Abstract
In an effort to identify a clinical biomarker for lung cancer, we used cDNA microarray and 2D protein analyses to demonstrate that increased Fas-associated death domain (FADD) mRNA and protein were significantly associated with poor survival. Analyses of copy number and sequence of the FADD gene in 24 independent tumors ruled out the existence of an amplified and/or mutated FADD gene in aggressive lung cancers. Immunohistochemistry-based tissue microarray analysis showed that nuclear localization of FADD and elevation of the phosphorylated form of FADD (p-FADD) correlated with poor outcome (P = 0.003). Tumors with increased p-FADD expression showed elevated NF-κB (P = 0.004) activation, a frequent molecular alteration associated with tumorigenesis and metastasis in a variety of cancers. To provide a link between p-FADD and NF-κB, cell culture studies demonstrated that overexpression of p-FADD leads to an increase in NF-κB activity and a decrease in the number of cells in the G2 phase of the cell cycle, compared with cells expressing the nonphosphorylatable form of FADD or the vector control. Furthermore, cDNA microarray analyses of lung tumor samples showed that increased levels of FADD transcripts were significantly correlated with overexpression of cyclins D1 (P < 0.01) and B1 (P < 0.01), genes that are involved in the regulation of cell cycle progression and are inducible by NF-κB. These studies demonstrate that induction of NF-κB activity and its effects on cell-cycle progression may represent a molecular basis underlying the aggressive tumor behavior associated with elevated p-FADD expression in lung adenocarcinoma.
Keywords: survival, proliferation, lung cancer
Lung malignancy remains the leading cause of cancer death in industrialized countries. The recent use of gene-expression profiling and proteomic analyses of human lung cancer has led to the identification of genes and proteins associated with reduced survival in lung adenocarcinomas (1, 2). In many instances, however, the exact role of these biomarkers in the biology and pathogenesis of lung cancers or the basis for their association with patient prognosis is unclear.
Fas-associated death domain (FADD)/Mort1 was first isolated in a yeast two-hybrid system as a protein that interacts with the cytosolic tail of the Fas receptor (3-5). Later, it was shown that this proapoptotic adaptor molecule recruits the initiator caspases 8 and 10 to instigate formation of the death-inducing signal complex (DISC), which mediates receptor-induced apoptosis (reviewed in refs. 6-8). FADD harbors a “death domain” (DD) at its carboxyl terminus which serves as a protein-protein interaction region with several death-domain-harboring members of the tumor necrosis factor receptor family, as well as a “death effector domain” (DED) at its amino terminus to recruit initiator caspases (9, 10). Recruitment of initiator caspases 8 and 10 to the DISC leads to intracellular processing and activation of these caspases, eventually resulting in cleavage of downstream targets and apoptosis (6-8). Expression of a dominant-negative form of FADD, consisting of the amino-terminal DD alone, impairs the relay of the apoptotic signal from the death receptors (11, 12).
Analysis of FADD-/- mice revealed that, apart from its role in apoptosis, FADD also plays a role in embryonic development, cell-cycle control, and proliferation of lymphoid cells (13-16). However, the role of DD or DED in these functions remains unclear. These activities have been partly attributed to a third domain in FADD consisting of 35 C-terminal amino acids that encompass the potential phosphorylation site at Ser194 in humans or Ser191, in the case of mouse FADD (17, 18). FADD has also been reported to be localized to the nucleus and shown to contain nuclear localization and export signals (19, 20). Furthermore, FADD was shown to interact with the nuclear proteins MBD4 (involved in the repair of GT mismatches in chromatin) or PIDD, a p53 downstream target regulated by DNA damage (20, 21). There is compelling evidence that phosphorylation of FADD plays an important role in its translocation to the nucleus and in cell-cycle progression, because the mutation of serine at position 194 to alanine leads to uniform localization throughout the cytoplasm and aberrant cell-cycle progression (17, 22). Although upstream kinases with the potential to phosphorylate FADD have been identified (18, 23) and unique FADD interacting partners discovered (20), the role of FADD in the induction of cellular proliferation and tumorigenesis remains poorly understood.
In this report, we provide evidence for a strong correlation between overexpression of FADD mRNA and protein in human lung adenocarcinoma with poor clinical outcome and show that such association is not because of amplification or mutation in FADD. We also show that phosphorylated FADD (p-FADD), which is predominantly localized to the nucleus in lung tumor tissues, is associated with poor survival in patients. Additionally, tumor samples with higher p-FADD showed significantly elevated levels of active NF-κB and correlated with increased cyclin D1 and B1 expression. Using FADD knockout tissue culture cells, we demonstrate that expression of phosphorylated, but not nonphosphorylated, FADD results in NF-κB activation and the associated changes in cell-cycle progression. Thus, dysregulated expression of p-FADD may be a key event in the progression of lung cancer and may provide a marker to differentiate indolent lung cancers from those that are more aggressive.
Materials and Methods
Tumors and Cell Lines. Patients undergoing resection for lung cancer in the General Thoracic Surgery Section of the University of Michigan Hospital from May 1991 to July 2000 were evaluated for inclusion in this study. All patient identifiers were coded to protect confidentiality. Consent was obtained from all patients and the protocol approved by the University of Michigan Institutional Review Board (Medicine). All lung tumors and adjacent normal lung tissues were obtained immediately at the time of surgery and transported to the laboratory in Dulbecco's modified Eagle's medium (Life Technologies, Gaithersburg, MD) on ice. Detailed patient medical records were available for each sample. A portion of each tumor and/or lung tissue was embedded in optimal cutting temperature (OCT) compound (Miles Scientific), frozen in isopentane cooled with liquid nitrogen for cryostat sectioning, and stored at -80°C until use. Hematoxylin-stained cryostat sections (5 μm), prepared from tumors used for protein or mRNA isolation, were evaluated by a study pathologist and compared with hematoxylin/eosin-stained sections made from paraffin blocks of the same tumors. Tumor cellularity of specimens was >70%, and no patient had received prior chemotherapy or radiotherapy. Mutational analysis of K-ras was determined as described in ref. 1.
Affymetrix Oligonucleotide Microarrays. RNA isolation and oligonucleotide microarray analysis are described in refs. 1 and 24.
Quantitative 2D PAGE Analysis and 2D Western Blotting. Analytical 2D PAGE protein quantification and 2D Western blotting were performed as described in ref. 25.
Immunohistochemical Staining of Tissue Microarray. Tissue microarray (TMA) construction and staining methods were performed as described in ref. 26. For immunohistochemical staining of TMA, the primary antibodies anti-FADD (BD Pharmingen) and p-FADD (Cell Signaling Technology, Beverly, MA), were incubated over-night at 4°C. The immunocomplex was visualized by the Ig enzyme bridge technique by using the ABC-peroxidase kit (Vector Laboratories), with 3, 3′ diaminobenzidine tetrachloride as a substrate.
Cell Culture, Cloning, Transfection, and Western Blotting Analysis. For information, see Supporting Methods, which is published as supporting information on the PNAS web site.
NF-κB Functional Assay. NF-κB activity was monitored by an ELISA-based colorimetric TransAM NF-κB human p50 kit, (Active Motif, Carlsbad, CA). In brief, 107cells were washed with 10 ml of cold PBS containing 125 mM NaF, 250 mM β-glycerophosphate, 250 mM p-nitrophenolphosphate, and 25 mM NaVO3. Nuclear lysates were prepared by using a CelLytic Nu-CLEAR extraction kit (Sigma) according to the manufacturer's protocol. Nuclear lysate (20 μg) was used for detection of NF-κB activity. For competition experiments, NF-κB WT and mutated consensus oligonucleotides were used at a concentration of 20 pmol per well. NF-κB activity was also monitored indirectly by detecting the amount and phosphorylation status of inhibitor of NF-κB (I-κB) by Western blotting.
Cell Proliferation and Cell Death Measurement. To induce cell death in the Jurkat cells by activation of Fas receptor, an anti-Fas antibody clone APO 1-3 (Kamiya Biomedical, Seattle, WA) was used at the concentration of 1 μg/ml in conjunction with 1 ng/ml protein A (Sigma) to increase cross-linking. Cell viability was monitored by calcein AM and propidium iodide (PI) (Molecular Probes) staining (see below). For analysis of cell death by cytotoxic agents in various Jurkat cell lines expressing the WT FADD or mutant FADDs, cells were monitored for cell viability 18 h after the addition of 10 μg/ml 5-fluorouracil. Percent cell death was monitored by using the dual-labeling method for live and dead cells. Live cells were stained by calcein AM (Molecular Probes), and dead cells were stained by either membrane impermeable dyes such as PI or Hoechst dye 33258. Briefly, calcein AM was added to a final concentration of 1 μM and PI at 5 μg/ml or Hoechst dye 33258 at 1 μg/ml were added directly to the medium at the end of treatment. After incubating for 5 min at 37°C, cells were visualized under a fluorescence microscope using filters with an excitation wavelength of 488 nm (calcein AM), 533 nm (PI) or 350 nm (Hoechst dye 33258). The dead cells are stained with DNA dyes, and live cells appear green. Photographs of random fields were taken, and the number of dead and live cells was counted. A minimum of 1,500 cells were counted for each treatment and plotted as percent cell death.
Small Interfering RNA (SiRNA) Transfection Analyses. SKLU-1 cells were plated onto 100-mm plates at a density of 1 × 106 cells. Cells were transfected with FADD or scramble siRNAs duplexes by using TransIT TKO reagent (Mirus), in accordance with the manufacturer's suggested protocol. Briefly, 50 μl of TransIT TKO reagent was added to 500 μl of OptiMEM medium and incubated at room temperature for 20 min. To this mixture, either FADD siRNA or scramble duplex (at a final concentration of 50 nM) and Cy3-luciferase GL2 duplex (at a final concentration of 0.5 nM) was added and left at room temperature for 20 min. The siRNA complex was then added to cells with 4.5 ml of media and twice the amount of serum. The medium was replaced 24 h later with fresh complete medium. The cells were harvested 72 h posttransfection, and either nuclei or whole-cell lysates were prepared for NF-κB binding assay or Western blotting. The FADD siRNA (SMART-pool reagent) against human FADD and other control siRNAs were purchased from Dharmacon Research (Lafayette, CO).
Immunostaining, Subcellular Fractionation, and FACS Analysis. For immunostaining, cells grown on a coverslip or in suspension were fixed with 3.5% paraformaldehyde in PBS for 15 min at room temperature. Chilled methanol was then added, and cells were incubated at -20°C for 10 min. Cells were then blocked by using 10% donkey serum and incubated with 1:50 anti-FADD antibodies (BD Pharmingen) for 1 h at 37°C in a humidified environment. After washing, cells were incubated with donkey anti-mouse antibodies coupled to Cy3 (Jackson ImmunoResearch) and counter-stained with Hoechst DNA dye. Staining was monitored with a fluorescence microscope using filters with an excitation wavelength of 533 nm (Cy3) or 350 nm (Hoechst DNA dye). For subcellular fractionation, N-XTRACT and nuclei isolation NucleiPure prep kit (Sigma) were used. Jurkat cells (10 × 106) were used for isolating nuclear and cytosolic extracts according to the protocol suggested by the manufacturer. For FACS analysis, 5 × 106 Jurkat cells were fixed in methanol, stained with 5 μg/ml PI, and delivered to the FACS Analysis Core at the University of Michigan.
Statistical Analysis. F tests were used for examining the difference between FADD protein and each clinical pathology group, except the tumor-normal comparison group, for which t tests were used; t tests were also used to examine differences between FADD mRNA expression and clinical variables. The Pearson correlation coefficient was used for the analysis of the correlation between FADD and Ki-67 protein expression. Univariate Cox regression analysis, Kaplan-Meier survival curve, and log-rank test were used for survival analysis.
Results
Elevated FADD mRNA Is Correlated with Poor Outcome in Lung Adenocarcinomas. Gene-expression analysis of lung adenocarcinomas using oligonucleotide microarrays revealed that increased FADD mRNA was significantly correlated with poor survival (log-rank test P = 0.008, Fig. 1A). This correlation with poor outcome was also observed by analysis of another independent data set of 84 lung adenocarcinomas from Harvard University (Cox P value = 0.02, β = 0.001) (27). Analysis with additional clinical variables revealed that FADD mRNA was increased in tumors with nodal metastasis, stage III (vs. stage I), poor differentiation status, and bronchial-derivation, and was up-regulated in tumors with 12th/13th codon K-ras mutation (see Table 1, which is published as supporting information on the PNAS web site). These associations are consistent with elevated FADD mRNA being involved in poor clinical outcome.
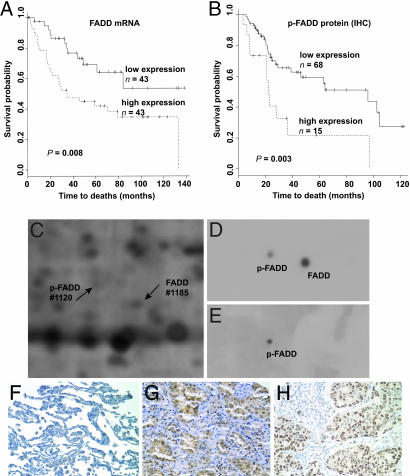
Fig. 1.
FADD mRNA and protein expression in lung adenocarcinomas. (A) Kaplan-Meier survival analysis showed a significantly different outcome between tumors with high and those with low levels of FADD mRNA expression (median value as cutoff value, long-rank test, P = 0.008). (B) Kaplan-Meier survival plot reveals that the 15 patients with highest p-FADD protein expression (score 3 vs. 0 -2) show a poor outcome (P = 0.003) in an independent set of lung adenocarcinomas. (C) Region of a 2D PAGE from a primary lung cancer, showing FADD (1185) and p-FADD (1120) proteins. (D) Western blot with anti-FADD antibody identified two spots representing FADD and p-FADD proteins. (E) Western blot with anti-p-FADD antibody identified only the phosphorylated form of FADD. (F) Immunohistochemical analysis of normal lung tissue, using the anti-FADD antibody on tissue arrays, displays a weak FADD staining. (G) Lung adenocarcinomas display strong cytoplasmic and nuclear immunoreactivity with the anti-FADD antibody. (H) By using the antibody to the p-FADD, immunostaining was predominantly detected in the nucleus of lung adenocarcinomas.
FADD Protein Is Increased in Aggressive Lung Tumors. To determine whether an increase in FADD mRNA results in increased FADD protein expression, quantitative 2D PAGE protein analyses were performed on the same lung adenocarcinomas as those used for mRNA analysis. To identify the location of the FADD protein spots on 2D gels (Fig. 1C), 2D Western blots were performed. Two protein spots, distinct in their pI but with similar molecular mass, were detected when anti-FADD antibodies were used. This finding suggested the existence of a posttranslationally modified form of FADD (presumably phosphorylated) in the tumor samples (Fig. 1D). To confirm this assumption, anti-p-FADD-specific antibodies were used that detected one of the two FADD protein spots on 2D Western blots of tumor lysates, indicating that a shift in mobility was due to the posttranslational phosphorylation of FADD (Fig. 1E). The locations of the two spots were matched to spots 1185 and 1120 on 2D PAGE gel (Fig. 1C) to enable quantitative analysis of both the nonphosphorylated FADD and p-FADD proteins, respectively.
Expression of FADD (1185) was elevated in node-positive tumors, stage III tumors (vs. stage I), tumors showing angiolymphatic invasion, and those lacking a lymphocytic response (Table 1). Because multiple overlapping protein spots were observed near the p-FADD spot (1120), quantitative analysis of p-FADD based on 2D PAGE gels was not possible.
FADD DNA Is Not Amplified or Mutated in Lung Cancers. To investigate whether increased FADD mRNA and protein expression was due to gene amplification, we performed differential PCR (for primer sets, see Table 2, which is published as supporting information on the PNAS web site) using 24 lung adenocarcinomas showing a high level of FADD expression. However, no evidence of gene-dosage changes was observed (see Fig. 6, which is published as supporting information on the PNAS web site). Based on the role of FADD in mediating death-receptor-initiated apoptosis, we hypothesized that the overexpressed form of FADD may be mutated and, thus, could act as a dominant negative to inhibit apoptosis. FADD gene DNA sequencing was performed on 24 of the high-FADD-expressing lung adenocarcinomas; however, no mutations in the coding region of this gene were detected (data not shown).
Nuclear Localization and Phosphorylation of FADD Is Associated with Poor Outcome in an Independent Set of Lung Adenocarcinomas. Tissue-microarray analysis of an independent set of 90 lung adenocarcinomas, including 69 stage I, 12 stage II, and 9 stage III tumor samples, using anti-FADD antibodies, revealed a variable staining pattern, with some tumors expressing little, and others very abundant, cytoplasmic and nuclear immunoreactivity (Fig. 1G). Anti-p-FADD reactivity was predominantly localized to the nucleus of lung adenocarcinoma cells, and, as with anti-FADD reactivity, the anti-p-FADD signal was variably expressed among lung cancer samples (Fig. 1H). Based on the intensity of the signal, p-FADD immunoreactivity was scored in this independent set of lung adenocarcinomas; higher levels of p-FADD were significantly correlated with reduced patient survival (Fig. 1B; n = 83, P = 0.003).
Increased p-FADD Is Correlated with Higher Cell Proliferation. To determine the relationship between cell proliferation and FADD/p-FADD expression, the same tumor samples as those used in the tumor tissue microarray analysis were examined by using the cell-proliferation marker Ki-67. Because the anti-FADD antibody recognized both FADD and p-FADD (Fig. 1D), it was not possible to investigate the correlation between nonphosphorylated FADD and cell proliferation. Staining for p-FADD (Fig. 1 E and H) was found to correlate with Ki-67 expression (Pearson correlation coefficient, n = 66 samples, r = 0.26, P = 0.04 or n = 4 groups, r = 0.98, P = 0.001), indicating that p-FADD overexpression is associated with increased proliferation. To examine the relationship between Ki-67 and nonphosphorylated FADD, quantitative 2D PAGE data (spot 1185) were used. Such analyses revealed no correlation between nonphosphorylated FADD and Ki-67 expression (n = 65, r = -0.02).
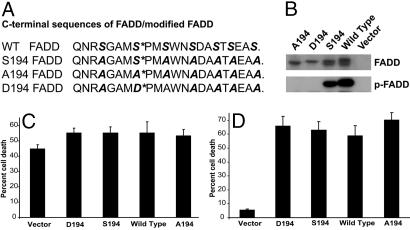
Ser194 Is the Key Phosphorylation Site in FADD. To elucidate the role of FADD phosphorylation in tumor progression and clinical outcome, we generated a series of mutant FADD constructs with a modified C-terminal domain. This domain harbors a serine residue at position 194 that has been reported to be phosphorylated and is implicated in the compartmentalization of FADD to the nucleus. Because this C-terminal domain of FADD harbors six additional serine residues, in anticipation of compensatory phosphorylation at alternate sites, we also generated mutants of FADD, wherein all the serine residues present in the C-terminal domain of FADD, except Ser194, were replaced with alanine (S194). Mutant A194 had a Ser-to-Ala substitution at position 194 (to mimic nonphosphorylated FADD), and D194 had a Ser-to-Asp mutation at position 194 (to mimic p-FADD; Fig. 2A). Stable expression of these constructs in a FADD-null cell line Jurkat-/- (28) followed by Western blot analysis using anti-FADD antibodies revealed that the nonphosphorylatable mutant A194-FADD had the highest gel mobility (Fig. 2B). Gel mobility of mutant D194 protein, which should mimic constitutively p-FADD, was slower and comparable with that of p-FADD (Fig. 2B). Mutant S194 showed both slow and high mobility bands (Fig. 2B) as did the WT FADD. Analysis of these lysates using antibodies specific to phosphorylated Ser194 revealed that S194 and WT had p-FADD. Similar results were seen with expression of FADD in COS (African green monkey kidney cells) and 293T cells (data not shown).
Fig. 2.
Generation and characterization of C-terminal FADD mutants. (A) The sequence of the C-terminal domain of FADD harbors key phosphorylation sites at Ser194, as shown by the asterisk. WT FADD contains seven serine residues (italicized) at the C-terminal domain, of which all except Ser194 were replaced by alanine in the mutant FADD constructs. S194, A194, and D194 FADD constructs have serine, alanine, and aspartic acid at position 194. (B) Whole lysates from Jurkat cells expressing the A194 FADD, D194 FADD, S194 FADD, WT FADD, or vector control were examined by Western blotting using either anti-FADD or anti-p-FADD antibodies. (C) Apoptosis induced by 5-fluorouracil in Jurkat cells expressing WT or FADD mutants, monitored by the live-dead assay using calcein AM and PI staining. (D) Cell death, in stable Jurkat cell lines of WT or FADD mutants, induced by activation of the Fas receptor with anti-Fas antibodies was assessed by using calcein AM and PI staining. All of the above-mentioned experiments were performed in at least triplicate and the results plotted as mean (±SEM) of percent cell death.
Phosphorylation of FADD Does Not Influence Death Through the Intrinsic or Extrinsic Pathways. By using the above cell lines expressing the WT and mutant forms of FADD, the role of p-FADD in mediating apoptosis was investigated. Cell death induced by cytotoxic agents such as 5-fluorouracil (Fig. 2C) and staurosporin (data not shown) was similar between the vector control and any of the stable lines expressing WT or mutant FADD. Induction of apoptosis through the Fas receptor by using the APO-1 agonist antibody revealed that vector control cells (FADD-deficient), had marginal cell death, when compared with any of the FADD-expressing lines (Fig. 2D). Interestingly, there was no significant difference in the cell death potential of A194, S194, D194, or WT, suggesting that phosphorylation of FADD may not be imperative for induction of cell death.
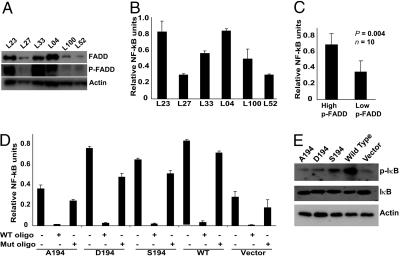
p-FADD Induces NF-κB in Lung Adenocarcinoma and Jurkat Cells. FADD has been reported to increase NF-κB activity (29). In an effort to mechanistically explain the correlation between tumors that have high p-FADD content and NF-κB activation status, we performed the following experiments. Ten lung adenocarcinoma tissues (five with high and five with low p-FADD, determined by immunohistochemistry) were selected. Western blot analysis confirmed the p-FADD levels of these tumor samples (6 of 10 are shown in Fig. 3A). Nuclear lysates prepared from these tissues were used to determine their NF-κB activation status. As shown in Fig. 3 B and C, activation of NF-κB was significantly higher in tissues with elevated levels of p-FADD, as compared with those having low levels of p-FADD (P = 0.004). To confirm the role of p-FADD in NF-κB activation, we used stable cell lines expressing WT or FADD mutants and monitored the activation status of NF-κB. WT, D194, and S194 showed higher NF-κB activation levels (>2-fold), when compared with A194 or vector control (Fig. 3D). These results suggest that the phosphorylation of FADD results in an increased NF-κB activation. To confirm the specificity of the NF-κB activation assay, we preincubated nuclear lysates with oligonucleotides containing the NF-κB-binding consensus sequences, or ones containing mutant sequences, and assessed NF-κB activity. Nuclear lysates preincubated with an excess of oligonucleotide having NF-κB-binding consensus sequences, showed a marked decrease in NF-κB-specific signal, whereas mutant oligonucleotides were ineffective in competing with the binding of NF-κB to its target sequences (Fig. 3D). To independently demonstrate NF-κB activation in these samples, the phosphorylation status of I-κB was evaluated, because I-κB interacts with NF-κB to silence its activity. In response to specific stimuli, I-κB is phosphorylated by I-κB kinase, leading to the liberation of NF-κB, which translocates to the nucleus and activates specific genes. When lysates were evaluated for phosphorylation of I-κB, cells expressing WT FADD showed an intense phospho-I-κB signal followed by S194 and D194, as compared with A194 or vector control cells (Fig. 3E). These results indicate that the phosphorylation of FADD is required for the efficient activation of NF-κB.
Fig. 3.
Induction of NF-κB activity by p-FADD. (A) Lysates from lung tumor samples expressing high (L23, L33, and L04) and low (L27, L100, and L52) p-FADD were subjected to Western blot analysis using anti-FADD, anti-p-FADD, and anti-actin antibodies. (B) Lysates from nuclei-enriched fractions were prepared from high (L23, L33, and L04) and low (L27, L100, and L52) p-FADD-expressing tumors samples, and NF-κB activity was monitored by using the functional NF-κB TransAM kit from Active Motif. Data shown is derived from mean (±SEM) of relative NF-κB of three separate experiments performed in triplicate. (C) NF-κB activity was significantly higher in tissues containing high levels of p-FADD, as compared with those having low levels of p-FADD (data derived from A and B; n = 10, P = 0.004). (D) Nuclear lysates from various Jurkat cell lines expressing FADD or mutant FADDs were prepared, and NF-κB activity was monitored. For competition experiments, lysates were preincubated with either WT NF-κB consensus oligonucleotide or mutant NF-κB oligonucleotide, and NF-κB activity was then assessed. (E) Western blot analysis of whole-cell lysates obtained from the WT FADD or various mutant FADD-expressing Jurkat cell lines by using anti-I-κB, anti-phospho I-κB, and anti-actin antibodies.
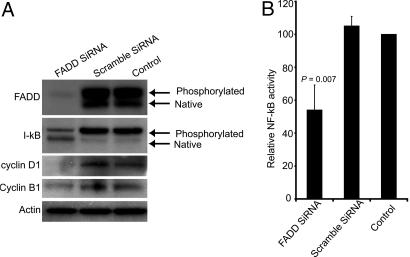
SiRNA-Mediated Down-Regulation of FADD Decreases NF-κB Activity. In an effort to demonstrate the effect of FADD on NF-κB activity in lung cancer cells, we used siRNA to knock down FADD transcripts. These cells were then monitored for NF-κB activity, both at the level of phosphorylation of I-κB and binding of NF-κB to its consensus sequences. SKLU-1, a lung adenocarcinoma cell line, was selected for this study because it showed a very high expression of p-FADD (Fig. 4A). SiRNA-mediated targeted down-regulation of FADD expression resulted in a significant decrease in levels of FADD protein (Fig. 4A). This decrease in FADD expression led to decrease in levels of phosphorylated I-κB and an increase in native I-κB, when compared with nontransfected or scrambled SiRNA-transfected SKLU-1 cells (Fig. 4A). Importantly, a 43% decrease in NF-κB activity was observed in FADD SiRNA-transfected cells, compared with controls (P = 0.007, Fig. 4B). Taken together, these results suggest a strong connection between FADD and NF-κB activity mediated by I-κB-kinase complex.
Fig. 4.
siRNA mediated down-regulation of FADD results in decrease in NF-κB activity. (A) SKLU-1 lysates from cells transfected with FADD siRNA and scramble siRNA and nontransfected control cells were subjected to Western blot analyses using antibodies to FADD, I-κB, and cyclins D1 and B1. Antibody to actin was used as a control. (B) Nuclear lysates were made from SKLU-1 cells transfected with FADD siRNA and scramble siRNA and from nontransfected control cells and were subjected to NF-κB activity by using the TransAM NF-κB kit from Active Motif. Data shown is derived from mean (±SD) of relative NF-κB activity to nontransfected controls of three separate experiments, performed in triplicate
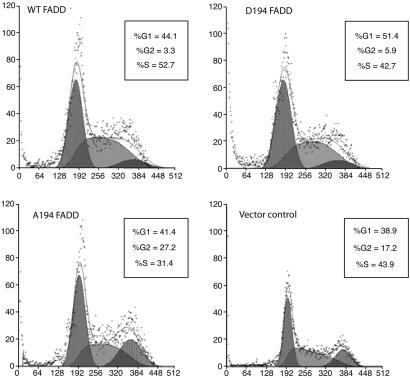
Cell-Cycle Regulation by p-FADD. To delineate the observation that p-FADD and Ki-67 expression was correlated, we directly tested the role of FADD and p-FADD in cell-cycle progression. We analyzed cell-cycle distribution of FADD-deficient vector control cells and cells expressing WT and mutant forms of FADD. Cells lacking FADD (vector control) had a G1:S:G2/M distribution of 38.3:43.9:17.2%, and cells expressing WT FADD had a distribution of 44.1:52.7:3.3%. In contrast, A194-expressing cells had a distribution of 41.4:31.4:27.2%, indicating that phosphorylation of Ser194 results in the abrogation of the G2/M peak. In support of this finding, D194-expressing cells had a distribution of 51.4:42.7:5.9% (Fig. 5).
Fig. 5.
Influence of FADD phosphorylation on cell-cycle progression. Analysis of cell-cycle progression of stable Jurkat cell lines expressing the WT FADD or FADD mutants was performed by using FACS. More cells were arrested in G2/M phase in A194 FADD (27.2%) or vector (17.2%), as compared with D194 FADD (5.9%) or WT FADD (3.3%).
We also investigated the expression of specific cyclins in tumors overexpressing FADD mRNA and examined the correlation between levels of FADD and expression of cyclins B1 and D1. Expression of cyclin B1 and phosphorylated cyclin D1 are G2/M phase markers (31, 32). Of the 86 tumor samples analyzed, cyclins B1 and D1 were strongly correlated with FADD expression (Spearman correlation coefficient r = 0.53 and 0.33, n = 86, P < 0.01). Interestingly, increased cyclin D1 expression was also related to a poor outcome in these patients (P = 0.02). However there was no correlation with other cyclins and FADD (see Table 3, which is published as supporting information on the PNAS web site). Furthermore, we checked the protein levels of cyclins D1 and B1 in SKLU-1 cells transfected with FADD or scramble siRNAs. Expression of cyclin D1 was decreased in cells transfected with FADD siRNA, when compared with scramble-transfected or nontransfected control cells (Fig. 4A). Cyclin B1 levels were not notably decreased by targeted down-regulation of FADD expression in SKLU-1 cells (Fig. 4A).
Discussion
Our studies demonstrate that, both at the mRNA and protein levels, overexpression of FADD, a protein thought to be involved in mediating death-receptor-induced apoptosis, is also an indicator of cell proliferation and poor survival of lung adenocarcinoma patients. FADD is a member of the death-inducing signaling complex (DISC) whose role as an adapter protein is key in transmitting death signals from cell-surface receptors (6, 7). Cells deficient in FADD show resistance to receptor-mediated cell death (ref. 28 and Fig. 2). A recent study using immunohistochemistry and subcellular fractionation analysis, however, showed that FADD is predominantly a nuclear protein in a number of cell lines (19, 20). This raises the possibility that the nuclear localization of FADD may be a regulatory mechanism wherein it is sequestered, thus preventing DISC formation. In addition, accumulating evidence suggests that FADD may have supplementary functions, at present not understood, that require it to reside in the nucleus (20, 22). FADD-null mutations in mice are embryonic-lethal, and defects in the proliferation and activation of FADD-/- T cells support this notion (13, 16, 32, 33). Adding to this functional intricacy, posttranslational modification of FADD has also been suggested to be important for some of these proposed functions. Recent reports have indicated that the phosphorylation status of FADD plays a role in nuclear or cytosolic distribution (17). Furthermore, FADD was shown to contain functional nuclear localization and export signals (19). Our studies not only provide evidence for a correlation between clinical outcome and the presence of nuclear-localized p-FADD but also provides substantial proof that p-FADD induces activation of NF-κB and abnormalities in cell proliferation. In contrast to our finding in lung cancer, a recent report in acute myeloid leukemia shows that the absence or low expression of FADD protein is a poor prognostic factor (34). One way to comprehend these contradictory results is by emphasis on cell-type-specific aberrations in dichotomous FADD-mediated pathways that regulate both cell growth and apoptosis.
Multiple cellular receptors and signaling pathways promote NF-κB activation, which plays a central role in tumor development, progression, and therapy (35). FADD was previously shown to induce NF-κB (29). Our data demonstrates a specific role for p-FADD in the efficient activation of NF-κB (Fig. 3). Upstream pathways and mechanistic events that trigger p-FADD-induced NF-κB activation are yet to be understood. Fas-interacting Serine/Threonine kinase (FIST)/homeodomain-interacting protein kinase 2 (HIPK2) has been implicated as a potential FADD kinase, although the regulation of this kinase is not well understood (23). Much is known about the downstream target genes that are under the direct effect of NF-κB. These include regulators of the cell cycle (cyclin D1) (36), growth promoters (c-myc and c-myb) (37, 38), and inhibitors of apoptosis (XIAP, IEX-1, twist, and the Bcl-2 homologues A1/Bfl-1, BCL-XL, and Nr13) (39, 40). Additionally, NF-κB regulates potent secreted mitogens, such as human macrophage-colony-stimulating factor (M-CSF), platelet-derived growth factor (PDGF), VEGF, and melanoma-growth-stimulating activity (MGSA)/Groα, which may serve in an autocrine manner to propagate growth stimuli (reviewed in ref. 35). Furthermore, NF-κB is reported to be involved in invasion and metastasis by regulating the expression of matrix metalloproteinases, plasminogen activator, and heparinase (reviewed in ref. 35). Thus, it appears that, in addition to its role in the survival of transformed cells, NF-κB has subsequent effects on the malignant behavior of tumors, perhaps partly explaining why the overexpression of p-FADD leads to poor survival. We are currently investigating the specific mediators that relay the signal from p-FADD to NF-κB.
Recent reports have suggested a role for FADD in cell-cycle progression (17, 43). These studies have also suggested that phosphorylation (at Ser191 in mice and Ser194 in humans) of FADD may play a role in the regulation of the cell cycle. T cells lacking FADD and MCF-10A cells overexpressing the C-terminal phosphorylation domain of FADD show alterations in cell-cycle progression (43). Our studies show striking differences in cells in the G2/M phase, as well as other changes in the cell-cycle distribution of cells expressing the phosphorylatable and nonphosphorylatable forms of FADD (Fig. 5). The correlation between cyclins D1 and B1 and FADD in tumor samples also indicates possible involvement of p-FADD in regulating the cell cycle. Cyclin B1 associates with cdc2 to form the maturation-mitosis-promoting factor, and cyclin D1 is phosphorylated and translocated to the nucleus in the G2/M phase (30, 31). Both cyclins play a key role in the G2/M phase of the cell cycle (30, 31). Cyclins D1 and B1 are overexpressed in many cancers, including lung cancer (44-46). Interestingly, the cyclin D gene promoter contains multiple NF-κB response elements that may link NF-κB induction with cyclin D1 overexpression in p-FADD-overexpressing tumors and cell lines. This finding suggests that the induction of NF-κB may be a relatively early event in lung adenocarcinoma development. Dysregulation of the cell cycle, resulting from elevated p-FADD expression, may represent one of the mechanisms for the poor survival observed in lung adenocarcinoma patients.
Supplementary Material
Acknowledgments
We thank Theodore Lawrence, Mats Ljungman, Dan Hamstra, Jonathan Maybaum, and Mukesh Nyati for useful discussion during these studies. The laboratories of D.G.B. and A.R. contributed equally to this work. This work was supported by National Institutes of Health/National Cancer Institute Grants P50CA01014 (to B.D.R. and A.R.), P01CA85878 (to B.D.R. and A.R.), National Cancer Institute Grant CA-71606 (to D.G.B.), and University of Michigan Cancer Center Support Grant 5P30CA46592.
Author contributions: G.C., M.S.B., D.G.B., and A.R. designed research; G.C., M.S.B., A.C.H., D.C.C., B.L., D.G.T., L.B.G., J.Y., J.M.C., T.J.G., and L.L. performed research; M.B.O., B.D.R., D.G.B., and A.R. contributed new reagents/analytic tools; G.C., M.S.B., D.G.B., and A.R. analyzed data; and G.C., M.S.B., D.G.B., and A. R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DD, death domain; FADD, Fas-associated DD; I-κB, inhibitor of NF-κB; PI, propidium iodide; SiRNA, small interfering RNA; p-FADD, phosphosylated FADD.
References
- 1.Beer, D. G., Kardia, S. L., Huang, C. C., Giordano, T. J., Levin, A. M., Misek, D. E., Lin, L., Chen, G., Gharib, T. G., Thomas, D. G., et al. (2002) Nat. Med. 8, 816-824. [DOI] [PubMed] [Google Scholar]
- 2.Chen, G., Gharib, T. G., Wang, H., Huang, C. C., Kuick, R., Thomas, D. G., Shedden, K. A., Misek, D. E., Taylor, J. M., Giordano, T. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13537-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnaiyan, A. M., O'Rourke, K., Tewari, M. & Dixit, V. M. (1995) Cell 81, 505-512. [DOI] [PubMed] [Google Scholar]
- 4.Boldin, M. P., Varfolomeev, E. E., Pancer, Z., Mett, I. L., Camonis, J. H. & Wallach, D. (1995) J. Biol. Chem. 270, 7795-7798. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, J. & Winoto, A. (1996) Mol. Cell. Biol. 16, 2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tibbetts, M. D., Zheng, L. & Lenardo, M. J. (2003) Nat. Immunol. 4, 404-409. [DOI] [PubMed] [Google Scholar]
- 7.Bhojani, M. S., Rossu, B. D. & Rehemtulla, A. (2003) Cancer Biol. Ther. 2, S71-S78. [PubMed] [Google Scholar]
- 8.Baker, S. J. & Reddy, E. P. (1996) Oncogene 12, 1-9. [PubMed] [Google Scholar]
- 9.Kischkel, F. C., Hellbardt, S., Behrmann, I., Germer, M., Pawlita, M., Krammer, P. H. & Peter, M. E. (1995) EMBO J. 14, 5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzio, M., Chinnaiyan, A. M., Kischkel, F. C., O'Rourke, K., Shevchenko, A., Ni, J., Scaffidi, C., Bretz, J. D., Zhang, M., Gentz, R., et al. (1996) Cell 85, 817-827. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan, A. M., Tepper, C. G., Seldin, M. F., O'Rourke, K., Kischkel, F. C., Hellbardt, S., Krammer, P. H., Peter, M. E. & Dixit, V. M. (1996) J. Biol. Chem. 271, 4961-4965. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, H., Shu, H. B., Pan, M. G. & Goeddel, D. V. (1996) Cell 84, 299-308. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, J., Cado, D., Chen, A., Kabra, N. H. & Winoto, A. (1998) Nature 392, 296-300. [DOI] [PubMed] [Google Scholar]
- 14.Newton, K., Harris, A. W., Bath, M. L., Smith, K. G. & Strasser, A. (1998) EMBO J. 17, 706-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh, C. M., Wen, B. G., Chinnaiyan, A. M., O'Rourke, K., Dixit, V. M. & Hedrick, S. M. (1998) Immunity 8, 439-449. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, J., Kabra, N. H., Cado, D., Kang, C. & Winoto, A. (2001) J. Biol. Chem. 276, 29815-29818. [DOI] [PubMed] [Google Scholar]
- 17.Hua, Z. C., Sohn, S. J., Kang, C., Cado, D. & Winoto, A. (2003) Immunity 18, 513-521. [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi, C., Volkland, J., Blomberg, I., Hoffmann, I., Krammer, P. H. & Peter, M. E. (2000) J. Immunol. 164, 1236-1242. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Angelats, M. & Cidlowski, J. A. (2003) Cell Death Differ. 10, 791-797. [DOI] [PubMed] [Google Scholar]
- 20.Screaton, R. A., Kiessling, S., Sansom, O. J., Millar, C. B., Maddison, K., Bird, A., Clarke, A. R. & Frisch, S. M. (2003) Proc. Natl. Acad. Sci. USA 100, 5211-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telliez, J. B., Bean, K. M. & Lin, L. L. (2000) Biochim. Biophys. Acta 1478, 280-288. [DOI] [PubMed] [Google Scholar]
- 22.Sheikh, M. S. & Huang, Y. (2003) Cell Cycle 2, 346-347. [PubMed] [Google Scholar]
- 23.Rochat-Steiner, V., Becker, K., Micheau, O., Schneider, P., Burns, K. & Tschopp, J. (2000) J. Exp. Med. 192, 1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano, T. J., Shedden, K. A., Schwartz, D. R., Kuick, R., Taylor, J. M., Lee, N., Misek, D. E., Greenson, J. K., Kardia, S. L., Beer, D. G., et al. (2001) Am. J. Pathol. 159, 1231-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, G., Gharib, T. G., Huang, C. C., Taylor, J. M., Misek, D. E., Kardia, S. L., Giordano, T. J., Iannettoni, M. D., Orringer, M. B., Hanash, S. M. & Beer, D. G. (2002) Mol. Cell. Proteomics 1, 304-313. [DOI] [PubMed] [Google Scholar]
- 26.Chen, G., Gharib, T. G., Huang, C. C., Thomas, D. G., Shedden, K. A., Taylor, J. M., Kardia, S. L., Misek, D. E., Giordano, T. J., Iannettoni, M. D., et al. (2002) Clin. Cancer Res. 8, 2298-2305. [PubMed] [Google Scholar]
- 27.Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juo, P., Woo, M. S., Kuo, C. J., Signorelli, P., Biemann, H. P., Hannun, Y. A. & Blenis, J. (1999) Cell Growth Differ. 10, 797-804. [PubMed] [Google Scholar]
- 29.Hu, W. H., Johnson, H. & Shu, H. B. (2000) J. Biol. Chem. 275, 10838-10844. [DOI] [PubMed] [Google Scholar]
- 30.Stacey, D. W. (2003) Curr. Opin. Cell Biol. 15, 158-163. [DOI] [PubMed] [Google Scholar]
- 31.Takizawa, C. G. & Morgan, D. O. (2000) Curr. Opin. Cell Biol. 12, 658-665. [DOI] [PubMed] [Google Scholar]
- 32.Beisner, D. R., Chu, I. H., Arechiga, A. F., Hedrick, S. M. & Walsh, C. M. (2003) J. Immunol. 171, 247-256. [DOI] [PubMed] [Google Scholar]
- 33.Kabra, N. H., Kang, C., Hsing, L. C., Zhang, J. & Winoto, A. (2001) Proc. Natl. Acad. Sci. USA 98, 6307-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tourneur, L., Delluc, S., Levy, V., Valensi, F., Radford-Weiss, I., Legrand, O., Vargaftig, J., Boix, C., Macintyre, E. A., Varet, B., et al. (2004) Cancer Res. 64, 8101-8108. [DOI] [PubMed] [Google Scholar]
- 35.Amit, S. & Ben-Neriah, Y. (2003) Semin. Cancer Biol. 13, 15-28. [DOI] [PubMed] [Google Scholar]
- 36.Cao, Y., Bonizzi, G., Seagroves, T. N., Greten, F. R., Johnson, R., Schmidt, E. V. & Karin, M. (2001) Cell 107, 763-775. [DOI] [PubMed] [Google Scholar]
- 37.Toth, C. R., Hostutler, R. F., Baldwin, A. S., Jr., & Bender, T. P. (1995) J. Biol. Chem. 270, 7661-7671. [DOI] [PubMed] [Google Scholar]
- 38.Duyao, M. P., Kessler, D. J., Spicer, D. B., Bartholomew, C., Cleveland, J. L., Siekevitz, M. & Sonenshein, G. E. (1992) J. Biol. Chem. 267, 16288-16291. [PubMed] [Google Scholar]
- 39.Zong, W. X., Edelstein, L. C., Chen, C., Bash, J. & Gelinas, C. (1999) Genes Dev. 13, 382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grumont, R. J., Rourke, I. J. & Gerondakis, S. (1999) Genes Dev. 13, 400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, C. Y., Guttridge, D. C., Mayo, M. W. & Baldwin, A. S., Jr. (1999) Mol. Cell. Biol. 19, 5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stehlik, C., de Martin, R., Kumabashiri, I., Schmid, J. A., Binder, B. R. & Lipp, J. (1998) J. Exp. Med. 188, 211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alappat, E. C., Volkland, J. & Peter, M. E. (2003) J. Biol. Chem. 278, 41585-41588. [DOI] [PubMed] [Google Scholar]
- 44.Schauer, I. E., Siriwardana, S., Langan, T. A. & Sclafani, R. A. (1994) Proc. Natl. Acad. Sci. USA 91, 7827-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soria, J. C., Jang, S. J., Khuri, F. R., Hassan, K., Liu, D., Hong, W. K. & Mao, L. (2000) Cancer Res. 60, 4000-4004. [PubMed] [Google Scholar]
- 46.Yoshida, T., Tanaka, S., Mogi, A., Shitara, Y. & Kuwano, H. (2004) Ann. Oncol. 15, 252-256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.