Abstract
Spore formation in Saccharomyces cerevisiae involves the sequential deposition of multiple spore wall layers between the prospore membranes that surround each meiotic product. The Smk1p mitogen-activated protein (MAP) kinase plays a critical role in spore formation, but the proteins that interact with Smk1p to regulate spore morphogenesis have not been described. Using mass spectrometry, we identify Gsc2p as a Smk1p-associated protein. Gsc2p is a 1,3-β-glucan synthase subunit involved in synthesizing an inner spore wall layer. We find that 1,3-β-glucan synthase activity is elevated in smk1 mutants, suggesting that SMK1 negatively regulates GSC2. Although deposition of the two inner spore wall layers is normal in smk1 mutants, deposition of the outer layers is aberrant. However, eliminating GSC2 activity restores normal deposition of the third spore wall layer in smk1 mutants, indicating that negative regulation of GSC2 by SMK1 is important for spore wall deposition. Our findings suggest a model for the coordination of spore wall layer deposition in which Smk1p facilitates the transition between early and late phases of spore wall deposition by inhibiting a spore wall-synthesizing enzyme important for early phases of spore wall deposition.
Keywords: cell wall, sporulation, yeast, chitosan
The coordination of sequential events is a regulatory challenge in biological processes from cell-cycle control to cell differentiation. Proper ordering of events can involve coupling the completion of one event to the initiation of the next. Spore formation in Saccharomyces cerevisiae involves the sequential deposition of four spore wall layers (1, 2), but the mechanisms that coordinate the deposition of the different spore wall layers are unknown.
Sporulation in S. cerevisiae occurs in an a/α diploid cell and involves meiosis followed by the packaging of the meiotic products during spore formation (reviewed in refs. 3 and 4). During spore formation, the four layers of the spore wall are sequentially deposited within the lumen of a double membrane structure (the prospore membrane) surrounding each meiotic product (1, 2), providing a system in which to examine the sequential deposition of a structure. The spore wall enables the spore to withstand harsh environmental conditions and is comprised of two inner spore wall layers, which resemble the vegetative cell wall, and two outer layers, which contain spore-specific materials (Fig. 1A). The inner layers are comprised of a mannan layer and a glucan polysaccharide layer, whereas the outer two layers consist of a chitosan layer and a dityrosine layer.
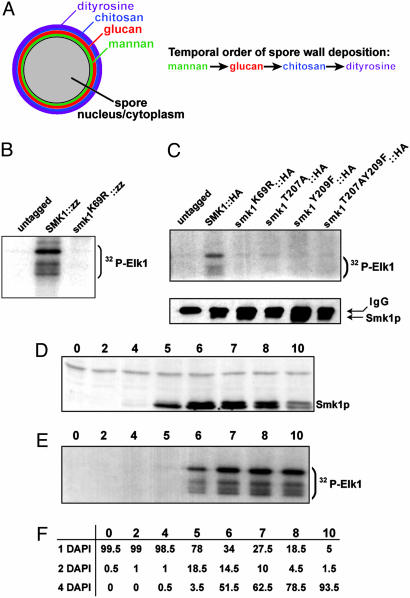
Fig. 1.
Smk1p is active during spore morphogenesis. (A) Diagram showing the spatial and temporal order of spore deposition. (B) In vitro kinase assay on sporulating cells. Smk1p was immunoprecipitated from untagged (LH177), SMK1::zz (LH414), and smk1(K69R)::zz (LH484) strains. (C) Kinase assay performed on strains carrying Smk1p untagged (LH177), tagged with HA (LH178), and HA-tagged smk1 mutations containing the following amino acid changes: K69R (LH415), T207A (LH416), Y209F (LH418), and T207A Y209F (LH417). Upper gel shows in vitro kinase assay. Lower gel is an immunoblot probed with anti-HA containing one-fifth of the immunoprecipitation reaction used for the kinase assay in the upper gel. (D–F) Sporulation time course examining Smk1p level and activity. Numbers at top of each panel indicate number of hours yeast were in sporulation media. All assays in D–F use cells from the same sporulating culture. (D) Anti-HA immunoblot. Blot contains extracts from SMK1::HA (LH178) strain undergoing sporulation. (E) Kinase assay using immunoprecipitated Smk1-HA. (F) DAPI staining to monitor progression through sporulation. A cell with a single DAPI staining body has not undergone meiosis, a cell with two DAPI staining bodies has completed meiosis I, and a cell with four DAPI staining bodies has completed meiosis II. Numbers listed are in percentages; a total of 200 cells for each time point were counted.
Proteins involved in the synthesis of the spore wall layers have been identified and include Gsc2p, a component of 1–3-β-glucan synthase (5, 6). Several proteins have been proposed to act as regulators of spore morphogenesis including Smk1p, a mitogen-activated protein (MAP) kinase essential for spore morphogenesis (7). Although Smk1p is not required for progression through meiosis, electron microscopic studies on smk1 mutant spores revealed perturbations to spore wall structure indicating that the nuclei are not properly packaged (7, 8).
We identify Gsc2p, a 1,3-β-glucan synthase subunit, as a protein that physically associates with Smk1p. We find that glucan synthase (GS) activity is elevated in smk1 mutants, but not in gsc2 smk1 double mutants, suggesting that SMK1 negatively regulates GSC2. We further examine spore wall deposition in smk1 mutants and find that, although deposition of the inner two spore wall layers is independent of SMK1, deposition of the third chitosan layer is highly aberrant in smk1 mutants. Elimination of Gsc2p suppresses the chitosan layer deposition defects of smk1 mutants. These results suggest that Smk1p promotes proper deposition of the spore wall through negative regulation of 1,3-β-glucan production via Gsc2p. We propose that Smk1p regulates a transition between early and late phases in spore wall deposition by regulating an enzyme important for early spore wall biosynthesis.
Materials and Methods
Yeast Strains and Growth Conditions. S. cerevisiae strains used are listed in the supporting information, which is published on the PNAS web site. All strains are in the SK1 genetic background and isogenic to LH177, which is essentially identical to KBY259 (9). All strains are homozygous diploid; only relevant genotypes are shown. Standard media and growth conditions were used (10). Synchronous sporulation was performed in 1% KOAc and 0.02% raffinose using a standard regimen (11), and the kinetics of sporulation were monitored by DAPI staining of cells withdrawn during sporulation (12). Cultures that did not sporulate with normal kinetics or to ≥85% efficiency were discarded. Yeast cell transformations were performed as described (13, 14). Gene disruptions and epitope tagging were carried out by using PCR-mediated techniques (15–17); details are provided in supporting information. All gene fusions were tested for function by analyzing their ability to undergo successful sporulation with kinetics and efficiencies similar to wild type and for their ability to support normal germination.
Protein Immunoblotting, Kinase Assays, and Immunoprecipitation. Denaturing protein lysates for immunoblotting were prepared by TCA precipitation as described (18). Native lysates for immunoprecipitation were prepared from 50 ml of cells lysed by using a MiniBeadBeater8 (Biospec) and glass beads into IP buffer (300 mM NaCl/5 mM EGTA/50 mM Tris, pH 7.4/0.5% Nonidet P-40) and a mix of protease inhibitors and phosphatase inhibitors as described (9). Lysates were clarified by centrifugation in a microcentrifuge or a Beckman TLX centrifuge using a TLA100.4 rotor at 23,000 × g. Smk1-HA was immunoprecipitated by using HA-11 beads (Covance). Smk1-zz was immunoprecipitated by using IgG Sepharose (GE Healthcare). Trichloroacetic acid (TCA)-precipitated proteins, native lysates, and immune complexes were resuspended in SDS/PAGE sample buffer and separated by SDS/PAGE. Gels were blotted onto nitrocelluose and probed with rabbit preimmune antiserum to detect the zz tag (gift from K. Benjamin), with α-HA (12CA5; Covance) or with α-myc (9E10; Covance).
Kinase assays were performed by using Smk1p immunoprecipitated from ≈1 × 109 yeast cells. Immunoprecipitated Smk1p was added to a 20-μl reaction containing 20 mM Tris (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 10 μM ATP, 5 μCi of [γ-32P]ATP plus 1 μg of Elk-1, presented as a recombinant protein of amino acids 307–428 of Elk-1 fused to GST (Cell Signaling Technologies). Kinase reaction was carried out at 30°C for 45 min, stopped by addition of SDS/PAGE sample buffer, run on a 12.5% polyacrylamide gel, and visualized by using Kodak X-Omat film or a PhosphorImager using imagequant software (Molecular Dynamics).
Protein Purification and Mass Spectrometry. Pilot purifications using 250 ml of synchronously sporulating cells per strain (≈1 × 1010 cells) were done by using the SMK1::zz strain LH414 and the untagged control strain LH177. Purifications were carried out as below, except cells were lysed in a MiniBeadBeater8 (Biospec) and clarified by using a TLA100.4 rotor. Proteins were separated by using a 4–15% gradient gel and visualized by silver staining. For large-scale purification, a total of 13 liters each of LH414 and LH177 were grown under conditions of synchronous sporulation. Given the high aeration requirements of sporulating cells, cells were grown as 250-ml cultures in Fernbach flasks. Cells were harvested 8 h into sporulation and stored at –80°C until enough cells were collected for purification of SMK1-zz. Yeast cells were lysed in IP buffer with protease and phosphatase inhibitors (see above) by using glass beads in a BeadBeater (Biospec) and checked microscopically for at least 80% cell breakage. Lysates were clarified with a Beckman ultracentrifuge using a 70Ti rotor at 23,000 × g at 4°C for 45 min. IgG Sepharose (GE Healthcare) was added to clarified lysates and incubated at 4°C for 2 h. Lysate and beads were applied to a column and washed with ≥10 bed volumes of IP buffer followed by 250 mM MgCl2 in IP buffer and 1M MgCl2 in IP buffer. Protein was eluted by using 0.5 M acetic acid, pH 3.4, collected into 1 M Tris (pH 7.4), concentrated by using a Centricon 30 (Millipore), and precipitated (19). Samples were electrophoresed in SDS/PAGE sample buffer on a 1.5-mm-thick 9% polyacrylamide gel. Bands were visualized by using Sypro Ruby Red (Bio-Rad) and excised. Sequence analysis was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ DECA quadrupole ion trap mass spectrometer. The MS/MS spectra were correlated with known sequences by using previously developed algorithms (20, 21).
GS Assay. GS was partially purified from sporulating yeast by membrane isolation essentially as described (22), with the addition of the protease and phosphatase inhibitor mixture described above. Membrane sedimentation was performed in a Beckman Opti-MAX centrifuge using a TLA100.2 rotor at 125,000 × g at 4°C for 40 min. GS activity was measured by using [14C]UDP-glucose (MP Biomedicals). Measurements were normalized to the number of cells collected because we found that the OD600, cell pellet weight per ml of culture, and amount of membrane protein purified per ml culture fluctuated over time during sporulation. Within a particular time point, experiments normalized to either cell weight or μg of membrane purified yielded similar results.
Microscopy and Spore Wall Assays. Visualization of mannan, glucan, and chitosan layers was performed as described (1), except that the described class 5 (mannan and chitosan) and class 6 (mannan and faint staining for chitosan) were grouped into a single class, as we found the classification of “not faint” versus “faint” staining subjective. At least 200 cells were counted per time point; each strain was grown in at least two independent cultures on different days. Only cells with four DAPI staining bodies were counted. Cells were scored by using a ×100, numerical aperture 1.45 objective on a Zeiss Axioskop Mot2 with an Orca-ER cooled charge-coupled device camera (Hammamatsu) and openlab (Improvision) software. Dityrosine fluorescence was performed as described (23). Don1-GFP and differential interference contrast (DIC) images were acquired from living cells by using the system described above. All other DIC microscopy was performed by using an Olympus BX-60 microscope with an attached SPOT camera and software (Diagnostic Instruments).
Results
The Smk1p MAP Kinase Is Active During Spore Morphogenesis. We determined when Smk1p was catalytically active by establishing an in vitro assay for Smk1p kinase activity during sporulation. The endogenous SMK1 gene was modified with a C-terminal addition of an IgG-binding zz epitope-tag (from protein A) or a HA epitope tag. Smk1p was isolated from cell extracts by immunoprecipitation and incubated with [γ-32P]ATP and Elk-1, a substrate of mammalian MAP kinases. Immunoprecipitates from both SMK1::zz--containing and SMK1::HA-containing yeast phosphorylated Elk-1 (Fig. 1 B and C). This kinase activity was associated with Smk1p, as no activity was detected in immunoprecipitates from cells that contained only untagged Smk1p or tagged forms of mutant Smk1p with the conserved catalytic lysine residue (K69) changed to arginine (Fig. 1 B and C). Kinase activity was also greatly reduced by mutations of sites in the conserved potential activating T loop of Smk1p, from phosphorylatable threonine (T207) and tyrosine (Y209) to nonphosphorylatable amino acids (Fig. 1C). These kinase-dead and nonphosphorylatable smk1 mutants failed to support sporulation.
The time course of Smk1p activity during sporulation was examined. Cells were induced to undergo synchronous sporulation. At 4 h, 98.5% of cells had not yet begun the meiotic divisions, whereas by 10 h, 93.5% of cells had completed meiosis (Fig. 1F). Smk1p was noticeably induced after 5 h in sporulation medium, reaching maximal levels between 6 and 8 h (Fig. 1D). Smk1p kinase activity was examined by using cells from the same time course and appeared strongly induced by 6 h after the initiation of sporulation, with detectable activity persisting through 10 h of sporulation, when mature spore walls appear (Fig. 1E). Thus, the Smk1p MAP kinase becomes active after completion of meiosis and remains active throughout the time when spore wall deposition occurs, consistent with a role in spore morphogenesis.
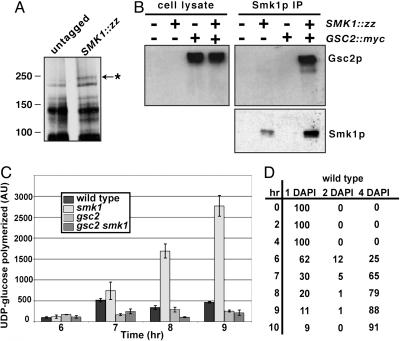
Smk1p Associates with Gsc2p, a 1,3-β-Glucan Synthase Subunit. To identify proteins that interact with Smk1p during the time when it is active, lysates were made from SMK1::zz cells and wild-type untagged control cells harvested after the meiotic divisions are completed, during the time of spore morphogenesis. Smk1-zz and associated proteins were isolated from extracts by using an affinity matrix. A protein with an apparent molecular mass of ≈250 kDa was enriched specifically in lysates from SMK1::zz cells, suggesting that it physically associated with Smk1p (Fig. 2A). The 250-kDa protein was analyzed by tryptic digestion followed by mass spectrometry analysis and identified as Gsc2p.
Fig. 2.
Smk1p and Gsc2p interact and SMK1 modulates GS activity. (A) Silver stained gel from pilot purification comparing an untagged control strain (LH177) with the SMK1::zz strain (LH414). Location of protein size standards denoted at left. Arrow with asterisk points to an ≈250-kDa protein that appears to bind the Smk1-zz protein specifically. (B) Immunoprecipitation experiment from strains containing untagged Smk1p (LH177), Smk1-zz (LH414), Gsc2-myc (LH565), and Smk1-zz Gsc2-myc (LH566). Immunoblot at left contains 5% of the native lysates used for immunoprecipitations shown at the right. The two immunoblots on the right are from the same gel showing immune complexes isolated by immunoprecipitation of Smk1-zz. The top blot shows the top of the gel, where Gsc2-myc migrates, whereas the bottom blot shows the middle of the gel, where Smk1-zz migrates. Blots were probed with a mouse monoclonal anti-myc IgG. Smk1-zz can be visualized by using this antibody, as the zz-tag binds IgG. (C) GS activity assay performed on membrane fractions isolated from sporulating cells. The wild-type strain is LH177, smk1Δ is LH185, and gsc2Δ is LH625, and gsc2Δsmk1Δ is LH701. Error bars show the standard error of the mean. (D) Kinetics of sporulation of the wild-type culture used for the GS assay in C, as monitored by DAPI staining. Numbers listed are in percentages; a total of 200 cells for each time point were counted. The other cultures used in C were chosen to have kinetics similar to the wild-type culture.
Gsc2p is required for proper sporulation (6) and encodes a subunit of 1,3-β-glucan synthase (5, 6), an enzyme responsible for synthesis of the 1,3-β-glucan layer of the spore wall. We confirmed the physical association of Smk1p and Gsc2p by coimmunoprecipitating the two proteins from sporulating cells (Fig. 2B). Endogenous Gsc2p was tagged with the myc epitope. When Smk1-zz was immunoprecipitated from sporulating cells, Gsc2-myc was detected in the immunoprecipitate (Fig. 2B), reflecting a specific interaction between Smk1p and Gsc2p. Although high levels of Gsc2-myc were detected in sporulating cells containing either untagged Smk1p or Smk1-zz (Fig. 2B), Gsc2-myc was detected in immunoprecipitates only in the presence of Smk1-zz. These results confirm that Smk1p physically associates with Gsc2p.
GS Activity Is Increased in Cells Lacking Smk1p. The physical interaction between Smk1p and Gsc2p raised the possibility that Gsc2p may be required to activate Smk1p or that Smk1p may regulate Gsc2p activity. These possibilities were explored by directly monitoring the biochemical activities associated with each protein. Robust Smk1p kinase activity was detected in the absence of Gsc2p (data not shown), indicating that Gsc2p was not required for the activation of Smk1p.
We examined Smk1p regulation of Gsc2p activity by using a GS assay (Fig. 2C) (22). In wild-type cells, GS activity is low at 6 h (when 26% of cells have completed the meiotic divisions) and peaks at 7 h (when 65% of cells have completed the meiotic divisions). Membrane preparations from gsc2 mutant cells did not show a significant increase between 6 and 7 h and polymerized ≈30% as much glucan as wild-type cells at 7 h. These data are consistent with the Gsc2p-dependent enzyme contributing the majority of the GS activity in sporulating cells. Residual GS activity in gsc2 mutants likely reflects the activity of Fks1p (5, 6, 24), a Gsc2p homolog used predominantly in vegetatively growing cells. Such Gsc2p-independent activity is neither necessary nor sufficient to support sporulation, as gsc2 mutants do not form mature spores (see below), whereas fks1 mutants sporulate and germinate with wild-type kinetics and efficiencies (data not shown).
Like wild-type cells, smk1 mutant cells showed an increase in GS activity between 6 and 7 h into sporulation. However, unlike wild-type cells, GS activity levels continue to increase after 7 h in smk1 mutants. smk1 mutant cells show a >5-fold increase in GS activity compared to wild-type cells at 8 and 9 h. This increase in GS activity depends on GSC2, as gsc2 smk1 double mutants do not show this dramatic increase in GS activity. These observations suggest Smk1p may negatively regulate Gsc2p activity.
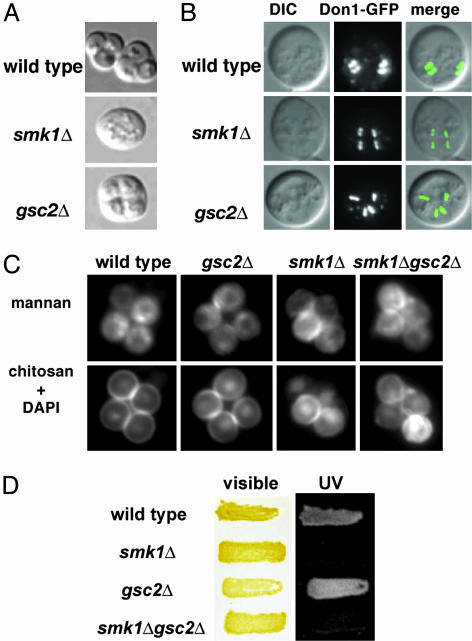
Spore Wall Deposition in smk1 and gsc2 Cells. Our biochemical data indicated that Smk1p negatively regulated GS activity and predicted that the underlying defects in smk1 and gsc2 mutants would differ, prompting us to analyze the spore morphology phenotypes of both mutants in greater detail. Sporulation of wild-type cells results in the creation of a compact ascus containing four tetrahedrally arranged spores. The majority of gsc2 mutant cells formed spores that appeared immature and failed to compact, whereas smk1 mutants failed to form any structures resembling spores (Fig. 3A). The observation that smk1 and gsc2 mutants generate morphologically distinct sporulation products is consistent with the existence of distinct underlying defects in the two mutants.
Fig. 3.
SMK1 negatively regulates GSC2.(A) DIC images of sporulating yeast. (B) Don1-GFP localization. Rings of Don1-GFP are shown from the side, such that all four rings in an ascus can be visualized in a single focal plane. The wild-type stain is LH790, smk1Δ is LH788, and gsc2Δ is LH789. (C) Immunofluorescence pictures showing mannan, the nuclei (DAPI), and chitosan. Note that, in smk1 mutants, the top two spores do not show detectable chitosan staining but do stain for mannan. For B–D, the wild type is LH177, smk1Δ is LH185, gsc2Δ is LH625, and gsc2Δsmk1Δ is LH701. (D) Dityrosine fluorescence. Cells were grown on a membrane and subjected to UV light to visualize the dityrosine layer. (Left) Patches of yeast on a membrane. (Right) The same patches under UV light.
Before deposition of the spore wall, the prospore membrane grows from the spindle pole body of each meiotic product to surround the associated nucleus. To determine whether SMK1 or GSC2 were required at early stages in spore wall morphogenesis, we examined Don1-GFP (a marker for the leading edge of the developing prospore membrane; ref. 25) localization in smk1 and gsc2 mutants. As in wild-type cells, Don1-GFP rings are found in smk1 and gsc2 mutants, suggesting that the leading edge complex is intact (Fig. 3B). Because prospore membrane growth precedes spore wall synthesis, these data suggested that SMK1 and GSC2 act at later stages in spore wall formation.
Spore wall deposition in smk1 and gsc2 mutants was assessed by using fluorescent probes to visualize the mannan, glucan, and chitosan layers (1). The innermost mannan layer binds the lectin concanavalin-A (ConA) and can be visualized by using FITC coupled to ConA. The overlying 1,3-β-glucan layer can be detected by using an anti-1,3-β-glucan antibody. The chitosan layer binds to Calcofluor White. Consistent with previous observations (1), the mannan layer appeared first in wild-type yeast, followed by the glucan layer and then the chitosan layer (Table 1, see also Fig. 1A). Thus, as spores progress in maturity, they begin with only a mannan layer, then have both mannan and glucan, followed by mannan, glucan, and chitosan. As spores continue to mature, the ability to detect the glucan layer with antibody staining is lost (why this occurs is unknown), resulting in cells that show staining for only mannan and chitosan. It is important to note that Calcofluor White staining does not allow the distinction between the deposition of and the assembly of the chitosan layer. The outermost layer, the dityrosine layer, can be detected by the inherent ability of dityrosine to fluoresce under UV light on patches of wild-type yeast grown on membranes (ref. 23; Fig. 3D). These methods were used to examine spore wall formation in smk1 and gsc2 mutants.
Table 1. Spore wall deposition analysis.
| Genotype/staining pattern, h | DAPI only | Mannan | Mannan + glucan | Mannan + glucan + chitosan | Mannan + chitosan | Mannan + glucan + <4 spores with chitosan* | Mannan + <4 spores with chitosan* |
|---|---|---|---|---|---|---|---|
| Wild type | |||||||
| 6 | 9.5 | 9 | 12.3 | 33.2 | 36.1 | - | - |
| 7 | 0.5 | 2.4 | 8 | 35.3 | 53.5 | - | - |
| 8 | - | - | 4.6 | 22.8 | 72.6 | - | - |
| 9 | - | - | 2.4 | 7.6 | 90.1 | - | - |
| smk1 | |||||||
| 6 | 36.8 | 51.5 | 11.7 | - | - | - | - |
| 7 | 18.8 | 54.5 | 17.4 | - | - | 8.5 | 0.9 |
| 8 | - | 32.3 | 33.6 | - | - | 31 | 3.1 |
| 9 | - | 4 | 52.6 | - | - | 38.5 | 4.9 |
| gsc2 | |||||||
| 6 | 14.5 | 45.4 | 2.4 | 8.2 | 29.5 | - | - |
| 7 | 1.4 | 38.3 | 1.4 | 11 | 47.8 | - | - |
| 8 | - | 18.3 | 2.4 | 11.5 | 67.7 | - | - |
| 9 | - | 7.7 | 2.9 | 6.2 | 83.3 | - | - |
| smk1 gsc2 | |||||||
| 6 | 21.2 | 71.7 | 3.8 | - | 3.3 | - | - |
| 7 | 4.4 | 45.7 | 4.4 | 2.9 | 42.7 | - | - |
| 8 | 0.5 | 23.6 | 4.2 | 10.8 | 60.8 | - | - |
| 9 | - | 18.4 | 5.7 | 4.2 | 68.4 | - | 3.3 |
Numbers are in percentages. Because of rounding, not all amounts for a particular experiment add up to exactly 100%.
One, two, or three spores within an ascus are surrounded by chitosan. An example of this is depicted in Fig. 3D.
As detailed in Table 1 and Fig. 3C, smk1 mutant cells produced the inner mannan and glucan layers, but did not deposit the outermost layers properly. Unlike in wild-type cells, where a chitosan layer surrounds all four meiotic products, only a subset of the meiotic products deposit chitosan in smk1 mutant cells. Although deposition of chitosan occurred in smk1 mutant cells, this deposition was aberrant and incomplete. Finally, the outermost dityrosine layer was not detected in smk1 mutant cells (Fig. 3D), consistent with previous biochemical studies demonstrating a lack of dityrosine in the spore wall of smk1 mutant cells (8). Thus, SMK1 appears specifically required for production of the outer spore wall layers.
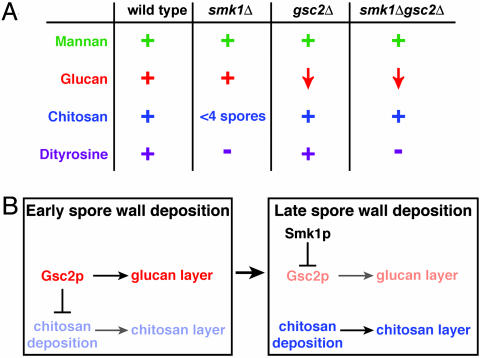
gsc2 mutants were severely compromised in the production of a different spore wall layer, the 1,3-β-glucan layer (Fig. 3C). The reduction in this layer is consistent with our biochemical measurement indicating that Gsc2p is required to support the majority of GS activity during sporulation (Fig. 2C). A few gsc2 spores had the glucan layer (Table 1), likely because of the activity of Fks1p (see above). All four spores in an ascus showed staining for glucan whenever staining was detected, consistent with our detection of refractile tetrads in gsc2 mutants (Fig. 3A). Despite the low frequency of glucan deposition in gsc2 mutants, deposition of the outer chitosan and dityrosine layers proceeded at frequencies comparable to wild-type cells (Table 1 and Fig. 3 C and D). Thus, normal deposition of the glucan layer was not required for deposition of the outer spore wall layers. Taken together, these observations demonstrated that smk1 and gsc2 mutants were defective in producing different layers of the spore wall. As summarized in Fig. 4A, the outer two layers of the spore wall were not produced normally in smk1mutants, whereas in gsc2 mutants, the inner glucan layer was selectively disrupted.
Fig. 4.
Summary. (A) Summary of spore wall deposition phenotypes shown quantitatively in Table 1. (B) Model for Smk1p regulation of Gsc2p activity during spore morphogenesis. See text for details.
SMK1 Inhibition of GSC2 Is Important for Spore Wall Deposition. Our biochemical data suggested that Smk1p negatively regulated Gsc2p activity. To test whether the inhibition of Gsc2p was an important aspect of Smk1p function, spore wall production was examined in gsc2 smk1 double mutants. We reasoned that if SMK1 inhibition of GSC2 were important for spore morphogenesis, the elimination of GSC2, might allow proper deposition of outer spore wall layers to proceed without SMK1. As predicted, the gsc2 smk1 mutant cells did not exhibit the chitosan deposition defect of the smk1 single mutant. In most gsc2 smk1 double mutant cells, all four meiotic products have detectable chitosan deposition (Table 1 and Fig. 3C). As in the gsc2 single mutant, gsc2 smk1 double mutant cells were compromised in their ability to produce the glucan layer, presumably due to the lack of GSC2 (Table 1). As summarized in Fig. 4A, the elimination of GSC2 allowed proper deposition of chitosan even in the absence of SMK1, consistent with SMK1 promoting chitosan deposition through the inhibition of GSC2.
Although inhibition of GSC2 is critical for proper chitosan deposition, SMK1 clearly has additional functions during spore morphogenesis. The other outer layer of the spore wall, the dityrosine layer, was not produced in the gsc2 smk1 double mutant (Fig. 3D), suggesting that SMK1 controls dityrosine deposition in a GSC2-independent fashion. Taken together, our biochemical and cell biological data demonstrate that SMK1 is required for proper deposition of the two outer layers of the spore wall, the chitosan and dityrosine layers, and suggest that inhibition of GSC2 is one essential aspect of SMK1 function.
Discussion
Cell division and differentiation involve the construction of structures specialized for particular tasks, from the mitotic spindle found in all dividing cells to the stereocilia of vertebrate hair cells. The spore wall that protects the meiotic products of S. cerevisiae is a complex structure comprised of multiple layers deposited in a spatial and temporal sequence. Our work provides insight into the regulatory strategies controlling spore wall formation and demonstrates the existence of a previously unknown role for the Smk1p MAP kinase in regulating spore morphogenesis.
In many multistep processes, the initiation of subsequent events depends on the successful completion of prior events. This is not the case for late steps in spore morphogenesis, as the outer spore wall layers are laid down normally in gsc2 mutants that are defective in inner wall synthesis. Nonetheless, deposition of inner and outer spore wall layers does appear coordinated. Our data indicate that appropriate deposition of the outer chitosan layer requires Smk1p-dependent inhibition of GS activity. The ability to suppress the chitosan deposition defect of smk1 by simply eliminating GSC2 points to Smk1p-dependent inhibition of Gsc2p as an important step for chitosan deposition.
These observations suggest a model in which SMK1 increases the fidelity of chitosan deposition by assisting the transition from exclusive deposition of inner spore wall material (mannan and glucan layers) to deposition of outer spore wall layers (chitosan and dityrosine layers; Fig. 4B). In this model, Smk1p is not active during early phases of spore wall production, whereas Gsc2p is fully active, producing the glucan layer and potentially inhibiting chitosan deposition. Later during sporulation, Smk1p kinase is activated by an unknown mechanism. Active Smk1p inhibits Gsc2p, both decreasing glucan layer production and facilitating chitosan deposition.
This model suggests that GS activity should normally rise early in spore wall production and then decrease in response to Smk1p activity. Our data (Fig. 2C) show a sharp increase in GS activity in wild-type cells between the 6- and 7-h time point, with no subsequent increases in GS activity at the 8- and 9-h time points. In interpreting these data, it is critical to note that even in the most highly synchronously sporulating culture, there is substantial asynchrony. Over the course of our assay, an increasing percentage of cells will have passed the point in sporulation when glucan deposition is initiated, and even at 8 and 9 h, some fraction of wild-type cells have not yet begun to deposit chitosan (Table 1). Thus, if GS activity remained constant once induced, total GS activity should continue to increase as the assay progresses. Instead, although the percentage of cells completing meiosis rises by ≈35% between 7 and 9 h of sporulation (Fig. 2D), GS activity remains roughly flat. Thus, GS activity per sporulating cell does not rise and may well decrease as sporulation progresses. In contrast, the level of GS activity in smk1 mutants continues to increase over time, rising >4-fold between 7 and 9 h of sporulation, consistent with a failure to inhibit GS activity in smk1 mutant cells.
Several genes required for the proper production of the outer spore wall layers have been identified, and include SSP2, CRR1, SPO77, OSW1, and MUM3 (2, 26, 27). These genes may be important in working with or in parallel to SMK1 to regulate the transition between inner and outer spore wall deposition. Thus, it is of interest to examine the relationship between SMK1 and these genes.
The transition from inner to outer spore wall layer deposition could be regulated in several ways. First, deposition of outer layers may depend on successful completion of inner layers, with the completion of one layer triggering the deposition of the next. Because our data show that the chitosan layer can be deposited even without proper glucan layer deposition in gsc2 mutants, this is likely not a major mechanism. Alternatively, deposition of different layers could be completely independent, perhaps relying on a timing mechanism that switches on the synthesis, deposition, and/or assembly of the next layer at the appropriate time, irrespective of the state of the previous layer. Because we do not see proper chitosan deposition in smk1 mutants and because mutants exist that block in sporulation after synthesis of the glucan layer (2), this is also likely not the case.
Instead, our data are consistent with a model where the deposition of inner and outer spore wall layers is interdependent, but where the deposition of one layer needs to be inhibited before the next layer can be deposited. We speculate that there may exist a Smk1p-dependent timing mechanism that is activated independently of the success of spore wall deposition and shuts down the synthesis of a layer, permitting the deposition of the next. How synthesis of one layer may disrupt the synthesis of a subsequent layer is unknown, but this inhibition could be as simple as the need for one completed layer to act as a template for the deposition of the next. In the current example, continued synthesis of glucan could preclude proper deposition of chitosan, whereas the complete absence of glucan may lead to the next layer being deposited onto the mannan layer.
Because we see heterogeneity of chitosan deposition within an ascus in smk1 mutants, we further speculate that individual spores may at some rate override the timing mechanism, perhaps analogous to the ability of cells to override the DNA damage checkpoint (28). A spore may have ways of overriding the need for GS inhibition, and whether this occurs is a decision made on a spore-by-spore basis. Alternatively, it is possible that GS activity varies from spore to spore in smk1 mutants; those spores that have less GS activity can support chitosan deposition, resulting in a heterogenous phenotype within an ascus.
Our results raise important issues for future investigation. We do not understand the mechanisms that activate Smk1p at the appropriate time. How glucan activity may interfere with chitosan deposition is also unknown. Understanding the biochemical mechanism of Gsc2p regulation by Smk1p is of interest, especially given the existence of SMK1 homologues in other fungal species and the importance of 1,3-β-glucan synthase as a target for antifungal agents (29–33). Nonetheless, the association of Smk1p with a spore wall biogenesis enzyme provides a clear opportunity for this MAP kinase to act directly on the machinery responsible for generating structure within the cell.
Supplementary Material
Acknowledgments
We thank B. Lane and the Harvard Microsequencing Facility for performing the mass spectrometry analysis; E. Cabib for advice regarding the GS assay; A. Neiman for sharing advice regarding spore wall assay; K. Benjamin (Molecular Sciences Institute, Berkeley, CA) and K. Irie (University of Tsukuba, Tsukuba, Japan) for gifts of reagents; M. Shiaris and K. Campbell for use of laboratory space, equipment, and reagents; A. Neiman, P. Garrity, and the Herskowitz laboratory for helpful discussions; and K. Benjamin, P. Garrity, K. Jaglo, and J. Urano for comments on the manuscript. This work was supported National Institutes of Health Grant GM59256 (to I.H.) and from start-up funds and a Faculty Scholarship Support Award from UMass Boston (to L.S.H.). L.S.H. was a Herbert W. Boyer Postdoctoral Fellow and a Special Fellow of the Leukemia and Lymphoma Society.
Author contributions: L.S.H. and I.H. designed research; L.S.H. and H.K.D. performed research; L.S.H. contributed new reagents/analytic tools; L.S.H., H.K.D., and I.H. analyzed data; and L.S.H. and I.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MAP, mitogen-activated protein; GS, glucan synthase.
References
- 1.Tachikawa, H., Bloecher, A., Tatchell, K. & Neiman, A. M. (2001) J. Cell Biol. 155, 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coluccio, A., Bogengruber, E., Conrad, M. N., Dresser, M. E., Briza, P. & Neiman, A. M. (2004) Eukaryot. Cell 3, 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kupiec, M., Byers, B., Esposito, R. E. & Mitchell, A. P. (1997) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology, eds. Pringle, J. R., Broach, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 889–1036.
- 4.Shimoda, C. (2004) J. Cell Sci. 117, 389–396. [DOI] [PubMed] [Google Scholar]
- 5.Inoue, S. B., Takewaki, N., Takasuka, T., Mio, T., Adachi, M., Fujii, Y., Miyamoto, C., Arisawa, M., Furuichi, Y. & Watanabe, T. (1995) Eur. J. Biochem. 231, 845–854. [DOI] [PubMed] [Google Scholar]
- 6.Mazur, P., Morin, N., Baginsky, W., el-Sherbeini, M., Clemas, J. A., Nielsen, J. B. & Foor, F. (1995) Mol. Cell. Biol. 15, 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krisak, L., Strich, R., Winters, R. S., Hall, J. P., Mallory, M. J., Kreitzer, D., Tuan, R. S. & Winter, E. (1994) Genes Dev. 8, 2151–2161. [DOI] [PubMed] [Google Scholar]
- 8.Wagner, M., Briza, P., Pierce, M. & Winter, E. (1999) Genetics 151, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamin, K. R., Zhang, C., Shokat, K. M. & Herskowitz, I. (2003) Genes Dev. 17, 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose, M. D., Winston, F. & Hieter, P. (1990) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 11.Padmore, R., Cao, L. & Kleckner, N. (1991) Cell 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- 12.Grether, M. E. & Herskowitz, I. (1999) Mol. Biol. Cell 10, 3689–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, H., Fukuda, Y., Murata, K. & Kimura, A. (1983) J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. (1995) Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- 15.Kitada, K., Yamaguchi, E. & Arisawa, M. (1995) Gene 165, 203–206. [DOI] [PubMed] [Google Scholar]
- 16.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 17.Puig, O., Rutz, B., Luukkonen, B. G., Kandels-Lewis, S., Bragado-Nilsson, E. & Seraphin, B. (1998) Yeast 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- 18.Philips, J. & Herskowitz, I. (1998) J. Cell Biol. 143, 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wessel, D. & Flugge, U. I. (1984) Anal. Biochem. 138, 141–143. [DOI] [PubMed] [Google Scholar]
- 20.Eng, J. K., McCormick, A. L. & Yates, J. R. I. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 21.Chittum, H. S., Lane, W. S., Carlson, B. A., Roller, P. P., Lung, F. D., Lee, B. J. & Hatfield, D. L. (1998) Biochemistry 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- 22.Mol, P. C., Park, H. M., Mullins, J. T. & Cabib, E. (1994) J. Biol. Chem. 269, 31267–31274. [PubMed] [Google Scholar]
- 23.Briza, P., Breitenbach, M., Ellinger, A. & Segall, J. (1990) Genes Dev. 4, 1775–1789. [DOI] [PubMed] [Google Scholar]
- 24.Douglas, C. M., Foor, F., Marrinan, J. A., Morin, N., Nielsen, J. B., Dahl, A. M., Mazur, P., Baginsky, W., Li, W., el-Sherbeini, M., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 12907–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knop, M. & Strasser, K. (2000) EMBO J. 19, 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar, P. K., Florczyk, M. A., McDonough, K. A. & Nag, D. K. (2002) Mol. Genet. Genomics 267, 348–358. [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Esquer, F., Rodriguez-Pena, J. M., Diaz, G., Rodriguez, E., Briza, P., Nombela, C. & Arroyo, J. (2004) Microbiology 150, 3269–3280. [DOI] [PubMed] [Google Scholar]
- 28.Toczyski, D. P., Galgoczy, D. J. & Hartwell, L. H. (1997) Cell 90, 1097–1106. [DOI] [PubMed] [Google Scholar]
- 29.Tzung, K. W., Williams, R. M., Scherer, S., Federspiel, N., Jones, T., Hansen, N., Bivolarevic, V., Huizar, L., Komp, C., Surzycki, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3249–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, S., Fares, M. A., Zimmermann, W., Butler, G. & Wolfe, K. H. (2003) Genome Biol. 4, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 32.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 33.Kurtz, M. B. & Rex, J. H. (2001) Adv. Protein Chem. 56, 423–475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.