Abstract
The human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr has previously been shown to bind to the cellular uracil DNA glycosylase UNG. We show here that the binding of Vpr to UNG and to the related enzyme SMUG induces their proteasomal degradation. UNG and SMUG were found to be encapsidated in Δvpr HIV-1 virions but were significantly less abundant in vpr+ virions. Δvpr virions contained readily detectable uracil-DNA glycosylase enzymatic activity, while the activity was reduced to undetectable levels in vpr+ virions. Consistent with proteasomal degradation, complexes that contained Vpr and the E3 ubiquitin ligase components Cul1 and Cul4 were detected in cell lysates. We hypothesized that the interaction of Vpr might be a means for the virus to reduce the frequency of abasic sites in viral reverse transcripts at uracil residues caused by APOBEC3-catalyzed deamination of cytosine residues. Although APOBEC3 is largely neutralized by the Vif accessory protein, residual enzyme could remain in virions that would generate uracils. In support of this, Δvif vpr+ HIV-1 produced in the presence of limited amounts of APOBEC3G was significantly more infectious than Δvif Δvpr virus. In Addition, vpr+ HIV-1 replicated more efficiently than vpr− virus in cells that expressed limited amounts of APOBEC3G. The findings highlight the importance of cytidine deamination in the virus replication cycle and present a novel function for Vpr.
Lentiviruses encode the accessory protein viral protein R (Vpr), the function of which is not entirely clear (18). Vpr is expressed late in infection from a spliced mRNA. The protein localizes to the nuclei of infected cells (16, 23), where it causes the cells to arrest in the G2/M phase of the cell cycle (14). Unlike the other regulatory proteins of human immunodeficiency virus type 1 (HIV-1), Vpr is encapsidated in significant quantity in virions. Encapsidation of Vpr is mediated by interaction with amino acids of the p6 region of the Gag polyprotein precursor, Pr55gag (8, 32). Vpr is not required for HIV-1 replication, but Δvpr virus replicates less efficiently in macrophages (1, 9). Vpr has a small effect on HIV-1 replication in primary CD4+ T cells that can be magnified by rapid passage of the virus (14). In experimentally infected rhesus macaques, Δvpr simian immunodeficiency virus reverted to wild type in some animals and was associated with more rapid progression to disease, demonstrating a role for Vpr in AIDS pathogenesis (20).
Several roles for Vpr in virus replication have been proposed (reviewed in references 29 and 45). An analysis of HIV-1 replication in nondividing cells suggested that Vpr, acting in conjunction with the viral matrix protein, mediates nuclear import of the preintegration complex. Vpr was also found to be a weak transactivator of cellular genes and of the HIV-1 long terminal repeat. Expression of Vpr by transfection or in infected cells arrests or delays progression through the cell cycle at G2/M. G2 arrest may result in more efficient expression of the viral genome as a result of cellular factors that are preferentially expressed late in the cell cycle (14). G2 arrest results in apoptosis of infected cells, and this is thought to be caused by activation of ATR, a protein involved in the response to DNA damage (38).
Vpr has been found to interact with several cellular proteins, although the relative biological importance of these remain to be determined (reviewed in reference 18). A yeast two-hybrid screen in which Vpr was used as bait to screen a human cDNA library identified the cellular uracil DNA glycosylase (UDG) UNG as a binding partner (3, 41). This interaction suggested that Vpr might serve to bring UNG into virions. In support of this hypothesis, Mansky et al. (25) reported that UNG was present in vpr+ but not Δvpr HIV-1 virions. Willetts et al. (50) also detected UNG in HIV-1 virions but found that its encapsidation was not dependent upon Vpr. Instead, using a panel of HIV-simian immunodeficiency virus chimeric viruses, they mapped the virion component required for encapsidation to integrase. This finding was unexpected in light of the two-hybrid results (35, 50). Mutational analysis identified a Vpr point mutant with a Trp54→Arg exchange that failed to bind UNG (41). Conversely, mutational analysis of UNG identified a carboxy-terminal WXXF motif that was required for interaction with Vpr (4). A rationale for Vpr-mediated encapsidation of UNG was provided by Chen et al. and Mansky et al., who found that vpr+ virus reverse transcripts contained fewer mutations than those of Δvpr HIV-1. This finding suggested that the encapsidated UNG serves to correct errors in reverse transcription, increasing the fidelity of HIV-1 replication (6, 25).
In humans, there are at least four known UDGs: UNG, SMUG1, TDG, and MBD4. The enzymes vary in substrate specificity and localize to different cellular compartments (19). Human UNG exists in two forms differing in the amino-terminal 44 amino acids: UNG1, which is mitochondrial, and UNG2, which is nuclear (31). UNG2 (here termed UNG) and SMUG1 (here termed SMUG) remove uracil from single- and double-stranded DNA, whereas TDG and MBD4 act only on double-stranded DNA (31). The enzymes repair DNA that contains uracil that results either from the misincorporation of dUMP during DNA synthesis or from C→U deamination. Upon excision of uracil from double-stranded DNA, the abasic position is corrected by short- or long-patch repair in which the mutant base is resynthesized using the complementary strand as a template (19).
In cells infected with Δvif HIV-1, APOBEC3 family cytidine deaminases are encapsidated into the virion (13, 26, 43). Encapsidated APOBEC3F or APOBEC3G blocks virus replication by catalyzing the C→U deamination of the reverse transcripts synthesized in newly infected cells (2, 15, 21, 22, 24, 26, 49, 54, 55). Most of the uracil-containing DNA is degraded prior to integration, and as a result, few proviruses are generated (24, 26). Degradation of the uracil-containing reverse transcripts is thought to be mediated by host DNA repair enzymes. In cells infected with wild-type HIV-1, Vif binds to APOBEC3F and APOBEC3G, inducing their polyubiquitination and proteasomal degradation (22, 27, 28, 44, 46, 49, 52).
The finding that cytidine deaminases generate C→U changes in HIV-1 reverse transcripts led us to further explore the role of the association of Vpr with UNG in virus replication. We show here that Vpr increases the infectivity of HIV-1 that contains small amounts of APOBEC3G and enhances HIV-1 replication in cells stably expressing APOBEC3G. Vpr prevented the encapsidation of UNG and SMUG by inducing their polyubiquitination and proteasomal degradation. These findings suggest that Vpr, like Vif, prevents the encapsidation of cellular enzymes that inhibit virus replication.
MATERIALS AND METHODS
Expression vectors.
UNG (corresponding to UNG2) was amplified with a hemagglutinin (HA) epitope tag from pAS1B.UNG2.HA (provided by S. Benichou) (42) using primers UNG2F and UNG2R containing HindIII and XhoI sites, respectively, and an antisense HA epitope tag and cloned into pcDNA3.1(+). SMUG.3HA was amplified from vector pGEX-3X (provided by C. Radom) with antisense primers encoding a C-terminal triple HA tag and cloned into pcDNA3.1(+). SMUG-myc-His6 was subcloned from pGEX-3X.SMUG1 into the EcoRI and BamHI sites of pcDNA 3.1(−) myc-His6 (Invitrogen). TDG.HA was amplified from phytohemagglutinin-activated peripheral blood mononuclear cell (PBMC) cDNA using a forward primer containing an NheI site and a reverse primer containing an HA sequence and a KpnI site and cloned into pcDNA3.1(+). The APOBEC3G expression vector has been described previously (26). pNL43-Luc-E− R− (HIV-1 Δvpr), pNL-Luc-E− R− V− (HIV-1 Δvif Δvpr), and pNL-Luc-E− R+ (HIV-1 wild type [WT]) have been described previously (9, 9a). pNL-Luc-E− R+ V− (HIV-1 Δvif) was generated by cloning vpr from pNL4-3 into the unique PflMI and NheI sites of pNL-Luc-E− R− V−. pNL-E− R− p6− and pNL− E− p6− have been described previously (32). Vpr expression vectors pcVpr and pcHA.Vpr have been described previously (32). The HA.VprW54R mutant was generated using the Gene-Tailor site-directed mutagenesis system (Invitrogen) and verified by sequencing. The ubiquitin expression vector Ub-myc-His6 was a gift from Heinrich Gottlinger. UbK48R-myc-His6 is encoded by pRbG4-His6-myc-Ub (48). myc-tagged Cul1-Cul5 expression vectors were provided by Peter Jackson.
Reporter virus assay.
Single-cycle luciferase reporter virus was generated by cotransfection of 293T cells with 4.5 μg of plasmid DNA consisting of a mixture of 2 μg reporter virus plasmid, 0 to 2 μg pcAPOBEC3G or control pcDNA3.1(+), and 0.5 μg pcVSV-G. When Vpr was tested, the transfection contained 2 μg of pNL-Luc− E− R− V−, 1 μg pcVpr, 0.5 μg pcAPOBEC3G or control pcDNA3.1(+), and 0.5 μg pcVSV-G. Two days posttransfection, virus was harvested and quantified by p24 enzyme-linked immunosorbent assay (ELISA). HOS.T4 cells or PBMCs were infected with virus containing 1 ng p24 in triplicate, and luciferase activity was measured 3 days later by using Luc-Lite Plus reagent (Packard). Data are presented as the average counts per second of the triplicates ± the standard error.
Viral replication kinetics.
HOS.CD4.X4 cells (105) that stably expressed APOBEC3G were infected with WT, Δvpr, Δvif, or Δvif Δvpr NL43 at a multiplicity of infection of 0.1. Culture supernatant was collected over 14 days, and virus was quantitated by p24 ELISA.
Encapsidation.
UNG and SMUG encapsidation was measured as previously described (26). Briefly, 293T cells were transfected with 6 μg of reporter virus plasmid and 4 μg pcUNG.HA or pcSMUG.3HA. Two days posttransfection, the virions were centrifuged through 20% sucrose for 1 h at 30,000 rpm in a Beckman SW40.1 rotor. UNG and SMUG were detected in cell lysates (30 μg) and solubilized virions (100 ng p24) on immunoblots probed with anti-HA monoclonal antibody (MAb) 16b12 (Covance), followed by horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin. The blots were developed with ECL reagents (Amersham), stripped, and reprobed with anti-Vpr MAb. To verify similar loading, the immunoblot was probed with anti-HIV-1 serum, followed by horseradish peroxidase-conjugated anti-human immunoglobulin.
Immunoblot analysis.
293T cells were cotransfected with 5 μg pcUNG.HA and 0 to 2 μg pcHA.Vpr and brought to a total of 7 μg with pcDNA3.1(+). Where indicated, the proteasome inhibitors MG132 (12.5 μM) and clasto-lactocystin β-lactone (10 μM) or dimethyl sulfoxide (DMSO) were added 24 h posttransfection. The cells were lysed 16 h later in buffer that contained 1% Triton X-100 and normalized for protein concentration. The lysates (50 μg) were analyzed on immunoblots probed with anti-HA MAb and with anti-tubulin MAb to control for equal loading.
Immunofluorescence.
Cells were transfected with Vpr expression vector and 3 days later were fixed in 1% paraformaldehyde. The cells were stained with 1:500 anti-UNG antiserum (provided by Geir Slupphaug) and 1:1,000 anti-HA.11 (Covance) as previously described (53). Images were collected on a Deltavision deconvolution microscope. Merges and montages were generated using ImageJ (National Institutes of Health).
Pulse-chase labeling and coimmunoprecipitation.
293T cells were cotransfected with pcHA.Vpr and pcSMUG-myc-his at a ratio of 1:3. Two days later, the cells were starved in methionine-cysteine-free medium for 30 min. The cells were then labeled in medium containing 500 μCi [35S]methionine for 30 min and chased in complete medium supplemented with unlabeled methionine and cysteine. The cells were lysed at increasing times, and the SMUG or Vpr was immunoprecipitated with anti-HA MAb. Labeled proteins were detected by phosphorimager analysis after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE).
For coimmunoprecipitation, 293T cells were cotransfected with pcUNG.HA, pcSMUG.3HA, or empty vector and pcVpr or pcDNA3.1(+) at a ratio of 1:1. After 24 h, 12.5 μM MG132 was added to the culture medium, and after another 16 h, the cells were lysed in CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer (5 μM CHAPS, 50 μM NaCl, 20 μM Tris, pH 7.5). Lysates (200 μg) were precleared with protein A-Sepharose for 30 min at 4°C and immunoprecipitated with anti-HA MAb for 1 h. Complexes were collected by addition of 15 μl protein A-Sepharose and analyzed on immunoblots probed with anti-Vpr or anti-HA MAb. Vpr-Cullin complexes were detected by cotransfection of the myc-tagged Cullin expression vector with pcHA.Vpr at a ratio of 1:1. The cells were lysed in CHAPS buffer, and complexes were immunoprecipitated with anti-myc MAb 9E10 (Roche). The complexes were detected on an immunoblot probed with anti-HA MAb or anti-myc-horseradish peroxidase MAb. To detect UNG-Vpr-Gag complexes, 293T cells were cotransfected with pcUNG.HA, pNL43-E−, or pNL43-E− R− or pcVpr. The next day, 1 μM nelfinavir and 12.5 μM MG132 were added to the medium, and 16 h later, the cells were lysed in CHAPS buffer. The complexes were immunoprecipitated with anti-HA MAb and analyzed on an immunoblot probed with anti-Vpr MAb, anti-HIV serum, and anti-HA MAb.
Detection of ubiquitinated proteins.
293T cells were transfected with pcUNG.HA or pcSMUG.3HA, pcVpr and pRbG4-His6-myc-Ub, or pRbG4-His6-myc-UbK48R at a ratio of 1:1:2. After 24 h, the cells were incubated with MG132 and lysed 16 h later in RIPA buffer. The lysates were precleared with protein A-Sepharose for 30 min and incubated with 15 μl Ni-nitrilotriacetic acid beads (QIAGEN) for 1.5 h. The beads were washed with three changes of RIPA buffer, and the bound protein was detected on an immunoblot probed with anti-HA MAb.
Uracil glycosylase assay.
Virus was produced in transfected HeLa cells or in infected CEMx174 5.25 indicator cells. Virions were harvested at times of peak virus production and banded on a 60%-20% sucrose step gradient by ultracentrifugation at 30,000 rpm for 1.5 h in an SW40 rotor (Beckman). The virions were pelleted by ultracentrifugation at 30,000 rpm for 30 min and solubilized in 100 μl of virion lysis buffer (50 mM Tris [pH 8.0], 40 mM KCl, 50 mM NaCl, 5 mM EDTA, 10 mM dithiothreitol, and 0.1% [vol/vol] Triton X-100). Lysate containing 10 ng p24 was mixed with 105 cpm of 5′-32P-labeled deoxyoligonucleotide ATT ATT ATT ATT ATT CCU AAT TAT TTA TTT ATT TAT TTA TTT in UDG buffer (20 mM Tris [pH 8.0], 1 mM dithiothreitol) and incubated for 1 h at 37°C. The reaction mixture was then brought to 0.15 M NaOH and incubated another 30 min. The cleaved product was detected on 15% Tris-borate-EDTA-urea PAGE by autoradiography.
Quantitative PCR quantification of HIV-1 cDNA.
HOS.T4 cells were infected, and total DNA was isolated 0 to 24 h postinfection. Early and late reverse transcription products were quantified using primers that hybridized 5′ to the primer-binding site and to the 5′ region of Gag, respectively, which have been previously described (5).
RESULTS
Vpr reduces UNG and SMUG encapsidation.
In light of the role of cytidine deaminases in the lentiviral life cycle and the reported interaction of Vpr and UNG, we tested whether Vpr might have a role in the protection of HIV-1 from cytosine deaminases. We initially sought to study the role of Vpr in mediating the encapsidation of UNG. In addition, we also tested the related enzyme SMUG. To test this, 293T cells were cotransfected with wild-type or Δvpr HIV-1 and HA-tagged UNG or SMUG. The virions produced were pelleted and normalized for p24, and the encapsidated UNG and SMUG were detected on an immunoblot probed with anti-HA MAb. To determine the amounts of UNG and SMUG produced in the transfection, cell lysates were analyzed in parallel. Analysis of the lysates confirmed that UNG and SMUG were expressed in the transfected cells. Vpr had only a small effect on the amounts of both proteins (Fig. 1A, left). Virions were found to contain readily detectable UNG and SMUG. Unexpectedly, Vpr was associated with a significant reduction in the amount of virion-associated UNG and SMUG (Fig. 1A, right). The UNG and SMUG were virion associated, as their detection required cotransfection of the viral DNA. Furthermore, UNG and SMUG encapsidation was not the result of nonspecific packaging, because two other proteins that were tested, UNG1 and TDG, were not detected in similarly prepared virions (not shown). Much of the virion-associated UNG and SMUG migrated on the gel as 17- to 14-kDa proteolytic fragments. These were likely to have resulted from proteolytic cleavage by the viral protease, and their presence further supports the conclusion that they are encapsidated. Vpr did not completely eliminate UNG and SMUG from the virions under these conditions, but this could be because of their relatively high expression levels.
FIG. 1.
Encapsidation of UNG and SMUG is reduced by Vpr. (A) Vpr reduces UNG and SMUG encapsidation. Wild-type and Δvpr virions were produced in 293T cells cotransfected with HA-tagged UNG or SMUG expression vector. The virions (50 ng p24) were analyzed on an immunoblot probed with anti-HA MAb to detect UNG and SMUG (top right), anti-Vpr MAb (middle left and right), and anti-HIV-1 serum (bottom right). The cell lysates were analyzed in parallel (top left). Controls lacking viral DNA are labeled “no virus.” (B) Vpr reduces intracellular UNG and SMUG. 293T cells were cotransfected with UNG expression vector and decreasing amounts of pcHA.Vpr (left). The proteins were detected in the cell lysates by immunoblot analysis with anti-HA MAb. 293T cells were cotransfected with HA-tagged SMUG or TDG expression vector, with or without 0.5 μg pcVpr (right), and detected with anti-HA MAb. The total amount of plasmid DNA in each transfection was kept constant. (C) Vpr reduces UNG more efficiently in the absence of p6. 293T cells were transfected with UNG.HA expression vector and the indicated proviral DNA. Cell lysates and virions were analyzed on immunoblots probed with anti-HA MAb (top left and right) and anti-tubulin (bottom left) or anti-HIV-1 serum (bottom right). (D) Vpr reduces the UDG enzymatic activity of HIV-1 virions. The uracil glycosylase activities in wild-type and Δvpr sucrose gradient-purified virions were measured using a 5′-32P-labeled deoxyoligonucleotide containing a single uracil. Controls included recombinant UDG and mock virions (no virus) in which viral DNA was omitted from the transfection. (E) 5.25 cells were infected with WT or Δvpr NL4-3, harvested 6 days postinfection, and purified through a sucrose gradient. The uracil glycosylase activities of the virions were measured as for panel D.
The finding that Vpr prevented the encapsidation of UNG and SMUG seemed to parallel the effect of Vif on APOBEC3. We therefore tested whether Vpr reduced UNG encapsidation by inducing its degradation. To do this, UNG was expressed over a range of Vpr concentrations, and its level in the cell lysate was measured on an immunoblot. As little as 0.25 μg pc-HA.Vpr was found to reduce the steady-state level of UNG by 50% (Fig. 1B, left). In the presence of increased Vpr, UNG became undetectable. Vpr also reduced the amount of SMUG in the cell but had no significant effect on TDG, a double-strand-specific UDG (Fig. 1B, right). The specificity of the effect demonstrated that the results were not caused by global effects of Vpr on cellular protein stability.
The Vpr-induced reduction in intracellular UNG was more pronounced when Vpr was expressed alone than in the context of the virus (Fig. 1A and B). This finding led to the hypothesis that when Vpr was produced in the context of the virus some of the Vpr molecules bound to Gag, reducing the number of molecules available for UNG binding. To test the hypothesis, the effect of Vpr on UNG was measured in cells that expressed wild-type HIV-1 or Δp6 HIV-1. The Δp6 virus expressed a Gag precursor truncated before p6 and therefore did not encapsidate Vpr (32). Vpr was found to more effectively reduce UNG in the cell (Fig. 1C, left) and exclude it from Δp6 virions than from the wildtype (Fig. 1C, right). These findings supported the hypothesis that binding of Gag to Vpr titrated out a portion of the molecules, reducing the efficiency of UNG degradation.
The experiments described above detected the transfected epitope-tagged UNG and SMUG but not the endogenous proteins. To determine whether the endogenous cellular UDGs were encapsidated, we measured virion-associated uracil-DNA glycosylase enzymatic activity with an in vitro assay. Virions were generated by transfection of HeLa cells with wild-type or Δvpr HIV-1, purified on a sucrose step gradient, and lysed in NP-40-containing buffer. The lysates were incubated with a 5′-32P-labeled oligonucleotide containing a single uracil and then exposed to high pH to cleave at abasic sites generated by UDG. The cleaved product was then quantitated by PAGE, followed by autoradiography. UDG activity in Δvpr virions was readily detectable, requiring only a brief assay duration and exposure to film (Fig. 1D). In contrast, UDG activity was not detected in wild-type virions. UDG activity was also measured in replication-competent virions produced by productive infection of T cells. CEMx174 5.25 indicator cells were infected with replication-competent wild-type or Δvpr NL4-3, and at the peak of virus production, virions were harvested. Measurement of intravirion UDG activity showed readily detectable activity in Δvpr but not wild-type virions (Fig. 1E). Taken together, the findings further supported the conclusion that Vpr prevents the encapsidation of UNG and SMUG.
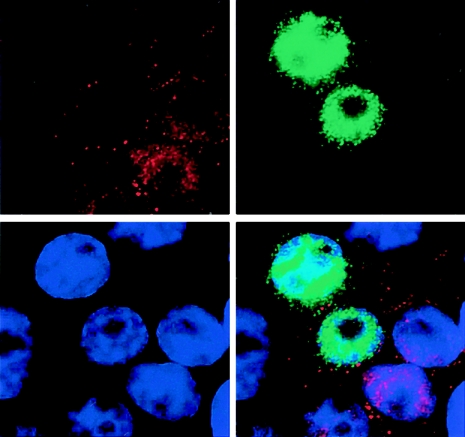
The effect of Vpr in single cells was assessed by two-color immunofluorescence of fixed, transfected cells. HeLa cells were transfected with HA-Vpr expression vector and then stained with anti-HA and anti-UNG sera. Vpr and UNG were localized to the nucleus, consistent with their known localization (23) (Fig. 2). Inspection of several fields showed that of the cells that expressed Vpr, 97% were negative for UNG. Conversely, nearly all of the cells that expressed UNG lacked Vpr. These observations were consistent with Vpr-induced degradation of intracellular UNG.
FIG. 2.
Immunofluorescence detection of Vpr and endogenous UNG. HeLa cells were transfected with HA-Vpr expression vector. The cells were fixed and stained with anti-HA MAb (green) and anti-UNG serum (red). Upper left, UNG staining; upper right, Vpr staining; lower left, DAPI nuclear staining; lower right, merged UNG, Vpr, and DAPI. The cells shown are representative of those contained in several fields. Secondary-antibody controls yielded black fields.
Vpr induces polyubiquitination and proteasomal degradation of UNG and SMUG.
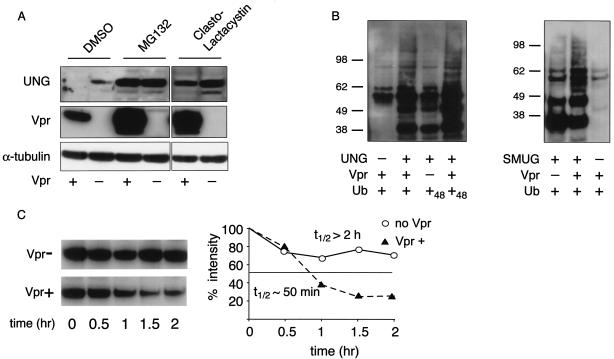
The reduction in intracellular UNG and SMUG could result from inhibition of synthesis or increased protein turnover. To test for proteasomal degradation, the effect of proteasome inhibitors on Vpr-induced reduction in UNG was measured. Transfected cells were treated with the proteasome inhibitor MG132, clasto-lactacystin, or DMSO alone. Vpr reduced UNG in cultures treated with DMSO but not in cells treated with MG132 or clasto-lactacystin (Fig. 3A). Proteasome inhibitors were associated with an overall increase in the levels of UNG and Vpr, consistent with proteasomal control of these proteins. These findings suggested that the Vpr-induced reduction in UNG is mediated through proteasomal degradation.
FIG. 3.
Vpr induces ubiquitination and proteasomal degradation of UNG and SMUG. (A) Proteasome inhibitor blocks the effect of Vpr on UNG. 293T cells were cotransfected with UNG expression vector with (+) or without (−) pcHA.Vpr and then cultured for 16 h with DMSO, MG132, or clasto-lactacystin. Cell lysates were analyzed on immunoblots probed with anti-HA MAb, anti-Vpr MAb, and anti-tubulin MAb. (B) Vpr induces polyubiquitination of UNG and SMUG. 293T cells were transfected with UNG (left) or SMUG (right) expression vector, Vpr, and Ub-His6-myc or UbK48R-His6-myc expression vector. The cell lysates were incubated with Ni-nitrilotriacetic acid beads, and coprecipitated UNG and SMUG were detected on an immunoblot probed with anti-HA MAb. (C) Vpr reduces the half-life of SMUG. The stability of SMUG in the presence and absence of Vpr was determined by pulse-chase metabolic labeling. Bands were quantified on a phosphorimager, and cpm at time zero was normalized to 100.
We next tested whether Vpr induced the ubiquitination of UDG and SMUG. 293T cells were cotransfected with His6-tagged ubiquitin, Vpr, and UNG.HA or SMUG.3HA expression vector. After 2 days, the ubiquitinated proteins were captured on nickel beads and the bound UNG and SMUG were detected on an immunoblot probed with anti-HA MAb. Vpr was found to cause an increase in the amount of heterogeneous higher-molecular-weight forms of UNG (Fig. 3B, left) and SMUG (Fig. 3B, right). To confirm the identity of the higher-molecular-weight forms as polyubiquitinated UNG, the experiment was repeated with cells transfected with K48R mutant ubiquitin, a variant that stabilizes polyubiquitinated substrates and results in increased amounts of ubiquitinated substrate (12, 47). The K48R mutant caused an increase in the intensity of the higher-molecular-weight forms, consistent with the identification of the smear as ubiquitinated UNG and SMUG.
To directly demonstrate Vpr-induced degradation, the half-life (t1/2) of SMUG in the presence or absence of Vpr was tested by pulse-chase metabolic labeling. In the absence of Vpr, SMUG was relatively stable, with a t1/2 of >2 h (Fig. 3C). In the presence of Vpr, the t1/2 of the protein was reduced to ∼50 min. A similar analysis of UNG was impractical due to poor metabolic labeling of the protein (not shown). The half-life of Vpr in the presence or absence of UNG was about 4 hours and was not affected by Vpr, suggesting that Vpr itself was not degraded (data not shown).
Vpr forms a complex with UNG, SMUG, and Cullins.
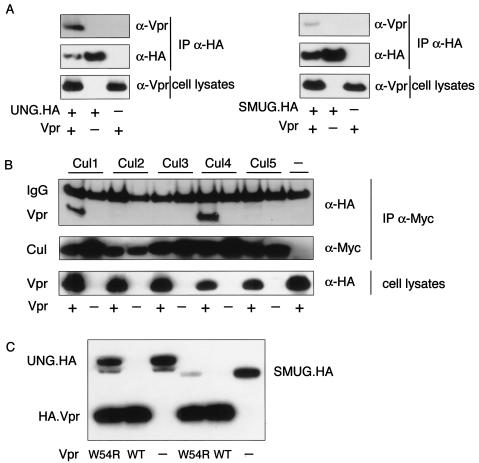
Vpr has been found to interact with UNG in yeast two-hybrid screens (3, 41). To detect complexes of Vpr with UNG and SMUG in mammalian cells, we immunoprecipitated the proteins from cell lysates made with nondenaturing detergent. HA-tagged UNG or SMUG and Vpr were expressed by transfection in 293T cells. The cells were lysed with CHAPS buffer, and UNG and SMUG were immunoprecipitated with anti-HA MAb. Coimmunprecipitated Vpr was detected by analysis of the complexes on an immunoblot probed with anti-Vpr MAb. In this analysis, Vpr was found to coimmunoprecipitate with UNG and, to a lesser extent, with SMUG, suggesting that the proteins were present as a complex in the cells (Fig. 4A).
FIG. 4.
Vpr interacts with UNG, SMUG, and Cullins. (A) Vpr interacts with UNG and SMUG. 293T cells were cotransfected with (+) UNG (left) or SMUG (right) expression vector with or without (−) Vpr expression plasmid and were then cultured for 16 h with MG132. Cell lysates were immunoprecipitated with anti-HA MAb, and the coimmunoprecipitated proteins were detected on an immunoblot probed with anti-Vpr MAb or anti-HA MAb. Vpr in the cell lysates was quantitated by immunoblot analysis with anti-Vpr MAb (bottom). (B) Vpr interacts with Cul1 and Cul4a. Vpr-Cullin complexes were detected by coimmunoprecipitation from 293T cells cotransfected with the indicated Cullin-myc and HA.Vpr expression vectors. Cell lysates were immunoprecipitated with anti-myc MAb, and the coimmunoprecipitated proteins were detected on an immunoblot probed with anti-HA MAb. The membrane was stripped and probed with anti-myc MAb to confirm Cullin expression. Vpr in the cell lysates was detected by probing with anti-HA MAb. (C) Vpr W54R does not decrease levels of UNG. 293T cells were cotransfected with UNG or SMUG expression vector and HA.Vpr, HA.VprW54R plasmid, or empty vector. Cell lysates were analyzed on an immunoblot probed with anti-HA MAb.
To extend the analogy with Vif (28, 52), we tested whether Vpr interacted with Cullin family E3 ligase components. To detect complexes of Vpr with Cullins, HA-tagged Vpr was coexpressed with myc-tagged Cul1 to -5. Cell lysates were prepared, and each Cullin was immunoprecipitated with anti-myc MAb. Coimmunoprecipitated Vpr was detected by immunoblot analysis with anti-HA MAb. Vpr was found to form a complex with Cul4a and, to a lesser extent, Cul1 (Fig. 4B). These findings further supported a role for proteasomal degradation as the mechanism by which Vpr induced UNG and SMUG polyubiquitination and degradation.
Vpr with a Trp→Arg mutation at amino acid 54 (W54R) does not associate with UNG (25). This mutant Vpr was used to further associate the interaction of Vpr with UNG and SMUG with its effects. UNG or SMUG was expressed with W54R Vpr by contransfection of 293T cells, and the proteins were detected on an immunoblot. Wild-type, but not W54R, Vpr caused a significant reduction in UNG (Fig. 4C) and a lesser reduction in SMUG. These findings suggested that binding of Vpr to UNG and SMUG was required for their degradation.
Vpr rescues the infectivity of Δvif HIV-1 virus produced in the presence of limited amounts of APOBEC3G.
The presence of UNG or SMUG in an HIV-1 virion could be deleterious to the virus, particularly if some amount of cytidine deamination occurred upon cDNA synthesis. Encapsidated UDG would remove the uracil generated by cytidine deamination, and the presence of even a single abasic site in a minus strand would impose a block to synthesis of the complementary plus strand. To test this model, we determined the infectivity of wild-type, Δvif, Δvpr, and Δvif Δvpr luciferase reporter viruses that were prepared in cells cotransfected with decreasing amounts of human APOBEC3G expression vector. For viruses prepared in the absence of APOBEC3G, the infectivities were similar (Fig. 5A). Δvif viruses were slightly more infectious than the wild type, a reproducible finding in this system that may be due to a slight toxicity of Vif in human cells (39). When produced in cells expressing a low level of APOBEC3G, Δvif vpr+ virus was substantially more infectious than Δvif Δvpr virus. As APOBEC3G levels increased, the infectivity of the Δvif Δvpr virus was reduced to near to background, while the vif+ viruses remained relatively stable. In peripheral blood mononuclear cells, vpr+ virus was also less sensitive to APOBEC3G than Δvpr virus (Fig. 5B). In these cells, Vpr was advantageous not only for Δvif virus but also slightly for vif+ virus.
FIG. 5.
Vpr partially rescues Δvif HIV-1 infectivity. (A) Wild-type, Δvif, Δvpr, and Δvif Δvpr luciferase reporter viruses were produced in 293T cells cotransfected with the indicated amounts of human APOBEC3G expression vector. Luciferase activities generated by infection of HOS cells with 1 ng of each virus are shown. The error bars indicate standard deviations. (B) Same as panel A, but infectivity was measured in PBMCs. (C) Reporter viruses were generated in cells cotransfected with wild-type or W54R Vpr in trans with or without human APOBEC3G, and their infectivities were determined by luciferase assay. The data shown are representative of three independent repetitions of the experiment. (D) HOS.CD4.X4 (left) or HOS.CD4.X4 APOBEC3G.HA (right) cells were infected with the indicated viruses at a multiplicity of infection of 0.1. Virus production was determined at the indicated time points by p24 ELISA.
To determine whether the increase was related to the interaction of Vpr with UNG and SMUG, Δvif Δvpr luciferase reporter viruses were generated in cells that expressed wild-type or W54R Vpr and a limited amount of APOBEC3G (0.5 μg plasmid). Vpr restored the infectivity of Δvif HIV-1 about sixfold, whereas W54R Vpr increased the infectivity twofold (Fig. 5C). These findings suggested that the increase in infectivity required an interaction of Vpr with UNG. The small increase in infectivity in the presence of W54R Vpr may have been caused by the weak interaction of SMUG with this mutant. A small increase in infectivity was found for Δvif Δvpr virus produced without APOBEC3G, probably because of a slight toxicity of Vif and Vpr in human cells.
Vpr has little effect on HIV-1 replication in transformed cell lines. To determine whether the effect of Vpr on virus replication is increased in the presence of APOBEC3G, cells that stably expressed a transduced POBEC3G were tested for their ability to support HIV-1 replication. HOS.CD4.X4.APO3G cells (26) were infected with wild-type, Δvpr, Δvif, or Δvif Δvpr NL4-3 HIV-1, and virus replication kinetics were measured by p24 production (Fig. 5D, right). In control HOS.CD4.X4 cells, all of the viruses replicated with similar kinetics (Fig. 5D, left). In the HOS.CD4.X4.APO3G cells (Fig. 5D, right), vpr+ viruses replicated significantly better than Δvpr HIV-1. In addition, Δvif Δvpr HIV-1 replicated poorly but vpr+ Δvif HIV-1 replicated to significantly higher levels. These findings suggested a cooperative effect of Vpr and Vif in cells that express APOBEC3G.
Vpr increases late HIV-1 reverse transcripts.
Small amounts of UDG in virions could be deleterious to the virus because removal of even rare minus-strand uracil bases would leave abasic sites that would block synthesis of the complementary strand. To determine whether Vpr had an effect on the relative amounts of early and late reverse transcripts, reverse transcripts were quantitated by quantitative PCR in newly infected cells. Early reverse transcripts increased in copy number until 12 h postinfection and then decreased in abundance, and this was similar for viruses generated with or without APOBEC3G (Fig. 6A and B). Late transcripts peaked around 12 h and decreased over time (Fig. 6C). In cells infected with APOBEC3G-containing virions, significantly fewer late transcripts were detected for the Δvif Δvpr virus. APOBEC3G had no effect on Δvif vpr+ virus (Fig. 6D). These data suggest that the removal of uracil by encapsidated UDG results in incomplete second-strand synthesis and that Vpr relieves this inhibition.
FIG. 6.
Vpr increases the copy number of late reverse transcription (RT) products. The relative copy number of reverse transcripts in cells newly infected with wild-type, Δvif, Δvpr, or Δvif Δvpr that had been generated in 293T cells cotransfected with or without APOBEC3G expression vector was determined by fluorescence-detected quantitative PCR. DNA was isolated from the infected cells at the indicated times postinfection, and the relative copy number of the reverse transcript was determined with primers specific for early or late products. The experiments shown are representative of at least two repetitions.
DISCUSSION
The primary means by which lentiviruses protect themselves against cytidine deamination by APOBEC3 is through Vif (reviewed in references 30, 37, and 40). However, recent insight into the importance of cytidine deaminases in HIV-1 replication coupled with earlier findings of an association between Vpr and UNG (7) led us to consider a role for Vpr as a secondary mechanism by which the virus protects its genome from damage caused by APOBEC3G. In transfection experiments, small amounts of APOBEC3G remained in wild-type viruses that expressed Vif, and a low level of G→A mutation was detected (26, 51). This suggested that in cells that express relatively large amounts of APOBEC3G, Vif may not be entirely efficient. This could account for a secondary viral mechanism that functions to neutralize the effects of cytidine deamination.
The interaction between UNG and Vpr was first detected by Bouhamdan et al. and Willetts et al. in a yeast two-hybrid screen (3, 50). Mansky et al. (25) subsequently showed that UNG was encapsidated and that this appeared to be mediated by the interaction with Vpr. Paradoxically, Priet et al. and Willetts et al. found that UNG encapsidation was independent of Vpr but dependent on integrase (35, 50). In our experiments, Vpr appeared not to increase but to reduce UNG encapsidation. The reason for this difference is not clear but could be due to differences in experimental conditions. Expression of large amounts of UNG relative to Vpr could overwhelm the negative effects of Vpr, or expression of large amounts of Vpr could overwhelm the capacity of the E3 ligase to induce UNG degradation. Both situations would result in the presence of UNG in vpr+ virus.
Herpesviruses and poxviruses encode UDG and nonprimate lentiviruses encode a dUTPase, which serve to augment their replication (7). Herpesvirus and poxvirus genomes are double stranded and therefore can repair abasic sites generated by UNG using the complementary strand as a template. In contrast, APOBEC3G-catalyzed cytidine deamination of HIV-1 occurs when the viral genome is single stranded (51), in which case the uracils generated by cytosine deamination cannot be repaired because there is no template strand from which to correct them. This difference could provide a rationale for why HIV-1 induces the degradation of UDG when other viruses use these enzymes to promote their replication. In addition, single-stranded DNA containing abasic sites is a poor template for plus-strand synthesis because of the difficulty in synthesizing over positions that lack a base. It has been reported that reverse transcriptase was able to repair a uracil-containing model substrate in which a single uracil was positioned at the site of chain extension (36). However, this activity cannot account for the repair of uracils present within single-stranded cDNA.
Because the viral minus strand is deaminated shortly after its synthesis (51), excision of even a single uracil by UDG could prevent synthesis of the complementary plus strand. Vpr could, therefore, serve to inhibit the encapsidation of UDG, preventing the formation of deleterious abasic sites. Although Vif effectively prevents APOBEC3G encapsidation, small amounts of the enzyme are found in vif+ virions (26, 39). This residual deaminase could generate a small number of uracils in the reverse transcripts.
Vpr induced UDG degradation more efficiently when expressed in the absence of Gag than in the context of a proviral vector. Furthermore, deletion of p6 from the proviral vector increased the efficiency of Vpr-induced UDG degradation. These findings suggested that free Vpr in the cell binds to UDG and causes its degradation. Binding of Vpr to p6 during virus assembly would prevent association with UDG. Consistent with this, we were unable to coimmunoprecipitate a trimolecular complex containing Vpr, Gag, and UDG (data not shown).
The interaction of Vpr with p6 in the mature virus is weak (17). We speculate that in the mature virus, Vpr is released from p6 and functions to bind target cell UDG in the next round of replication. This would distinguish Vpr from Vif, which appears to act only in the producer cell to bind its cellular partner. That Vpr from virions is biologically active in the target cell has been shown by Poon et al., who found that incoming virus could cause G2 cell cycle arrest (33).
Priet et al. recently showed that RNA interference knockdown of UNG in macrophages blocked HIV-1 replication (34). They further showed that in vitro, on model substrates, UNG and reverse transcriptase acted in concert to remove uracils that had been misincorporated during reverse transcription. In our study, removal of UNG by Vpr did not appear to interfere with virus infectivity. It is not clear how to reconcile these findings, but we cannot rule out the possibility that small amounts of UNG are required and that these are not removed by Vpr.
APOBEC3 proteins appear to constitute an arm of the innate immune system that functions to interfere with retrovirus replication and transposition of endogenous retrotransposons (10, 11). The single-strand-specific UDGs, UNG and SMUG, appear to also be involved. This proposed model further points out the need for HIV-1 to inhibit the interference of cellular DNA-modifying enzymes with virus replication.
Acknowledgments
We thank Peter Jackson, C. Radom, Heinrich Gottlinger, Serge Benichou, and Geir Slupphaug for reagents.
S.G.Z. is supported by an NIH F32 fellowship. This work was supported by NIH grants DA14494 and AI58864 to N.R.L. N.R.L. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
REFERENCES
- 1.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 3.Bouhamdan, M., S. Benichou, F. Rey, J. M. Navarro, I. Agostini, B. Spire, J. Camonis, G. Slupphaug, R. Vigne, R. Benarous, and J. Sire. 1996. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J. Virol. 70:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.BouHamdan, M., Y. Xue, Y. Baudat, B. Hu, J. Sire, R. J. Pomerantz, and L. X. Duan. 1998. Diversity of HIV-1 Vpr interactions involves usage of the WXXF motif of host cell proteins. J. Biol. Chem. 273:8009-8016. [DOI] [PubMed] [Google Scholar]
- 5.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R., E. Le Rouzic, J. A. Kearney, L. M. Mansky, and S. Benichou. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 279:28419-28425. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R., H. Wang, and L. M. Mansky. 2002. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83:2339-2345. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, E. A., G. Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J. Virol. 64:3097-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:936-944. [DOI] [PubMed] [Google Scholar]
- 9a.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, S. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 10.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 12.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaddis, N. C., E. Chertova, A. M. Sheehy, L. E. Henderson, and M. H. Malim. 2003. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 77:5810-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65-71. [DOI] [PubMed] [Google Scholar]
- 15.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 16.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins, Y., O. Pornillos, R. L. Rich, D. G. Myszka, W. I. Sundquist, and M. H. Malim. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6(Gag). J. Virol. 75:10537-10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kino, T., and G. N. Pavlakis. 2004. Partner molecules of accessory protein Vpr of the human immunodeficiency virus type 1. DNA Cell Biol. 23:193-205. [DOI] [PubMed] [Google Scholar]
- 19.Krokan, H. E., F. Drablos, and G. Slupphaug. 2002. Uracil in DNA—occurrence, consequences and repair. Oncogene 21:8935-8948. [DOI] [PubMed] [Google Scholar]
- 20.Lang, S. M., M. Weeger, C. Stahl-Hennig, C. Coulibaly, G. Hunsmann, J. Muller, H. Muller-Hermelink, D. Fuchs, H. Wachter, M. M. Daniel, et al. 1993. Importance of vpr for infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 67:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 22.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 23.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 25.Mansky, L. M., S. Preveral, L. Selig, R. Benarous, and S. Benichou. 2000. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J. Virol. 74:7039-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 27.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 28.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 29.Muthumani, K., B. M. Desai, D. S. Hwang, A. Y. Choo, D. J. Laddy, K. P. Thieu, R. G. Rao, and D. B. Weiner. 2004. HIV-1 Vpr and anti-inflammatory activity. DNA Cell Biol. 23:239-247. [DOI] [PubMed] [Google Scholar]
- 30.Navarro, F., and N. R. Landau. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16:477-482. [DOI] [PubMed] [Google Scholar]
- 31.Nilsen, H., M. Otterlei, T. Haug, K. Solum, T. A. Nagelhus, F. Skorpen, and H. E. Krokan. 1997. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 25:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type-1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon, B., K. Grovit-Ferbas, S. A. Stewart, and I. S. Chen. 1998. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science 281:266-269. [DOI] [PubMed] [Google Scholar]
- 34.Priet, S., N. Gros, J. M. Navarro, J. Boretto, B. Canard, G. Querat, and J. Sire. 2005. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol. Cell 17:479-490. [DOI] [PubMed] [Google Scholar]
- 35.Priet, S., J. M. Navarro, N. Gros, G. Querat, and J. Sire. 2003. Differential incorporation of uracil DNA glycosylase UNG2 into HIV-1, HIV-2, and SIV(MAC) viral particles. Virology 307:283-289. [DOI] [PubMed] [Google Scholar]
- 36.Priet, S., J. M. Navarro, N. Gros, G. Querat, and J. Sire. 2003. Functional role of HIV-1 virion-associated uracil DNA glycosylase 2 in the correction of G:U mispairs to G:C pairs. J. Biol. Chem. 278:4566-4571. [DOI] [PubMed] [Google Scholar]
- 37.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291-297. [DOI] [PubMed] [Google Scholar]
- 38.Roshal, M., Y. Zhu, and V. Planelles. 2001. Apoptosis in AIDS. Apoptosis 6:103-116. [DOI] [PubMed] [Google Scholar]
- 39.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrofelbauer, B., Q. Yu, and N. R. Landau. 2004. New insights into the role of Vif in HIV-1 replication. AIDS Rev. 6:34-39. [PubMed] [Google Scholar]
- 41.Selig, L., S. Benichou, M. E. Rogel, L. I. Wu, M. A. Vodicka, J. Sire, R. Benarous, and M. Emerman. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 71:4842-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selig, L., J. C. Pages, V. Tanchou, S. Preveral, C. Berlioz-Torrent, L. X. Liu, L. Erdtmann, J. Darlix, R. Benarous, and S. Benichou. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J. Virol. 73:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 44.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 45.Sherman, M. P., C. M. De Noronha, S. A. Williams, and W. C. Greene. 2002. Insights into the biology of HIV-1 viral protein R. DNA Cell Biol. 21:679-688. [DOI] [PubMed] [Google Scholar]
- 46.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 47.Tsirigotis, M., M. Zhang, R. K. Chiu, B. G. Wouters, and D. A. Gray. 2001. Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J. Biol. Chem. 276:46073-46078. [DOI] [PubMed] [Google Scholar]
- 48.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121-127. [DOI] [PubMed] [Google Scholar]
- 49.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willetts, K. E., F. Rey, I. Agostini, J. M. Navarro, Y. Baudat, R. Vigne, and J. Sire. 1999. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J. Virol. 73:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 52.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 53.Zeitlin, S. G., C. M. Barber, C. D. Allis, K. F. Sullivan, and K. Sullivan. 2001. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114:653-661. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]