Abstract
Background
The active copy of the imprinted gene H19 is turned off by inappropriate methylation in several pediatric tumors including Wilms' Tumour and embryonal rhabdomyosarcoma. H19 controls in cis the linked Insulin-like Growth Factor 2 (IGF2) gene, encoding an important growth factor. Recent work has suggested that methylation of a gene may lead to deacetylation of its associated histones and that silenced genes can be reactivated by increasing histone acetylation levels.
Results
Treatment of a rhabdomyosarcoma cell line which has a silent, methylated H19 gene with histone deacetylase (HDAC) inhibitors under conditions which gave maximal hyperacetylation of histone 4, both globally and at the H19 gene itself could not reactivate H19 or affect the active Insulin-like Growth Factor 2 (IGF2) gene, but caused clear up-regulation of the Tissue-type Plasminogen Activator (TPA) gene, a non-imprinted gene known to respond to changes in histone acetylation. In contrast, mild treatment of the cells with the methylation inhibitor 5-AzaC-2'-deoxycytidine (AzaC) on its own was able to reactivate H19. Combining AzaC treatment with HDAC inhibitors gave a reduced rather than enhanced reactivation. These findings were confirmed in mouse primary liver and kidney explants which maintain normal imprinting, where we also found that the silent Igf2 gene could not be reactivated by HDAC inhibitors.
Conclusion
These results suggest that DNA methylation rather than histone acetylation is the primary determinant of silencing of H19 in rhabdomyosarcoma.
Background
Inactive genes in mammals often have particular features which differentiate them from their active neighbours. High levels of DNA methylation have been found to be associated with a large number of inactive endogenous genes in normal tissues and also with many genes which are aberrantly silenced in tumour cells [1]. In contrast, high levels of histone acetylation are associated with transcriptionally active genes [2]. DNA methylation and histone deacetylation have also been mechanistically linked, since it has been found that proteins which bind to methylated cytosine such as MeCP2 can recruit HDACs [3]. Treatment of cancer cell lines with demethylating agents can cause reactivation of certain silenced tumor suppressor genes and this effect was augmented for some by the addition of HDAC inhibitors such as Trichostatin A (TSA) or the chemotherapeutic agent sodium butyrate [4]. This raises the possibility that a combination of agents which inhibit DNA methylation and promote histone acetylation may lead to efficient reactivation of growth-inhibitory genes in certain cancers, including those which currently have a poor response to chemotherapy such as rhabdomyosarcoma.
Two genes which are known to be effected by changes in DNA methylation and which are implicated in rhabdomyosarcoma, are the imprinted genes H19 and IGF2, which are tightly linked on human chromosome 11p15 [5]. IGF2 encodes a fetal growth factor and is expressed only from that copy of chromosome 11 inherited from the father, while H19 is expressed exclusively from the maternally inherited allele. The inactive paternal copy of H19 is heavily methylated and is less accessible to nucleases, while the maternal copy is unmethylated and in a more open conformation [6]. H19 and IGF2 compete for shared enhancer elements: methylation of the paternal H19 allows IGF2 to gain control of the enhancers and become expressed [7]. The importance of methylation has been demonstrated in mice with decreased levels of methylation, which show expression of H19 from both alleles and a loss of Igf2 expression [8]. Besides rhabdomyosarcoma, H19 and IGF2 have been implicated in a number of other human cancers [9]. In many cases, the primary lesion appears to be not a deletion or point mutation, but rather a change in methylation, usually an aberrant methylation of the maternal H19 allele [10-12]. Since this renders both alleles methylated, it has the effect of silencing H19 and results in a concomitant biallelic expression of IGF2. In Wilms' tumors, a pediatric tumor of the kidney, inactivation of H19 by methylation is almost as common as loss of heterozygosity (LOH) and thus represents a significant pathway for inactivation of a growth-inhibiting gene in tumors [12].
In order to assess the possibility for reactivating H19 in rhabdomyosarcoma, we set out to determine whether treatment of a recently-derived line of rhabdomyo-sarcoma cells with histone deacetylases, on their own, or in combination with inhibitors of DNA methylation, could cause a significant reactivation of a silenced H19 allele. We found that the silenced allele is unresponsive to inhibitors of deacetylation, while it could be reactivated readily using an inhibitor of methylation. A combination of the two treatments carried out simultaneously resulted in less reactivation than seen for the methylation inhibitor on its own, rather than more. These experiments indicate that methylation and not acetylation is the primary determinant of aberrant H19 silencing in this tumor type.
Results
Treatment with HDAC inhibitors increases acetylation levels globally and at the H19 locus without relieving silencing of the gene
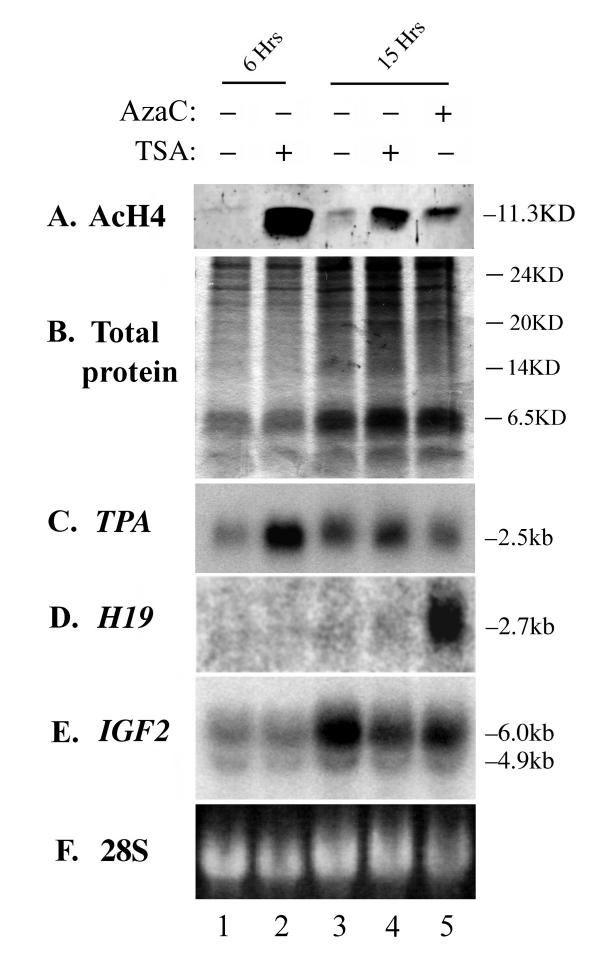
In order to investigate the effects of increasing histone acetylation levels in RD cells on the silenced H19 gene, cells were grown in culture medium supplemented with a range of concentrations of TSA from 25 nM to 1 mM for periods ranging from 3 hrs to 2 weeks and sodium butryate (3 mM) for various times from 3 hrs to 24 hours. Proteins were then extracted and analyzed by Western blotting using an antibody against acetylated histone 4. Histone acetylation levels are maximally effected by inhibitors at very early time points, with the increase in histone acetylation dropping off rapidly in the first day of treatment and protein samples taken on subsequent days of treatment showed no difference between treated and untreated cells. This effect can be seen in Figure 1, where RD cells were treated with 500 nM TSA for 6 or 15 hours, after which proteins were harvested and analysed by Western hybridisation with an antibody to the acetylated form of histone 4. At 6 hr, acetylation levels have strongly increased (Fig 1A, lanes 1 and 2), but this effect has diminished significantly at 15 hr (Fig 1A, lanes 3 and 4): by 48 hrs, no difference in acetylation levels between treated and untreated samples can be detected (data not shown). Increases in histone acetylation levels were equivalent at 3 hr and 6 hr (data not shown). These data agree with recent studies showing cells can homeostatically adjust to maintain a steady-state level of histone acetylation [13]. Treatment with sodium butyrate (3 mM), another inhibitor of histone deacetylases (HDACs) showed similar effects to 500 nM TSA and results are shown for TSA alone.
Figure 1.
Effects of histone deacetylase inhibitor on rhabdomyosarcoma cells. Cells were treated for 6 hr (lanes 1, 2) or 15 hr (lane 3–5) with 500 nM of either Trichostatin A (TSA) or 5'-aza-2'deoxycytidine (AzaC) as indicated at top. (A) Western analysis of protein derived from the cells using the anti-acetylated histone 4 (AcH4) antibody shows that treatment with TSA markedly increases the amount of acetylated histone in the cells at 6 hr (lanes 1 and 2). While treatment for 15 hr also increases acetylation levels, the effect is less marked, presumably due to a compensatory mechanism in the cell (lanes 3 and 4). (B) Coomasie stained total protein loading control for the Western. (C) Northern hybridization of RNA derived from the same cells to a probe for Tissue-type Plasminogen Activator (TPA). Transcription levels can be seen to increase in parallel with the marked increase in histone acetylation (lanes 1 and 2), indicating that acetylation plays an important role in determining levels of TPA transcript in the cell. The less marked increase in acetylation seen at 15 hr has only a slight effect on transcript levels (lanes 3 and 4). (D) The membrane used in (C) was stripped and rehybridized with a probe for H19, which is normally silent in these cells. No signal was detected in the normal or TSA-treated samples (lanes 1–4), but some reactivation could be seen after even a brief treatment with a low level of AzaC (lane 5). (E) Rehybridization with an IGF2 probe shows no difference between the TSA treated and untreated samples at 6 hr (lanes 1 and 2) or 15 hr (lanes 3 and 4). IGF2 transcripts accumulate in the cells during the log phase of growth in culture and at 15 hr, basal levels of IGF2 have increased from those seen at 6 hr (compare lanes 3 and 1). The two major IGF2 transcripts of 6 kb and 4.9 kb are visible. (F) 28S rRNA loading control for the Northern.
In order to ensure that the alterations in histone acetylation were meaningful in terms of gene transcription, we examined the transcription levels of TPA, which has previously been shown to be up-regulated by hyperacetylation [14]. Levels of transcription of the TPA gene increase in proportion to the level of acetylation seen at histone 4 (Fig 1C): at 6 hr post-treatment, TPA mRNA levels are significantly increased, while at 15 hr only a slight increase in transcript levels can be seen (Fig 1C, lanes 2 and 4). Taken together with the observations above, this indicates that the cell can compensate for the presence of HDAC inhibitors on prolonged treatment, possibly by up-regulating the enzymes. This is a significant finding, since many previous studies using these drugs have looked at gene transcription after treatments of 15 hr or more. However, robust effects on acetylation levels and gene transcription are achieved at shorter timepoints.
We examined expression of H19 in the cells treated with 500 nM TSA for 6 hr (which show hyperacetylation of histones) by stripping the membrane used to probe for TPA and rehybridising with an H19 probe. No reactivation of the gene could be seen (Fig 1D, lanes 1 and 2). Treatment with concentrations of TSA from 25 nm to 1 mM for periods up to 12 days or with sodium butyrate at 3 mM for up to 15 hrs also failed to cause any detectable increase in H19 transcription. However, treatment for as little as 15 hr with 500 nM AzaC, which causes decreases in DNA methylation, resulted in detectable reactivation of the gene (Fig 1D, lane 5), indicating that even low levels of transcription can be detected in this way. We also examined mRNA levels for the neighbouring IGF2 gene. IGF2 is normally active in RD cells and no increase or decrease in transcript levels was seen at 6 hr post-treatment, when acetylation levels are highest (Fig 1E, lanes 1 and 2). Expression of IGF2 increases during the log phase of cell growth, so that levels in untreated cells are higher at 15 hr (Fig 1C, lane 3) than they were at 6 hr: cells grown for 15 hours in the presence of 500 nM TSA or 500 nM AzaC show a slight decrease in IGF2expression which is most likely to be due to the differentiating effect of these two drugs.
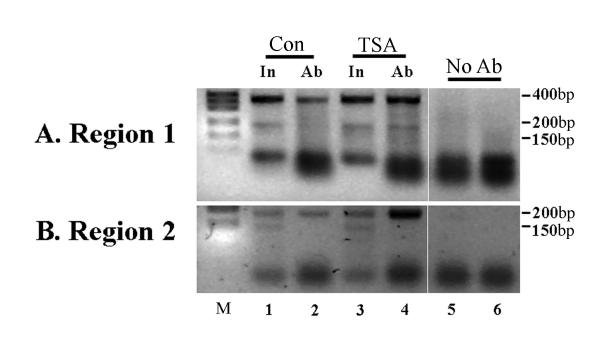
It is possible that histones at the H19 and IGF2 locus are particularly refractory to inhibitors of histone deacetylases. In order to test this possibility, we carried out chromatin immunoprecipitation experiments to assay whether changes in acetylation levels occur at the H19 locus on treatment with TSA. Chromatin was cross-linked to DNA using formaldehyde, sonicated to reduce the fragment size, then immunoprecipitated with the anti-acetylated H4 antibody. PCR using primers located in the transcribed region (Region 1) or in the upstream insulator region (Region 2) of the H19 gene were used to determine the fraction of total templates which are associated with acetylated histone 4. Input DNA (Fig 2, lanes 1 and 3, In) which has not been treated was used as a control between samples, while a sample of DNA which has been mock-immunoprecipitated without antibody was used as a negative control (Fig 2, lanes 5 and 6, No Ab). We found that after treatment with TSA there was a reproducible increase in the amount of acetylated H4 associated with both the transcribed and the upstream regions (Fig 2A and 2B). Each experiment was carried out independently at least three times. These results indicate that H4 acetylation levels are being increased at the H19 locus in these experiments, but with no detectable effect on H19 transcription.
Figure 2.
Chromatin immunoprecipitation assay of histone acetylation at the H19 locus in treated rhabdomyosarcoma cells. Cells were either treated (TSA) or not treated (Con; control) with histone deacetylase inhibitor (500 nM TSA for 6 hrs). DNA and proteins were cross-linked and precipitated with an antibody to acetylated histone 4, prior to reversing cross-links, amplifying DNA with specific primers and separating the products on a gel. (A) Results obtained using primers specific for the transcribed region of the H19 gene (Region 1), which give an expected band size of 355 bp. Equal amounts of input DNA (In) were used in each immunoprecipitation (lanes 1 and 3) and a no antibody control was run for each sample (lanes 5 and 6). Treatment with TSA gives an increase in signal for the 355 bp target, indicating an increase in acetylated histones associated with the transcribed region (lanes 2 and 4). (B) Primers for a region in the insulator (Region 2) upstream of H19 and downstream of IGF2 give a specific band of 161 bp and this also shows a marked increase after TSA treatment of the cells. Controls are as for (A) above. M; 1 kb DNA size marker (Life Tech.). Minor bands present in some lanes represent non-specific artifacts: primers are visible at the bottom of the gel. Negative images of ethidium-stained gels are shown.
Reactivation of H19 by demethylation is inhibited by TSA
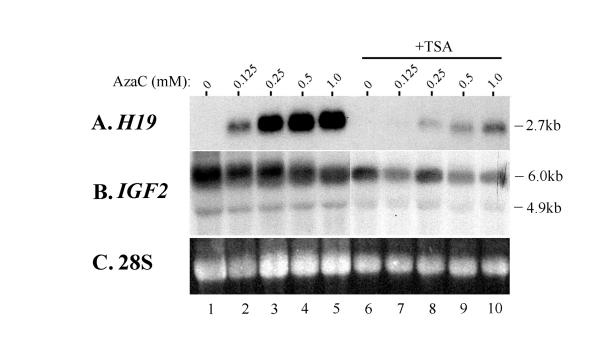
It has been suggested that some epigenetically silenced genes can only be reactivated by treatment with a combination of histone deacetylase inhibitors and methylation. In order to test whether such a combination treatment might enhance the reactivation of H19 in rhabdomyosarcoma seen using methylation inhibitor alone, we grew RD cells for long or short periods in various combinations of AzaC and TSA. Both long and short-term cultures gave similar results and a typical experiment can be seen in Figure 3 (treatment for 1 week). Here, the silenced H19 allele shows a dose-dependent reactivation in response to AzaC treatment, with higher levels of the drug (0.5 mM to 1 mM) resulting in greater activation (Fig 3A, lanes 1–5). Addition of 500 nM TSA to the culture medium containing various amounts of AzaC resulted in a significant decrease in the level of H19 transcript produced (Fig 3A, lanes 6–10). A decrease in IGF2 transcript levels on addition of TSA was also seen in these cells when compared to treatment with AzaC alone (Fig 3B). These results indicate that a combination of drug treatments do not result in an increase in H19 expression, but rather inhibit reactivation of the gene by demethylation.
Figure 3.
Simultaneous treatment of cells to increase histone acetylation and decrease DNA methylation. Rhabdomyosarcoma cells were treated with the indicated amounts of AzaC in the absence (lanes 1–5) or presence (+TSA; lanes 6–10) of 500 nM TSA. RNA was extracted and separated on an agarose gel before transferring to a nylon membrane and hybridizing with the indicated probes. (A)H19 transcription is reactivated by AzaC in a dose-dependent fashion with higher doses resulting in greater reactivation (lanes 1–5). Combining the AzaC treatment with 500 nM TSA however results in lower levels of reactivation of the gene (compare lanes 6–10 with lanes 1–5). (B) The same membrane stripped and rehybridized with a probe for IGF2. Levels of IGF2 show a slight decrease as H19 transcription increases in the cells treated with AzaC alone (lanes 1–5). H19 and IGF2 compete for common enhancer elements, which would explain this effect. The effect is diminished in cells treated with both TSA and AzaC (lanes 6–10), which also show less H19 expression. (C) 28S rRNA loading control for the Northern.
Igf2 and H19 transcript levels in primary cells are not effected by inhibitors of histone acetylation
H19 and IGF2 expression are highest in embryonal tissues, and silencing of the active allele has been seen in normal fetal tissues prior to tumor formation. In order to extend our findings from the embryonal rhabdomyosarcoma-derived cell line RD to primary cells and to provide for sensitive detection of reactivation of a silent Igf2 allele, we developed an assay based on explanting fetal tissues from normal mice or mice with a homozygous deletion of the Igf2 gene. Each liver or kidney from an embryonic day 16.5 fetal mouse was dissected out and dissociated into a single-cell suspension prior to splitting between four wells of a six-well tissue-culture plate. Cells from both kidney and liver formed large patches of cuboidal epithelia interspersed with fibroblast cells. Two of the four wells were treated with a histone deacetylase inhibitor for 6 hr and two were left untreated. Protein extract was prepared from one treated and one untreated sample and the remaining treated and untreated sample were used to prepare RNA.
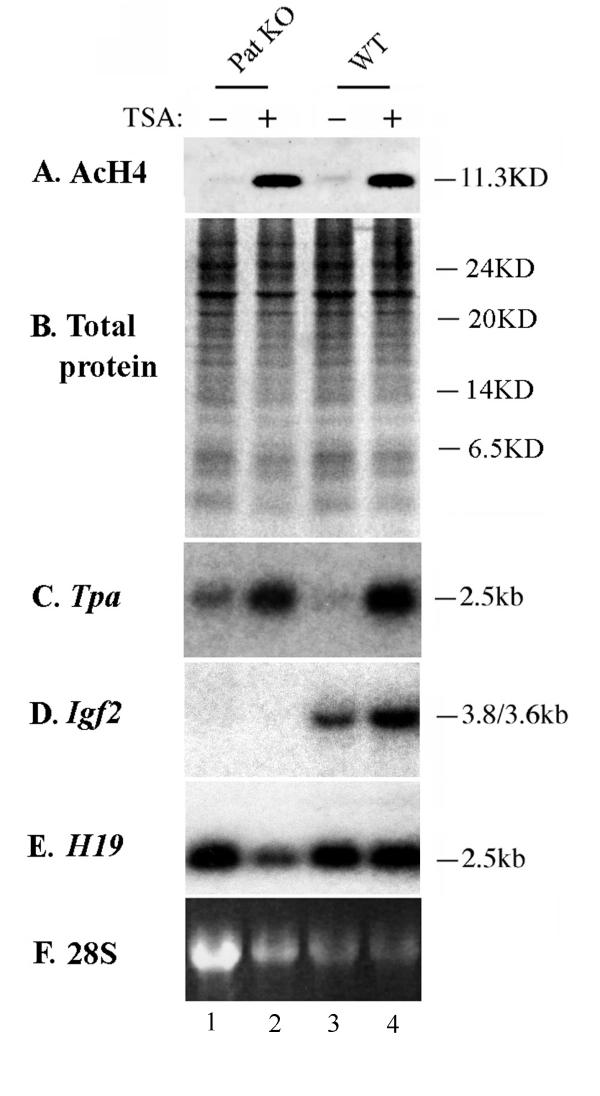
Treatment of primary liver or kidney cells which had never been passaged with 500 nM TSA or 3 mM Sodium butyrate for 6 hr resulted in a marked increase in H4 acetylation, as seen previously in RD cells: results are shown for liver cell cultures (Fig 4A). This also resulted in an up-regulation of the mouse tissue-type plasminogen activator gene (Tpa) (Fig 4C), indicating that histone acetylation levels were again being altered to a degree significant enough to effect gene transcription. We found that Igf2 imprinting was maintained in these primary cells, as shown by the lack of expression of the gene in the cultures derived from a mouse with a paternally-inherited Igf2 knockout (Fig 4D, lanes 1–2), whereas Igf2 transcripts were detected in liver explants from wild-type littermates (Fig 4D, lanes 3–4). However, no significant reactivation of the silent maternal Igf2 allele from any of the three embryonic promoters was seen in the samples, even on long exposure of autoradiographs (Fig 4D and data not shown). There was no significant effect seen on the active Igf2 or H19 alleles in this system. Results for sodium butryrate were identical to TSA and no effect of inhibitors on transcription was seen on longer-term culturing of cells (2–15 days) or in primary kidney cells (data not shown).
Figure 4.
Effects of histone deacetylase inhibitor on Igf2 knockout cells. Primary embryonic liver cells were derived from mice which carried a paternally-inherited deletion of Igf2 (Pat KO; lanes 1 and 2) or their wild-type littermates (WT; lanes 3 and 4). First passage cells were either treated with 500 nM TSA for 6 hr or left untreated as indicated. (A) Western analysis of protein derived from the cells using the anti-acetylated histone 4 (AcH4) antibody shows that treatment with TSA markedly increases the amount of acetylated histone in the primary cells (lanes 2 and 4). (B) Coomasie stained total protein loading control for the Western. (C) Northern hybridization of RNA derived from the same cells to a mouse probe for Tissue-type plasminogen activator (Tpa). Transcription levels can be seen to increase on TSA treatment (lanes 2 and 4) as in RD cells. (D) The membrane used in (C) was stripped and rehybridized with a probe for Igf2. Mice which carry a deletion of the gene on the paternally inherited allele (Pat KO) are missing the allele which is normally active but retain the silenced maternal allele and show no expression of the gene (lane 1). Reactivation of the silent maternal allele was not seen in the cells treated with TSA (lane 2). The two major transcripts ran as a single band here. (E) Rehybridization with an H19 probe shows no significant difference between the TSA treated and untreated samples, allowing for differences in loading. (F) 28S rRNA loading control for the Northern.
Discussion
There is growing evidence for the existence of a pathway to gene inactivation in cancer which does not involve the loss or alteration of the primary sequence of DNA, but rather a change in the activation status of the gene through epigenetic means [1]. Alterations in DNA methylation have been the type of epigenetic change most closely studied and it has been found that hypermethylation is associated with the silencing of key genes in many cancers [15,16]. Inactivation of the H19 gene by methylation is implicated in a number of childhood cancers, including Wilms' tumour and embryonal rhabdomyosarcoma [12]. Wilms' tumors (WT) often occur in association with Beckwith-Weidemann Syndrome (BWS) and it is the subset of BWS patients with methylation of the maternal allele of H19 which usually develop WT [17,18]. In some cases of BWS and of Wilms' tumors the methylation and inactivation of the H19 gene is seen in preneoplastic kidney tissue, indicating that it is an early event in tumorigenesis [10-12]. The H19 gene is part of a cluster of imprinted genes on human chromosome 11p15 which includes the IGF2 gene and several others [12]. In normal tissues, methylation on the paternal allele of H19 inactivates the promoter of the gene. In addition, there is a region upstream which acts as an insulator, preventing Igf2 from interacting with the enhancers downstream of H19 on the maternal allele. On the paternal allele, the insulator, like the H19 promoter, is also methylated and inactivated, allowing IGF2 to interact with the enhancers. The maternal LOH commonly seen in tumors such as WT and RD eliminates the active maternal H19 gene and usually duplicates the active paternal IGF2 gene. Methylation of the normally unmethylated maternal H19 allele in these tumors has a similar effect, as both copies of H19 are now silent and IGF active biallelically. Deletion of the H19 locus or of the insulator upstream [7] by homologous recombination in mice causes the silent maternal allele of Igf2 to become active. In contrast, removal of methylation from the maternal H19 gene by inactivation of the major methyltransferase activity in the cell causes activation of the silent paternal H19 allele and a concomitant decrease in Igf2 expression [8]. Treatment of RD cells by AzaC has also been shown previously to have this effect [5].
Recent work has indicated that DNA methylation and histone deacetylation may act along the same pathway of inactivation. Three proteins which bind to methylated DNA, MeCP2, MBD1 and MBD2 have been shown repress transcription through a mechanism which involves histone deacetylases [3]. Additionally, both the major DNA methyltransferase enzyme DNMT1 [19] and the CTCF protein [20] which binds the upstream insulator region between H19 and IGF2 may cause histone deacetylation either directly or by interacting with HDACs. The inactive alleles of H19 and Igf2 have also recently been shown to have lower levels of histone acetylation than their active counterparts [21], as would be expected if methylation and deacetylation were coupled. If methylation must be followed by histone deacetylation to inactivate a gene, then it is possible that reacetylation of histones associated with a silenced gene may be sufficient to relieve repression and allow transcription. Alternatively, it may be necessary to use a combined treatment which both demethylates the gene and acetylates its associated histones to reactivate a silenced gene.
We found in the work described here that the HDAC inhibitors TSA and sodium butyrate did cause clear increases in H4 acetylation both globally and at the H19 locus in RD cells treated for 6 hrs with these drugs. These increases, while insufficient to reactivate the silent H19 gene, caused up-regulation of the already active TPA gene, which has previously been shown to be responsive to changes in histone acetylation levels [14]. This is similar to the findings of Cameron et al, who showed that TSA alone could up-regulate the basally transcribed CDKN2B and CDKN1A (p21) genes, but not reactivate the silent MLH1 and TIMP3 genes in the colorectal carcinoma cell line RKO [4]. Our results also agree with previous work in mice [22], where it was found that TSA did not reactivate the silenced H19 allele in the vast majority of cells in the mouse embryo. TSA failed to reactivate H19 in RD cells at any concentration from one low enough to show no effect on histone acetylation (25 nM) to a level high enough to have significant cytotoxic side-effects (1 mM) and for any length of time tested from 6 hr to 12 days. In contrast to these results, treatment of RD cells with as little as 500 nM AzaC for a period as short as 15 hrs was sufficient to reactivate H19 in some RD cells, indicating that methylation is a far more important determinant of H19 inactivation in RD cells. These results agree with earlier findings from treatment of RD cells [5] or cells from uniparental embryos [23] with AzaC and also with studies on mice deficient in Dnmt1 activity [8,24]. H19 seems particularly sensitive to changes in methylation levels compared to other imprinted genes in the same cluster as shown by our previous work [12] and that of other laboratories [24,25].
Cameron et al had also found that treatment of cells with a combination of HDAC and methylation inhibitors could reactivate silent genes which were unresponsive to the HDAC inhibitors alone [4]. While Pedone and co-workers found that H19 could be reactivated in this way in cultured mouse fibroblasts when RT-PCR assays were used for detection [26], our own results from RD cells presented here and those of El Kharroubi et al from cultured mouse fibroblasts [23] show that treatment of cells with both types of inhibitor together in fact decreases the levels of H19 mRNA detected relative to those seen with AzaC treatment alone. We found this to be the case at all timepoints and concentrations of inhibitors tested. The evidence therefore suggests that under most circumstances the silent paternal H19 allele cannot be reactivated by treatment with HDAC inhibitors on their own or in combination with demethylating agents.
We also found that the silent Igf2 allele could not be significantly reactivated on treatment with HDAC inhibitors in mouse primary cells. Again, this was true of all drug concentrations and times used, while we could show clear effects on histone acetylation and on the control gene, Tpa in this system. The use of cells which were derived from embryos carrying an Igf2 deletion allowed for sensitive assaying for reactivation against a background of zero expression, similar to the situation for H19 in RD cells. The results agree with those from RD cells, where no increase in transcription from the active IGF2 allele is seen on treatment with acetylase inhibitors and are in agreement with some earlier reports, which have either found no effect [21,27] or some inhibitory effect [28] of HDAC inhibitors on Igf2expression. In order to be certain that a significant reactivation of the silenced Igf2 or H19 is occurring, we chose to examine expression using Northern blots against a zero expressing background rather than RT-PCR assays, which are prone to artifact nonlinearity. It can be noted that levels of reactivation detected by RT-PCR for p57Kip2 or Peg3 in some experiments using acetylase inhibitors are only a small fraction of the wild-type levels [23]. We have previously successfully used Northern blots to detect two-fold differences in expression of H19 and Igf2 in Dnmt1 mutant mice [29] and in AzaC-treated RD cells [5] and here we could detect very low levels of H19 expression which are a result of 15 hr treatment with AzaC (Fig 1B). We also confined our studies to a very recently-derived cell line with no H19 expression or to primary mouse cells which have never been passaged and have a Igf2 null background, to increase sensitivity. Using these methods, we found no evidence that hyperacetylation caused significant reactivation of Igf2 or H19 in either RD cells or primary mouse cells.
Conclusions
We find that silencing by DNA methylation in rhabdomyosarcoma can be uncoupled from histone acetylation for the H19 gene. Likewise, methylation and acetylation can be altered independently in mouse cells carrying a silenced mouse Igf2 gene. These experiments confirm that methylation is the primary determinant of silencing of the H19-Igf2 pair in tumour cells and in normal tissues and that histone hyperacetylation does not reproduce the effects of demethylation.
Materials and Methods
Human cell line culture
Cells derived from an embryonal rhabdomyosarcoma cell line termed RD [5] were grown in DMEM medium containing L-glutamine (Life technologies) supplemented with 10% fetal bovine serum (Hyclone) and 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma). Stock solutions of 5-Aza-2'-deoxycytidine (50 mM in DMEM, single-use; Sigma), TSA (5 mM in ethanol; Wako chemicals) or sodium butyrate (50 mM in deionised water; Sigma) were stored at -85°C and diluted in tissue culture medium to give the required final concentrations (see text and figure legends) before being added to cells in the exponential phase of growth. For experiments in which TSA and AzaC treatment was carried out simultaneously, TSA was added directly to medium which contained AzaC.
Mouse breeding and primary tissue explants
For primary tissue explants, male mice heterozygous for an inactivating mutation at the Igf2 gene (kind gift of A. Efstratiadis)were crossed to WT C57B6/J females. Pregnant females were sacrificed at e16.5 and liver or kidney collected into prewarmed DMEM medium containing 10% fetal bovine serum and antibiotics as above. Each tissue was triturated through a 23 gauge needle and the cell suspension used to seed four wells of a six-well plate which had been pre-treated with fibronectin (Sigma) at 0.5 μg/cm2. After 2 days, the medium was changed to remove unattached cells and dead cells – at this stage the remaining cells had formed extensive cuboidal epithelia interspersed with pockets of fibroblast cells. For these first passage cells, two wells of the four were then treated with histone deacetylase inhibitors (see text and figure legends) prior to harvesting protein or DNA.
Protein harvesting and Western analysis
Tissue culture cells were harvested by washing twice in ice-cold, sterile PBS, then scraped directly in 1:1 dilution of protein loading buffer (125 mM Tris-OH, 20% glycerol, 6% SDS and 1.4 M 2-mercaptoethanol) in PBS, boiled for 5 mins and chilled on ice/water prior to separation on a 15% SDS/PAGE gel. Western blotting was carried out essentially as described [30]. The primary antibody was anti-tetraacetylated histone H4 antibody (Upstate Biotech), used at a dilution of 1:2000 at room temperature for 2 hrs and the secondary antibody was goat anti-rabbit alkaline phosphatase (Sigma) used for one hour at room temperature.
RNA isolation and Northern analysis
RNA was harvested according to standard methods [31]. Cells were initially washed twice with ice-cold, sterile PBS then lysed and scraped directly in solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% sarcosyl, 7 μl/ml 2-mercaptoethanol) before phenol:chloroform extracting and precipitating twice with isopropanol. Purified RNA was resuspended in sterile deionised water. 15 μg of each sample was fractionated by electrophoresis on formaldehyde containing gels and transferred to nylon membranes exactly as previously described [29].
Probes for RNA blot hybridization analysis
Human Tissue-type plasminogen activator (TPA) exon 13 (Genbank accession no. L00152) was amplified using primers F1 TGC GGC CTG AGA CAG TAC AG and R1 TCT CCT GGA AGC AGT GGG CG and cloned into the plasmid vector pCR2.1 (Invitrogen). A probe for mouse Tpa (Genbank accession no. J03520) was generated and cloned in a similar fashion using the primers F1 GGA TGA AGG TCT GGC TTT GG and R1 TGG TGC TGT GAT TCG GCC AG. Human IGF2 and H19 probes have been described previously [12]. A rat Igf2 probe used to detect mouse Igf2 and the mouseH19 probe were as detailed in [29]. Hybridizations were carried out as in [29].
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation was carried out with the anti-tetraacetylated histone 4 antibody (Upstate biotechnology) as per manufacturer's instructions but with the following modifications. Chromatin was cross-linked to DNA in RD cells by adding formaldehyde to the medium to a final concentration of 1% and incubating at 37°C, 30 mins. Sonication to reduce the size of the cross-linked DNA was for 3 x 10 sec with cooling on ice between cycles. The chromatin solution was pre-cleared by adding 60 μl of a 50% protein A sepharose slurry (Upstate) containing 20 micrograms sonicated salmon sperm DNA and 1 mg/ml BSA in TE (10 mM Tris; 1 mM EDTA, pH 8.0) and rocking overnight at 4°C. 5 μl anti-acetyl Histone H4 was added to the resulting supernatant which was then incubated for 2 h at 4°C with rotation before adding 60 μl of 50% protein A sepharose/ssDNA slurry and incubating overnight as before. Proteinase K treatment was done overnight at 55°C. DNA was recovered by phenol/chloroform extraction and ethanol precipitation at room temperature using 20 μg tRNA as a carrier before resuspending in 50 μl (immunoprecipitated) or 200 μl (input) of water. Specific sequences were detected using 3 μl input or 5 μl immunoprecipitated DNA using PCR with the primers H19F11 GCTCCCAGAA CCCACAACA and H19R10 GTGTCTTTGATGTTGGGCT for the transcribed region (Region 1: expected size 355 bp) and H19F5 GCCCTGATGGCGCAGAATC and H19R3 ATGAGTGTCTATCTCTGAC for the insulator region (Region 2: expected size 161 bp). PCR conditions were as follows: an initial deannealing at 94°C, 3 mins, was followed by 35 cycles of 91°C, 10 sec; 55°C, 30 sec; 68°C, 1 min and one final extension at 72°C, 7 min.
Authors' contributions
CL set up and carried out much of the ChiP analysis and some of the cotreatment experiments. BT carried out the cotreatment experiments in Fig 3. CPW carried out the primary cell work and initial RD work. CPW, BT and THB participated in the design of the experiments. CPW wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work has been supported by grants from the NIH (to THB and BT), and from Action Cancer, Combat Cancer and the Cancer Recognised Research Group of Northern Ireland (to CPW).
Contributor Information
Catherine A Lynch, Email: c.lynch@ulster.ac.uk.
Benjamin Tycko, Email: bt12@columbia.edu.
Timothy H Bestor, Email: thb12@columbia.edu.
Colum P Walsh, Email: cp.walsh@ulster.ac.uk.
References
- Tycko B. Epigenetic gene silencing in cancer. J Clin Invest. 2000;105:401–7. doi: 10.1172/JCI9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr Top Microbiol Immunol. 2000;249:55–74. doi: 10.1007/978-3-642-59696-4_4. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Chung WY, Yuan L, Feng L, Hensle T, Tycko B. Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms' tumors. Hum Mol Genet. 1996;5:1101–8. doi: 10.1093/hmg/5.8.1101. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–73. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang SP, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–95. [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Rainier S, Johnson L, Dobry C, Ping A, Grundy P, Feinberg A. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- Frevel MA, Sowerby SJ, Petersen GB, Reeve AE. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. J Biol Chem. 1999;274:29331–40. doi: 10.1074/jbc.274.41.29331. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Morison IM, Taniguchi T, Reeve AE. Epigenetic changes at the insulin-like growth factor II/H19 locus in developing kidney is an early event in Wilms tumorigenesis. Proc Natl Acad Sci U S A. 1997;94:5367–71. doi: 10.1073/pnas.94.10.5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao D, Walsh CP, Yuan L, Gorelov D, Feng L, Hensle T, Nisen P, Yamashiro DJ, Bestor TH, Tycko B. Multipoint analysis of human chromosome 11p15/mouse distal chromosome 7: inclusion of H19/IGF2 in the minimal WT2 region, gene specificity of H19 silencing in Wilms' tumorigenesis and methylation hyper-dependence of H19 imprinting. Hum Mol Genet. 1999;8:1337–52. doi: 10.1093/hmg/8.7.1337. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–52. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts J, Lansink M, Grimbergen J, Toet KH, Kooistra T. Stimulation of tissue-type plasminogen activator gene expression by sodium butyrate and trichostatin A in human endothelial cells involves histone acetylation. Biochem J. 1995;310:171–6. doi: 10.1042/bj3100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, Palazzo JP, Fishel R, Goodfellow PJ. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–6. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet. 2002;70:604–11. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JR, Smallwood A, Harper A, Higgins MJ, Oshimura M, Reik W, Schofield PN, Maher ER. Epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. J Med Genet. 2000;37:921–6. doi: 10.1136/jmg.37.12.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, Schultheiss H, Brehm A, Kouzarides T, Lobanenkov V, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–13. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V Grandjean, O'Neill L, T Sado, B Turner, A Ferguson-Smith. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 2001;488:165–9. doi: 10.1016/S0014-5793(00)02349-8. [DOI] [PubMed] [Google Scholar]
- Svensson K, Mattsson R, James TC, Wentzel P, Pilartz M, MacLaughlin J, Miller SJ, Olsson T, Eriksson UJ, Ohlsson R. The paternal allele of the H19 gene is progressively silenced during early mouse development: the acetylation status of histones may be involved in the generation of variegated expression patterns. Development. 1998;125:61–9. doi: 10.1242/dev.125.1.61. [DOI] [PubMed] [Google Scholar]
- El Kharroubi A, Piras G, Stewart CL. DNA demethylation reactivates a subset of imprinted genes in uniparental mouse embryonic fibroblasts. J Biol Chem. 2001;276:8674–80. doi: 10.1074/jbc.M009392200. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–35. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–74. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone PV, Pikaart MJ, Cerrato F, Vernucci M, Ungaro P, Bruni CB, Riccio A. Role of histone acetylation and DNA methylation in the maintenance of the imprinted expression of the H19 and Igf2 genes. FEBS Lett. 1999;458:45–50. doi: 10.1016/S0014-5793(99)01124-2. [DOI] [PubMed] [Google Scholar]
- Eversole-Cire P, Ferguson-Smith AC, Sasaki H, Brown KD, Cattanach BM, Gonzales FA, MA Surani, PA Jones. Activation of an Imprinted Igf 2 Gene in Mouse Somatic Cell Cultures. Mol Cell Biol. 1993;13:4928–4938. doi: 10.1128/mcb.13.8.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG, Yakovleva T, Hartmann W, Tally M, Bakalkin G, Ekstrom TJ. IGF-II enhances trichostatin A-induced TGFbeta1 and p21(Waf1, Cip1, sdi1) expression in Hep3B cells. Exp Cell Res. 1999;253:618–28. doi: 10.1006/excr.1999.4661. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–97. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Chomczynski , Sacchi A single-step method for isolation of RNA. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]