Abstract
The human immunodeficiency virus type 1 (HIV-1) Vpr protein has important functions in advancing HIV pathogenesis via several effects on the host cell. Vpr mediates nuclear import of the preintegration complex, induces host cell apoptosis, and inhibits cell cycle progression at G2, which increases HIV gene expression. Some of Vpr's activities have been well described, but some functions, such as cell cycle arrest, are not yet completely characterized, although components of the ATR DNA damage repair pathway and the Cdc25C and Cdc2 cell cycle control mechanisms clearly play important roles. We investigated the mechanisms underlying Vpr-mediated cell cycle arrest by examining global cellular gene expression profiles in cell lines that inducibly express wild-type and mutant Vpr proteins. We found that Vpr expression is associated with the down-regulation of genes in the MEK2-ERK pathway and with decreased phosphorylation of the MEK2 effector protein ERK. Exogenous provision of excess MEK2 reverses the cell cycle arrest associated with Vpr, confirming the involvement of the MEK2-ERK pathway in Vpr-mediated cell cycle arrest. Vpr therefore appears to arrest the cell cycle at G2/M through two different mechanisms, the ATR mechanism and a newly described MEK2 mechanism. This redundancy suggests that Vpr-mediated cell cycle arrest is important for HIV replication and pathogenesis. Our findings additionally reinforce the idea that HIV can optimize the host cell environment for viral replication.
Human immunodeficiency virus type 1 (HIV-1), like other viruses, has evolved mechanisms that alter the physiology of the host cell during viral replication. These alterations presumably enhance viral replication or blunt the host response to viral infection in ways that advance viral pathogenesis. Vpr, encoded by an HIV accessory gene, has many distinct, profound effects upon host cells. It is a 96-amino-acid 14-kDa virion-associated protein, expressed primarily from a singly spliced Rev-dependent mRNA (4). While not required for HIV replication in cell culture systems, Vpr plays an important role in HIV replication and pathogenesis in vivo (16, 25), and Vpr aids viral replication in primary monocytes/macrophages (5, 8, 36). Some animal studies, however, suggest that simian immunodeficiency virus can at times remain pathogenic while lacking Vpr (14).
Vpr's known activities include nuclear import of the HIV preintegration complex (12, 13, 27, 31), induction of host cell apoptosis (21, 28, 29, 37), and, in some instances, antiapoptotic activity (6, 7, 11). Vpr arrests the cell cycle at the G2 stage (17, 22, 42). This may help increase HIV gene expression (16). Vpr's transcriptional activities may also be mediated through effects of the p300 coactivator (9, 23). However, Vpr's many varied effects on the host suggest that there may be additional, previously unappreciated effects due to Vpr. In vitro studies indicated that cell cycle arrest enables HIV-1 to maximize viral replication and to suppress immune responses by preventing T-cell clonal expansion, facilitating viral persistence. Elucidation of the detailed mechanisms of G2/M cell cycle arrest induced by Vpr should enhance understanding of an important aspect of HIV biology.
Earlier studies demonstrated that Vpr induces G2 arrest by inhibiting activation of Cdc2, which is necessary for the G2-to-M transition (15, 17, 33, 41). In response to Vpr, the Cdc2-specific phosphatase, Cdc25C, is hyperphosphorylated in a pattern consistent with inactivation. Additional evidence suggests that p21 may also play a role (2). More recently, one mechanism responsible for Vpr-mediated cell cycle arrest involving activation of the ATR DNA repair pathway has been characterized in additional detail (34, 46). However, since the regulation of the cell cycle is complex, and since Vpr-mediated cell cycle arrest activity appears to be important for Vpr's actions to advance HIV replication and HIV pathogenesis, we undertook an expression profiling-based screen to identify cellular genes showing altered expression associated with wild-type (wt) Vpr expression and expression of Vpr mutated in key sites related to cell cycle arrest. We constructed a series of mammalian cell lines that express wild-type and mutant vpr genes under the control of a tetracycline-inducible promoter. Using flow cytometry, oligonucleotide microarray analysis, and phosphoprotein expression profiling, we found that wild-type Vpr overexpression was associated with the up- or down-regulation of several genes in the mitogen-activated protein kinase (MAPK) pathway, including MAP2K2 (mitogen-activated protein kinase kinase 2) (MEK2) and MKNK2 (MAP kinase-interacting serine/threonine kinase 2). Overexpression of MEK2 abolished cell cycle G2 arrest induced by Vpr. We further found that the phosphorylation of the MEK2 effector protein ERK is down-regulated in cells expressing wild-type-Vpr. MAPK proteins are a family of kinases, including MEK2, that phosphorylate specific substrates at serine and/or threonine residues in response to intrinsic or extrinsic stimuli. They regulate signaling cascades that relay signals mediating critical cellular responses, including proliferation or cell cycle arrest and survival or death. Our data suggest that Vpr affects two disparate cell signaling cascades to induce cell cycle arrest, the previously described pathway acting through ATR and the newly identified MEK2 pathway. These signaling cascades may play important roles in HIV-1 pathogenesis and may represent new antiretroviral therapeutic targets.
MATERIALS AND METHODS
Construction of plasmids expressing wild-type Vpr.
The Vpr gene was originally cloned from HIV-1 NL4-3 by PCR amplification using the forward primer 5′-ACGGATCCATGGAACAAGCCCCAGA-3′ and the reverse primer 5′-CTAATCTACTGGCTCCATT-3′. The amplified PCR product was subcloned in a plasmid with a tetracycline-responsive DNA element, pcDNA5/FRT/TO/TOPO (Invitrogen, Carlsbad, CA). Sequences encoding the FLAG immunoepitope were added at the N terminus of vpr cDNA by PCR amplification using the following primers: 5′-GACGATGACGACAAGATGGAACAAGCCCCA-3′ and 5′-TTTGTAGTCCATGGTGGATCCGTAAGGGCG-3′. All constructs were sequenced to verify their integrity.
PCR-mediated mutagenesis.
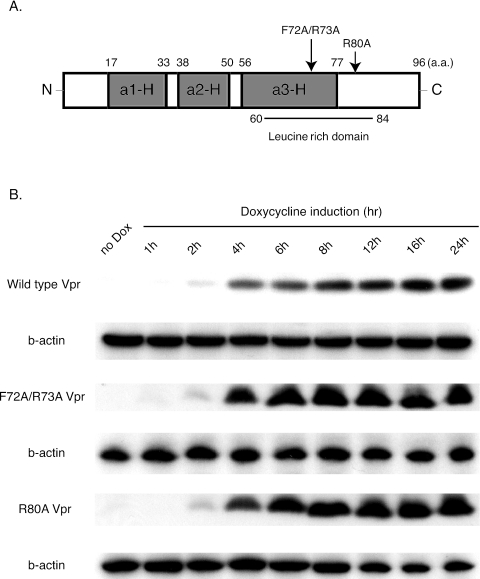
To generate the substitution mutants designated F72A/R73A (23, 35) and R80A (9, 41), we introduced site-specific mutations into the pcDNA5/FRT/TO vector containing wild-type Vpr using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's protocol. The primers to introduce the F72A/R73A mutations were 5′-CCATGCCGCAATTGGGTGTCGACATAGCAGAATAGGCGTTACTCG-3′ and 5′-CGAGTAACGCCTATTCTGCTATGTCGACACCCAATTGCGGCATGG-3′. The primers to introduce the R80A mutation were 5′-CGACATAGCGCAATAGGCGTTACTCGACAGAGGAGAGCAAGA-3′ and 5′-TCTTGCTCTCCTCTGTCGAGTAACGCCTATTGCGCTATGTCG-3′. All the constructs were sequenced to verify the integrity of the additional mutations. The locations of the mutations are schematically shown in the context of the vpr gene in Fig. 1A.
FIG. 1.
Construction and analysis of the Vpr-expressing cell lines. (A) Diagram of the primary structure of wild-type Vpr representing α-helical domains I (a1-H), II (a2-H), and III (a3-H) and the leucine-rich domain. Mutation sites (F72A/R73A and R80A) are indicated by arrows. (B) Protein immunoblot analysis of Vpr protein expression regulated by doxycycline. Protein lysate was prepared from cells not treated with doxycycline (no Dox) and at 1, 2, 4, 6, 8, 12, 16, and 24 hours after doxycycline addition. Mouse anti-FLAG M2 monoclonal antibody was used to detect FLAG-tagged Vpr protein expression. As an internal control, mouse anti-human β-actin (b-actin) monoclonal antibody was used. a.a., amino acids.
Cells.
The Flp-In TREx 293 cell line (Invitrogen), which contains a Flp recombination target (FRT) site in its genome, was maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Biosource, Camarillo, CA), 1% penicillin and streptomycin (Sigma, St. Louis, MO), 2 mM l-glutamine (Sigma), 200 μg/ml zeocin (Invitrogen), and 15 μg/ml blasticidin S (Invitrogen). To establish a tetracycline-inducible system in 293 cells, pcDNA5/FRT/TO vectors encoding wild-type or mutant Vpr and pOG44 vector (Invitrogen), which expresses Flp recombinase, were cotransfected at a molar ratio of 1:9 into Flp-In TREx 293 cells using Superfect lipofection reagent (QIAGEN, Valencia, CA), according to the manufacturer's protocol. pcDNA5/FRT vector only or encoding chloramphenicol acetyltransferase was also used for transfection as a negative control. After 48 h posttransfection, hygromycin B (100 μg/ml; Invitrogen) was added to the culture for selection. After 2 weeks, hygromycin-resistant cells were harvested without cloning and used for the further analysis described below.
Immunoblotting.
Cells were lysed with triple-detergent buffer (50 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 0.5% sodium deoxycholate) containing Complete Mini Protease Inhibitor Cocktail (Roche, Basel, Switzerland). The cell lysates were then centrifuged at 14,000× g for 15 min at 4°C to remove the cell debris. The supernatants were used for immunoblotting. The protein concentration in the lysate was determined using the BCA protein assay reagent kit (Pierce, Rockford, IL) with pooled bovine serum albumin as the standard. Immunoblotting was performed following denaturing of samples and gel separation by SDS-polyacrylamide gel electrophoresis using the enhanced-chemiluminescence detection system. Briefly, 50 μg of protein lysates was loaded on NuPAGE 4 to a 12% Bis-Tris Gel (Invitrogen) and electroblotted onto a polyvinylidene difluoride membrane (Invitrogen). The filters were blocked in blocking buffer containing 5% nonfat dry milk and 0.05% Tween 20 in Tris-buffered saline. After being blocked for 2 h at room temperature, the filters were incubated with mouse anti-FLAG monoclonal antibodies (Sigma) in incubation buffer containing 0.1% bovine serum albumin-0.05% Tween 20 in Tris-buffered saline for 90 min at room temperature with occasional shaking. After being washed, the filters were incubated with 1:10,000 horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia, Piscataway, NJ). After the final washing, immunoreactivity was visualized using the enhanced-chemiluminescence system (Amersham Pharmacia).
Cell cycle analysis.
Vpr-mediated cell cycle arrest was evaluated by propidium iodide (PI) staining and flow cytometry analysis. Briefly, cells were trypsinized, washed once with standard growth medium, and fixed in 70% ethanol. After an additional wash with phosphate-buffered saline, the cells were resuspended in PI staining solution (20 μg/ml of propidium iodide [Sigma], 200 μg/ml of RNase A [Roche] in 1 ml of 0.1% Triton X-100-phosphate-buffered saline) and incubated at 37°C for 30 min. Flow cytometric analysis was performed in a FACScalibur instrument (Becton Dickinson, San Jose, CA). Cell cycle analysis was performed using CellQuest software (Becton Dickinson).
RNA preparation.
Doxycycline-induced and uninduced Flp-In TREx 293/Vpr cells were collected 0, 1, 2, 4, 6, 8, 12, 16, and 24 h postinduction (p.i.). Doxycycline-induced and uninduced Flp-In TREx 293/Blank cells were also collected as a reference for the following experiments. Total RNA was isolated using an RNAeasy Midi kit (QIAGEN) according to the manufacturer's protocol.
Oligonucleotide microarray analysis.
Human long oligonucleotide microarrays were obtained from the National Cancer Institute Microarray Facility, Advanced Technology Center (Gaithersburg, MD). The microarrays (Hs Operon V2) contained 22,434 oligonucleotide (60- to 70-mer) spots on a glass slide. Twenty-five micrograms of total RNAs isolated from doxycycline-induced Flp-In TREx 293 stably transfected blank- and FLAG-tagged Vpr cells were reverse transcribed using an oligo(dT) primer and aminoallyl-deoxynucleoside triphosphate (Amersham Pharmacia). Unincorporated deoxynucleoside triphosphates were removed with a QIAquick PCR purification kit (QIAGEN). Purified cDNA was processed for coupling reactions with Cy3 or Cy5 ester dye, respectively. Uncoupled fluor dye was removed with a QIAquick PCR purification kit (QIAGEN). Purified fluor probes were mixed with 20 μg of Cot-1 DNA (Roche), 20 μg of poly(dA)20-60 (Amersham Pharmacia), and 4 μg of Saccharomyces cerevisiae tRNA (Sigma) and then applied to the microarray for hybridization at 42°C for 12 to 16 h. Following hybridization, the slides were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS briefly, followed by washes in 1× SSC-0.2× SSC for 2 min. The slides were scanned using an Axon 4000A scanner. The scanned images were analyzed first using GenePix 5.0 software (Axon instruments), and spots of poor quality as determined by visual inspection were also removed before further data analysis using Micro Array Database databases (Center for Information Technology, National Institutes of Health, Bethesda, MD). For each time point, there were at least three biological replicates. The spot-filtering criteria were as follows. A median target signal-over-background ratio was required to be at least 1.5, a spot diameter was required to be at least 90 μm, each gene was required to have values in at least 66% of the arrays, and each array was required to have values for at least 66% of the gene spots. Detailed methods of statistical analysis were described previously (24). Briefly, comparison of the expression profiles of Vpr-positive and Vpr-negative cells, or wild-type- and mutant-expressing cells, at a given time point was performed with univariate parametric and multivariate permutation tests based on the one-sample random-variance statistic in BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools). Statistical significance was based on a P value of <0.001 for a parametric one-sample random-variance t test. For evaluation of differential expression, a multivariate permutation test based on the one-sample random-variance t statistic was used in which the proportion of false discoveries was limited to 0.10 with 90% confidence. Beyond the statistical analysis, we chose to arbitrarily consider twofold changes in the expression of a cellular gene a threshold level for a plausible biological effect, and here we report the results mainly on that basis. The microarray data sets included in this study have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE2296. Hierarchical clustering analyses of the resulting data sets were done with the Micro Array Database system, as well as Cluster and TreeView software programs provided by the Eisen Laboratory (http://rana.lbl.gov/EisenSoftware.htm).
Transient transfection of MAP2K2 and determination of MAPK activity.
MAP2K2 cDNA was obtained from I.M.A.G.E consortium clone ID 2961198, digested by XhoI and EcoRI, and directionally subcloned into pcDNA3.1(+). To introduce constitutively active mutations (Ser222Glu and Ser226Asp) (26, 32, 43), we performed PCR-based site-directed mutagenesis using the primers 5′-GCCATTTCGTCTATGAGCTGGCCG-3′ and 5′-CAACGACTTCGTGGGCACGC-3′, as described previously (19). The mutant was sequenced to verify the integrity of the additional mutations. The wild-type and constitutively active forms of MAP2K2 (MEK2) were transiently transfected into Flp-In TREx 293/Vpr cells using Superfect (QIAGEN). Plasmid encoding enhanced green fluorescent protein (GFP) (pEGFP-N1; Clontech, Palo Alto, CA) was cotransfected with MAP2K2-encoding plasmids at a molar ratio of 1:10. Vpr expression was induced using 1 μg/ml of doxycycline. For immunoblotting, samples were collected at 0, 0.5, and 1 h p.i, and lysed with PhosphoSafe Protein Extraction Buffer (Novagen, Darmstadt, Germany) containing Complete Mini Protease Inhibitor Cocktail (Roche). The activation of ERK1/2 and MEK1/2 was determined by immunoblotting with antibodies specific for phosphorylated, activated forms of these kinases (Cell Signaling, Beverly, MA). To detect endogenous ERK1/2 and endogenous or exogenous MEK2, antibodies specific for these kinases (Cell Signaling, Beverly, MA) were also used for the immunoblotting.
Samples were also collected at 24 h p.i. for cell cycle analysis. The cells were fixed in 1% paraformaldehyde to preserve GFP fluorescence and stained with the PI staining solution. GFP-positive cells were gated so as to examine only the transfected fraction of the cells.
RESULTS
Establishment of a doxycycline-inducible Vpr expression system in Flp-In TREx 293 cells.
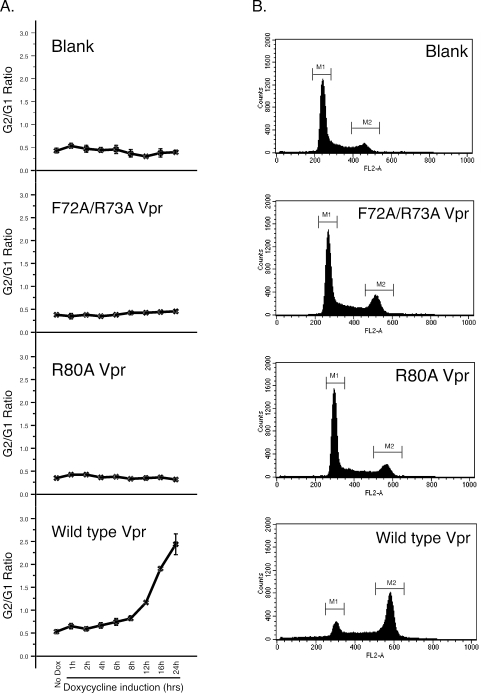
We first established Flp-In TREx cells, in which Vpr expression is regulated by doxycycline addition. The Flp-In TREx system streamlines the generation of stable, tetracycline-inducible mammalian cell lines by taking advantage of a yeast DNA recombination system that uses Flp recombinase and site-specific recombination to facilitate integration of the gene of interest into the genome. The established cell lines contain one copy per cell of the inducibly expressed gene of interest. Figure 1B shows the expression kinetics of the FLAG-Vpr protein following induction by doxycycline. Vpr was quickly induced within 2 h p.i after doxycycline addition and reached a maximum level by 4 to 6 h p.i. To examine whether the inducibly expressed Vpr produced G2 cell cycle arrest, we performed cell cycle profiles of the Flp-In TREx 293 cells inducibly expressing wild-type Vpr after treatment with doxycycline. Doxycycline-induced cells were fixed and stained with propidium iodide as whole cells, and flow cytometry was used to assess the DNA content. As shown in Fig. 2A, G 2 cell cycle arrest induced by Vpr was induced by 8 h p.i and reached a maximum level at 16 to 24 h p.i.
FIG. 2.
Effects of induced wild-type and mutant Vpr expression on the cell cycle. (A) Kinetic analyses of G2/M cell cycle arrest after doxycycline treatment. Vpr-induced G2/M cell cycle arrest was measured by flow cytometry after DNA was stained with propidium iodide. The extent of G2/M arrest was evaluated by the G2/G1 ratio. The results represent the mean of at least three experiments. The standard deviations of the values are shown as error bars. (B) Representative data for Vpr-induced G2/M cell cycle arrest measured by flow cytometry at 24 h after doxycycline addition. The y axis represents the cell count, and the x axis represents the DNA content. The experiments were conducted at least three times.
Production of cell lines expressing mutant versions of Vpr protein and demonstration of impaired function.
To further characterize the mechanisms through which Vpr produced G2 cell cycle arrest, we established cell lines that inducibly expressed two mutants of Vpr, F72A/R73A and R80A (Fig. 1A), which fail to induce G2 cell cycle arrest activity (9, 23, 35, 41). As shown in Fig. 2A, induced expression of wild-type Vpr yielded a dramatic increase in the proportion of cells in the G2/M phase. However, while the cell lines stably transfected with the F72A/R73A and R80A Vpr mutants exhibited good expression of the mutant Vpr proteins, similar to that of the cell lines expressing the wild type (Fig. 1B), the overexpression of the Vpr mutants F72A/R73A and R80A yielded significantly decreased cell cycle arrest (Fig. 2).
Large-scale expression profiling of cellular genes following overexpression of wild-type and mutant Vpr.
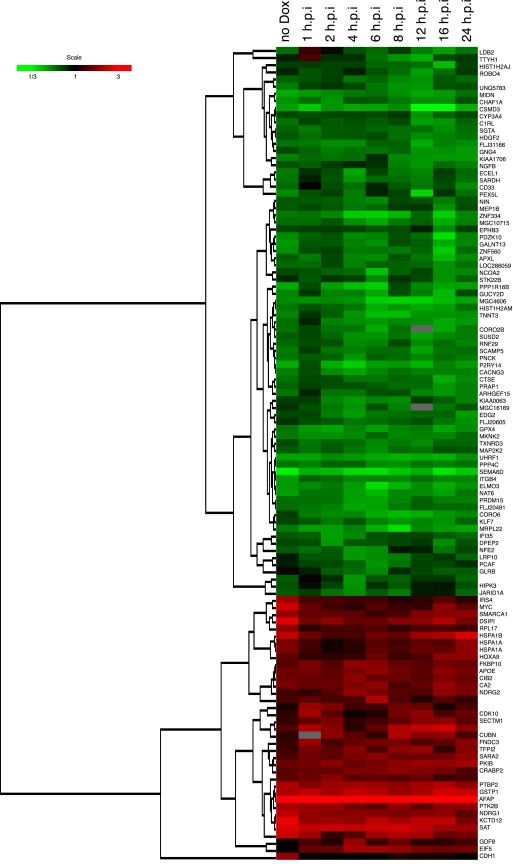
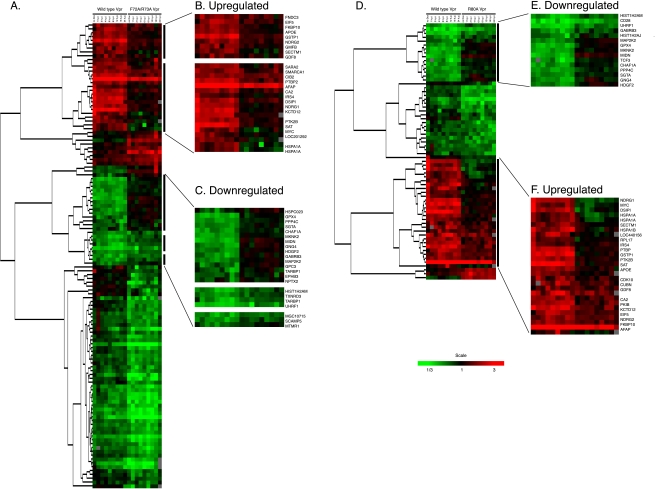
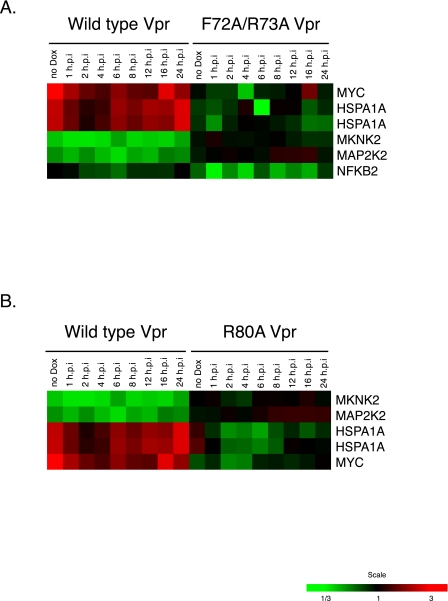
To study whether certain host cell genes showed alterations in expression following induced expression of Vpr, we conducted a large-scale host cell gene expression profiling study using microarray technologies. To determine whether the differential expression of certain genes was associated with Vpr-induced cell cycle arrest, we compared the gene expression pattern of cells expressing wild-type Vpr with the pattern of cells expressing Vpr mutant F72A/R73A or R80A Vpr. Using long oligonucleotide microarrays containing 22,000 elements in each array, we identified 565 genes that showed statistically significant differential regulation by wt Vpr (P < 0.001) (see Table S1 in the supplemental material). Among them, 113 genes were differentially regulated by twofold or more, a change we arbitrarily considered likely to be a biologically significant effect (Fig. 3 and Table 1 ). When we compared the expression profiles of the cell lines expressing wt and F72A/R73A Vpr, we found that 494 genes showed statistically significant differential regulation by either wt or F72A/R73A Vpr (P < 0.001). Among them, 121 genes were differentially regulated by twofold or more (Fig. 4A). The differential expression of 47 of the 121 genes was a wild-type-specific event (26 were up-regulated, and 21 were down-regulated) (Fig. 4B and C and Table 2). When we compared the expression profiles of the cell lines expressing wt and R80A Vpr, we found that 479 genes showed statistically significant differential regulation by either wt or R80A Vpr (P < 0.001). Among them, 66 genes were differentially regulated by twofold or more (Fig. 4D and Table 2). The differential expression of 42 of the 66 genes was a wild-type-specific event (27 were up-regulated, and 15 were down-regulated) (Fig. 4E and F and Table 2). Among the genes with wild-type-specific differential expression, 31 genes were commonly differentially expressed in comparison to both mutants (F72A/R73A and R80A) (Table 2). Some genes were differentially expressed only with respect to one mutant: 16 were differentially expressed with respect to the F72A/R73A mutant Vpr, and 11 genes were differentially expressed only with respect to the R80A mutant Vpr (Table 2). Interestingly, wild-type Vpr specifically altered the expression of several host cell genes involved in the control of cell growth and cell division (Tables 1 and 2). We noted that among these genes, wild-type (but not mutant) Vpr specifically down-regulated expression of genes involved in the MEK2-ERK pathway, notably including MAP2K2 (MEK2) and MKNK2, before the induction of cell cycle arrest (<6 h p.i) (Table 3 and Fig. 5). The observation that the expression of genes in the MEK2-ERK pathway was down-regulated by wild-type Vpr, coupled with the known activity of the MEK2-ERK pathway in controlling the cell cycle, suggested that this pathway might be important in mediating Vpr-dependent cell cycle arrest and suggested a possible additional mechanism responsible for the cell cycle arrest function of Vpr. Wild-type Vpr was also specifically associated with the up-regulation of Myc and HSPA1A (Table 1), which were up-regulated after the induction of cell cycle arrest (>8 h p.i.) (Table 3 and Fig. 5). Interestingly, we noted that the gene for cyclin-dependent kinase inhibitor 1A (CDKN1A; p21) (2), a possible cell cycle effector, was among the genes that we found to be statistically significantly differentially expressed in Vpr-expressing cells, and this gene was also observed to be up-regulated in HIV-infected macrophages (39) (see Table S1 in the supplemental material). However, we found that the up-regulation in our system was quantitatively small and was not a wild-type-specific alteration.
FIG. 3.
Hierarchical clustering of differentially expressed cellular genes altered by the inducible expression of wild-type Vpr protein. The figure shows the hierarchical clustering of the cellular genes that showed statistically significant differences in expression (P < 0.001) and twofold changes in the expression of a cellular gene at least at one time point. Genes shown in red showed up-regulation, and those in green were down-regulated, while those that did not show any change with respect to a normalized matched control are shown in black (see color scale). The gray areas indicate missing data.
TABLE 1.
Cellular genes differentially expressed with wild-type Vpr overexpression
| Regulation (n) | UniGene | Gene | Description |
|---|---|---|---|
| Up-regulated (36) | Hs.109438 | KCTD12 | Potassium channel tetramerization domain containing 12 |
| Hs.127428 | HOXA9 | Homeo box A9 | |
| Hs.129867 | CIB2 | Calcium and integrin binding family member 2 | |
| Hs.152292 | SMARCA1 | SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 1 | |
| Hs.155097 | CA2 | Carbonic anhydrase II | |
| Hs.159609 | IRS4 | Insulin receptor substrate 4 | |
| Hs.166206 | CUBN | Cubilin (intrinsic factor-cobalamin receptor) | |
| Hs.202453 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | |
| Hs.269895 | PTBP2 | Polypyrimidine tract binding protein 2 | |
| Hs.274402 | HSPA1B | Heat shock 70-kDa protein 1B, mRNA | |
| Hs.279582 | SARA2 | SAR1a gene homolog 2 (S. cerevisiae) | |
| Hs.28491 | SAT | Spermidine/spermine N1-acetyltransferase | |
| Hs.349570 | CDNA clone IMAGE:4328048; partial cds | ||
| Hs.372914 | NDRG1 | N-myc downstream regulated gene 1 | |
| Hs.374588 | RPL17 | Ribosomal protein L17 | |
| Hs.405662 | CRABP2 | Cellular retinoic acid binding protein 2 | |
| Hs.406858 | CDNA: FLJ21966 fis, clone HEP05644 | ||
| Hs.41565 | GDF8 | Growth differentiation factor 8 | |
| Hs.433702 | EIF5 | Eukaryotic translation initiation factor 5 | |
| Hs.438231 | TFPI2 | Tissue factor pathway inhibitor 2 | |
| Hs.461086 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | |
| Hs.463035 | FKBP10 | FK506 binding protein 10; 65 kDa | |
| Hs.464404 | CDNA: FLJ23438 fis, clone HRC13275 | ||
| Hs.486354 | PKIB | Protein kinase (cAMP-dependent; catalytic) inhibitor beta | |
| Hs.491322 | PTK2B | PTK2B protein tyrosine kinase 2 beta | |
| Hs.508010 | FNDC3 | Fibronectin type III domain containing 3 | |
| Hs.515465 | APOE | Apolipoprotein E | |
| Hs.520028 | HSPA1A | Heat shock 70-kDa protein 1A | |
| Hs.522074 | DSIPI | Delta sleep-inducing peptide; immunoreactor | |
| Hs.523836 | GSTP1 | Glutathione S-transferase pi | |
| Hs.525205 | NDRG2 | NDRG family member 2 | |
| Hs.529095 | LOC440156 | ||
| Hs.529369 | AFAP | Actin filament-associated protein | |
| Hs.532368 | Clone 24587 mRNA sequence | ||
| Hs.77313 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | |
| Hs.95655 | SECTM1 | Secreted and transmembrane 1 | |
| Down-regulated (77) | Hs.108106 | UHRF1 | Ubiquitin-like, containing PHD and RING finger domains 1 |
| Hs.12308 | PDZK10 | PDZ domain containing 10 | |
| Hs.126667 | EDG2 | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor 2 | |
| Hs.129910 | NAT6 | N-Acetyltransferase 6 | |
| Hs.131819 | SUSD2 | Sushi domain containing 2 | |
| Hs.134999 | HIST1H2AM | Histone 1, H2am | |
| Hs.1355 | CTSE | Cathepsin E | |
| Hs.143046 | CORO6 | Coronin 6 | |
| Hs.147041 | ZNF334 | Zinc finger protein 334 | |
| Hs.15951 | PRAP1 | Proline-rich acidic protein 1 | |
| Hs.159711 | GNG4 | Guanine nucleotide binding protein (G protein) gamma 4 | |
| Hs.194777 | MEP1B | Meprin A beta | |
| Hs.198003 | SARDH | Sarcosine dehydrogenase | |
| Hs.201918 | HIPK3 | Homeodomain-interacting protein kinase 3 | |
| Hs.203910 | SGTA | Small glutamine-rich tetratricopeptide repeat (TPR)-containing alpha | |
| Hs.207603 | CDNA FLJ30190 fis, clone BRACE2001312 | ||
| Hs.226422 | FLJ31166 | Hypothetical protein FLJ31166 | |
| Hs.232614 | ZNF560 | Zinc finger protein 560 | |
| Hs.23748 | LDB2 | LIM domain binding 2 | |
| Hs.2465 | P2RY14 | Purinergic receptor P2Y, G-protein coupled 14 | |
| Hs.24907 | CORO2B | Coronin, actin binding protein 2B | |
| Hs.2561 | NGFB | Nerve growth factor, beta polypeptide | |
| Hs.268728 | TTYH1 | Tweety homolog 1 (Drosophila) | |
| Hs.26880 | ECEL1 | Endothelin-converting enzyme-like 1 | |
| Hs.2913 | EPHB3 | EPH receptor B3 | |
| Hs.292986 | MGC16169 | Hypothetical protein MGC16169 | |
| Hs.306673 | LOC286059 | Hypothetical protein LOC286059 | |
| Hs.3094 | KIAA0063 | KIAA0063 gene product (KIAA0063); mRNA. | |
| Hs.309958 | GUCY2D | Guanylate cyclase 2D, membrane (retina specific) | |
| Hs.310429 | NIN | Ninein (GSK3B-interacting protein) | |
| Hs.32973 | GLRB | Glycine receptor beta | |
| Hs.336954 | CDNA clone IMAGE:4110850; partial cds | ||
| Hs.368486 | APXL | Apical protein-like (Xenopus laevis) | |
| Hs.369042 | FLJ20605 | Hypothetical protein FLJ20605 | |
| Hs.370255 | ITGB4 | Integrin beta 4 | |
| Hs.372633 | DPEP2 | Dipeptidase 2 | |
| Hs.374180 | SCAMP5 | Secretory carrier membrane protein 5 | |
| Hs.377416 | ELMO3 | Engulfment and cell motility 3 (ced-12 homolog; Caraorhabditis elegans) | |
| Hs.406691 | HIST1H2AJ | Histone 1, H2aj | |
| Hs.43071 | HDGF2 | Hepatoma-derived growth factor-related protein 2 | |
| Hs.433951 | GPX4 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) | |
| Hs.436667 | PNCK | Pregnancy-up-regulated nonubiquitously expressed CaM kinase | |
| Hs.442527 | CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | |
| Hs.443109 | ARHGEF15 | Rho guanine nucleotide exchange factor (GEF) 15 | |
| Hs.446678 | NCOA2 | Nuclear receptor coactivator 2 | |
| Hs.45719 | PPP1R16B | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | |
| Hs.460568 | MGC4606 | Hypothetical protein MGC4606 | |
| Hs.460857 | FLJ20481 | Hypothetical protein FLJ20481 | |
| Hs.462080 | UNQ5783 | DTFT5783 | |
| Hs.465529 | MIDN | PREDICTED: Homo sapiens midnolin | |
| Hs.465627 | MAP2K2 | Mitogen-activated protein kinase kinase 2 | |
| Hs.470277 | GALNT13 | UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) | |
| Hs.471221 | KLF7 | Kruppel-like factor 7 (ubiquitous) | |
| Hs.473893 | PRDM15 | PR domain-containing 15 | |
| Hs.477475 | TXNRD3 | PREDICTED: Homo sapiens thioredoxin reductase 3 | |
| Hs.478393 | PEX5L | Peroxisomal biogenesis factor 5-like | |
| Hs.483924 | MRPL22 | Mitochondrial ribosomal protein L22 | |
| Hs.487994 | KIAA1706 | KIAA1706 protein | |
| Hs.505323 | STK22B | Serine/threonine kinase 22B | |
| Hs.50842 | IFI35 | Interferon-induced protein 35 | |
| Hs.511265 | SEMA6D | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D | |
| Hs.515032 | MKNK2 | MAP kinase-interacting serine/threonine kinase 2 | |
| Hs.516846 | MGC10715 | Novel C2H2-type zinc finger protein | |
| Hs.524121 | ROBO4 | Roundabout homolog 4, magic roundabout (Drosophila) | |
| Hs.525232 | LRP10 | Low-density lipoprotein receptor-related protein 10 | |
| Hs.525264 | C1RL | Complement component 1, r subcomponent-like | |
| Hs.531766 | CDNA: FLJ22840 fis, clone KAIA4709 | ||
| Hs.533055 | PCAF | p300/CBP-associated factor | |
| Hs.534338 | PPP4C | Protein phosphatase 4 (formerly-X), catalytic subunit | |
| Hs.7235 | CACNG3 | Calcium channel, voltage-dependent, gamma subunit 3 | |
| Hs.73454 | TNNT3 | Troponin T3, skeletal, fast | |
| Hs.75643 | NFE2 | Nuclear factor (erythroid-derived 2), 45 kDa | |
| Hs.76272 | JARID1A | Jumonji, AT-rich interactive domain 1A (RBBP2-like) | |
| Hs.79018 | CHAF1A | Chromatin assembly factor 1, subunit A (p150) | |
| Hs.83731 | CD33 | CD33 antigen (gp67) (CD33), mRNA. | |
| Hs.85524 | RNF29 | Ring finger protein 29 | |
| Hs.91381 | CSMD3 | CUB and Sushi multiple domains 3 |
FIG. 4.
Differential expression of cellular genes following induction of wild-type and mutant Vpr expression. Comparison of differentially expressed cellular genes altered after Vpr expression between cells expressing (A) wild-type Vpr and F72A/R73A Vpr and (D) wild-type Vpr and R80A Vpr. Black bars on the right side of panels A and D indicate the clusters, which showed wild-type-specific differential regulation of gene expression. These clusters are highlighted in larger images with gene symbols. (B) Up- and (C) down-regulated in a comparison with F72A/R73A Vpr-expressing cells. (E) Down- and (F) up-regulated in a comparison with R80A Vpr-expressing cells. The genes shown in this figure passed two filtering criteria: (i) statistical significance (P < 0.001) and (ii) twofold changes in the expression of a cellular gene at least at one time point.
TABLE 2.
Cellular genes differentially expressed specifically in association with wild-type Vpr overexpression (genes not affected by overexpression of mutant Vprs)
| Comparison | Regulation (a) | UniGene ID | Gene | Gene description |
|---|---|---|---|---|
| Botha | Up (19) | Hs.529369 | AFAP | Actin filament-associated protein |
| Hs.515465 | APOE | Apolipoprotein E | ||
| Hs.155097 | CA2 | Carbonic anhydrase II | ||
| Hs.522074 | DSIPI | Delta sleep-inducing peptide; immunoreactor | ||
| Hs.433702 | EIF5 | Eukaryotic translation initiation factor 5 | ||
| Hs.463035 | FKBP10 | FK506 binding protein 10; 65 kDa | ||
| Hs.41565 | GDF8 | Growth differentiation factor 8 | ||
| Hs.523836 | GSTP1 | Glutathione S-transferase pi | ||
| Hs.520028 | HSPA1A | Heat shock 70-kDa protein 1A | ||
| Hs.159609 | IRS4 | Insulin receptor substrate 4 | ||
| Hs.109438 | KCTD12 | Potassium channel tetramerization domain containing 12 | ||
| Hs.202453 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | ||
| Hs.372914 | NDRG1 | N-myc downstream regulated gene 1 | ||
| Hs.525205 | NDRG2 | NDRG family member 2 | ||
| Hs.269895 | PTBP2 | polypyrimidine tract binding protein 2 | ||
| Hs.491322 | PTK2B | PTK2B protein tyrosine kinase 2 beta | ||
| Hs.28491 | SAT | Spermidine/spermine N1-acetyltransferase | ||
| Hs.95655 | SECTM1 | Secreted and transmembrane 1 | ||
| Hs.464404 | CDNA: FLJ23438 fis, clone HRC13275 | |||
| Down (12) | Hs.79018 | CHAF1A | Chromatin assembly factor 1, subunit A (p150) | |
| Hs.302352 | GABRB3 | Gamma-aminobutyric acid (GABA) A receptor beta 3 | ||
| Hs.159711 | GNG4 | Guanine nucleotide binding protein (G protein) gamma 4 | ||
| Hs.433951 | GPX4 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) | ||
| Hs.43071 | HDGF2 | Hepatoma-derived growth factor-related protein 2 | ||
| Hs.134999 | HIST1H2AM | Histone 1, H2am | ||
| Hs.465627 | MAP2K2 | Mitogen-activated protein kinase kinase 2 | ||
| Hs.465529 | MIDN | PREDICTED: Homo sapiens midnolin | ||
| Hs.515032 | MKNK2 | MAP kinase-interacting serine/threonine kinase 2 | ||
| Hs.534338 | PPP4C | Protein phosphatase 4 (formerly X); catalytic subunit | ||
| Hs.203910 | SGTA | Small glutamine-rich tetratricopeptide repeat (TPR)-containing alpha | ||
| Hs.108106 | UHRF1 | Ubiquitin-like, containing PHD and RING finger domains 1 | ||
| F72A/R73Ab | Up (7) | Hs.129867 | CIB2 | Calcium and integrin binding family member 2 |
| Hs.508010 | FNDC3 | Fibronectin type III domain containing 3 | ||
| Hs.151413 | GMFB | Glia maturation factor beta | ||
| Hs.189823 | LOC201292 | Hypothetical protein LOC201292 | ||
| Hs.279582 | SARA2 | SAR1a gene homolog 2 (S. cerevisiae) | ||
| Hs.152292 | SMARCA1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 1 | ||
| Hs.281434 | CDNA clone IMAGE: 3831740, partial cds | |||
| Down (9) | Hs.2913 | EPHB3 | EPH receptor B3 | |
| Hs.435036 | GPC3 | Glypican 3 | ||
| Hs.231616 | HSPC023 | HSPC023 protein | ||
| Hs.516846 | MGC10715 | Novel C2H2-type zinc finger protein | ||
| Hs.347187 | MTMR1 | Myotubularin-related protein 1 | ||
| Hs.3281 | NPTX2 | Neuronal pentraxin II | ||
| Hs.374180 | SCAMP5 | Secretory carrier membrane protein 5 | ||
| Hs.498115 | TARBP1 | TAR (HIV) RNA binding protein 1 | ||
| Hs.477475 | TXNRD3 | PREDICTED: Homo sapiens thioredoxin reductase 3 | ||
| R80Ac | Up (8) | Hs.77313 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 |
| Hs.166206 | CUBN | Cubilin (intrinsic factor-cobalamin receptor) | ||
| Hs.274402 | HSPA1B | Heat shock 70-kDa protein 1B. | ||
| Hs.486354 | PKIB | Protein kinase (cAMP-dependent, catalytic) inhibitor beta | ||
| Hs.374588 | RPL17 | Ribosomal protein L17 | ||
| Hs.529095 | LOC440156 | |||
| Hs.406858 | CDNA: FLJ21966 fis, clone HEP05644 | |||
| Hs.523491 | LOC440030 | |||
| Down (3) | Hs.1987 | CD28 | CD28 antigen (Tp44) | |
| Hs.406691 | HIST1H2AJ | Histone 1, H2aj | ||
| Hs.371282 | TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) |
Genes with differential expression associated with wild-type Vpr. in comparison with both F72A/R73A and R80A mutant Vprs.
Differentially expressed genes observed only in the comparison of wild-type Vpr-expressing cells with F72A/R73A-Vpr-expressing cells.
Differentially expressed genes observed only in the comparison of wild-type Vpr-expressing cells with R80A-Vpr-expressing cells.
TABLE 3.
Temporal relationship of cell cycle arrest and changes in cellular gene expression associated with Vpr overexpression
| Time | Comparison | UniGene | Gene | Description | WT |
|---|---|---|---|---|---|
| Beforea | Bothb | Hs.159609 | IRS4 | Insulin receptor substrate 4 | Up |
| Hs.41565 | GDF8 | Growth differentiation factor 8 | Up | ||
| Hs.463035 | FKBP10 | FK506 binding protein 10; 65 kDa | Up | ||
| Hs.464404 | CDNA: FLJ23438 fis, clone HRC13275 | Up | |||
| Hs.465627 | MAP2K2 | Mitogen-activated protein kinase kinase 2 | Down | ||
| Hs.515032 | MKNK2 | MAP kinase-interacting serine/threonine kinase 2 | Down | ||
| F72A/R73Ac | Hs.129867 | CIB2 | Calcium and integrin binding family member 2 | Up | |
| Hs.151413 | GMFB | Glia maturation factor beta | Up | ||
| Hs.189823 | LOC201292 | Hypothetical protein LOC201292 | Up | ||
| Hs.231616 | HSPC023 | HSPC023 protein | Down | ||
| Hs.347187 | MTMR1 | Myotubularin-related protein 1 | Down | ||
| R80Ad | Hs.374588 | RPL17 | Ribosomal protein L17 | Up | |
| Aftere | Both | Hs.202453 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | Up |
| Hs.520028 | HSPA1A | Heat shock 70-kDa protein 1A | Up | ||
| Hs.95655 | SECTM1 | Secreted and transmembrane 1 | Up | ||
| Hs.159711 | GNG4 | Guanine nucleotide binding protein (G protein) gamma 4 | Down | ||
| Hs.203910 | SGTA | Small glutamine-rich tetratricopeptide repeat (TPR)-containing alpha | Down | ||
| Hs.43071 | HDGF2 | Hepatoma-derived growth factor-related protein 2 | Down | ||
| F72A/R73A | Hs.152292 | SMARCA1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 1 | Up | |
| Hs.279582 | SARA2 | SAR1a gene homolog 2 (S. cerevisiae) | Up | ||
| Hs.2913 | EPHB3 | EPH receptor B3 | Down | ||
| Hs.3281 | NPTX2 | Neuronal pentraxin II | Down | ||
| Hs.374180 | SCAMP5 | Secretory carrier membrane protein 5 | Down | ||
| R80A | Hs.274402 | HSPA1B | Heat shock 70-kDa protein 1B | Up | |
| Hs.523491 | LOC440030 | Up | |||
| Hs.371282 | TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | Down | ||
| Hs.406691 | HIST1H2AJ | Histone 1, H2aj | Down | ||
| Independentf | Both | Hs.109438 | KCTD12 | Potassium channel tetramerization domain-containing 12 | Up |
| Hs.155097 | CA2 | Carbonic anhydrase II | Up | ||
| Hs.269895 | PTBP2 | Polypyrimidine tract binding protein 2 | Up | ||
| Hs.28491 | SAT | Spermidine/spermine N1-acetyltransferase | Up | ||
| Hs.372914 | NDRG1 | N-myc downstream regulated gene 1 | Up | ||
| Hs.433702 | EIF5 | Eukaryotic translation initiation factor 5 | Up | ||
| Hs.491322 | PTK2B | PTK2B protein tyrosine kinase 2 beta | Up | ||
| Hs.515465 | APOE | Apolipoprotein E | Up | ||
| Hs.522074 | DSIPI | Delta sleep-inducing peptide; immunoreactor | Up | ||
| Hs.523836 | GSTP1 | Glutathione S-transferase pi | Up | ||
| Hs.525205 | NDRG2 | NDRG family member 2 | Up | ||
| Hs.529369 | AFAP | Actin filament-associated protein | Up | ||
| Hs.108106 | UHRF1 | Ubiquitin-like, containing PHD and RING finger domains 1 | Down | ||
| Hs.134999 | HIST1H2AM | Histone 1; H2am | Down | ||
| Hs.302352 | GABRB3 | Gamma-aminobutyric acid (GABA) A receptor beta 3 | Down | ||
| Hs.433951 | GPX4 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) | Down | ||
| Hs.465529 | MIDN | PREDICTED: Homo sapiens midnolin | Down | ||
| Hs.534338 | PPP4C | Protein phosphatase 4 (formerly X), catalytic subunit | Down | ||
| Hs.79018 | CHAF1A | Chromatin assembly factor 1, subunit A (p150) | Down | ||
| F72A/R73A | Hs.281434 | CDNA clone IMAGE: 3831740, partial cds | Up | ||
| Hs.508010 | FNDC3 | Fibronectin type III domain-containing 3 | Up | ||
| Hs.435036 | GPC3 | Glypican 3 | Down | ||
| Hs.477475 | TXNRD3 | PREDICTED: Homo sapiens thioredoxin reductase 3 | Down | ||
| Hs.498115 | TARBP1 | TAR (HIV) RNA binding protein 1 | Down | ||
| Hs.516846 | MGC10715 | Novel C2H2-type zinc finger protein | Down | ||
| R80A | Hs.166206 | CUBN | Cubilin (intrinsic factor-cobalamin receptor) | Up | |
| Hs.406858 | CDNA: FLJ21966 fis, clone HEP05644 | Up | |||
| Hs.486354 | PKIB | Protein kinase (cAMP-dependent, catalytic) inhibitor beta | Up | ||
| Hs.529095 | LOC440156 | Up | |||
| Hs.77313 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | Up | ||
| Hs.1987 | CD28 | CD28 antigen (Tp44) | Down |
Genes differentially expressed before the induction of cell cycle arrest (<6 h p.i.).
Genes differentially expressed in comparison with both mutant Vpr-expressing cells.
Genes differentially expressed only in comparison with F72A/R73A-Vpr expressing cells.
Genes differentially expressed only in comparison with R80A-Vpr expressing cells.
Genes differentially expressed after the induction of cell cycle arrest (>8 h p.i.).
Genes differentially expressed gene independent of cell cycle arrest.
FIG. 5.
Differentially regulated genes related to the MAPK pathway. Assignment of these genes was performed with the Cancer Genome Anatomy Project pathway databases (http://cgap.nci.nih.gov/). Pathway information was provided by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/).
Vpr-mediated changes in the phosphorylation status of ERK.
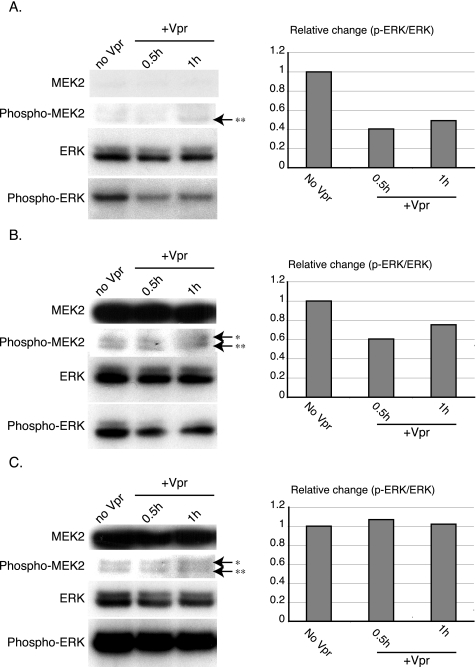
Since we hypothesized that one mechanism by which Vpr arrests the cell cycle is through down-regulating the expression of genes in the MEK2-ERK pathway, those encoding MAP2K2 (MEK2) and MKNK2, it would be reasonable to hypothesize that overexpression of Vpr might be associated with decreased phosphorylation of ERK. We therefore examined the ERK phosphorylation status in the cells before and after Vpr expression was induced with doxycycline. We found that inducing overexpression of Vpr with doxycycline was associated with a decrease in phosphorylated ERK (Fig. 6A). To confirm that the down-regulation of the MEK2-ERK pathway may help mediate G2 cell cycle arrest in the cells expressing wild-type Vpr, we constructed eukaryotic expression vectors encoding wild-type or a constitutively active form of MEK2. The activation of MAPKKs may be mediated by an increased charge at the phosphorylation site (32). We therefore constructed a constitutively active mutation by producing a mutated protein with increased negative charge at the phosphorylation site by replacing two phosphorylation targets of MEK2, Ser-222 and Ser-226 (43), with Glu and Asp, respectively. To investigate whether providing extra exogenous MEK2 would block the decrease in ERK phosphorylation presumably caused by Vpr, we transfected plasmids encoding either extra wild-type MEK2 (Fig. 6B) or a constitutively active form of MEK2 into the Vpr-expressing cells (Fig. 6C). Providing extra MEK2 to overcome the presumed Vpr-mediated decrease in MEK2 expression prevented the decrease in ERK phosphorylation seen with Vpr overexpression. We also examined the amounts of phosphorylated MEK2 and found that Vpr overexpression and the cell cycle arrest associated with Vpr overexpression were not related to changes in the phosphorylation state of MEK2 (Fig. 6A) and that the ability to phosphorylate MEK2 was intact, because providing extra MEK2 (wild type and the constitutively active mutant) also increased MEK2 phosphorylation (Fig. 6B and C), even when Vpr was overexpressed.
FIG. 6.
Analysis of ERK2 phosphorylation status following induced expression of Vpr and abrogation of the effect by MEK2. Flp-In TREx 293 cells stably transfected with wild-type Vpr were transiently transfected with either a blank vector as a control plasmid (A), wild-type MEK2 (MEK2 WT) (B), or constitutively active mutant MEK2 (MEK2 CA) (C) and treated with 1 μg/ml doxycycline for the times indicated. The cells were lysed and analyzed for ERK phosphorylation using an anti-phospho-p44 (ERK1)/phospho-p42 (ERK2) phosphospecific antibody. Endogenous ERK1/2 expression levels were detected by immunoblotting using anti-ERK1/2 antibody. The higher band of the doublet detected by anti-ERK1/2 antibody is p44 (ERK1), and the lower band is p42 (ERK2). The signal due to phospho-ERK protein was quantitated using Image J software (http://rsb.info.nih.gov/ij/) and normalized to the signal due to endogenous ERK protein (p-ERK/ERK ratio). The relative changes in the p-ERK/ERK ratio (y axis) at each time point (+Vpr) were calculated by comparing the p-ERK/ERK ratio at each time point to the p-ERK/ERK ratio without doxycycline treatment (no Vpr). Endogenous or exogenous MEK2 expression levels were detected by immunoblotting using anti-MEK2 antibody. The phosphorylated forms of MEK1/2 were detected by immunoblotting using anti-phospho-MEK1/2 antibodies. * indicates the band for the phosphorylated form of MEK1/2, while ** indicates a cross-reacting band (1). We repeated the experiments at least three times. Representative data are shown.
Overexpression of MEK2 reverts G2/M arrest induced by Vpr protein.
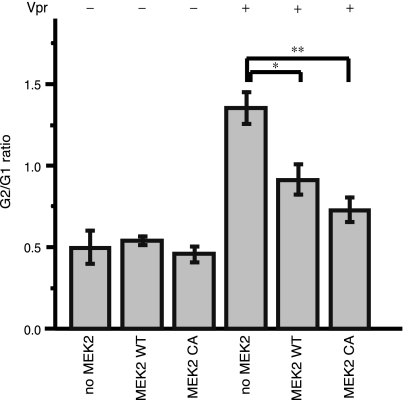
To verify whether providing extra MEK2 via transfection could overcome the effect of the Vpr protein in G2 cell cycle arrest, we transiently transfected plasmids encoding the wild-type or the constitutively active form of MEK2 into Flp-In TREx 293 cells inducibly expressing wild-type Vpr and analyzed cell cycle profiles by flow cytometry. As shown in Fig. 7, supplying MEK2 exogenously abolished the G2 cell cycle arrest induced by Vpr, suggesting that one mechanism used by Vpr to arrest the cell cycle involves decreasing the expression of MEK2 and reducing the phosphorylation of ERK2.
FIG. 7.
Inhibition of Vpr-induced G2/M arrest by wild-type MEK2 and a constitutively active MEK2 mutant. Flp-In TREx 293 cells stably transfected with wild-type Vpr were transiently transfected with either a blank vector as a control plasmid, a wild-type MEK2 construct (MEK2 WT), or a constitutively active mutant of MEK2 (MEK2 CA). Twenty-four hours after transfection, the cells were treated with 1 μg/ml doxycycline for 24 h to induce Vpr (Vpr +) or left untreated (Vpr -). The results represent the mean of four experiments. The standard deviations of the values are shown as error bars. Differences between groups were examined for statistical significance using the unpaired t test. *, P < 0.02; **, P < 0.005.
DISCUSSION
One of Vpr's several important functions is to arrest the cell cycle at G2/M. Arresting the cell cycle likely aids viral replication in many ways, including increasing expression from the HIV long terminal repeat (10, 16, 18, 45). One clear mechanism responsible for Vpr-mediated cell cycle arrest involves the activation of the ATR DNA damage response pathway (34, 46). Inhibiting ATR function with drugs or with a dominant-negative mutant or knocking down its expression with small interfering RNA abrogates cell cycle arrest associated with Vpr. The Rad17 and Hus1 proteins involved in the ATR pathway are required for cell cycle arrest by Vpr, which involves the phosphorylation of Chk1 and histone 2A variant X. Ultimately, Vpr leads to the inactivation by phosphorylation of Cdc25c, the Cdc2 phosphatase, resulting in the subsequent hyperphosphorylation and inactivation of Cdc2 (17, 22, 33). The Cdc2-cyclin B kinase activity is needed for the progression of the cell cycle from G2 to M. Other recent data show that Vpr can bind Cdc25c directly, inhibiting the Cdc25c phosphatase activity (15), perhaps blocking cyclin B1-Cdc2 redundantly with the ATR pathway. Other cellular proteins can block Vpr's cell cycle arrest activity. For example, the chaperone protein heat shock protein 70 (Hsp70), when overexpressed, can partially prevent Vpr from arresting the cell cycle (20). The studies implicating the involvement of the ATR pathway in Vpr-mediated cell cycle arrest, however, contained some intriguing hints that the ATR pathway may not be the only mechanism responsible for Vpr-mediated cell cycle arrest. For example, knocking down ATR, Chk1, or Rad17 expression does not completely inhibit cell cycle arrest by Vpr (34, 46). Our finding that Vpr also arrests the cell cycle through a signal transduction pathway involving MEK2 offers some additional insight into these observations.
While the ATR mechanism can clearly result in cell cycle arrest by Vpr, the Vpr cell cycle arrest function is apparently an important enough activity that HIV has evolved two distinct mechanisms, both mediated by Vpr, to arrest the cell cycle: the previously described ATR-mediated pathway and the new pathway that we now describe, acting through the down-regulation of MEK2. Why might HIV Vpr target the cell cycle through two distinct mechanisms? The mechanisms of radiation-induced cell cycle arrest may offer some insight.
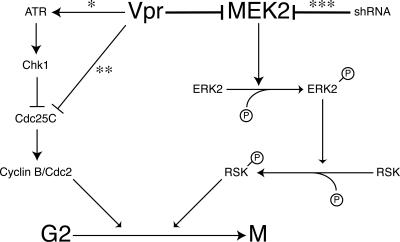
Once ionizing radiation exposure occurs, G2/M checkpoint cascades are activated through the activation of ATR for the repair of damaged DNA (Fig. 8). In addition to this activation, ionizing radiation also activates MEK1 and MEK2 to control the extent of G2/M checkpoint activity and to promote survival in response to the stress (1). However, cells expressing dominant-negative MEK2 showed a decreased ability to recover from G2/M checkpoint arrest once activated (1). Accumulating evidence, including our findings, suggest that the activation of ATR-mediated G2/M checkpoint arrest by Vpr acts as an initiator, while the down-regulation of the MEK2-ERK pathway by Vpr augments or maintains the G2/M cell cycle arrest induced by ATR activation.
FIG. 8.
Schematic representation of the putative signaling cascades mediating G2/M cell cycle arrest induced by Vpr protein. Previously described pathways, including those described by Zimmerman et al. (*) (46), who reported the involvement of the ATR-Chk1-mediated checkpoint pathway; Goh et al. (**) (16), who reported the direct inhibition of Cdc25C by Vpr; and Ussar and Voss (***) (38), who reported that the MEK2 knockdown results in centrosome amplification and multipolar spindle formation through reduced phosphorylation of RSK, are indicated. In our model, blockade of MEK2 activity also leads to cell cycle arrest at the G2/M checkpoint by decreasing ERK phosphorylation and abolishing the ability of the cells to recover from the G2/M checkpoint arrest.
Our results show that Vpr overexpression is associated with the down-regulation of genes in the MEK2-ERK pathway and that overexpression of MEK2 can ablate Vpr's ability to arrest the cell cycle. While the tetracycline-inducible promoter driving the Vpr gene can be slightly “leaky,” the cell cycle arrest that we observed to accompany treatment of the cells with doxycycline clearly shows that overexpression of Vpr alters the phenotype of the cells. The identification of MEK2 and the ability of MEK2 supplied in trans to abrogate the Vpr-mediated cell cycle arrest phenotype support the idea that the pathways identified using the cells have physiological significance. Since Vpr overexpression had no appreciable effect on MEK2 phosphorylation in the absence of extra MEK2, and since overexpression of MEK2 resulted in increased amounts of phosphorylated MEK2 even when Vpr was overexpressed (Fig. 6), it is unlikely that Vpr's effects result from inhibition of MEK2 phosphorylation by Vpr. These findings also help confirm the model that Vpr overexpression is associated with decreases in MEK2 RNA, as observed in the microarray data. The overexpression of the constitutively active MEK2 increased MEK2 phosphorylation (Fig. 6C), although the phosphorylation targets, Ser-222 and Ser-226 (43), were replaced by Glu and Asp, respectively. This finding suggests that constitutively active MEK2 may phosphorylate endogenous MEK2 before the induction of Vpr expression via auto- or transphosphorylation mechanisms (44).
How might Vpr act through the down-regulation of MEK2 to arrest the cell cycle? One plausible mechanism might involve the centrosome. Ussar and Voss (38) reported that the MEK2 knockdown by short hairpin RNA resulted in deregulated centrosome duplication, followed by multipolar spindle formation. Their work showed that the MEK2 knockdown-mediated decrease in ERK phosphorylation decreases, in turn, the phosphorylation of RSK, which is required for the mitotic exit, providing a possible explanation for MEK2 down-regulation leading to the accumulation of the cells at G2/M (Fig. 8).
Some of the changes in cellular gene expression that accompany overexpression of Vpr may reflect a viral strategy aimed at optimizing the host cell for viral replication, but other changes may instead reflect a cellular response aimed at blunting the effects of Vpr. For example, the finding that Hsp70 is overexpressed following HIV infection (40) and with overexpression of Vpr (20), findings that we confirmed in our study, may represent such an attempt by the cell to subvert the pathogenesis strategies of the virus.
Although the de novo expression of Vpr occurs at late stages of the HIV replication cycle, large amounts of Vpr are efficiently packaged into progeny viral particles (3). Virion-associated Vpr acts as an immediate-early protein after virus entry and helps establish G2/M arrest and increases viral gene expression (18, 30). Since we examined the effects of Vpr within 24 h after induction of Vpr expression, our findings should reflect the immediate host cell response. Our results suggest that inhibition of the MEK2-ERK pathway may be elicited by virion-associated Vpr in vivo soon after virus entry.
Although p21, which was up-regulated in HIV-1-infected macrophages (39), was also up-regulated in our expression data, this up-regulation was not wild type specific. The previous report also showed that p21 was predominantly up-regulated beginning 7 days after infection, perhaps to enhance the next cycle of viral replication. Since our expression data reflect an immediate host cell response to Vpr overexpression, this discrepancy may be due to differences in the experimental model and our focus on the effects of a single HIV protein. During active viral replication, there may be several different confounding effects that make it difficult to appreciate the isolated effects of a single viral protein on the host cell.
While the focus of this report is the host cell cycle effects of Vpr, the data set developed in the course of this study is a rich one, and we expect that careful examination of the data may reveal other implications of Vpr's effects on host cell gene expression. Our findings also lend some additional support to the hypothesis that viruses have evolved ways of changing the host cell milieu to facilitate viral replication and that a careful study of those changes can illuminate important viral replication and pathogenesis strategies that can be investigated at a mechanistic level and that may suggest additional avenues for the development of novel antiretroviral therapeutics.
Supplementary Material
Acknowledgments
We thank Michael Lu for his helpful assistance, Rich Simon for his advice on statistics, careful readings of the manuscript, and helpful suggestions, and Mike Kruhlak for helpful suggestions.
This work was supported in part by the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abbott, D. W., and J. T. Holt. 1999. Mitogen-activated protein kinase kinase 2 activation is essential for progression through the G2/M checkpoint arrest in cells exposed to ionizing radiation. J. Biol. Chem. 274:2732-2742. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury, I. H., X. F. Wang, N. R. Landau, M. L. Robb, V. R. Polonis, D. L. Birx, and J. H. Kim. 2003. HIV-1 Vpr activates cell cycle inhibitor p21/Waf1/Cip1: a potential mechanism of G2/M cell cycle arrest. Virology 305:371-377. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, E. A., G. Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J. Virol. 64:3097-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, E. A., E. F. Terwilliger, Y. Jalinoos, J. Proulx, J. G. Sodroski, and W. A. Haseltine. 1990. Identification of HIV-1 vpr product and function. J. Acquir. Immune Defic. Syndr. 3:11-18. [PubMed] [Google Scholar]
- 5.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 6.Conti, L., P. Matarrese, B. Varano, M. C. Gauzzi, A. Sato, W. Malorni, F. Belardelli, and S. Gessani. 2000. Dual role of the HIV-1 vpr protein in the modulation of the apoptotic response of T cells. J. Immunol. 165:3293-3300. [DOI] [PubMed] [Google Scholar]
- 7.Conti, L., G. Rainaldi, P. Matarrese, B. Varano, R. Rivabene, S. Columba, A. Sato, F. Belardelli, W. Malorni, and S. Gessani. 1998. The HIV-1 vpr protein acts as a negative regulator of apoptosis in a human lymphoblastoid T cell line: possible implications for the pathogenesis of AIDS. J. Exp. Med. 187:403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckstein, D. A., M. P. Sherman, M. L. Penn, P. S. Chin, C. M. De Noronha, W. C. Greene, and M. A. Goldsmith. 2001. HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J. Exp. Med. 194:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felzien, L. K., C. Woffendin, M. O. Hottiger, R. A. Subbramanian, E. A. Cohen, and G. J. Nabel. 1998. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc. Natl. Acad. Sci. USA 95:5281-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forget, J., X. J. Yao, J. Mercier, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 vpr protein transactivation function: mechanism and identification of domains involved. J. Mol. Biol. 284:915-923. [DOI] [PubMed] [Google Scholar]
- 11.Fukumori, T., H. Akari, S. Iida, S. Hata, S. Kagawa, Y. Aida, A. H. Koyama, and A. Adachi. 1998. The HIV-1 Vpr displays strong anti-apoptotic activity. FEBS Lett. 432:17-20. [DOI] [PubMed] [Google Scholar]
- 12.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs, J. S., A. A. Lackner, S. M. Lang, M. A. Simon, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1995. Progression to AIDS in the absence of a gene for vpr or vpx. J. Virol. 69:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh, W. C., N. Manel, and M. Emerman. 2004. The human immunodeficiency virus Vpr protein binds Cdc25C: implications for G2 arrest. Virology 318:337-349. [DOI] [PubMed] [Google Scholar]
- 16.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65-71. [DOI] [PubMed] [Google Scholar]
- 17.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrimech, M., X. J. Yao, F. Bachand, N. Rougeau, and E. A. Cohen. 1999. Human immunodeficiency virus type 1 (HIV-1) Vpr functions as an immediate-early protein during HIV-1 infection. J. Virol. 73:4101-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iordanskiy, S., Y. Zhao, L. Dubrovsky, T. Iordanskaya, M. Chen, D. Liang, and M. Bukrinsky. 2004. Heat shock protein 70 protects cells from cell cycle arrest and apoptosis induced by human immunodeficiency virus type 1 viral protein R. J. Virol. 78:9697-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacotot, E., L. Ravagnan, M. Loeffler, K. F. Ferri, H. L. Vieira, N. Zamzami, P. Costantini, S. Druillennec, J. Hoebeke, J. P. Briand, T. Irinopoulou, E. Daugas, S. A. Susin, D. Cointe, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2000. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 191:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kino, T., A. Gragerov, O. Slobodskaya, M. Tsopanomichalou, G. P. Chrousos, and G. N. Pavlakis. 2002. Human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr induces transcription of the HIV-1 and glucocorticoid-responsive promoters by binding directly to p300/CBP coactivators. J. Virol. 76:9724-9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan, V., and S. L. Zeichner. 2004. Host cell gene expression during human immunodeficiency virus type 1 latency and reactivation and effects of targeting genes that are differentially expressed in viral latency. J. Virol. 78:9458-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lum, J. J., O. J. Cohen, Z. Nie, J. G. Weaver, T. S. Gomez, X. J. Yao, D. Lynch, A. A. Pilon, N. Hawley, J. E. Kim, Z. Chen, M. Montpetit, J. Sanchez-Dardon, E. A. Cohen, and A. D. Badley. 2003. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J. Clin. Investig. 111:1547-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansour, S. J., W. T. Matten, A. S. Hermann, J. M. Candia, S. Rong, K. Fukasawa, G. F. Vande Woude, and N. G. Ahn. 1994. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265:966-970. [DOI] [PubMed] [Google Scholar]
- 27.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muthumani, K., D. S. Hwang, B. M. Desai, D. Zhang, N. Dayes, D. R. Green, and D. B. Weiner. 2002. HIV-1 Vpr induces apoptosis through caspase 9 in T cells and peripheral blood mononuclear cells. J. Biol. Chem. 277:37820-37831. [DOI] [PubMed] [Google Scholar]
- 29.Patel, C. A., M. Mukhtar, and R. J. Pomerantz. 2000. Human immunodeficiency virus type 1 Vpr induces apoptosis in human neuronal cells. J. Virol. 74:9717-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon, B., and I. S. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raingeaud, J., A. J. Whitmarsh, T. Barrett, B. Derijard, and R. J. Davis. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278:25879-25886. [DOI] [PubMed] [Google Scholar]
- 35.Sawaya, B. E., K. Khalili, J. Gordon, A. Srinivasan, M. Richardson, J. Rappaport, and S. Amini. 2000. Transdominant activity of human immunodeficiency virus type 1 Vpr with a mutation at residue R73. J. Virol. 74:4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, M. P., C. M. de Noronha, L. A. Eckstein, J. Hataye, P. Mundt, S. A. Williams, J. A. Neidleman, M. A. Goldsmith, and W. C. Greene. 2003. Nuclear export of Vpr is required for efficient replication of human immunodeficiency virus type 1 in tissue macrophages. J. Virol. 77:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, S. A., B. Poon, J. Y. Song, and I. S. Chen. 2000. Human immunodeficiency virus type 1 vpr induces apoptosis through caspase activation. J. Virol. 74:3105-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ussar, S., and T. Voss. 2004. MEK1 and MEK2, different regulators of the G1/S transition. J. Biol. Chem. 279:43861-43869. [DOI] [PubMed] [Google Scholar]
- 39.Vazquez, N., T. Greenwell-Wild, N. J. Marinos, W. D. Swaim, S. Nares, D. E. Ott, U. Schubert, P. Henklein, J. M. Orenstein, M. B. Sporn, and S. M. Wahl. 2005. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J. Virol. 79:4479-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wainberg, Z., M. Oliveira, S. Lerner, Y. Tao, and B. Brenner. 1997. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology 233:364-373. [DOI] [PubMed] [Google Scholar]
- 41.Yao, X. J., N. Rougeau, D. Ghislaine, J. Lemay, and E. A. Cohen. 2004. Analysis of HIV-1 Vpr determinants responsible for cell growth arrest in Saccharomyces cerevisiae. Retrovirology 1:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, Y., J. Cao, M. R. O'Gorman, M. Yu, and R. Yogev. 1996. Effect of human immunodeficiency virus type 1 protein R (vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J. Virol. 70:5821-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, C. F., and K. L. Guan. 1994. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 13:1123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, C. F., and K. L. Guan. 1993. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J. Biol. Chem. 268:23933-23939. [PubMed] [Google Scholar]
- 45.Zhu, Y., H. A. Gelbard, M. Roshal, S. Pursell, B. D. Jamieson, and V. Planelles. 2001. Comparison of cell cycle arrest, transactivation, and apoptosis induced by the simian immunodeficiency virus SIVagm and human immunodeficiency virus type 1 vpr genes. J. Virol. 75:3791-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmerman, E. S., J. Chen, J. L. Andersen, O. Ardon, J. L. Dehart, J. Blackett, S. K. Choudhary, D. Camerini, P. Nghiem, and V. Planelles. 2004. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol. Cell. Biol. 24:9286-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.