate of ∼0.14 μm/sec. This rate was consistent with rapid axonal transport.
The rate was inconsistent with spread from glial cell to glial cell, outside of retinal ganglion cell axons (1, 23). There are about 2.4 astrocytes/33-μm length of optic pathway; the length of the pathway from ON1 to the OC is about 6 mm (11, 15). A chain of about 465 astrocytes would be needed to span the distance from the optic nerve head to the beginning of the OC. To reach the beginning of the OC by 48 h, the HSV would have only about 6.2 min/astrocyte to enter, replicate, and spread to the next astrocyte.
EM localization of viral protein and DNA.
In the most proximal portion of ganglion cell axons, we found a variety of organelles containing viral antigens (Fig. 3D) (see above). In the most distal portions of the same axons, i.e., in the optic tracts, we found two sizes of immunostained particles at 5 dpi. One class of particle was about 150 to 180 nm in diameter and was composed of a host membrane surrounding viral antigenic contents (Fig. 6B through D).
FIG. 6.
EM immunohistochemistry of cross-sections of axons in the optic tracts of mice 5 dpi with wt strain. Two types of HSV-positive particles were seen in retinal ganglion cell axons (a). (A) Small (∼110-nm-wide), round, immunostained particles (enlarged in upper inset). Bars = 1 μm (A) and 200 nm (inset). (B to D) Larger membrane-bound organelles surrounded an immunostained substructure. Bars = 400 nm.
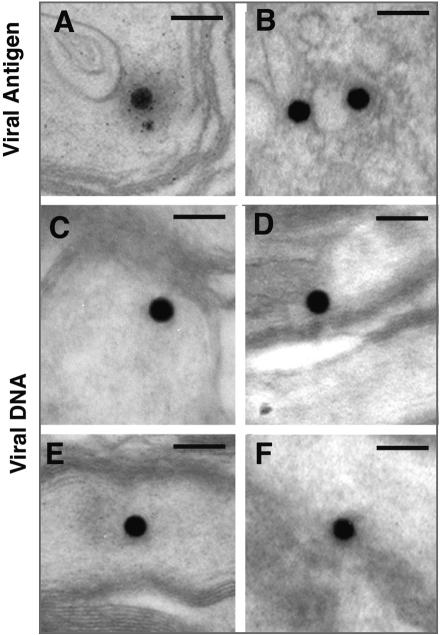
The other class of particles was small (∼110 nm in diameter), capsid-like, and often had a diffuse halo surrounding the particle (Fig. 7A). The capsid-like particles resembled structures immunostained with a monoclonal antibody to VP5 that were found within the nuclei of infected astrocytes distributed along the OT in tissues from animals not given valacyclovir (Fig. 7B). Localization of viral DNA within the capsid-like structures was confirmed by examination of sections of optic axons from infected mice that had been hybridized with a DIG-labeled probe for the Us6 (gD) gene (Fig. 7C through F). Negative controls included tissue from uninfected mice or tissue from infected mice in which the primary antibody or DIG label was omitted. Similar structures were never observed in the control tissues (data not shown). These results demonstrate directly that the major structure in which viral DNA is axonally transported is a nucleocapsid.
FIG. 7.
Immunohistochemical and DNA in situ labeling of the small, round particles in the optic tracts of mice 5 dpi. (A) Polyclonal antiserum to human HSV labeled a small particle in an axon, similar to that illustrated in Fig. 6A. (B) Similar particles are immunolabeled with a monoclonal antibody to VP5 in the cytoplasm of an infected astrocyte in the OT. (C to F) The DIG-HRP reaction product is concentrated on similar small, round particles in the axons. Bars = 200 nm.
DISCUSSION
Genetic analysis of HSV assembly in situ.
We have used a genetic approach to test if viral-DNA replication is needed for viral DNA to enter the axon in an infected neuron in situ. Because viral-DNA synthesis is absolutely necessary for viral replication, viruses defective for DNA synthesis are unable to spread from cell to cell (32). Thus, we needed to deliver these viruses directly to the host neuron by injection. Although previous studies have used viruses that are defective for nonessential gene products, such as viral glycoproteins that determine neuronal targeting in vivo (38), this is the first attempt to examine the role of an essential viral-replication process in viral assembly in vivo through the use of replication-defective mutants. Despite the presence of viral particles around retinal ganglion cell axons in the optic fiber layer, these replication-defective virions failed to reach the proximal portions of the optic nerve.
Neurons can internalize a variety of macromolecules, including trophic factors, lectins, toxins, and other pathogens. After uptake at the dendritic or cell soma membrane, these macromolecules can be moved into and down the axon. For example, wheat germ agglutinin is transcytosed by chick retinal ganglion cells (10). Subsequent release after anterograde transport results in transcytosis across the neuron and release at the synapse (for a review, see reference 40). It has not been clear whether viruses can be similarly transcytosed across a neuron or whether transneuronal transfer occurs by the replication of virus in each cell body in a synaptic chain. Our studies demonstrate that viral-DNA replication must occur for HSV to move to and be transported anterograde down the length of the axon in detectable amounts. Virus that is incapable of replication, as a result of either UV irradiation or genetic mutation, fails to be delivered to the axon compartment.
Several explanations might account for why mutant virus is not detected in the optic pathway. The concentration of virus that reaches the axon compartment may be too low to be detectable. The cellular-transport machinery of an infected neuron may not be able to recognize nucleocapsids that enter the retinal ganglion cell. Alternatively, the host machinery may be able to recognize nucleocapsids, but the nucleocapsid itself may lack the necessary recognition signals. Whatever the failure of recognition, transcytosis of virions does not play a significant role in the delivery of HSV DNA to the axon.
The recognition mechanism that allows encapsulated viral DNA to be sorted to the axon compartment remains unknown. New nucleocapsids are associated with a complement of tegument proteins which may provide some sort of signal for recognition between the viral particle and the motor-associated transport proteins (see below) (for a review, see reference 18). Diefenbach et al. have suggested that conventional kinesin heavy chain and the US11 tegument protein are associated during capsid movement (3).
Rate of movement of viral DNA in the axon in situ.
We and others have previously tried to identify particles found at the EM level in infected axons as enveloped virons (2, 9, 12, 23). We have faced the difficulty of distinguishing between dense core vesicles which are characteristic of sensory and autonomic neurons and viral particles (28). Without the advantage of immunostaining, this is problematic; the vesicles and particles are about the same size, and both contain an electron dense core. EM immunostaining with antibodies to viral proteins and DNA probes resolve the difficulty; viral particles are stained, and dense core vesicles are not.
In this study, we also estimated the rate of transport of HSV DNA in mature, infected central nervous system neurons in vivo. Specifically, we found HSV DNA in the most distal axonal segment (the OT) by 2.5 dpi. Viral capsid protein VP5 was detected in this distal optic segment at 3 to 4 dpi. This difference is likely due to the difference in sensitivities of PCR for viral DNA and Western blotting for viral protein, although other explanations are possible.
An estimate of the rate of axonal transport of the viral DNA can be made if one makes several assumptions. First, we assume that the viral DNA is transported at about an equivalent rate in all infected axons. Potential variability is reduced but not eliminated by valacyclovir treatment, which limits the period of infection to the first 24 h. Second, we assume that the average length of the nerve between the OC to the point where the axon reaches the lateral geniculate nucleus of the thalamus is about 6 mm. Third, we assume that the appearance of DNA in a segment is not limited to the most proximal limit or the most distal limit of that segment. Given these assumptions, we found that DNA is transported about 6 mm in 12 h (between 48 and 60 h postinfection). This suggests that the viral DNA moves at a rate of about 0.5 mm/hr or 0.14 μm/sec.
This estimated rate is less than 10% of the rate estimated for instantaneous axonal transport of viral capsids in embryonic neurons in vitro, which is about 1.97 μm/sec and with an average run length of about 13 μm (34). Several factors can account for this difference between in vivo and in vitro estimates. First, the in vitro estimates are done for single particles moving small distances interrupted by pauses. The pauses between movements were not included in the rate of movement. Our in vivo measurements are done for populations of particles moving over significantly longer distances, and most measurements certainly include the time of delays between saltatory jumps and temporary reversals in direction. Second, the difference in rate in vivo may relate to the steric interference of moving DNA (in capsid) with other organelles in axons of larger caliber. Last, the composition of axoplasm in mature axons and the stability of microtubules in mature axons are known to be different than those of relatively immature neurites (24).
Structures containing viral DNA in the OT.
By 5 dpi, we found viral proteins localized in at least two types of particles in the axons of the OT: small, round 110-nm particles and larger 150- to 180-nm particles. The smaller, round particles were immunopositive with a polyclonal antiserum to HSV and positive for viral DNA by EM in situ hybridization. The capsid identification was further supported by the similarities in size and VP5 immunostaining of capsids in infected astrocytes in the OT. Simultaneous labeling for identification at the EM level of specific viral proteins and DNA is currently under investigation.
In some cases, the capsids were surrounded by a halo of diffuse, electron-dense material, presumably tegument proteins. Several tegument proteins have been found to associate with newly synthesized capsids of HSV and pseudorabies virus (8, 14, 18, 20, 35). The proteins may also be associated with host cell motor adaptor molecules to allow the particle to move anterograde in the cytoplasm (3).
The other class of particles was larger, and the contents were immunopositive for HSV. The surrounding membrane did not react with the HSV antiserum. The identity of these organelles remains to be determined, and the use of monoclonal antibodies should clarify the particular viral proteins that are intrinsic to the particles. One possibility is that the structures are late endosomal organelles that have picked up viral proteins at the cell surface and are in the process of retrograde transport from the axon surface back toward the cell body (26). However, since we see no immunolabel on the axon surface, this seems unlikely.
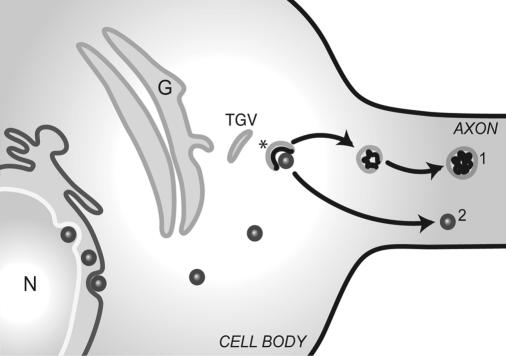
Alternatively, they may contain newly synthesized viral envelope components. Mettenleiter has proposed a model in which trans-Golgi vesicles partially envelop nucleocapsids in the cytosol (17). The glycoproteins within the trans-Golgi vesicle may be segregated to one pole of the vesicle by interaction with proteins of the tegument that surround the nucleocapsid. The portion of the vesicle that contains viral glycoproteins invaginates into the lumen, resulting in a double-membrane vesicle: the host cell membrane surrounding the viral envelope membrane (Fig. 8). The capsid is transported independently of the vesicle (Fig. 8). The union of the capsid and envelope components in the axon may occur by budding of capsid, DNA, and tegument into the larger immunopositive organelle that we have found (39). However, viral egress from an axon may depend on more than one mechanism, and our sample is small.
FIG. 8.
Model of derivation and axonal transport of two classes of particles. In the neuron cell body, a membrane vesicle derived from the trans-Golgi network and containing envelope proteins partially enfolds a maturing capsid and associated tegument proteins (asterisk). In the cell body-proximal axon, the capsid is separated from the vesicle (denoted by the number 2). The capsid containing the DNA is transported independently of the host cell vesicle containing envelope components (denoted by the number 1). N, nucleus; G, Golgi cisternae; TGV, trans-Golgi vesicle.
In summary, after the entry of HSV into neurons and the delivery of viral DNA to the neuronal nucleus, new viral components and DNA must be produced before the viral DNA moves anterograde from the cell soma to nerve terminals in quantities we can detect. A critical question has been the identity of the package in which the DNA is transported within the axon. Using EM immunohistochemistry, we identified two types of HSV-immunopositive organelles in the OT by 5 dpi: small, round organelles and larger, membrane-bound vesicles. Both contain viral proteins, as evidenced from the immunostaining. Using EM in situ hybridization, we provided new evidence that the smaller, ∼100-nm capsid-like structures contain viral DNA. These results provide the first definitive evidence for viral-DNA transport in the capsids. They provide support for the subassembly model of viral maturation in situ and show that movement of viral DNA into the axon requires new synthesis of viral DNA.
Acknowledgments
This work was supported by PHS grants P01 NS35138 (D.K.), P01 HL-24136, EY-08773, and P30 EY02162 (J.L.) and by funds from That Man May See, Inc., and Fight for Sight.
We thank Gary Cohen and Roselyn Eisenberg for gifts of monoclonal antibodies, Peter Ohara for discussion, and Suling Wang for graphic assistance.
REFERENCES
- 1.Brown, A. 2003. Axonal transport of membranous and nonmembranous cargoes: a unified perspective. J. Cell Biol. 160:817-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Card, J. P., L. Rinaman, R. B. Lynn, B.-H. Lee, R. P. Meade, R. R. Miselis, and L. W. Enquist. 1993. Pseudorabies virus infection of the rat central nervous system: ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci. 13:2515-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao, M., and D. M. Knipe. 1989. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J. Virol. 63:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giuditta, A., B. B. Kaplan, J. van Minnen, J. Alvarez, and E. Koenig. 2002. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 25:400-404. [DOI] [PubMed] [Google Scholar]
- 6.Harrabi, O., A. N. Tauscher, and J. H. LaVail. 2004. Temporal expression of herpes simplex virus type 1 mRNA in murine retina. Curr. Eye Res. 29:191-194. [DOI] [PubMed] [Google Scholar]
- 7.Hayat, M. A. 2002. Microscopy, immunohistochemistry and antigen retrieval methods for light and electron microscopy, p. 218-219. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 8.Holland, D. J., M. Miranda-Saksena, R. A. Boadle, P. Armati, and A. L. Cunningham. 1999. Anterograde transport of herpes simplex virus proteins in axons of peripheral human fetal neurons: an immunoelectron microscopy study. J. Virol. 73:8503-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensson, K., B. Ghetti, and H. Wisniewski. 1974. Study on the propagation of herpes simplex virus (type 2) into the brain after intraocular inoculation. Brain Res. 69:189-201. [DOI] [PubMed] [Google Scholar]
- 10.LaVail, J. H., I. K. Sugino, and D. M. McDonald. 1983. Localization of axonally transported 125I-wheat germ agglutinin beneath the plasma membrane of chick retinal ganglion cells. J. Cell Biol. 96:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaVail, J. H., A. N. Tauscher, E. Aghaian, O. Harrabi, and S. S. Sidhu. 2003. Axonal transport and sorting of herpes simplex virus components in mature mouse visual system. J. Virol. 77:6117-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaVail, J. H., K. S. Topp, P. A. Giblin, and J. A. Garner. 1997. Factors that contribute to the transneuronal spread of herpes simplex virus. J. Neurosci. Res. 49:485-496. [PubMed] [Google Scholar]
- 13.Lee, S.-K., and P. Hollenbeck. 2003. Organization and translation of mRNA in sympathetic axons. J. Cell Sci. 116:4467-4478. [DOI] [PubMed] [Google Scholar]
- 14.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabuchi, F., M. Aihara, M. R. Mackey, J. D. Lindsey, and R. N. Weinreb. 2003. Optic nerve damage in experimental mouse ocular hypertension. Investig. Ophthalmol. Vis. Sci. 44:4321-4330. [DOI] [PubMed] [Google Scholar]
- 16.Meignier, B., R. Longnecker, P. Mavromara-Nazos, A. Sears, and B. Roizman. 1988. Virulence and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251-254. [DOI] [PubMed] [Google Scholar]
- 17.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 18.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, D. R. Thompsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 22.Ohara, P. T., M. S. Chin, and J. H. LaVail. 2000. The spread of herpes simplex virus type 1 from trigeminal neurons to murine cornea: an immunoelectron microscopy study. J. Virol. 74:4776-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohara, P. T., A. N. Tauscher, and J. H. LaVail. 2001. Two paths for dissemination of herpes simplex virus from infected trigeminal ganglion to the murine cornea. Brain Res. 899:260-263. [DOI] [PubMed] [Google Scholar]
- 24.Owen, R., and P. R. Gordon-Weeks. 2003. Inhibition of glycogen synthase kinase 3beta in sensory neurons in culture alters filopodia dynamics and microtubule distribution in growth cones. Mol. Cell. Neurosci. 23:626-637. [DOI] [PubMed] [Google Scholar]
- 25.Pauli, U., K. Wright, A. van Wijnen, G. Stein, and J. Stein. 1991. DNA footprinting techniques: applications to eukaryotic nuclear proteins, p. 228-249. In J. D. Karam, L. Chao, and G. W. Warr (ed.), Methods in nucleic acids research. CRC Press, Boca Raton, La.
- 26.Pelkmans, L., and A. Helenius. 2002. Endocytosis via caveolae. Traffic 3:311-320. [DOI] [PubMed] [Google Scholar]
- 27.Penfold, M. E., P. Armati, and A. L. Cunningham. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. USA 91:6529-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters, A., S. L. Palay, and H. d. Webster. 1991. The fine structure of the nervous system: neurons and their supporting cells, 3rd ed. Oxford University Press, New York, N.Y.
- 29.Potel, C., K. Kaelin, L. Danglot, A. Triller, C. Vannier, and F. Rozenberg. 2003. Herpes simplex virus type 1 glycoprotein B sorting in hippocampal neurons. J. Gen. Virol. 84:2613-2624. [DOI] [PubMed] [Google Scholar]
- 30.Rajcåni, J., V. Andrea, and R. Ingeborg. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293-310. [DOI] [PubMed] [Google Scholar]
- 31.Revzin, A. 1991. Gel retardation assays for nucleic acid-binding proteins, p. 206-226. In J. D. Karam, L. Chao, and G. W. Warr (ed.), Methods in nucleic acids research. CRC Press, Boca Raton, La.
- 32.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 33.Sarkar, G., and S. S. Sommer. 1991. Parameters affecting susceptibility of PCR contamination to UV inactivation. BioTechniques 10:591-593. [PubMed] [Google Scholar]
- 34.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci USA. 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 37.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 38.Tomishima, M. J., and L. W. Enquist. 2001. A conserved α-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomishima, M. J., and L. W. Enquist. 2002. In vivo egress of an alphaherpesvirus from axons. J. Virol. 76:8310-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Bartheld, C. S. 2003. Axonal transport and neuronal transcytosis of trophic factors, tracers and pathogens. J. Neurobiol. 58:295-314. [DOI] [PubMed] [Google Scholar]