Abstract
Glycosylated natural products are reliable platforms for the development of many front-line drugs, yet our understanding of the relationship between attached sugars and biological activity is limited by the availability of convenient glycosylation methods. When a universal chemical glycosylation method that employs reducing sugars and requires no protection or activation is used, the glycorandomization of digitoxin leads to analogs that display significantly enhanced potency and tumor specificity and suggests a divergent mechanistic relationship between cardiac glycoside-induced cytotoxicity and Na+/K+-ATPase inhibition. This report highlights the remarkable advantages of glycorandomization as a powerful tool in glycobiology and drug discovery.
Keywords: carbohydrate, natural product, sugar
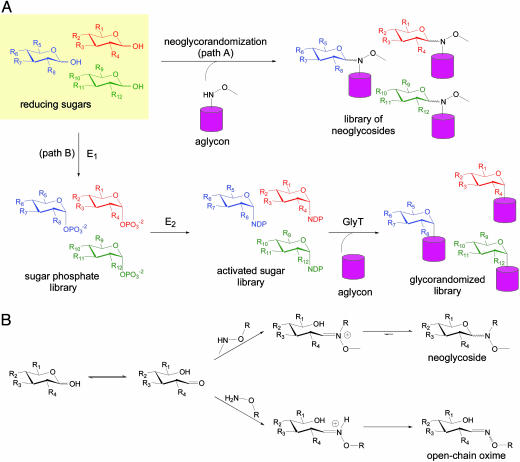
Carbohydrates mediate many essential biological processes (1–4). For example, the saccharide-containing macromolecules that decorate cell surfaces are vital to a variety of cellular functions, including cell–cell recognition, apoptosis, differentiation, and tumor metastasis. In a similar fashion, glycosylated natural products contain sugar attachments essential for their activity and continue to serve as reliable platforms for the development of many of our existing front-line drugs (5, 6). While the diverse chemical space accessible by carbohydrates contributes to a remarkably vast array of biological function (7), a precise understanding of the relationship between sugars and biological activity remains limited by the availability of convenient and effective glycosylation tools (8). In an effort to explore the contribution of the sugar constituents of pharmaceutically relevant glycosylated natural products, we have developed chemoenzymatic “glycorandomization” methods (Fig. 1A, path B) to rapidly convert a single aglycon structure into a library of analogs with a broad array of sugar attachments (9, 10). Despite these advances, chemoenzymatic glycorandomization currently excludes a number of essential glycoconjugates because it is limited to natural products for which promiscuous glycosyltransferases are available and can operate in vitro. Herein we describe a complementary robust chemical approach, referred to as “neoglycorandomization,” that accomplishes a one-step sugar ligation that does not require any prior sugar protection or activation (Fig. 1 A, path A). Using digitoxin as a simple pharmaceutically relevant model, we demonstrate that neoglycorandomization leads to the discovery of digitoxin analogs that are much more potent and/or tumor-specific cytotoxins, but less potent Na+/K+-ATPase inhibitors, relative to the parent natural product. This report reveals the remarkable utility of neoglycosylation as a general tool for glycobiology and drug discovery and highlights a potentially divergent relationship between Na+/K+-ATPase inhibition and cytotoxicity invoked by cardiac glycosides.
Fig. 1.
Glycorandomization is a tool used to convert a single aglycon molecule into a library of analogs with a diverse array of sugar attachments. (A) Neoglycorandomization involves the chemoselective formation of glycosidic bonds between reducing sugars and a secondary alkoxylamine to form a library of neoglycosides (path A). The utility of neoglycorandomization is limited only by the ease of installation of the reactive secondary alkoxylamine group onto a complex natural product aglycon. Chemoenzymatic glycorandomization (path B) exploits nucleotide sugar activation enzymes (E1 and E2) and glycosyltransferase enzymes (GlyT) that display natural or engineered promiscuity to glycosylate secondary metabolites. This method is limited to natural products for which promiscuous glycosylation machinery is available. (B) Whereas primary alkoxylamines react with reducing sugars to form open-chain oximes, secondary alkoxylamines react to form closed-ring neoglycosides.
Neoglycorandomization is based upon the chemoselective formation of glycosidic bonds between reducing sugars and a secondary alkoxylamine-containing aglycon to form “neoglycosides” (Fig. 1 A, path A). The notable advantage of this approach is, unlike most traditional chemical glycosylation reactions, unprotected and nonactivated reducing sugars are used as sugar donors in the reaction under mild conditions (11). In early examples of this chemoselective reaction, sugars and peptides that contain secondary alkoxylamines were reacted with d-glucose, d-mannose, d-galactose, lactose, and N-acetyl-d-glucosamine to generate oligosaccharide and glycopeptide mimics, respectively (12, 13). These pioneering studies revealed that, unlike primary alkoxylamines, which provide open-chain oxime isomers (14), secondary alkoxylamines react to form closed-ring neoglycosides (Fig. 1B). Although the stability of these model neoglycosides was not examined, the distribution of pyranose, and occasionally furanose, anomers in neoglycosides was found to be dependant on the identity of the sugar (15), and equilibration between the product isomers was sometimes observed (J.M.L. and J.S.T., unpublished data). Closed-ring neoglycosides were found to display conformational behavior similar to natural O-glycosides by NMR studies, molecular dynamics simulations, and ab initio calculations (16).
Digitoxin (1) was selected as our model platform to test the potential of this neoglycosylation reaction toward natural product glycorandomization. In addition to its well known cardiac activity, which is mediated by inhibition of the plasma membrane Na+/K+-ATPase (17), digitoxin has demonstrated in vitro anticancer properties (18), and patient profiling suggests the survival rate of cancer patients taking digitoxin is statistically enhanced (19, 20). Cardiac glycosides were also recently noted to inhibit the expression of four genes that are overexpressed in prostate cancer cells, including transcription factors and the apoptosis inhibitor survivin (21), and to provide protective effects against polyglutamine-based diseases (22). Digitoxin also inhibits activation of the NF-κB signaling pathway in cystic fibrosis (CF) cells, suppressing hypersecretion of IL-8, a protein implicated in lung inflammation, from CF lung epithelial cells (23). Given that the attached sugars are implicated as mediators of the unique spectrum of biological properties exhibited by cardiac glycosides (18, 24), digitoxin provides an excellent model both to examine the general utility of neoglycosylation to efficiently construct a glycorandomized library and to directly assess the biological impact of varying the sugars attached to a given natural product-based drug.
Materials and Methods
Aglycon Synthesis. Compounds 2a,b, 3β, and 3α were synthesized according to procedures described in Supporting Text, which is published as supporting information on the PNAS web site.
Library Synthesis and Purification. Aglycon 3β or 3α (≈40 μmol) was added to 4-ml glass vials equipped with stirring fleas. The appropriate sugar (2 eq) was added to each vial, followed by 3:1 (vol/vol) dimethylformamide (DMF)/AcOH (final concentration of aglycon = 90 mM). The reaction mixtures were stirred at 40°C by using a stirplate equipped with a 48-well reaction block and a contact thermometer. After 2 days, the reaction mixtures were concentrated by means of a SpeedVac (Savant) and suspended in 5% EtOH/CHCl3. The crude suspensions were purified in parallel on disposable SiO2 solid-phase extraction columns by using a 24-port vacuum manifold. Library members (β and α) 5–16, 23, 24, 30–36, and 41 were purified on 1,000-mg columns, eluting first with 5 ml of 5% EtOH/CHCl3 to remove the remaining aglycon and second with 5 ml of 15% EtOH/CHCl3 to collect the product neoglycosides. Library members (β and α) 4, 17–22, 25–29, 37–40, and 42 were purified on 500-mg columns, eluting first with 4 ml of 5% EtOH/CHCl3 to remove the remaining aglycon and second with 5 ml of 25% EtOH/CHCl3 to collect the product neoglycosides. The product solutions were concentrated by SpeedVac, weighed, and dissolved in DMSO to make 20 or 30 mM stock solutions. The stock solutions were characterized by liquid chromatography/mass spectrometry (LCMS) using reverse-phase HPLC on a Zorbax Eclipse XDB-C8 column (4.6 × 150 mm; Agilent Technologies, Palo Alto, CA) with a flow rate of 0.8 ml/min and a linear gradient of 45% CH3OH/H2O to 85% CH3OH/H2O over 20 min and electrospray ionization. Library member purities were estimated by dividing the sum of the peak areas at 220 nm of peaks corresponding to the desired product mass by the total area of all peaks. For mass information, the purity of specific library members, and a tabulation of which members display >90% of a single product isomer as judged by LCMS, see Table 1, which is published as supporting information on the PNAS web site. Average library purity was 91%.
Hydrolytic Stability of 4α. The chemical stability of the neoglycosidic linkage was examined by monitoring the hydrolytic degradation of neoglycoside 4α in a 3 mM solution of 1:1 DMSO/buffer. Three buffers were used, 50 mM sodium acetate buffer (pH 5), 50 mM sodium phosphate buffer (pH 7), or 50 mM Tris·HCl buffer (pH 9). Neoglycoside degradation was monitored by reverse-phase HPLC on an Agilent Technologies Zorbax Eclipse XDB-C8 column (4.6 × 150 mm) with a flow rate of 0.8 ml/min and a linear gradient of 49% CH3OH/H2Oto89% CH3OH/H2O over 20 min. At t = 0, neoglycoside 4α in 500 μl of DMSO was added to 500 μl of buffer, and the resulting solution was Vortex mixed for 40 sec, then immediately injected onto the HPLC column. Peak areas at 220 nm were used to estimate the neoglycoside/aglycon ratio, which is reported as percent neoglycoside remaining [Aneoglycoside/(Aneoglycoside + Aaglycon)] for each of the three buffer systems (see Fig. 5, which is published as supporting information on the PNAS web site).
Cytotoxicity Assays. All cell lines except NmuMG were maintained in RPMI medium 1640 supplemented with 10% (wt/vol) FBS and penicillin-streptomycin (PS) (100 units/ml and 100 μg/ml). NmuMG cells were maintained in DMEM supplemented with 10% wt/vol FBS (FBS), 10 μg/ml insulin, and penicillin/streptomycin (PS) (100 units/ml and 100 μg/ml, respectively). Cells were harvested by trypsinization using 0.25% trypsin and 0.1% EDTA and then counted in a hemocytometer in duplicate with better than 10% agreement in field counts. Cells were plated at a density of 10,000–15,000 cells per well of each 96-well black tissue culture treated microtiter plate. Cells were grown for 1 h at 37°C, with 5% CO2/95% air in a humidified incubator to allow cell attachment to occur before compound addition. Library members were stored at –20°C under desiccating conditions before the assay. Library member stocks (100×) were prepared in 96-well V-bottom polypropylene microtiter plates. Five serial 1:2 dilutions were made with anhydrous DMSO at 100× the final concentration used in the assay. The library member-containing plates were diluted 1:10 with complete cell culture medium. The 10× stocks (10 μl) were added to the attached cells by using a Biomek FX liquid handler (Beckman Coulter). Library member stocks (10 μl) were added to 90 μl of cells in each plate to ensure full mixing of stocks with culture media by using a Biomek FX liquid handler with 96-well head. Cells were incubated with the library members for 72 h before fluorescence reading. Test plates were removed from the incubator and washed once in sterile PBS to remove serum containing calcium esterases. Calcein AM (acetoxymethyl ester) reagent (30 μl, 1 M) was added and the cells were incubated for 30 min at 37°C. Plates were read for emission by using a fluorescein filter (excitation 485 nm, emission 535 nm).
IC50 Calculations. For each library member, at least six dose–response experiments were conducted. Within each experiment, percent inhibition values at each concentration were expressed as a percentage of the maximum fluorescence emission signal observed for a 0 nM control. To calculate IC50, percent inhibitions were plotted as a function of log[concentration] and then fit to a four-parameter logistic model that allowed for a variable Hill slope by using xlfit 4.1 (ID Business Solutions, Emeryville, CA) (see Table 2 and Fig. 6, which are published as supporting information on the PNAS web site).
Na+/K+-ATPase Assays. Inhibition of Na+/K+-ATPase on HEK-293 cells and CHO-K1 cells by the library hits was determined by Aurora Biomed (Vancouver) by using a high-throughput nonradioactive rubidium ion uptake assay. Experiments were conducted in duplicate at three different concentrations. Within each experiment, percent inhibition values at the three concentrations were expressed as the percent reduction of the maximum absorption signal observed for a 0 nM control. IC50 values were determined by using the following formula: IC50 = [(50 – low %)/(high % – low %)] × (high conc. – low conc.) + low conc. (see Table 3, which is published as supporting information on the PNAS web site).
Results and Discussion
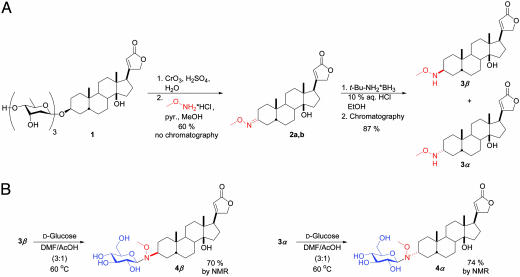
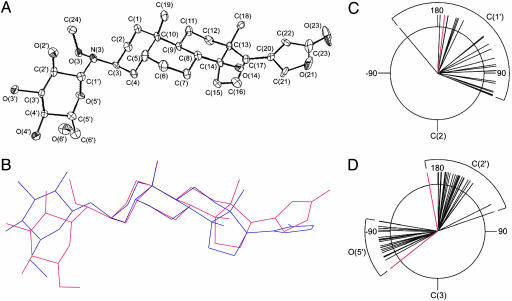
As highlighted in Fig. 2A, the requisite methoxylamine functional group was installed at the C3 of digitoxin (the natural position of sugar attachment) in three simple chemical steps. Specifically, digitoxin was oxidized under acidic conditions to simultaneously hydrolyze the O-glycoside and provide digitoxigenone, which was then converted to the corresponding set of oxime diastereomers (2a,b). We found that treatment of 2a,b with tert-butylamine·borane resulted in a 1:1 mixture of stereoisomers, which were easily resolved by standard column chromatography and assigned as 3β and 3α by x-ray crystallography (see Supporting Text). The accessibility of both digitoxigenin-like isomers 3β and 3α set the stage to explore the importance of the C3 stereochemistry on biological activity. Pilot reactions of aglycons 3β and 3α with d-glucose were first explored in an attempt to generate the corresponding neoglycosides (Fig. 2B). Aglycons 3β and 3α reacted with d-glucose in DMF/acetic acid to form neoglycosides 4β and 4α in good yields (>70%). Both reactions proceeded stereoselectively, providing the β-anomer exclusively as determined by 1H NMR. An x-ray crystal structure of neoglycoside 4β was obtained (Fig. 3A) and compared to the crystal structures of related O-glycosides available from the Cambridge Crystal Database (Fig. 3 B–D). The observed orientations about the C2–C3–N3–C1′ torsion (Fig. 3C) and the C3–N3–C1′–C2′ torsion in the neoglycoside structure (Fig. 3D) fall on the periphery of the narrow range of orientations displayed in the solid-state structures of 23 known cardiac O-glycosides. The chemical stability of the neoglycosidic linkage was examined by monitoring the hydrolytic degradation of neoglycoside 4α in a 3 mM solution in 1:1 DMSO/buffer using buffers at three different pHs. Compound 4α was completely stable over the period of 1 month under neutral or basic conditions but slowly hydrolyzed under acidic conditions over this same time period. When identical acidic conditions were used, aglycon 3α and d-glucose did not react to form neoglycoside 4α, ruling out equilibrium as a complicating factor in this analysis. Library member 27β, derived from aglycon 3β, also displayed no hydrolytic degradation under the same conditions at neutral and basic pHs, demonstrating that aglycon C3 stereochemistry does not significantly influence neoglycoside stability. In conjunction with our neoglycoside structural analyses and the previously reported NMR and molecular dynamics studies (16), these hydrolytic studies suggest the neoglycoside nitrogen to be predominately charge-neutral at physiological pH.
Fig. 2.
The synthesis of cardiac neoglycosides. (A) Aglycon 3β and its C3 epimer 3α were generated in three simple steps from the parent natural product, digitoxin. pyr., pyridine. (B) Aglycons 3β and 3α reacted with d-glucose (2 eq) in 3:1 DMF/acetic acid at 60°C to form neoglycosides 4β and 4α, respectively, in >70% yield by 1H NMR.
Fig. 3.
The crystal structure of a neoglycoside. (A) Solid-state structure of 4β shown with 50% thermal probability ellipsoids. Hydrogen atoms are omitted for clarity. Crystallographic structure refinement data for 4β and for 3β (structure not shown) can be found, respectively, in Tables 4 and 5, which are published as supporting information on the PNAS web site. (B) The solid-state structure of 4β (red) superimposed on the solid-state structure of a homologous O-glucoside, actodigin (blue). (C) A Newman projection along the C2–C3–N3–C1′ torsion of neoglycoside 4β (red) and the corresponding torsion of 23 known cardiac glycosides (black) reveals that the neoglycoside torsion falls within the range of torsions displayed in the solid state by the known cardiac glycosides. (D) A Newman projection along the C3–N3–C1′–C2′ torsion of 4β (red) and the corresponding torsions of 23 known cardiac glycosides (black) reveals that the neoglycoside torsion falls on the periphery of the narrow range of orientations displayed by natural O-glycosides (citations for the 23 structurally characterized cardiac glycosides can be found in the Supporting Text).
A library of 78 digitoxin derivatives was synthesized in parallel from 39 reducing sugars and aglycons 3β and 3α. The reaction mixtures were stirred for 2 days at 40°C, concentrated, and then submitted to solid-phase extraction in parallel to remove unreacted aglycon and sugar. The concentrated products were characterized by liquid chromatography/mass spectrometry to assess purity and to confirm product identity. Even though a diverse array of reducing sugars were used, including l-sugars, deoxy sugars, dideoxy sugars, disaccharides, and uronic acids, in every case neoglycosides were successfully generated. The average purity of the library members was 91%, and the liquid chromatography chromatograms suggested that ≈50% of the library members contained >90% of a single product isomer. Although combinatorial methods have been extensively applied to steroidal derivatives and cardenolides in particular (25, 26), the results reported herein represent the largest and most diverse glycorandomized library generated to date.
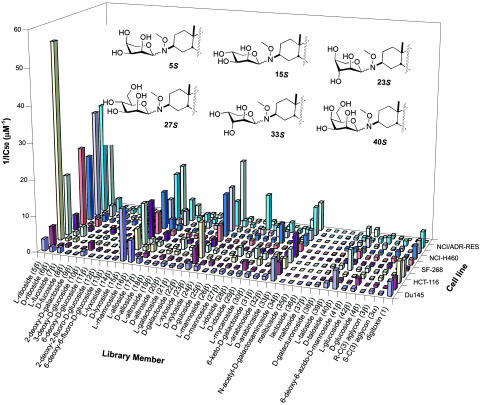
The activity of the library members was assessed by using a high-throughput cytotoxicity assay on nine human cancer cell lines representing a broad range of carcinomas, including breast, colon, CNS, liver, lung, and ovary, and a mouse mammary normal epithelial control line (Fig. 4). The cytotoxicities of digitoxin and aglycons 3β and 3α were also examined. Digitoxin was a modest cytotoxin toward the nine human cancer cell lines (average IC50 ≈ 440 nM) but was nonspecific because it affected these cancer cells with similar potencies. One library member (33β) closely mimicked this activity. Several hits identified from the neoglycoside library exhibited enhanced activities relative to the parent natural product, digitoxin (1), in terms of both potency and specificity. The two most significant hits, library members 5β and 27β, displayed striking potency and excellent selectivity, respectively. Specifically, library member 5β was a potent cytotoxin against six cancer cell lines (18 ± 2 nM in the case of HCT-116, >9-fold more potent than digitoxin), and also was modestly selective, because three of the nine cancer cell lines tested were much less affected. In contrast, library member 27β was a less potent cytotoxin than 5β, but 27β exhibited dramatic selectivity because it was 4 times more cytotoxic toward NCI/ADR-RES cells (IC50 = 100 ± 10 nM) than any other cell line. This result is especially significant because NCI/ADR-RES is a multidrug-resistant line that has high levels of MDR-1 and P-glycoprotein expression (27, 28). Given that cardiac glycosides are substrates for P-glycoprotein (29), such tumor specificity suggests that 27β may no longer serve as a P-glycoprotein substrate or may be interacting with a unique target. Other neoglycoside library members, although not as potent as 5β or as selective as 27β, also were significantly active. For example, library member 40β exhibited notable selectivity, with modest cytotoxicity toward only Du145 and Hep3B cells (IC50 = 200 ± 30 nM and 180 ± 30 nM, respectively), whereas library members 15β and 23β were significantly more potent than digitoxin against some cell lines, but were somewhat nonselective like digitoxin.
Fig. 4.
Summary of IC50 data from the high-throughput cytotoxicity assay. Reciprocal IC50 values are displayed for clarity, with the current illustration representing an IC50 range of 18 nM (5β, HCT-116) to >25 μM (e.g., 35β, all cell lines). In the assay, live cells were distinguished by the presence of a ubiquitous intracellular enzymatic activity that converts the nonfluorescent cell-permeant molecule calcein AM to the intensely fluorescent molecule calcein, which is retained within live cells. The IC50 value for each library member represents at least six replicates of dose–response experiments conducted over five concentrations at 2-fold dilutions. For the entire panel of 81 compounds in 10 cell lines, the average error was 17%. IC50 values and corresponding error values can be found in Tables 4 and 5, which are published as supporting information on the PNAS web site. The six library member “hits” are depicted in pyranose 4C1 conformations to facilitate structural comparisons. Du145, human colon carcinoma; MCF7, human breast carcinoma; HCT-116, human colon carcinoma; Hep3B, human liver carcinoma; SF-268, human CNS glioblastoma; SK-OV-3, human ovary adenocarcinoma; NCI-H460, human lung carcinoma; A549, human lung adenocarcinoma; NCI/ADR-RES, human breast carcinoma; NmuMG, mouse mammary normal epithelial cells.
In contrast to the 3β-derived analogs, the 38 neoglycosides derived from aglycon 3α uniformly displayed low cytotoxicities in the assay (IC50 > 25 μM, data not shown), as did aglycon 3α itself, establishing the importance of the natural β configuration of the C3 stereocenter. Aglycon 3β was only weakly cytotoxic against the cell lines (average IC50 ≈ 1.10 μM), consistent with the influence sugars have on library member cytotoxicity. Interestingly, the six hits described above all contain sugars with a common structural feature, an S-configured C2′ sugar stereocenter. This C2′ stereochemistry appears to be of critical importance for compound activity. For example, the C2′ epimer of the extremely potent library member 5β (l-arabinose-containing member 32β) was relatively inactive toward the 10 cell lines examined. Likewise, d-glucose-containing library member 4β was relatively inactive and displayed none of the cell line specificity observed for its C2′ epimer 27β. It should also be noted that neoglycosides with sugars containing reactive handles were successfully generated. For example, 41β contains the C2′ stereochemistry shared by the library hits and a reactive azido group, which is amenable to further diversification by Huisgen 1,3-dipolar cycloaddition (9).
To assess how these structural modifications affect the ability of library members to inhibit Na+/K+-ATPases, a fundamental activity of cardiac glycosides (17), library hits 5β, 15β, 23β, 27β, 33β, 40β, and digitoxin (1) were submitted to a nonradioactive rubidium uptake assay to gauge Na+/K+-ATPase inhibition in both HEK-239 human embryonic kidney cells and CHO-K1 hamster ovary cells (30). In HEK-239 cells, digitoxin displayed an IC50 of 75.4 ± 0.5 μM, whereas none of the library hits showed 50% inhibition even at the highest concentration tested (300 μM for 5β, 15β, 23β, and 33β; 200 μM for 27β and 40β). A similar trend was observed in the CHO-K1 cells. Thus, not only did hits identified from the neoglycoside library display enhanced cytotoxic properties toward human cancer cells, but the rubidium uptake assays revealed our neoglycoside hits to be less potent Na+/K+-ATPase inhibitors in a human cell line than digitoxin.
The growing body of epidemiological (19, 20), in vitro (18), and in vivo (31) evidence supporting the anticancer benefits of cardenolides has prompted the search for noncardioactive analogs that retain anticancer activity. The specific mechanism of cardenolide-induced cytotoxicity remains controversial. For example, a preferred ligand for cardenolides, the Na+/K+-ATPase, belongs to the “Na+/K+-ATPase signalosome,” the activation of which by certain cardenolides can lead to NF-κB pathway inactivation (32). Constitutive activation of the NF-κB pathway protects a large group of cancer cells against apoptosis, whereas suppression of this transcription factor can restore normal levels of apoptosis in cancer cells and also potentially block tumorigenesis and inflammation (33, 34). Yet digitoxin-mediated inhibition of the NF-κB signaling pathway in cystic fibrosis lung epithelial cells has been demonstrated to be mechanistically distinct from Na+/K+-ATPase inhibition (23). With respect to other implicated cellular players, the nonlethal cardenolide concentrations that inhibit breast cancer cell proliferation also activate Src kinase, stimulate the interaction between Na+/K+-ATPase, the activated Src kinase, and epidermal growth factor (EGFR), and lead to the activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) and subsequent cell cycle arrest caused by increased levels of p21Cip1 (35). Cardiac glycosides have also been demonstrated to initiate apoptosis via the classical caspase-dependent pathways in malignant T lymphoblasts (36) and prostate cancer cells (37). In the latter, cardiac glycosides also inhibit testosterone production in vivo (38). Thus, while the cytotoxicity of some cardiac glycosides may correlate with Na+/K+-ATPase inhibition (18, 21), the present study reveals a unique class of noncardioactive tumor-specific and potent cytotoxins, for which the mechanism of action remains to be elucidated.
Conclusions
The neoglycorandomization of digitoxin illustrates the remarkable utility of this simple, mild, and robust reaction with unprotected and nonactivated reducing sugars to rapidly survey the influence of differential glycosylation upon natural product scaffolds. In this prototype example, we show that subtle sugar modifications can dramatically, and independently, modulate both the cytotoxic properties and the Na+/K+-ATPase-inhibitory properties of cardiac glycosides. The potential of neoglycorandomization is further augmented by its compatibility with chemical handles (e.g., azido groups) for additional elaboration (8, 9). Neoglycorandomization is limited solely by the efficiency and specificity of alkoxylamine handle installation and the availability of reducing sugar donors; thus, these studies highlight the unique potential of neoglycosylation and/or neoglycorandomization as a universally powerful tool for glycobiology and drug discovery. Transformations paralleling those described in Fig. 2 will provide easy access to a wide array of bioactive small molecules and glycoconjugates, and the incorporation of alkoxylamine-bearing unnatural amino acids (39) will easily extend this approach into biologically relevant macromolecules such as glycoproteins and proteoglycans. Moreover, a wide range of reducing sugars are available commercially or by means of elegant transformations from simple precursors, presenting broad access to the only building blocks essential to this approach (40, 41).
Supplementary Material
Acknowledgments
We thank Megan K. Fitzgerald (Keck University of Wisconsin Comprehensive Cancer Center Small Molecule Screening Facility) and Dr. Lesley M. Liu (University of Wisconsin School of Pharmacy) for technical assistance. This research was supported in part by National Institutes of Health Grants AI52218, CA84374, and GM70637 (to J.S.T.) and National Cooperative Drug Discovery Group Grant U19 CA113297 from the National Cancer Institute. J.M.L. was supported in part by National Institutes of Health Postdoctoral Fellowship AI56652. We are grateful to the University of Wisconsin–Madison School of Pharmacy Analytical Facility for analytical support.
Author contributions: J.M.L. and J.S.T. designed research; J.M.L., N.R.P., and I.A.G. performed research; J.M.L., N.R.P., I.A.G., F.M.H., and J.S.T. analyzed data; J.M.L. and J.S.T. wrote the paper; I.A.G. performed crystallography and analysis only; and F.M.H. performed cytotoxicity analysis and participated in discussion.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: DMF, dimethylformamide.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CCDC reference nos. 269298 and 269299).
References
- 1.Sears, P. & Wong, C.-H. (2001) Science 291, 2344–2350. [DOI] [PubMed] [Google Scholar]
- 2.Bertozzi, C. R. & Kiessling, L. L. (2001) Science 291, 2357–2364. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, Z., Gildersleeve, J., Yang, Y.-Y., Xu, R., Loo, J. A., Uryu, S., Wong, C.-H. & Schultz, P. G. (2004) Science 303, 371–373. [DOI] [PubMed] [Google Scholar]
- 4.Prescher, J. A., Dube, D. H. & Bertozzi, C. R. (2004) Nature 430, 873–877. [DOI] [PubMed] [Google Scholar]
- 5.Clardy, J. & Walsh, C. (2004) Nature 432, 829–837. [DOI] [PubMed] [Google Scholar]
- 6.Thorson, J. S., Hosted, T. J., Jr., Jiang, J., Biggins, J. B. & Ahlert, J. (2001) Curr. Org. Chem. 5, 139–150. [Google Scholar]
- 7.Dobson, C. M. (2004) Nature 432, 824–865. [DOI] [PubMed] [Google Scholar]
- 8.Langenhan, J. M. & Thorson, J. S. (2005) Curr. Org. Synth. 2, 59–81. [Google Scholar]
- 9.Fu, X., Albermann, C., Jiang, J., Liao, J., Zhang, C. & Thorson, J. S. (2003) Nat. Biotechnol. 21, 1467–1469. [DOI] [PubMed] [Google Scholar]
- 10.Yang, J., Hoffmeister, D., Liu, L., Fu, X. & Thorson, J. S. (2004) Bioorg. Med. Chem. 12, 1577–1584. [DOI] [PubMed] [Google Scholar]
- 11.Van Vranken, D. L. & Chisolm, J. D. (2000) J. Org. Chem. 65, 7541–7553. [DOI] [PubMed] [Google Scholar]
- 12.Peri, F., Dentman, A., LaFerla, B. & Nicotra, F. (2002) Chem. Commun., 1504–1505. [DOI] [PubMed]
- 13.Carrasco, M. R. & Brown, R.T. (2003) J. Org. Chem. 68, 8853–8858. [DOI] [PubMed] [Google Scholar]
- 14.Cervigni, S. E., Dumy, P. & Mutter, M. (1996) Angew. Chem. Int. Ed. 35, 1230–1232. [Google Scholar]
- 15.Peri, F., Dumy, P. & Mutter, M. (1998) Tetrahedron 54, 12269–12278. [Google Scholar]
- 16.Peri, F., Jimenez-Barbero, J., Garcia-Aparicio, V., Tvaroska, I. & Nicotra, F. (2004) Chem. Eur. J. 10, 1433–1444. [DOI] [PubMed] [Google Scholar]
- 17.Paula, S., Tabet, M. R. & Ball, W. J., Jr. (2005) Biochemistry 44, 498–510. [DOI] [PubMed] [Google Scholar]
- 18.Johansson, S., Lindholm, P., Gullbo, J., Larsson, R., Bohlin, L. & Claeson, P. (2001) Anti-Cancer Drugs 12, 475–483. [DOI] [PubMed] [Google Scholar]
- 19.Stenkvist, B. (2001) Anti-Cancer Drugs 12, 635–636. [DOI] [PubMed] [Google Scholar]
- 20.Haux, J., Klepp, O., Spigset, O. & Tretli, S. (2001) BMC Cancer 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, P. H., Walker, R. P., Jones, S. W., Stephens, K., Meurer, J., Zajchowski, D. A., Luke, M. M., Eeckman, F., Tan, Y., Wong, L., et al. (2002) Mol. Cancer Ther. 1, 1293–1304. [PubMed] [Google Scholar]
- 22.Piccioni, F., Roman, B. R., Fischbeck, K. H. & Taylor, J. P. (2004) Hum. Mol. Genet. 13, 437–446. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava, M., Eidelman, O., Zhang, J., Paweletz, C., Caohuy, H., Yang, Q., Jacobson, K. A., Heldman, E., Huang, W., Jozwik, C., et. al. (2004) Proc. Natl. Acad. Sci. 101, 7693–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathore, H., From, A. H. L., Ahmed, K. & Fullerton, D. S. (1986) J. Med. Chem. 29, 1945–1952. [DOI] [PubMed] [Google Scholar]
- 25.Maltais, R., Tremblay, M. R., Ciobanu, L. C. & Poirier, D. (2004) J. Comb. Chem. 6, 443–456. [DOI] [PubMed] [Google Scholar]
- 26.Schneider, G. & Wolfling, J. (2004) Curr. Org. Chem. 8, 1381–1403. [Google Scholar]
- 27.Fairchild, C. R., Ivy, S. P., Kao-Shan, C. S., Whang-Peng, J., Rosen, N., Israel, M. A., Melera, P. W., Cowan, K. H. & Goldsmith, M. E. (1987) Cancer Res. 47, 5141–5148. [PubMed] [Google Scholar]
- 28.Scudiero, D. A., Monks, A. & Sausville, E. A. (1998) J. Natl. Cancer Inst. 90, 862 (lett.). [DOI] [PubMed] [Google Scholar]
- 29.Tanigawara, Y., Okumura, N., Hirai, M., Yasuhara, M., Ueda, K., Kioka, N., Komano, T. & Hori, R. (1992) J. Pharmacol. Exp. Ther. 263, 840–845. [PubMed] [Google Scholar]
- 30.Gill, S., Gill, R., Wicks, D., Despotovski, S. & Liang, D. (2004) Assay Drug Dev. Technol. 2, 535–542. [DOI] [PubMed] [Google Scholar]
- 31.Svensson, A., Azarbayjani, F., Backman, U., Matsumoto, T. & Christofferson, R. (2005) Anticancer Res. 25, 207–212. [PubMed] [Google Scholar]
- 32.Dmitrieva, R. I. & Doris, P. A. (2002) Exp. Biol. Med. 227, 561–569. [DOI] [PubMed] [Google Scholar]
- 33.Quanquebeke, E. V., Simon, G., Andre, A., Dewelle, J., El Yazidi, M., Bruyneel, F., Tuti, J., Nacoulma, O., Guissou, P., Decaestecker, C., et al. (2005) J. Med. Chem. 48, 849–856. [DOI] [PubMed] [Google Scholar]
- 34.Sreenivasan, Y., Sarkar, A. & Manna, S. K. (2003) Biochem. Pharmacol. 66, 2223–2239. [DOI] [PubMed] [Google Scholar]
- 35.Kometiani, P., Liu, L. & Askari, A. (2005) Mol. Pharmacol. 67, 929–936. [DOI] [PubMed] [Google Scholar]
- 36.Daniel, D., Susal, C., Kopp, B., Opelz, G. & Terness, P. (2003) Internatl. Immunopharmacol. 3, 1791–1801. [DOI] [PubMed] [Google Scholar]
- 37.Lin, H., Juang, J.-L. & Wang, P. S. (2004) J. Biol. Chem. 279, 29302–29307. [DOI] [PubMed] [Google Scholar]
- 38.Lin, H., Wang, S.-W., Tsai, S.-C., Chen, J.-J., Chiao, Y.-C., Lu, C.-C., Huang, W. J.-S., Wang, G.-J., Chen, C.-F. & Wang, P. S. (1998) Br. J. Pharmacol. 125, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin, J. W., Cropp, A. J., Anderson, J. C., Mukherji, M., Zhang, Z. & Schultz, P. G. (2003) Science 301, 964–967. [DOI] [PubMed] [Google Scholar]
- 40.Northrup, A. B. & MacMillan, D. W. C. (2004) Science 305, 1752–1755. [DOI] [PubMed] [Google Scholar]
- 41.Andreana, P. R., McLellan, J. S., Chen, Y. & Wang, P. G. (2002) Org. Lett. 4, 3875–3878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.