Abstract
Homologues of the UL7 gene of herpes simplex virus type 1 are conserved in alpha-, beta-, and gammaherpesviruses. However, little is known about their functions. Using a monospecific rabbit antiserum raised against a bacterial fusion protein, we identified the UL7 gene product of the neurotropic alphaherpesvirus pseudorabies virus (PrV). In Western blot analyses of infected cells and purified PrV particles the serum specifically detected a 29-kDa protein, which matches the calculated mass of the 266-amino-acid translation product of PrV UL7. For functional analysis, UL7 was deleted by mutagenesis of an infectious full-length clone of the PrV genome in Escherichia coli. The obtained recombinant PrV-ΔUL7F was replication competent in rabbit kidney cells, but maximum virus titers were decreased nearly 10-fold and plaque diameters were reduced by ca. 60% compared to wild-type PrV. Electron microscopy of infected cells revealed that in the absence of UL7, formation and nuclear egress of nucleocapsids were not affected, whereas secondary envelopment of cytoplasmic nucleocapsids appeared to be delayed and release of mature virions was less efficient. The observed replication defects were corrected by repair of the viral UL7 gene or by propagation of PrV-ΔUL7F in UL7-expressing cells. PrV-ΔUL7F was moderately attenuated in mice. Compared to wild-type virus, mean survival times were prolonged from 2 to 3 days after intranasal infection. However, neuroinvasion and transneuronal spread of PrV were not abolished in the absence of UL7. Thus, UL7 encodes a virion protein of PrV, which plays a role during virion maturation and egress both in vitro and in vivo.
Pseudorabies virus (PrV, suid herpesvirus 1) is the causative agent of Aujeszky's disease in pigs. However, it is also highly pathogenic for many other mammalian species. Whereas infection leads to fatal neurological disorders in most animals, adult pigs usually survive and become virus carriers due to latent infection of the central nervous system (38). PrV is classified as a member of the genus Varicellovirus within the Alphaherpesvirinae subfamily of the Herpesviridae (40) and possesses a class D herpesvirus genome which consists of long and short unique regions (UL and US) and of inverted repeat sequences (IRS and TRS) flanking the US region (Fig. 1A) (46). The complete nucleotide sequence of the 143-kbp PrV genome has been determined recently (29), and all 72 identified open reading frames (ORFs) were shown to possess homologues in at least one of the previously characterized genomes of other mammalian alphaherpesviruses, including herpes simplex virus type 1 (HSV-1) (36), varicella-zoster virus (12), and equine herpesvirus 1 (EHV-1) (48). Gene arrangement also proved to be widely colinear, except for an inversion within the UL genome region of PrV spanning the homologues of the HSV-1 UL27 to UL44 genes (3, 6).
FIG. 1.
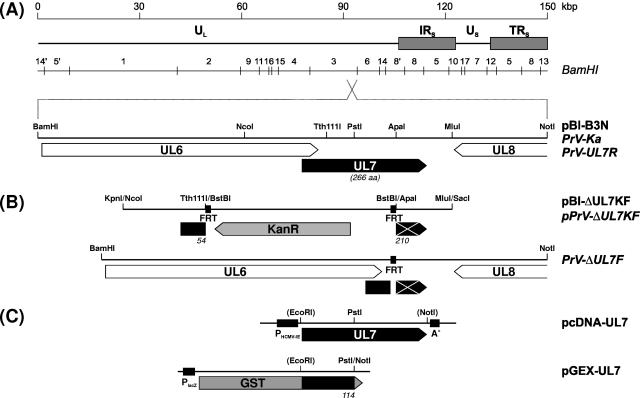
Construction of plasmids and PrV recombinants. (A) The PrV genome consists of two unique regions (UL and US) and of inverted repeat sequences (IRS and TRS) flanking the US region. The positions of BamHI restriction sites are indicated. An enlarged section shows the analyzed genome part which was cloned in pBl-B3N. Open reading frames (ORFs) are drawn as pointed rectangles. (B) In plasmid pBl-ΔUL7KF the UL7 ORF was replaced by a kanamycin resistance gene (KanR) flanked by Flp-recombinase target sites (FRT). The insert of pBl-ΔUL7KF was amplified by PCR and used for mutagenesis of an infectious full-length clone of the PrV-genome in E. coli. The resistance gene was removed by Flp-mediated recombination, and PrV-ΔUL7F was isolated after transfection of rabbit kidney cells. The UL7 rescuant PrV-UL7R was obtained after transfection of cells with genomic DNA of PrV-ΔUL7F and pBl-B3N. (C) Plasmid pcDNA-UL7 permits constitutive expression of UL7 in eucaryotic cells under control of the HCMV immediate-early promoter (PHCMV-IE) and was used for generation of the trans-complementing cell line RK13-UL7. The bacterial UL7 fusion protein with glutathione S-transferase (GST) expressed from pGEX-UL7 was used for rabbit immunization followed by antiserum preparation. Codon ranges of expressed or retained UL7 gene fragments as well as relevant restriction sites are indicated (artificial sites in parentheses). aa, amino acids.
Several gene clusters are conserved throughout mammalian and avian alpha-, beta-, and gammaherpesviruses and are therefore suggested to encode proteins which are required for fundamental steps of the viral life cycle (46). One of these clusters includes the UL7 gene of the alphaherpesvirus HSV-1 (36), whose homologues were named UL103 in the betaherpesvirus human cytomegalovirus (HCMV) (8) and BBRF2 in the gammaherpesvirus Epstein-Barr virus (EBV) (2). UL7 is surrounded by four essential genes. The UL5 and UL8 open reading frames encode two subunits of the helicase-primase complex required for viral DNA replication, whereas the UL9 gene codes for the origin binding protein (10, 43). The essential UL6 protein of HSV-1 has recently been shown to form the portal for entry and presumably also for release of viral DNA into and from capsids (41). However, the function of UL7 is still unclear, although it is also considered to be essential for replication of HSV-1 (45). The UL7 gene products of HSV-1 and -2 were recently identified using a cross-reacting antiserum raised against the HSV-2 protein (42). The HSV-2 gene product was characterized as a temporally nuclear localizing protein which is associated with A, B, and C capsids and is also present in mature virions (42). In contrast, the homologous UL7 gene product of another alphaherpesvirus, bovine herpesvirus 1 (BHV-1), has been described as a nonstructural protein which is predominantly localized in the cytoplasm of infected cells (47). Successful isolation of a deletion mutant revealed that UL7 is dispensable for replication of BHV-1 in cell culture, although virus release appeared to be impaired, and maximum virus titers were significantly reduced in the absence of the UL7 protein (47).
Previously, we identified and functionally characterized several PrV-encoded proteins which are not essential but nevertheless are relevant for virion formation and egress (reviewed in reference 39). Few of these proteins, like the UL31 and UL34 gene products, are absent from mature virus particles but involved in the export of newly synthesized nucleocapsids from the host cell nucleus by consecutive envelopment and de-envelopment at the inner and outer leaflets of the nuclear membrane (17, 27). Many more proteins have been shown to be involved in tegumentation and secondary envelopment of cytoplasmic nucleocapsids. Most of these viral gene products are either components of the tegument, like the UL36, UL37, and UL46 to UL49 proteins (18, 19, 20, 21, 28, 31), or are associated with the envelope of mature virus particles, like the UL11 gene product or glycoproteins gE and gM (4, 5, 32). Two other viral membrane proteins, gK and the interacting UL20 gene product, were demonstrated to be required for efficient release of finally enveloped virions from the cell (13, 16, 25). Among these proteins, only the UL36 gene product proved to be absolutely essential for productive replication in cell culture (21). However, several “nonessential” proteins play important roles during neuroinvasion and transneuronal spread of PrV in vivo. Neurovirulence in rodents was significantly affected in the absence of glycoprotein gE, the membrane-associated US9 and UL11 gene products, and the UL37, UL47, and UL48 tegument proteins (1, 7, 24).
Since the published results about function and virion localization of the UL7 proteins of herpes simplex viruses and BHV-1 are controversial (see above), we decided to investigate the respective gene product of PrV. The UL7 gene product of PrV was identified using a monospecific rabbit antiserum raised against a bacterial fusion protein. We further addressed the question whether the UL7 protein is relevant for in vitro virus replication and/or neurovirulence of PrV. For that purpose, an UL7-negative virus recombinant was generated by mutagenesis of an infectious plasmid clone of the PrV genome (32) in Escherichia coli. The growth properties of the obtained deletion mutant and a corresponding rescuant were analyzed in noncomplementing and in UL7-expressing cells. Virion morphogenesis of PrV-ΔUL7F was investigated by electron microscopy, and survival times of experimentally infected mice were determined.
MATERIALS AND METHODS
Viruses and cells.
The virus mutants used in this study were derived from PrV strain Kaplan (PrV-Ka) (23), whose genome has been cloned as a bacterial artificial chromosome (BAC) in E. coli (pPrV-ΔgB) (32). Viruses were propagated on rabbit kidney (RK13) cells in minimum essential medium (MEM) supplemented with 10% fetal calf serum (Invitrogen). A UL7-expressing cell line (RK13-UL7) was established to permit trans-complementation of UL7-negative PrV. For that purpose the UL7 ORF was amplified from genomic PrV DNA by PCR using custom-made (MWG Biotech) primers PUL7-F (CAGAATTCGGCTCCGCGATGGAGG), PUL7-R (CAGCGGCCGCTCACTCGTAGCGCACAAAC), and Pfx DNA polymerase (Invitrogen). The primers correspond to the reverse of nucleotides 87473 to 87489 (PUL7-F) and to nucleotides 86679 to 86697 (PUL7-R) of the PrV genome sequence (GenBank accession no. BK001744) (29) and contain the UL7 start and stop codons, respectively (underlined). At the 5′ primer ends, artificial EcoRI and NotI restriction sites (printed in italics) were added for convenient cloning. The 828-bp PCR product was digested with these enzymes and inserted into the similarly cleaved plasmid vector pcDNA3 (Invitrogen), which contained a neomycin/Geneticin resistance gene and also permitted constitutive expression of the UL7 protein under control of the HCMV immediate-early promoter. The correct insert sequence of the obtained plasmid pcDNA-UL7 (Fig. 1C) was verified by DNA sequencing. Two days after calcium-phosphate-mediated transfection (22) with this plasmid, RK13 cells were trypsinized, diluted in medium containing 500 mg/ml Geneticin (Invitrogen), and seeded into 96-well microtiter plates. Resistant cell clones were tested for UL7 expression by Western blot analyses with a UL7-specific antiserum (see below), and one stable cell line (RK13-UL7) was further propagated.
Generation of virus mutants.
A 3,335-bp BamHI/NotI subfragment of the genomic BamHI fragment 3 of PrV-Ka was cloned in pBluescript SK(−) (Stratagene) to yield plasmid pBl-B3N (Fig. 1A). For construction of a UL7 deletion plasmid, the insert of pBl-B3N was first shortened to 1,335 bp by consecutive MluI/SacI and KpnI/NcoI double digestions, followed by Klenow treatment and religation. From the resulting plasmid a 463-bp Tth111I/ApaI fragment spanning UL7 codons 55 to 209 was deleted and replaced by a 1,258-bp BstBI fragment of pKD13 (11), which contained a kanamycin resistance gene flanked by Flp-recombinase recognition target sites (FRT). The insert fragment of pBl-ΔUL7KF (Fig. 1B) was amplified by PCR using vector-specific M13/pUC (−47) and M13/pUC reverse (−48) primers (New England Biolabs). The obtained PCR product was used for Red recombinase-mediated mutagenesis of the BAC pPrV-ΔgB in E. coli as described previously (11, 32). After isolation of kanamycin-resistant clones, the resistance gene was removed by mutagenesis with the FRT site-specific Flp-recombinase, which was provided by transformation with helper plasmid pCP20 (9). Finally, the mini-F-plasmid vector sequences were removed from the gB gene locus of PrV, and the essential gB gene was restored by cotransfection of RK13 cells with BAC DNA and plasmid pUC-B1BclI (32). A single plaque isolate of the virus progeny was further propagated and designated PrV-ΔUL7F (Fig. 1B). The UL7 rescue mutant PrV-UL7R was isolated after cotransfection of RK13 cells with genomic DNA of PrV-ΔUL7F and plasmid pBl-B3N (Fig. 1A). Virion DNA of both PrV mutants was characterized by restriction analyses and Southern blot hybridization as well as by PCR amplification and sequencing of the UL7 gene region (results not shown).
In vitro growth studies.
For analysis of one-step growth kinetics, confluent monolayers of RK13 or RK13-UL7 cells were infected with 10 PFU per cell of wild-type PrV-Ka, PrV-ΔUL7F, or PrV-UL7R and incubated on ice for 1 h. Prewarmed medium was then added, and incubation was continued at 37°C. After 1 h, nonpenetrated virus was inactivated by low-pH treatment (37), and 1, 4, 8, 12, 24, 36, and 48 h after the temperature shift cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). For separate analysis of intra- and extracellular virions the suspensions were centrifuged for 5 min at 5,000 × g prior to lysis. The supernatants were transferred to new tubes, and the pellets were resuspended in the same volume of fresh medium. Progeny virus titers were determined by plaque assay in RK13-UL7 cells, which were overlaid with MEM containing 5% fetal calf serum and 6 g/liter methyl cellulose. After 2 days, cells were fixed for 1 h with 2% formaldehyde and stained for 15 min with 1% crystal violet in 50% ethanol. The mean virus titers of three independent experiments were calculated. Furthermore, the average diameters of 30 plaques per virus in RK13 and RK13-UL7 cells were converted into percent values of wild-type sizes, and standard deviations were determined.
Animal experiments.
The relevance of UL7 for virulence and neuronal spread of PrV was investigated in the mouse model essentially as described recently (24). Briefly, 8-week-old CD1 mice were infected by bilateral intranasal instillation of 106 PFU of PrV-ΔUL7F and PrV-Ka. Animals were observed three times a day, and mean survival times as well as standard deviations were determined. Viral spread in the trigeminal circuit was analyzed by immunohistochemistry of paraffin sections of the brains of animals which were necropsied 24, 48, and 72 h postinfection (p.i.). Viral antigen was detected by a monospecific rabbit serum against the major capsid protein of PrV (27).
Procaryotic expression and preparation of an UL7-specific antiserum.
The 817-bp EcoRI/NotI insert fragment of pcDNA-UL7 (Fig. 1C) was recloned into pGEX-4T-1 (Amersham) to permit expression of a UL7 fusion protein with glutathione S-transferase (GST) in E. coli. However, since the protein yield obtained after transformation of E. coli XL1-blue MRF′ (Stratagene) with the original construct was too low, the viral insert was shortened to 355 bp by double digestion with PstI and NotI, Klenow treatment, and religation. The resulting plasmid pGEX-UL7 (Fig. 1C) contained UL7 codons 1 to 114 which permitted abundant expression of a 40-kDa GST fusion protein. This protein was isolated and used for immunization of a rabbit as described previously (17). Sera collected before and after four applications of 100 mg protein in 4-week intervals were analyzed.
Western blot analyses.
RK13 cells were infected with PrV-Ka, PrV-ΔUL7F, or PrV-UL7R at a multiplicity of 5 PFU per cell and incubated at 37°C for 1 to 18 h. Samples of infected and noninfected RK13 cells, of RK13-UL7 cells, as well as of PrV virions were prepared as described previously (13), separated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (34), and electrotransferred to nitrocellulose membranes (Transblot SD cell; Bio-Rad). Blots were blocked with 5% low-fat milk in Tris-buffered saline (TBS-T; 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 0.25% Tween 20) and incubated for 1 h with rabbit antisera against the UL7 (this work) and UL34 (27) gene products or glycoprotein gH of PrV (26) at dilutions of 1:50,000 in TBS-T. Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
Electron microscopy.
RK13 cells were infected with 1 PFU per cell of PrV-ΔUL7F or PrV-UL7R. After 1 h on ice and an additional hour at 37°C, the inoculum was replaced by fresh medium, and incubation was continued for 13 h at 37°C. Fixation and embedding were done as described previously (27), and counterstained ultrathin sections were analyzed in an electron microscope (Tecnai 12; Philips).
RESULTS
Identification of the UL7 protein of PrV.
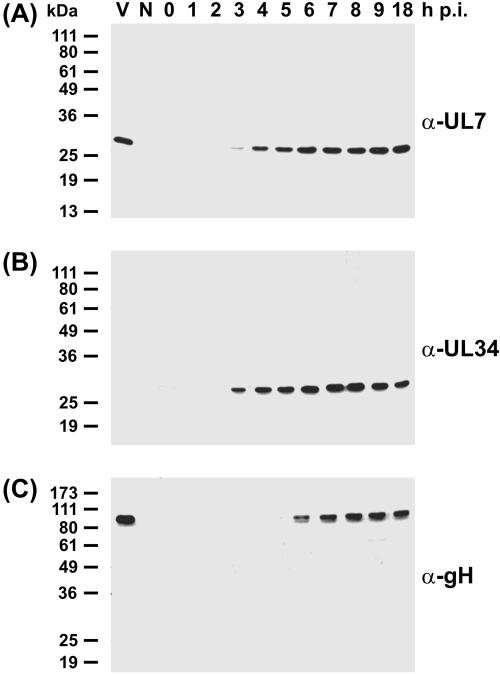
The deduced UL7 gene product of PrV strain Ka consists of 266 amino acids and possesses a calculated molecular mass of 29,046.41 Da (14). A monospecific rabbit antiserum raised against a bacterial fusion protein containing the UL7 amino acids 1 to 114 (Fig. 1C) specifically reacted with a protein of the expected size in Western blot analyses of PrV-infected RK13 cells (Fig. 2A). This protein was not found in uninfected cells (Fig. 2A) and also was not detected by the respective preimmune serum (data not shown). The amount of the UL7 protein increased from 3 to 9 h after infection, indicating early-late expression kinetics. The UL34 gene product of PrV (Fig. 2B) (27) exhibited similar expression kinetics, whereas envelope glycoprotein gH (Fig. 2C) (26) was first detected 6 h after infection. The 29-kDa UL7 protein was also expressed in RK13-UL7 cells which contain a genomic insertion of the PrV UL7 gene under control of the HCMV immediate-early promoter (Fig. 1C and 3A).
FIG. 2.
Expression kinetics and virion incorporation of the PrV UL7 protein. Rabbit kidney cells were infected with PrV-Ka at a multiplicity of infection of 5 and incubated at 37°C for 0 to 18 h. Lysates of infected and noninfected cells (N) and purified virions (V) were separated by SDS-PAGE. Western blots were incubated with monospecific antisera against the UL7 gene product (A), the nonstructural UL34 protein (B), or envelope glycoprotein gH (C). Molecular masses of marker proteins are indicated.
FIG. 3.
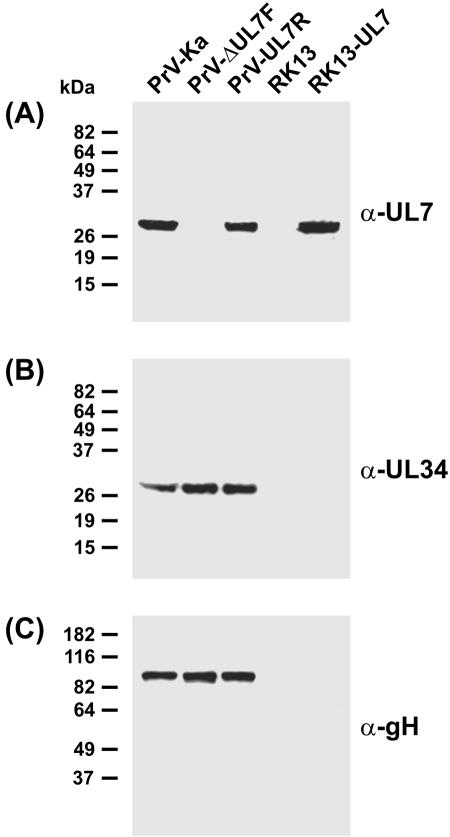
Protein expression of UL7 mutants. Western blots of rabbit kidney cells harvested 24 h after infection (multiplicity of infection, 5) with PrV-Ka, PrV-ΔUL7F, or PrV-UL7R of noninfected cells (RK13) and of RK13-UL7 cells were incubated with monospecific antisera against the UL7 protein (A), the UL34 protein (B), or gH (C). Molecular masses of marker proteins are indicated.
The UL7 protein was detectable not only in infected cells but also in sucrose gradient-purified PrV particles (Fig. 2A). The purity of the analyzed virion preparations was demonstrated by the absence of the nonstructural UL34 protein of PrV (Fig. 2B), which has been characterized as a constituent of primary enveloped intracellular, but not extracellular, virus particles (27). Thus, like glycoprotein gH (Fig. 2C), the UL7 gene product of PrV represents a structural component of mature virions.
The UL7 gene is dispensable for in vitro replication of PrV.
For functional characterization of the UL7 gene a deletion mutant was constructed by mutagenesis of a BAC clone of the PrV genome in E. coli (Fig. 1B). The resulting recombinant PrV-ΔUL7F exhibited a deletion of UL7 codons 55 to 209 and contained a 36-bp FRT site insertion as a rudiment of the mutagenesis reaction. The 5′ end of UL7 was not removed, since it overlaps the preceding UL6 gene (Fig. 1A). In contrast, expression of the retained 3′ end of UL7 was prevented by stop codons within the inserted FRT site. No UL7 gene product was found in Western blot analyses of RK13 cells infected with PrV-ΔUL7F (Fig. 3A), indicating that the N-terminal protein fragment might be either unstable or too small for detection. Wild-type-like protein expression was restored after repair of the viral UL7 gene in PrV-UL7R (Fig. 3A). Control blots were incubated with antibodies against other PrV proteins including the UL34 gene product and gH (Fig. 3B and C), which demonstrated that viral gene expression is not generally affected in the absence of UL7.
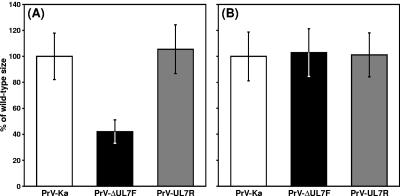
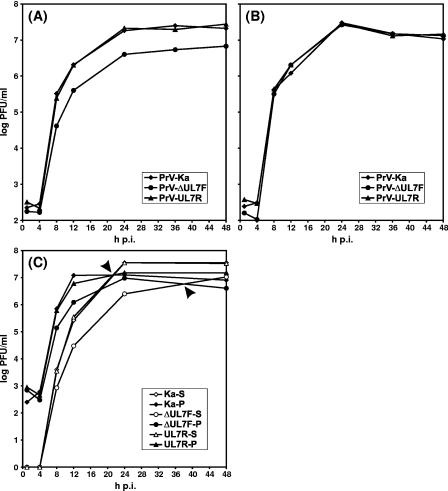
Since PrV-ΔUL7F was isolated from noncomplementing cells, the respective gene cannot be essential for in vitro virus replication. However, the UL7 deletion mutant exhibited significant growth defects in RK13 cells. Compared to wild-type PrV-Ka, plaque diameters of PrV-ΔUL7F were reduced by ca. 60% (Fig. 4A), and maximum virus titers were reduced 5- to 10-fold (Fig. 5A). Separate analysis of cell pellets and supernatants in one-step growth studies further indicated a release defect of PrV-ΔUL7F, since the titer of free virions exceeded the amount of cell-associated infectivity only as late as 40 h after infection, whereas in cells infected with PrV-Ka this already occurred after ca. 20 h (Fig. 5C). The observed replication defects could be corrected by repair of the viral UL7 gene in PrV-UL7R (Fig. 4A and 5A) or by propagation of PrV-ΔUL7F in trans-complementing RK13-UL7 cells (Fig. 4B and 5B). The latter finding indicates that the observed phenotype depends on the UL7 protein and not on cis-acting regulatory DNA sequences within the UL7 coding region.
FIG. 4.
Plaque sizes of UL7 mutants. RK13 (A) or RK13-UL7 (B) cells were infected with PrV-Ka, PrV-ΔUL7F, or PrV-UL7R and incubated for 48 h under semisolid medium. For each virus the average diameters of 30 plaques were determined and calculated in percentages compared to the plaques induced by PrV-Ka. Standard deviations are also shown.
FIG. 5.
One-step growth kinetics of UL7 mutants. RK13 (A and C) or RK13-UL7 (B) cells were infected with PrV-Ka, PrV-ΔUL7F, or PrV-UL7R at a multiplicity of infection of 5 and incubated at 37°C for 1, 4, 8, 12, 24, 36, and 48 h. Cells and medium were then harvested either together (A and B) or separately (C). After freeze-thawing, progeny virus titers (PFU/ml) in whole-cell lysates or cell pellets (P) and supernatants (S) were determined by plaque assays in RK13-UL7 cells. Crossover points of intra- and extracellular virus titers are indicated by arrowheads. The mean results of at least three independent experiments are shown.
The UL7 protein is involved in virus release.
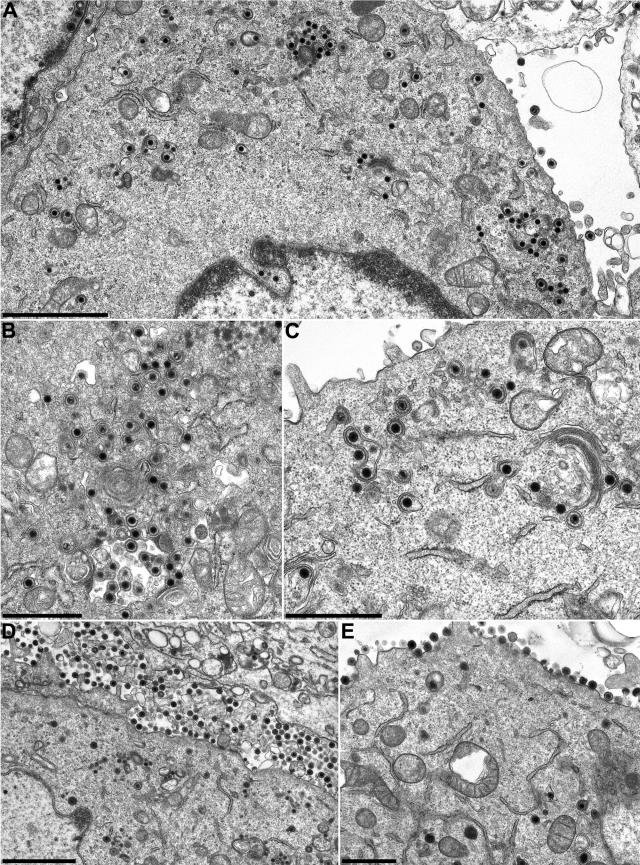
To investigate the role of UL7 during replication of PrV in more detail, RK13 cells were fixed 14 h after synchronized infection with PrV-ΔUL7F and analyzed by electron microscopy (Fig. 6A to C). These studies revealed that the intranuclear steps of virion formation, including capsid formation, DNA encapsidation, and budding of nucleocapsids through the nuclear membrane, were not detectably affected. Numerous DNA-filled capsids were present in the cytoplasm (Fig. 6A and B), and budding of nucleocapsids into Golgi-derived membrane vesicles was also observed (Fig. 6B and C). However, only very few enveloped virions were detectable in the extracellular space (Fig. 6A and C). In contrast, from RK13 cells which had been infected in parallel with similar doses of PrV-UL7R (Fig. 6D and E) or wild-type PrV-Ka (not shown), a majority of the virus progeny was already released after 14 h and detected in the extracellular space lining the plasma membrane. Thus, the UL7 protein is involved in a late step of virion morphogenesis.
FIG.6.
Egress of PrV-ΔUL7F. RK13 cells were fixed 14 h after infection with PrV-ΔUL7F (A, B, and C) or PrV-UL7R (D and E) at a multiplicity of infection of 1 and stained with uranyl acetate. Ultrathin sections were analyzed by electron microscopy. In the absence of UL7, nucleocapsids were formed and released from the host cell nucleus (A). However, secondary envelopment of cytoplasmic nucleocapsids appeared to be delayed (B and C), and only a few mature virus particles were detectable in the extracellular space (A and C). In contrast, virions of PrV-UL7R were efficiently released from cells (D and E). Bars represent 2.0 mm (A and D) or 1.0 mm (B, C, and E).
UL7 modulates neurovirulence of PrV.
Intranasal infection of 8-week-old mice with PrV-ΔUL7F was lethal for all animals, as was infection with parental wild-type PrV (Table 1). However, PrV-ΔUL7F-infected mice died at an average time of 70 h p.i., compared to 50 h after infection with PrV-Ka. At day 2 after infection with PrV-ΔUL7F only minor clinical signs (depression and anorexia) could be observed, whereas animals infected with PrV-Ka showed severe excitation and convulsions, heavy dyspnoea, and extensive scratching of the facial and nasal skin causing severe hemorrhagic dermal erosions (“mad itch” syndrome). One day later, the mice infected with PrV-ΔUL7F developed similar symptoms.
TABLE 1.
Virulence of PrV-ΔUL7F in infected mice
| Criterion | Virus
|
|
|---|---|---|
| PrV-Ka | PrV-ΔUL7F | |
| Mean time to death (h p.i.)a | 50.0 (1.9) | 70 (3.19) |
| Clinical symptomsb | ||
| 1 d p.i. | 0 | 0 |
| 2 d p.i. | +++ | + |
| 3 d p.i. | † | +++ |
| Immunohistochemistryc | ||
| Nasal cavity (d p.i.) | 1 | 1 |
| 1st order neuron (d p.i.) | 1 | 2 |
| 2nd order neuron (d p.i.) | 2 | 3 |
| 3rd order neuron (d p.i.) | ND | 3 |
Average times to death after intranasal infection with 106 PFU of the indicated virus were calculated for 10 animals each. Standard deviations are indicated in parentheses.
Clinical symptoms were scored as follows: 0, clinically inconspicuous; +, slight depression, hunched position, ruffled hair coat; ++, apathy, anorexia, moderate dyspnoea, slight facial pruritus; +++, severe attacks of excitation, self-mutilation, skin erosions, heavy dyspnoea; †, animals moribund or dead.
Time in days after infection (d p.i.) of the first UL19 antigen detection at the following levels of the trigeminal pathway: nasal cavity (respiratory mucosal epithelium), 1st order neurons (trigeminal ganglion), 2nd order neurons (spinal trigeminal nucleus, Sp5), 3rd order neurons (ectorhinal cortex). ND, not detected.
One day after infection with either virus, immunohistochemistry revealed the presence of the major capsid protein in clusters of respiratory epithelial cells of the nasal mucosa (Table 1). Infected first-order neurons in the trigeminal ganglion were detectable as early as 24 h after infection with PrV-Ka, whereas infection of the trigeminal ganglion by PrV-ΔUL7F was not observed before 48 h p.i. Viral antigen could be also detected in second-order neurons of the spinal trigeminal nuclei 48 h after infection with PrV-Ka or 72 h after infection with PrV-ΔUL7F (Table 1). In addition, PrV-ΔUL7F, but not PrV-Ka, could be detected in third-order cortical neurons (mainly in the ectorhinal cortex) at day 3 after infection. Taken together, these findings indicate that neuroinvasion is delayed in the absence of the UL7 protein.
DISCUSSION
The UL7 gene is conserved in all three subfamilies of mammalian and avian herpesviruses. However, besides the UL7 proteins of HSV-1 and -2, only the respective gene product of BHV-1 has been identified previously (42, 47). In the present study, we identified the UL7 gene product of PrV as a 29-kDa protein which matches the calculated molecular mass of the deduced 266-amino-acid translation product. Since the protein expressed in cells transfected with a UL7 expression plasmid and the in vitro translation product of the ORF (not shown) exhibited the same apparent mass, there is no evidence for posttranslational modifications of the viral gene product. Correlating with first detection of the UL7 mRNA of PrV at 3 h after infection (14), the protein was also found in the cells from this point. Increasing amounts of the protein until 9 h p.i. indicated early-late expression kinetics similar to those described for the homologous gene product of BHV-1 (47). Whereas the BHV-1 protein was not detected in purified virions (47), the UL7 gene products of HSV-2 (42) and PrV proved to be structural proteins. This difference may point to a different role of these gene products in the different viruses or may simply be due to insufficient sensitivity of the assays in the case of BHV-1.
UL7 is considered to be essential for replication of HSV-1 in cell culture (45), although the experimental evidence for this assumption is unknown to us. It has been speculated that the UL7 protein might be involved in cleavage and packaging of herpesvirus DNA, since the best conserved domain in all UL7 homologues [G-F-x(8)-E-D-x-V-x(12)-R] was also found in topoisomerase III of fission yeast (42). However, this domain is not perfectly conserved in topoisomerases of other species, and its functional relevance is unclear. We now demonstrate that a UL7-negative PrV mutant is replication competent in cell culture, although major parts of the respective ORF including the codons of the conserved domain (amino acids 71 to 97) had been deleted from the virus genome. Furthermore, electron microscopy of cells infected with PrV-ΔUL7F revealed that formation of DNA-filled capsids in the cell nucleus was not detectably affected. Our results are in good agreement with previous studies, which showed that the UL7 gene of BHV-1 is also dispensable for virus replication in vitro (47). Thus, despite their high degree of sequence homology the UL7 proteins might possess different functions in the Simplexvirus (HSV-1) and Varicellovirus (BHV-1, PrV) subfamilies of Alphaherpesvirinae. Possibly, other gene products of BHV-1 and PrV, but not of HSV-1, are able to compensate for lack of UL7. On the other hand, mutations introduced into the UL7 gene of HSV-1 might also affect expression of other genes, e.g., of UL6 which overlaps and forms a 3′-coterminal transcription unit with UL7 (36, 44). Remarkably, the UL6 gene encodes the portal protein which is essential for cleavage and encapsidation of virion DNA (35, 41). Overlapping and coterminally transcribed UL6 and UL7 ORFs were also found in PrV (14), and to avoid damage of UL6, codons 1 to 54 of UL7 were retained in our deletion mutant PrV-ΔUL7F.
Deletion of PrV UL7 resulted in significantly reduced plaque sizes and virus titers in cell culture. These defects were exclusively caused by the absence of the UL7 protein, since they were corrected in UL7-expressing cells. As mentioned, electron microscopy revealed that the defects of PrV-ΔUL7F were not a consequence of inefficient nucleocapsid formation. Apparently, nuclear egress of capsids by consecutive envelopment and de-envelopment at the nuclear membrane was also not impaired, since a large number of DNA-containing nucleocapsids was detected in the cytoplasm. Budding of these capsids into Golgi-derived vesicles was also observed. However, release of finally enveloped virions from the cells seemed to be inefficient in the absence of PrV UL7. Coinciding with these results of ultrastructural analyses, a major portion of infectious PrV-ΔUL7F remained cell associated, even at late times after infection. A similar phenotype was described for UL7-negative BHV-1 (47), which indicates similar functions of the investigated gene in both viruses. Most other deletions of “nonessential” genes which are involved in maturation and egress of PrV exhibited distinct ultrastructural phenotypes. In the absence of the tegument proteins encoded by UL37 or UL48, secondary envelopment of cytoplasmic nucleocapsids was almost completely abolished, whereas enveloped but capsidless L-particles were still formed in the trans-Golgi region and released from the cells (18, 28). In contrast, after simultaneous deletion of the nonessential envelope glycoproteins gE and gM, budding into membrane vesicles was inhibited, leading to aggregation of tegumented nucleocapsids in the cytoplasm (4, 5). In the absence of the membrane-associated UL11 protein, secondary envelopment of PrV was also impaired and accompanied by pronounced dilatations of trans-Golgi membranes (32, 33). Neither membrane proliferation, accumulation of capsids, electron-dense tegument material, nor L-particles were observed in the absence of UL7. Despite the severe replication defects, electron microscopy revealed similar unspectacular phenotypes of PrV mutants lacking the tegument protein encoded by UL51 (30) or the multiple-membrane-spanning proteins encoded by UL53 (gK) and UL20 (16, 25). The snapshots of the electron microscope cannot elucidate whether the absence of extracellular virions in the presence of all known maturation intermediates is due to delayed envelopment, impaired fusion of the virion-containing vesicles with the cytoplasmic membrane, or reentry of released virions into infected cells. The UL7 gene product exhibits no characteristics of membrane-anchored proteins and thus most likely represents a component of the herpesvirus tegument. Therefore, it appears more likely that the UL7 gene product is relevant during secondary envelopment of tegumented nucleocapsids than during later steps when it is located inside the enveloped virus particle. In transiently transfected cells expressing UL7 from the strong heterologous HCMV immediate-early promoter/enhancer, our antiserum showed strong immunoreactivity only in the cytoplasm (data not shown), which would correlate with the acquisition of the UL7 protein as a tegument component during virion morphogenesis in the cytoplasm. Unfortunately, in virus-infected cells only background staining was observed, which was probably due to the low abundance of the UL7 protein in wild-type PrV infection.
Deletion of UL7 affected not only in vitro replication of PrV but also neurovirulence in mice. After intranasal infection with PrV-ΔUL7F the animals survived for ca. 70 h, which is 20 h longer than observed after administration of the parental wild-type strain PrV-Ka. The effect of UL7 deletion was more pronounced than those found after deletion of several other nonessential genes, including UL11, UL46, UL47, and UL51 (24, 30). However, neuroinvasion was only delayed but not blocked in the absence of UL7. Thus, unlike the membrane protein encoded by US9 (7) or the tegument protein encoded by UL37 (24), the UL7 protein does apparently not possess functions which are absolutely required for replication or spread in neurons. Remarkably, PrV-ΔUL7F was demonstrated to infect not only first- and second-order neurons but also third-order neurons in the ectorhinal cortex, which is usually not affected by wild-type PrV (24). These findings indicate that delayed occurrence of disease in animals infected with PrV-ΔUL7F might coincide with delayed destruction of infected neurons and facilitate virus spread into higher regions of the brain. Due to its circuit-specific spread in the central nervous system, PrV has become a powerful tool for neurophysiology (15). Possibly, the viruses used as neuronal tracers can be further refined by mutations conferring a moderate attenuation, e.g., by deletion of UL7.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me854/5-2).
We thank Jutta Veits and Dorothee Wiesner for help with antiserum preparation and Jens P. Teifke for advice on the evaluation of animal experiments. The technical and photographical assistance of Charlotte Ehrlich, Mandy Jörn, Petra Meyer, Diana Werner, and Elke Zorn is greatly appreciated.
REFERENCES
- 1.Babic, N., B. Klupp, A. Brack, T. C. Mettenleiter, G. Ugolini, and A. Flamand. 1996. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology 219:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Ben Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology 127:194-204. [DOI] [PubMed] [Google Scholar]
- 4.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 7.Brideau, A. D., M. G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1 54:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Crute, J. J., T. Tsurumi, L. Zhu, S. K. Weller, P. D. Olivo, M. D. Challberg, E. S. Mocarski, and I. R. Lehman. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. USA 86:2186-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 13.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkstra, J. M., W. Fuchs, T. C. Mettenleiter, and B. G. Klupp. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17-35. [DOI] [PubMed] [Google Scholar]
- 15.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186:S209-S214. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., B. G. Klupp, H. Granzow, A. Mundt, C. Hengartner, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 24.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., H. Granzow, R. Klopfleisch, W. Fuchs, M. Kopp, M. Lenk, and T. C. Mettenleiter. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 36.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis-state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 39.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minson, A. C., A. Davison, R. Eberle, R. C. Desrosiers, B. Fleckenstein, D. J. McGeoch, P. E. Pellet, B. Roizman, and D. M. J. Studdert. 2000. Family Herpesviridae, p. 203-225. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, San Diego, Calif.
- 41.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:109923-110932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nozawa, N., T. Daikoku, Y. Yamauchi, H. Takakuwa, F. Goshima, T. Yoshikawa, and Y. Nishiyama. 2002. Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes 24:257-266. [DOI] [PubMed] [Google Scholar]
- 43.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1988. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc. Natl. Acad. Sci. USA 85:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel, A. H., and J. B. MacLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-468. [DOI] [PubMed] [Google Scholar]
- 45.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 46.Roizman, B., and P. E. Pellet. 2001. The family herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 47.Schmitt, J., and G. M. Keil. 1996. Identification and characterization of the bovine herpesvirus 1 UL7 gene and gene product which are not essential for virus replication in cell culture. J. Virol. 70:1091-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]