Abstract
Light is an essential environmental factor, and many species have evolved the capability to respond to it. Blue light is perceived through three flavin-containing photoreceptor families: cryptochromes, light-oxygen-voltage, and BLUF (sensor of blue light using flavin adenine dinucleotide, FAD) domain proteins. BLUF domains are present in various proteins from Bacteria and lower Eukarya. They are fully modular and can relay signals to structurally and functionally diverse output units, most of which are implicated in nucleotide metabolism. We present the high resolution crystal structure of the dark resting state of BlrB, a short BLUF domain-containing protein from Rhodobacter sphaeroides. The structure reveals a previously uncharacterized FAD-binding fold. Along with other lines of evidence, it suggests mechanistic aspects for the photocycle that is characterized by a red-shifted absorbance of the flavin. The isoalloxazine ring of FAD binds in a cleft between two helices, whereas the adenine ring points into the solvent. We propose that the adenine ring serves as a hook mediating the interaction with its effector/output domain. The structure suggests a unique photochemical signaling switch in which the absorption of light induces a structural change in the rim surrounding the hook, thereby changing the protein interface between BLUF and the output domain.
Keywords: blue light sensing, photochemistry, protein function, flavin, reaction mechanism
The ability to perceive light is crucial for the survival of most organisms, enabling them to adapt to changing environmental conditions. Consequently, the capacity to sense and respond to light is widespread among prokaryotes and eukaryotes. To date, six photoreceptor families have been identified: the rhodopsins, phytochromes, xanthopsins (photoactive yellow protein family), cryptochromes, light-oxygen-voltage (LOV) domain-containing proteins, and the sensors of blue light using flavine adenine dinucleotide (BLUF) domain-containing proteins (1). The latter three families are blue light receptors that use flavin chromophores. The primary photochemistry produces conformational changes in the cofactor, which induces the formation of a signaling state that communicates photon absorption to a downstream domain or protein partner. BLUF domain proteins represent the third class of flavin-containing blue-light receptors (2). Their identification in the protein AppA marks the latest piece of the puzzle in understanding how organisms respond to blue light. AppA is involved in the regulation of photosynthesis gene expression in the anoxygenic phototrophic α-proteobacterium Rhodobacter sphaeroides (3–6). AppA not only responds to blue light but also senses redox signals and has been shown to integrate these two stimuli (7, 8). The BLUF domain has thereupon been found in proteins from several branches of the phylogenetic tree of Bacteria and in photosynthetic lower Eukarya (Pfam protein domain database; www.sanger.ac.uk/Software/Pfam). In the photoactivated adenylyl cyclase (PAC) of the unicellular flagellate Euglena gracilis (9), BLUF mediates a photophobic response. Very recently, Klug and coworkers (10) showed that BLUF domains function modularly: the AppA blue light sensing module was replaced by the BLUF domain of PAC without disturbing light-dependent gene expression.
Great efforts have been undertaken to study the photochemistry and primary signaling processes of the BLUF domain. Blue light illumination of the AppA BLUF domain and the Synechocystis BLUF protein Slr1694 induces small but distinct and reversible red shifts of the flavine adenine dinucleotide (FAD) absorption spectrum (11, 12). Although intramolecular proton transfer processes and/or alterations in hydrogen bonding have been suggested to underlie this red shift (13, 14), the molecular mechanism of the photocycle is largely unknown.
The predicted secondary structure of BLUF domains resembles neither that of LOV domains nor that of the flavin-binding domains in cryptochromes (2). In fact, the BLUF domain has been predicted to establish a new FAD-binding fold because it could not be assigned to any of the existing FAD-binding protein families (15). Based on an analysis of domain architecture, Gomelsky and Klug divided the BLUF protein family into short and complex BLUF proteins (2). Short proteins contain, besides BLUF, an additional stretch of ≈50 amino acid residues at their C terminus but no additional identifiable protein domains. They were proposed to communicate the light signal to effector/output modules through protein–protein interactions. In complex BLUF proteins, sensor and effector modules reside on the same polypeptide chain. So far, the three-dimensional structure of BLUF domains, the photochemistry that underlies the activation of intraor intermolecular receptor domains, and, thus, the signal transduction process are unknown.
Here, we present the crystal structure of a full-length BLUF protein, BlrB from R. sphaeroides, at 1.9 Å resolution in its dark-adapted state. The structure reveals a FAD-binding fold and provides insights into the photochemical reaction. Moreover, the structure suggests a mechanism for how the BLUF domain communicates the blue light signal to either an effector domain within the same polypeptide chain or to an independent effector protein. This model is consistent with the idea of modularity.
Experimental Procedures
Protein Preparation and Quality Assurance. DNA encoding the full-length BlrA and BlrB proteins was PCR amplified from R. sphaeroides genomic DNA by using gene-specific primers and cloned into the expression vectors pET23a and pHis-Gβ1-Parallel1, where pHis-Gβ1-Parallel1 is a derivative of the pHis-Parallel1 vector (16). BlrB was purified from Escherichia coli cells grown in LB medium or M9 media containing 3 g/liter 13C6 glucose (for NMR samples), as described in supporting information, which is published on the PNAS web site. The protein was analyzed for cofactor content by reverse phase chromatography and subsequently reconstituted with its physiological cofactor, FAD, to nearly 100% (see supporting information). The protein was stored in 25 mM Tris·HCl, pH 8.0/40 mM NaCl/10 mM KCl/2 mM EDTA/5 mM DTT/5% glycerol at –80°C.
Spectroscopy. UV-visible absorption spectra were recorded on 0.05 mM protein samples in a buffer containing 50 mM Tris (pH 8.0) and 30 mM NaCl, using a Hewlett Packard 8452 A Diode Array Spectrophotometer at 4°C. Spectra on the putative signaling active state were acquired after exciting samples with a photographic flash. NMR spectra were obtained as described in supporting information.
Crystallization. Crystals of BlrB were grown by the hanging drop vapor diffusion method at 4°C in the dark by mixing 1 μl of protein (9 mg/ml) and reservoir (0.1 M acetate buffer pH 4.5/0.2 M Ca2+ acetate/0.05 M DTT/28% polyethylene glycol 400) solutions. Crystallization setups were inspected with a 2-mm-thick RG 630 filter (ITOS, Mainz, Germany) shielding the microscope bulb. Crystals were rinsed in cryoprotection solution (0.1 M acetate buffer, pH 4.5/0.2 M Ca2+ acetate/0.05 M DTT/35% polyethylene glycol 400) before flash-cooling in liquid nitrogen. Heavy-atom derivatives were prepared by soaking crystals in reservoir solutions containing the heavy atom compounds (EMP, KAu(CN)2, and uranylacetate, respectively, see Table 2, which is published as supporting information on the PNAS web site). Before flash-cooling in liquid nitrogen, the crystals soaked with EMP and KAu(CN)2 were rinsed briefly in the final cryoprotection solution.
Data Collection, Structure Determination, and Refinement. Diffraction data were collected under standard cryogenic conditions (100 K) at the European Synchrotron Radiation Facility or Swiss Light Source (see supporting information for details), and processed with the xds suite of programs (17). The crystals (space group P212121) contain two molecules per asymmetric unit that are related by a 2-fold rotation and a self-Patterson vector at 0.5, 0.5, and 0.007. This finding, together with noticeable nonisomorphism, complicated the structure determination by multiple isomorphous replacement with anomalous scattering. Heavy atom positions could only be identified by sharp (18). After density modification (dm in sharp), clearly interpretable electron density maps were obtained (supporting information). An initial partial model was built automatically by arp/warp (19) and extended manually in o (20). During several cyclic rounds of refinement with cns (21) and manual rebuilding, FMN and solvent molecules were included in the model. The final refinement step was done with refmac and included tls refinement (22) (for statistics, see supporting information). Figures were made with pymol (23).
Results and Discussion
Recombinant BlrB Contains a Functionally Active BLUF Domain. The R. sphaeroides genome (http://genome.ornl.gov/microbial/rsph) encodes three BLUF-domain containing proteins: AppA and two short BLUF proteins, RSP4060 (134 aa) and RSP1261 (140 aa). The latter two proteins were designated BlrA and BlrB, respectively, for putative blue-light receptors. The functions of these proteins are unknown. Upon induction of expression, BlrB, but not BlrA, made the E. coli host yellow. When purified, BlrB was found to contain noncovalently bound flavins. BlrB was therefore used for subsequent characterization. Full-length BlrB was expressed in E. coli and purified by affinity and size exclusion chromatography. Because cofactor analysis by HPLC showed ≈ 42% FAD, 26% FMN, and 32% riboflavin, the protein was reconstituted with FAD in vitro to its physiologically active form (see supporting information).
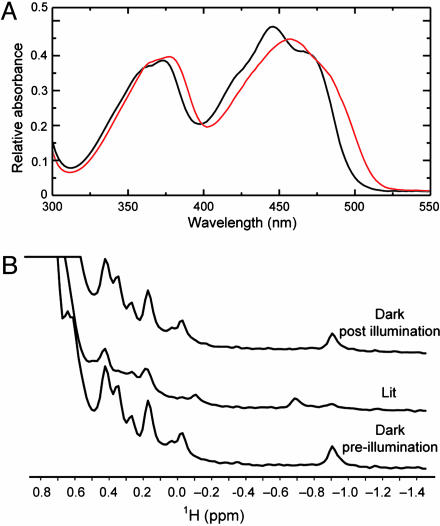
We used a combination of visible absorbance and NMR spectroscopy to characterize the effects of blue light illumination on BlrB. Visible absorbance spectra of dark state BlrB samples show a characteristic pattern of flavin-binding proteins, including two major absorbance bands centered near 365 and 450 nm (Fig. 1A). As observed with other BLUF domains (6, 12, 14), illumination leads to a red shift of these bands by ≈10 nm. These changes are reversible, reverting back to the original dark state spectra with an apparent first-order exponential time constant of ≈5 s at room temperature, pH 8.0. The kinetics for this regeneration process are similar to those observed for the short BLUF domain protein Synechocystis Slr1694 protein (t1/2 ≈ 5 s) (12) but are notably much faster than that observed for the isolated BLUF domain of AppA (t1/2 ≈ 15 min) (6).
Fig. 1.
Spectroscopic characterization of BlrB dark and signaling active states. (A) UV-visible absorption spectra of dark state (black line) and signaling state (red line) of BlrB recorded at 4°C. (B) 1H NMR spectra for the methyl regions of BlrB recorded under dark, signaling (lit), and postillumination dark states.
Although visible absorbance spectroscopy provides insights into the electronic environment of the FAD cofactor in BlrB, we sought to complement this finding by using NMR spectroscopy to obtain information about the surrounding protein environment. One-dimensional 1H NMR spectra acquired on BlrB in the dark show excellent chemical shift dispersion, including a number of upfield-shifted peaks (δ < 0.5 ppm) typically associated with methyl groups involved in stable tertiary interactions near aromatic rings (Fig. 1B). Illumination with blue light at 457 nm leads to the disappearance of several signals and the concomitant appearance of several new peaks, consistent with light-induced changes in the electronic environments near these protons and also with prior observations from AppA (14). After ceasing illumination and allowing ≈1 min for recovery, the original dark state spectrum is regenerated, critically establishing the reversibility of these changes at the levels of both the chromophore and protein.
A comparison of 2D 13C-1H HSQC spectra recorded on BlrB revealed a series of changes in peak locations and intensities that are reversible in the dark (supporting information). Notably, a much lower fraction of peaks are perturbed by illumination of BlrB than previously observed for the Avena sativa phototropin LOV2 domain (24), suggesting that light induces much smaller conformational changes within BLUF domains compared to LOV domains.
BLUF Domains Represent a Previously Uncharacterized FAD Binding Fold. The crystal structure of BlrB was determined through multiple isomorphous replacement (supporting information). The structure determination was complicated by the symmetry of the arrangement of the two molecules in the asymmetric unit of the orthorombic crystals of BlrB. The electron density indicates that the two BlrB molecules differ slightly in length. Molecule A encompasses residues 1 to 136, whereas molecule B ranges from residue 1 to 131. We determined the structure of BlrB in its dark state to a resolution of 1.9 Å (see Experimental Procedures and supporting information). The overall fold reveals a very compact protein that consists of a five-stranded mixed β-sheet with two α-helices running parallel to the β-strands on one side and a helix–turn–helix unit on the other side of the sheet. The dimer interface is made up by β-strands of each molecule (residues Phe-76 to Pro-83, respectively), running antiparallel to each other (Fig. 2A). The first 75 amino acid residues of the protein form an αβ sandwich with a typical ferredoxin-like β1 α1 β2 β3 α2 β4 topology. This structural motif is very similar to the fold adopted by acylphosphatase (PDB ID code 2ACY), a member of the ACT (aspartate kinase, chroismate mutase, TyrA) domain family (25) and one of the smallest enzymes known. However, the primary structure of BlrB does not reveal any significant similarity with the ACT consensus sequence and therefore represents a previously uncharacterized domain family.
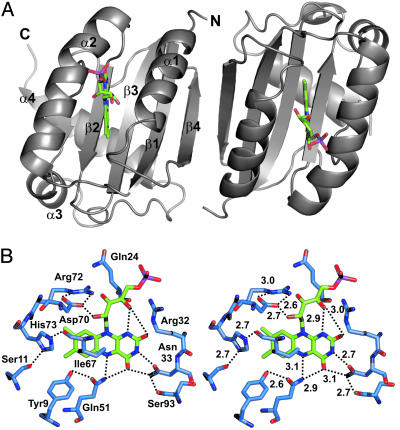
Fig. 2.
Structure and active site of BlrB. (A) The asymmetric unit of BlrB crystals contains two BlrB molecules with ferredoxin-like topology (apoproteins in gray and flavin cofactors in green). Secondary structure elements are assigned in molecule A (Left). (B) The flavin-binding pocket, shown in stereoview, is made up by the highest conserved residues of the BLUF domain. They position the cofactor by mostly hydrophobic interactions around the dimethylbenzene moiety and by hydrogen bonds to heteroatoms of the light absorbing ring system and the ribityl chain. Compared to most residues building up the cofactor pocket, the side chain of Arg-32 is less well defined. There is only a clear interaction with the O4′ and O2 atoms of the flavin in molecule A. Hydrogen bonds are indicated by dotted lines, and distances are given in angstroms.
While this manuscript was under revision, structures of two other BLUF domains were reported. These include a short BLUF domain protein (Tll0087; ref. 26) and a BLUF domain from the AppA protein (residues 17–133; ref. 27). All three of these structures adopt similar α/β folds immediately around the FAD isoalloxazine ring, emphasizing the structural conservation of the BLUF sequence homology region identified by Gomelsky and Klug (2). The key difference among these structures seems to be the presence of two C-terminal helices to the core BLUF domain, as observed in the short BLUF proteins (BlrB and Tll0078). AppA has a single short α-helix after the BLUF domain, but it is solvent exposed and appears to be stabilized by the presence of detergent in the crystallization mixture.
The FAD Binding Site. Chromatographic analysis indicated that the majority of protein molecules within the BlrB crystals bind FAD, which is consistent with our expectations (see supporting information for details). However, there is clear electron density for only the FMN moiety of FAD, but not for the adenosine monophosphate, AMP. The isoalloxazine moiety of the FAD cofactor is bound tightly between the helixes α1 and α2, above the β-sheet plane in a pocket that is built up by side chains originating from almost all secondary structure elements of the BLUF core, α1, α2, β1, β2, β3, and two loop regions (α2-β4 and β4-α3) (Fig. 2B). Because the ribityl side chain and phosphate moiety point toward the domain surface, the adenine moiety must be entirely solvent exposed and is probably highly flexible. This observation explains numerous biochemical findings, such as the instability of FAD bound to BLUF domains and the module's tolerance in binding diverse flavin derivatives (28). In particular, it explains the fact that the nature of the flavin chromophore hardly affects the capacity of the AppA-BLUF domain to photocycle (28, 29). As assumed in ref. 2, the FAD-binding mode described here establishes a previously uncharacterized FAD-fold family. The flavin-binding pocket of BlrB is similar to the FMN-binding pocket of phototropin LOV domains (30, 31) in being mainly polar on the pyrimidine side of the isoalloxazine ring and nonpolar around the dimethylbenzene moiety. The latter is surrounded predominantly by hydrophobic side chains (Ile-67, Phe-49, Ile-25, and Leu-42) and His-73, whose side chain forms hydrogen bonds with the backbone carbonyl oxygen atom of Ile-67 and the hydroxyl group of Ser-11.
The heteroatoms of the isoalloxazine ring form hydrogen bonds with residues Gln-51, Asn-33, Arg-32, and with the 4′-hydroxyl group of the cofactor's ribityl side chain (Fig. 2B). The terminal amine of Asn-33 forms two hydrogen bonds with the O4/N3 atoms of the isoalloxazine ring. Asn-33 and Gln-51 are oriented by interactions with the side chains of Ser-93 and Tyr-9, respectively. The hydroxyl groups of the ribityl side chain interact with side chains of residues Arg-32 and Asp-70. The side chain of Asp-70 is kept in position by forming two hydrogen bonds with Arg-72. In Fig. 2B, distances are given for BlrB MolA. Within the coordinate error, these distances are the same for both molecules in the asymmetric units of two data sets collected (data not shown). Although the conformations of the majority of residues within the flavin-binding pocket are well defined, the side chain of Arg-32 is somewhat more flexible. Consequently, distances between Arg-32 hydrogen bond donors and the FMN O2 and O4′ positions vary by up to 0.7 Å, as observed in the two molecules in the asymmetric unit and in different data sets. In contrast to the flavin-binding pocket of LOV domains, there are no interactions between the flavin phosphate group and the protein moiety of the BLUF domain, rendering the surface exposed phosphate comparatively flexible (Fig. 2B).
Structural Changes upon Light Excitation. As shown above, blue light illumination of BlrB generates a slightly red-shifted intermediate state as previously identified in the BLUF domains from AppA and Slr1694. This species is thought to be the signaling state responsible for signal propagation to downstream partners (2). Proton transfer processes and/or alterations of hydrogen–bond interactions of the flavin cofactor with the surrounding protein have been suggested to underlie the observed red shift (12, 14). These proposals are consistent with the fact that small conformational changes are likely to cause minor spectroscopic effects, including the visible absorbance and NMR spectroscopy data for BlrB. In contrast to phototropin LOV2 domains (24, 32), we observe no significant light-induced changes in the C-terminal helix–turn–helix span of BlrB; all of the sites clearly demonstrating light-induced NMR chemical shift changes in Fig. 1B and related 2D spectra map to the immediate vicinity of the FAD binding site.
Mechanistic Aspects/Photochemistry. BLUF proteins represent a previously undescribed class of blue light photoreceptors. Upon photon absorption, they perform a photocycle, which involves formation of a transient red-shifted intermediate believed to represent the signaling active state. It is thought to communicate the blue light information further downstream before it relaxes back to the receptor state on the timescale of seconds (BlrB and Slr1694) or minutes (AppA BLUF). What are the light-induced processes that lead to the reversible formation of this signaling state, and what is the mechanism of the BLUF photocycle? To address these questions, a great number of mainly spectroscopic experiments have been carried out, and several suggestions have been put forward based on the results. In light of the BlrB crystal structure presented here, we can eliminate some of these proposals and suggest mechanistic alternatives.
Based on mutagenesis data and spectroscopic considerations, the highly conserved tyrosine (Tyr-21 in AppA corresponding to Tyr-9 in BlrB) was assigned a prominent role in the photocycle (14). However, geometrically, Tyr-9 cannot interact with the isoalloxazine ring through hydrogen bonding or π stacking (Fig. 2B) and its location in the central β-sheet makes a light-induced approach to the isoalloxazine ring highly unlikely. Therefore, we exclude a hydrogen bond between the tyrosine hydroxyl group and N5 of the flavin (14), and a role of this tyrosine as transient acceptor of a proton donated by N5 (13). We can also exclude π–π stacking interactions between this tyrosine and the isoalloxazine ring (14) and between the isoalloxazine and the adenine moieties of the cofactor as proposed by Masuda et al. (12). Even though potentially indirectly involved in the receptor's primary photochemistry through an electron transfer-induced fluorescence quenching (29), the BlrB crystal structure suggests an important structural role for Tyr-9. It is part of the hydrophobic cavity surrounding the dimethylbenzene portion of the cofactor and its hydroxyl group orients Gln-51, which is 100% conserved in the BLUF domains, such that it forms a hydrogen bond with N5 of the isoalloxazine ring. The absence of this interaction in the AppA Y21I and Y21F mutants (13, 14) may be the reason for the mutants' deficiency in photocycling and decreased cofactor affinity.
Recent publications propose that the BLUF photocycle is rooted in alterations in hydrogen bonding between heteroatoms of the isoalloxazine ring and the protein (12, 33, 34). The underlying Fourier transform infrared studies show that illumination strengthens the N1C10a and/or C4aN5 bonding and weakens the C4 O and C2
O and C2 O bonding in the isoalloxazine ring. The latter was interpreted as a light-dependent strengthening of hydrogen bonding of C4
O bonding in the isoalloxazine ring. The latter was interpreted as a light-dependent strengthening of hydrogen bonding of C4 O and C2
O and C2 O. Based on comparisons with FTIR experiments carried out at –35°C and under dehydrating or deuterating conditions, Hasegawa and colleagues (12, 33, 34) proposed that the light-induced changes in hydrogen bonding of C4
O. Based on comparisons with FTIR experiments carried out at –35°C and under dehydrating or deuterating conditions, Hasegawa and colleagues (12, 33, 34) proposed that the light-induced changes in hydrogen bonding of C4 O trigger further rearrangements of the hydrogen bonding network, leading to alterations of the interactions between FAD and the protein matrix, which are suppressed by low temperature or dehydration. The BlrB crystal structure, together with high-level, nonempirical, quantum chemical calculations, allow extension of this model to fill in missing (structural) details and to propose a new model of the photocycle that links light-induced changes in the flavin ring to structural changes in the protein, resulting in signaling state formation.
O trigger further rearrangements of the hydrogen bonding network, leading to alterations of the interactions between FAD and the protein matrix, which are suppressed by low temperature or dehydration. The BlrB crystal structure, together with high-level, nonempirical, quantum chemical calculations, allow extension of this model to fill in missing (structural) details and to propose a new model of the photocycle that links light-induced changes in the flavin ring to structural changes in the protein, resulting in signaling state formation.
Our ab initio quantum chemical calculations on lumiflavin (unpublished data; see supporting information) show that a red shift of the spectrum of the excited flavin can be caused by (i) a protonation of N5, which, however, leads to very strong red shifts of 1.64 eV for both adsorption bands and is not structurally possible in BlrB (Fig. 2B), (ii) a protonation of O2 (calculated red shifts are 0.22 and 0.76 eV), and (iii) a protonation of N1 (calculated red shifts are 0.10 and 1.01 eV). Both (ii) and (iii) are structurally possible through Arg-32 and are consistent with the experimental findings (12, 33). In the ground state, the N1H and O2H cations have comparable energies, which are >10 kcal/mol lower than the energy of N5H cation. Thus, most likely N1 and O2 are proton acceptors in a flavin in the ground electronic state.
To test whether light excitation stimulates protonation, we compared the energies of the neutral lumiflavin and the cations in both the ground and excited states, respectively. The O2H and N5H cations were found to have higher deprotonation energies in the excited state than in the ground state, indicating a light-stimulated increase of the O2 and especially N5 basicity (31), whereas the deprotonation energy of the N1H cation does not change upon excitation. In the first excited state, the energies of N5H and O2H cations are significantly lower compared to the N1H cation (21 and 19 kcal/mol, respectively).
The negative charge developing on the N5 atom in the exited state would influence the hydrogen bond between N5 and the NE2 atom of Gln-51 (Fig. 2B). Because NE2 also forms a hydrogen bond with C4 O of the flavin, the changes from N5 could be transferred to C4
O of the flavin, the changes from N5 could be transferred to C4 O, which would explain the experimentally observed change in the corresponding stretch vibration (12, 33). Although the results of the calculations clearly suggest that chemical reactivity of a flavin associated with the N5 atom is significantly stimulated by light, structural analysis shows that in a BLUF domain, it would be rather difficult to transfer this chemical alteration into changes in the protein structure. The residues interacting with the portion of the cofactor containing N5 and C4
O, which would explain the experimentally observed change in the corresponding stretch vibration (12, 33). Although the results of the calculations clearly suggest that chemical reactivity of a flavin associated with the N5 atom is significantly stimulated by light, structural analysis shows that in a BLUF domain, it would be rather difficult to transfer this chemical alteration into changes in the protein structure. The residues interacting with the portion of the cofactor containing N5 and C4 O are part of a structurally rigid protein core. For example, light-induced flipping of Gln-51 is highly unlikely because of its interaction with the backbone carbonyl oxygen atom of Ser-93. Moreover, there is no prominent proton donor or acceptor that can be activated by flavin excitation. On the other hand, protonation of O2, which is light stimulated according to the calculations, may explain how excitation of the flavin ring is translated into a structural signal.
O are part of a structurally rigid protein core. For example, light-induced flipping of Gln-51 is highly unlikely because of its interaction with the backbone carbonyl oxygen atom of Ser-93. Moreover, there is no prominent proton donor or acceptor that can be activated by flavin excitation. On the other hand, protonation of O2, which is light stimulated according to the calculations, may explain how excitation of the flavin ring is translated into a structural signal.
We propose that excitation stimulates changes in the hydrogen bonding network around O2 possibly involving a proton transfer from Arg-32 to O2, resulting in a conformational change. This proposal is consistent with the fact that the C2 O stretch vibration is deuteration sensitive and suppressed at low temperature (33). The proposal of a light-induced reversible rate-limiting proton transfer from Arg-32 to the flavin O2 would also explain the observation that the rate of the back reaction from the light-induced signaling state to the ground state varies considerably in AppA [≈15 min; (14), Slr1694 (≈5 s; ref. 12)], and BlrB (≈5 s). These kinetic differences must reflect structural differences in close proximity to the light-absorbing isoalloxazine ring. In fact, all residues interacting directly with the flavin cofactor are identical in AppA, Slr1694, and BlrB, with one key exception, Arg-32 in BlrB (His in AppA and Asn in Slr1694). Proton transfer from this residue (or a bound water molecule in the case of Asn) could therefore be the rate-determining factor for the back reaction.
O stretch vibration is deuteration sensitive and suppressed at low temperature (33). The proposal of a light-induced reversible rate-limiting proton transfer from Arg-32 to the flavin O2 would also explain the observation that the rate of the back reaction from the light-induced signaling state to the ground state varies considerably in AppA [≈15 min; (14), Slr1694 (≈5 s; ref. 12)], and BlrB (≈5 s). These kinetic differences must reflect structural differences in close proximity to the light-absorbing isoalloxazine ring. In fact, all residues interacting directly with the flavin cofactor are identical in AppA, Slr1694, and BlrB, with one key exception, Arg-32 in BlrB (His in AppA and Asn in Slr1694). Proton transfer from this residue (or a bound water molecule in the case of Asn) could therefore be the rate-determining factor for the back reaction.
The question remains: How is the absorption of blue light by the FAD chromophore translated into a protein structural change that can be used in signal transduction? We propose that excitation stimulates changes in the hydrogen bonding network involving O2 and Arg-32. The light-induced changes in the flavin would be mediated through Arg-32 to the surface of the protein, which would form part of the interaction interface with an effector/output protein (module). In addition, the light-induced changes in charge at N1 could modify the interaction with the O4′ hydroxyl group of the ribityl side chain (Fig. 2B). This interaction could represent a fulcrum for the ribityl chain that acts as a lever arm resulting in a different location of the AMP moiety of FAD. We believe the latter to be essential for the interaction between the FAD-containing sensor domain and its regulated effector, as detailed in the next paragraph.
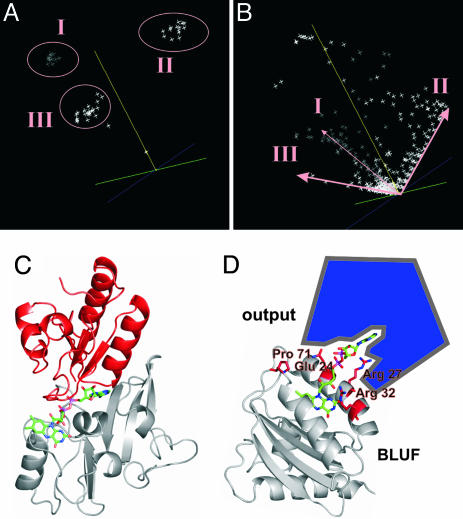
Model for BLUF Signal Transduction. For translation of the light signal into biologically useful information, any BLUF domain has to interact specifically with one or more output modules on the same polypeptide chain or with (an) independent effector unit(s). How does this communication take place? To identify potential interaction sites on the BLUF domain surface, we performed a sequence space analysis (35). This analysis allows us to define domain subfamilies and, moreover, to identify single functional residues that are specific for these subfamilies. It has been applied successfully to Ras-like proteins that constitute molecular switches, analogously to BLUF domains (35). With a multiple sequence alignment of >50 prokaryotic and eukaryotic BLUF domains as input (see supporting information), we identified three BLUF subfamilies I, II, and III (Fig. 3A), the latter of which can be subdivided into two classes, III A and III B (see supporting information), and residues specific for these subfamilies (Fig. 3B). The BLUF domain of BlrB is a representative of subfamily III, class A. Most residues that are conserved throughout all members of the BLUF family (Tyr-9, Ser-11, Asn-33, Gln-51, Leu-63, Phe-64, Ile-67, Asp-70, Arg-72, and His-73) serve in cofactor binding and probably stabilization of the signaling state (Fig. 2B). Strikingly, the majority of residues that are generally less conserved, but are highly conserved within subfamily III or even within class A of this subfamily, cluster around the entrance of the flavin pocket on the domain surface. For BlrB (subfamily III, class A), this group of residues encompasses Arg-27, Pro-71, Glu-24, and Arg-32. By analogy to the family of Ras-like proteins, this observation indicates that the interaction with effector modules takes place at the mouth of the flavin pocket where the entirely solvent-exposed adenine moiety of the FAD cofactor could serve as a recognition site or hook. In fact, the majority of complex BLUF proteins are light-driven enzymes involved in nucleotide metabolism. Their effector domains, C-terminally attached to the BLUF domain through flexible linkers, are represented by adenylyl cyclases or EAL (DUF2) domains encoding diguanylate-specific phosphodiesterases (36). These output units exhibit at least one nucleotide binding site. By interacting with one of these sites, the AMP moiety of the FAD cofactor could therefore function as a sort of competitive inhibitor that is released upon light absorption. A similar model can be developed for the communication of short BLUF proteins, like BlrB, with downstream effector proteins. Consistent with this proposal, in several instances, the genes located next to the BLUF-domain encoding genes encode EAL domains (2). The phenomenon of FAD holding together two subunits of a protein, one that mostly interacts with its isoalloxazine ring and another that binds the AMP moiety, is not unusual. It is observed in all members of a specific FAD-binding fold family, exemplified by p-cresol methylhydroxylase (PCMH, PDB ID code 1DII) (Fig. 3C).
Fig. 3.
A cofactor-mediated interaction/communication between BLUF domains and output modules is suggested by the analogy with the PCMH (p-cresol-methylhydroxylase) FAD-fold family and by sequence analysis of >50 BLUF domains. (A) Sequence space representation of >50 BLUF domains allows identification of three BLUF subfamilies, I, II, and III. BlrB belongs to subfamily III. (B) Sequence space representation of the entirety of residues covered by the 50 BLUF domains analyzed. Amino acid residues specific for these families are arranged along the edges of the space, respectively. The length of a residue vector in sequence space is proportional to its degree of conservation throughout the domain family. (C) In p-cresol-methylhydroxylase (PDB ID code 1DII), FAD is bound to two distinct subdomains of the protein, shown in gray and red, that interact predominantly with the isoalloxazine or the adenine moiety of the cofactor, respectively. (D) The surface exposed adenine unit of the BLUF-FAD could function as recognition site for effector/output domains. Residues that are unique in BlrB or specific for all members of the BLUF subfamily III and that are potentially involved in the signaling process as predicted by sequence space analysis (supporting information) cluster around the entrance of the flavin pocket (red). These residues (Glu-24, Arg-27, Arg-32, and Pro-71) are predestined to serve light-dependent modulation of the BLUF domain–output domain interaction.
The suggested mode of interaction explains the fact that FAD is the physiological cofactor of BLUF domains, although FMN is obviously sufficient to perform the photoreceptor's photocycle. However, binding of FMN to BLUF domains in a cellular environment might exert a regulatory function by rendering the domain inactive in signal propagation, similarly to the GDP-bound form of G proteins. The proposed mode of interaction is also consistent with the findings that the BLUF domain can function in trans with the C-terminal domains of AppA (10). Han et al. (10) also showed that a heterologous BLUF domain is able to communicate with the C-terminal domains of AppA in cis. The coincidence of highly conserved and less conserved, specific residues in the surface area near the cofactor cavity could also reflect an overlap of functions in this region. The surface residue Arg-32, for example, is involved in cofactor binding, but it is not highly conserved, suggesting a potential role in the photocycle (see above). In addition, this residue is specific for the class of BLUF domains exemplified by BlrB, which indicates an involvement in signal propagation and/or interaction with effectors. In conclusion, we propose the blue light signal to be propagated from the place of photon absorption in the flavin binding pocket along the neck of the pocket up to the domain's surface, where exposed amino acid side chains trigger the interaction with effector modules (Fig. 3D).
Supplementary Material
Acknowledgments
We thank Samuel Kaplan for initiating the collaboration and Ingrid Vetter for advice and support of the crystallographic software. Crystallographic data were collected at the Swiss Light Source, beamline X06SA, Paul Scherrer Institute, (Villigen, Switzerland), and the European Synchrotron Radiation Facility (Grenoble, France), and we are grateful to the beamline staff for their support. The Boehringer Ingelheim Fonds (to A.J.), the Robert A. Welch Foundation (Grant I-1424 to K.H.G.), the National Institutes of Health National Center for Research Resources (Centers of Biomedical Research Excellence) (Grant P20 RR15640 to M.G.), and Deutschen Forschungsgemeinschaft (to I.S.) are acknowledged for generous funding.
Author contributions: T.D., M.G., K.H.G., and I.S. designed research; A.J., T.D., M.T., Q.W., W.-h.K., R.L.S., M.G., K.H.G., and I.S. performed research; A.J., T.D., M.T., Q.W., R.L.S., M.G., K.H.G., and I.S. analyzed data; and A.J., T.D., Q.W., M.G., K.H.G., and I.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FAD, flavin adenine dinucleotide; BLUF, blue light sensing using FAD; LOV, light-oxygen-voltage.
Data deposition: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2BYC).
References
- 1.van der Horst, M. A. & Hellingwerf, K. J. (2004) Acc. Chem. Res. 37, 13–20. [DOI] [PubMed] [Google Scholar]
- 2.Gomelsky, M. & Klug, G. (2002) Trends Biochem. Sci. 27, 497–500. [DOI] [PubMed] [Google Scholar]
- 3.Gomelsky, M. & Kaplan, S. (1995) J. Bacteriol. 177, 4609–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomelsky, M. & Kaplan, S. (1997) J. Bacteriol. 179, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomelsky, M. & Kaplan, S. (1998) J. Biol. Chem. 273, 35319–35325. [DOI] [PubMed] [Google Scholar]
- 6.Masuda, S. & Bauer, C. E. (2002) Cell 110, 613–623. [DOI] [PubMed] [Google Scholar]
- 7.Braatsch, S., Gomelsky, M., Kuphal, S. & Klug, G. (2002) Mol. Microbiol. 45, 827–836. [DOI] [PubMed] [Google Scholar]
- 8.Braatsch, S., Moskvin, O. V., Klug, G. & Gomelsky, M. (2004) J. Bacteriol. 186, 7726–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iseki, M., Matsunaga, S., Murakami, A., Ohno, K., Shiga, K., Yoshida, K., Sugai, M., Takahashi, T., Hori, T. & Watanabe, M. (2002) Nature 415, 1047–1051. [DOI] [PubMed] [Google Scholar]
- 10.Han, Y., Braatsch, S., Osterloh, L. & Klug, G. (2004) Proc. Natl. Acad. Sci. USA 101, 12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer, C., Elsen, S., Swem, L. R., Swem, D. L. & Masuda, S. (2003) Philos. Trans. R. Soc. Lond., B, Biol. Sci. 358, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda, S., Hasegawa, K., Ishii, A. & Ono, T. A. (2004) Biochemistry 43, 5304–5313. [DOI] [PubMed] [Google Scholar]
- 13.Laan, W., van der Horst, M. A., van Stokkum, I. H. & Hellingwerf, K. J. (2003) Photochem. Photobiol. 78, 290–297. [DOI] [PubMed] [Google Scholar]
- 14.Kraft, B. J., Masuda, S., Kikuchi, J., Dragnea, V., Tollin, G., Zaleski, J. M. & Bauer, C. E. (2003) Biochemistry 42, 6726–6734. [DOI] [PubMed] [Google Scholar]
- 15.Dym, O. & Eisenberg, D. (2001) Protein Sci. 10, 1712–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheffield, P., Garrard, S. & Derewenda, Z. (1999) Protein Expression Purif. 15, 34–39. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch, W. (1993) J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- 18.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003) Acta Crystallogr. D. 59, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 19.Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003) Methods Enzymol. 374, 229–244. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjelgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 21.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999) Acta Crystallogr. D. 55, 247–255. [DOI] [PubMed] [Google Scholar]
- 23.DeLano, W. L. (2002) in The PYMOL User's Manual (DeLano Scientific, San Carlos, CA).
- 24.Harper, S. M., Neil, L. C. & Gardner, K. H. (2003) Science 301, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 25.Aravind, L. & Koonin, E. V. (1999) J. Mol. Biol. 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 26.Kita, A., Okajima, K., Morimoto, Y., Ikeuchi, M. & Miki, K. (2005) J. Mol. Biol. 349, 1–9. [DOI] [PubMed] [Google Scholar]
- 27.Anderson, S., Dragnea, V., Masuda, S., Ybe, J., Moffat, K. & Bauer, C. (2005) Biochemistry 44, 7998–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laan, W., Bednarz, T., Heberle, J. & Hellingwerf, K. J. (2004) Photochem. Photobiol. Sci. 3, 1011–1016. [DOI] [PubMed] [Google Scholar]
- 29.Zirak, P., Penzkofer, A., Schiereis, T., Hegemann, P., Jung, A. & Schlichting, I. (2005) Chem. Phys. 315, 142–154. [Google Scholar]
- 30.Crosson, S. & Moffat, K. (2001) Proc. Natl. Acad. Sci. USA 98, 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedorov, R., Schlichting, I., Hartmann, E., Domratcheva, T., Fuhrmann, M. & Hegemann, P. (2003) Biophys. J. 84, 2474–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper, S. M., Neil, L. C., Day, I. J., Hore, P. J. & Gardner, K. H. (2004) J. Am. Chem. Soc. 126, 3390–3391. [DOI] [PubMed] [Google Scholar]
- 33.Hasegawa, K., Masuda, S. & Ono, T. A. (2004) Biochemistry 43, 14979–14986. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa, K., Masuda, S. & Ono, T. A. (2005) Plant Cell Physiol. 46, 136–146. [DOI] [PubMed] [Google Scholar]
- 35.Casari, G., Sander, C. & Valencia, A. (1995) Nat. Struct. Biol. 2, 171–178. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, A. J., Ryjenkov, D. A. & Gomelsky, M. (2005) J. Bacteriol. 187, 4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.