Abstract
RPE65 is an abundant protein in the retinal pigment epithelium. Mutations in RPE65 are associated with inherited retinal dystrophies. Although it is known that RPE65 is critical for regeneration of 11-cis retinol in the visual cycle, the function of RPE65 is elusive. Here we show that recombinant RPE65, when expressed in QBI-293A and COS-1 cells, has robust enzymatic activity of the previous unidentified isomerohydrolase, an enzyme converting all-trans retinyl ester to 11-cis retinol in the visual cycle. The initial rate for the reaction is 2.9 pmol/min per mg of RPE65 expressed in 293A cells. The isomerohydrolase activity of RPE65 requires coexpression of lecithin retinol acyltransferase in the same cell to provide its substrate. This enzymatic activity is linearly dependent on the expression levels of RPE65. This study demonstrates that RPE65 is the long-sought isomerohydrolase and fills a major gap in our understanding of the visual cycle. Identification of the function of RPE65 will contribute to the understanding of the pathogenesis for retinal dystrophies associated with RPE65 mutations.
Keywords: isomerase, LRAT, retinal dystrophy, retinyl ester, 11-cis retinal
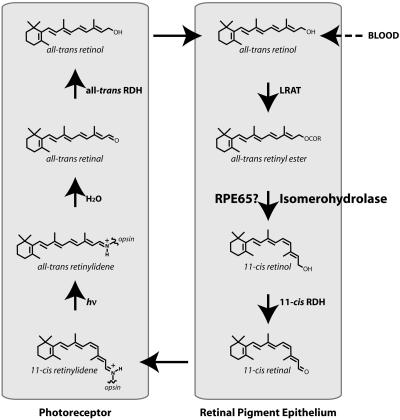
In vertebrates, vision is initiated in rod and cone photoreceptors. The photosensitive entities in these cells are the visual pigments which consist of an apoprotein, opsin, and a chromophore, 11-cis retinal, which is attached to the opsin by a Schiff's base bond (1). Upon absorption of light by the pigments, 11-cis retinal is isomerized to all-trans retinal, which leads to the conformational changes of opsin and subsequently activates G protein transducin, initiating vision (1, 2). Efficient regeneration of 11-cis retinal, referred to as the visual retinoid cycle (see Fig. 1) is critical for the regeneration of the visual pigments (for reviews, see refs. 3–5) and maintenance of visual function. Disruption of the visual cycle by mutation or dysfunction of one of the numerous enzymes involved in this process leads to a number of blinding disorders (6).
Fig. 1.
Scheme of retinoid visual cycle. RDH, retinol dehydrogenase.
The enzyme in the visual cycle that has eluded identification is the isomerohydrolase, which is responsible for the isomerization and hydrolysis of all-trans retinyl ester to 11-cis retinol. Rando and colleagues (7) first proposed that all-trans retinol is first acylated to retinyl esters by lecithin retinol acyltransferase (LRAT). The generated all-trans retinyl ester is then directly isomerized and hydrolized into 11-cis retinol in the retinal pigment epithelium (RPE). This hydrolysis-isomerization process has been proposed to be catalyzed by a single enzyme, referred to as isomerohydrolase, which is associated with the membrane in RPE microsomes (8). Although isomerohydrolase activity had been demonstrated in the RPE almost 20 years ago, identification of the enzyme catalyzing this reaction has been difficult, because this activity is associated with the membrane and is abolished by solubilization in all of the detergents investigated (9). As a result, the isomerohydrolase has not been identified despite intensive efforts of several groups over the past two decades. This missing enzyme represents a major gap in the elucidation of the visual cycle (Fig. 1).
RPE65 is an abundant protein in the RPE and is associated with the microsomal membrane (10, 11). A number of mutations of the RPE65 gene are associated with inherited retinal dystrophies in humans (12, 13) and in dogs (14, 15), indicating that RPE65 is essential for normal vision. Although physiologically significant, the exact function of RPE65 has remained elusive. The RPE65 gene knockout in mice resulted in a lack of 11-cis retinoids in the retina and RPE and an overaccumulation of all-trans retinyl ester in the RPE (16), suggesting an interrupted isomerization process. Recent studies demonstrated that purified RPE65 specifically binds all-trans retinyl palmitate, and these workers suggest that RPE65 is a retinyl ester-binding protein that is required for the isomerization reaction (17, 18).
RPE65 shares significant sequence homology only with the β-carotene monooxygenases, which cleaves β-carotene to generate all-trans retinal (19, 20). However, RPE65 itself does not cleave β-carotene (19), and no other enzymatic activities have ever been reported for RPE65.
Here, we show that RPE65, when coexpressed with LRAT in QBI-293A or COS-1 cells, efficiently generates 11-cis retinol from all-trans retinyl ester, suggesting that it is an enzyme responsible for the isomerohydrolase activity.
Methods
Cell Culture. The QBI-293A cell line for the adenovirus packaging and transient transfection was purchased from Qbiogene (Irvine, CA). The QBI-293A cells were cultured in DMEM (Invitrogen) containing 5% FBS (Invitrogen) supplemented with 100 units/ml penicillin G, 100 μg/ml streptomycin, and 250 ng/ml amphotericin. COS-1 cells (a generous gift from M. Kono, Medical University of South Carolina), a cell line derived from African Green Monkey kidney cells, were grown in DMEM with 10% newborn calf serum and the antibiotic mixture listed above.
Construction of Adenovirus Expressing RPE65 (Ad-RPE65) and Plasmid Expressing LRAT. To construct Ad-RPE65, the full-length coding region of the human RPE65 cDNA (from the translation start codon to the termination codon) was cloned into the pShuttle vector (Qbiogene). The purified plasmid DNA was digested with PmeI, then cotransformed with pAdEasy-1 vector (Qbiogene) into BJ5183-competent cells. The recombinant adenovirus DNA was digested with PacI, purified, and transfected into QBI-293A cells to generate virus particles. The resulting Ad-RPE65 was propagated in QBI-293A cells. Adenovirus infectious titer was determined by plaque formation assay as recommended by the manufacturer and expressed as plaque-forming units per ml.
For the LRAT expression plasmid, the human LRAT cDNA (from the translation start codon to the termination codon) was subcloned into an expression vector, pcDNA6 (Invitrogen) between EcoRI and XbaI sites. The plasmid DNA for transfection was prepared by QIAfilter Plasmid Maxi prep kit (Qiagen, Valencia, CA).
Transient Expression of LRAT, RPE65, and Coexpression. QBI-293A cells were cultured until 70–75% confluency. The LRAT expression plasmid was transiently transfected into the cells by using the calcium phosphate method. Twenty-four hours after the transfection, the cells were infected with Ad-RPE65 at a desired multiplicity of infection (MOI, i.e., ratio of virus plaque-forming units to cell number) and then incubated for another 24 h. The cells were harvested by cell scraper, washed twice by PBS, and stored at –80°C until use.
For RPE65 expression alone, QBI-293A cells were cultured in DMEM with 5% FBS until 70–75% confluent. Ad-RPE65 was incubated with the cells in 5 ml of DMEM for 2–3 h followed by addition of 5 ml of fresh medium. The infected cells were then cultured at 37°C for another 24 h. Cells were harvested with the cell scraper, rinsed twice with PBS, and stored at –80°C until use.
Western Blot Analysis. Protein concentrations in the cell lysates were determined by using the Bradford method (21). The same amount of total protein (40 μg) was blotted with an antibody (1:1,000 dilution) for RPE65 (22) or for LRAT (23) as described (24). The antibodies were then stripped and reblotted with a monoclonal antibody for β-actin (Novus Biologicals, Littleton, CO). The protein level of RPE65 was semiquantified by densitometry using the program genetools (Syngene, Cambridge, U.K.).
Immunocytochemistry. The cells were blocked with 3% BSA in 10% goat serum and then incubated with a polyclonal rabbit anti-LRAT antibody for 1 h. After three washes, a FITC-conjugated anti-rabbit antibody was added and incubated with the cells for 1 h. After five washes, the cells were incubated overnight with rabbit serum. The cells were incubated with an excess of monoclonal unconjugated anti-rabbit Fab antibody, followed by thorough washes. Then the cells were incubated overnight with a polyclonal rabbit anti-RPE65 antibody and 1 h with a Texas red-conjugated anti-rabbit antibody. The cells were washed, mounted, and covered.
Slides were visualized with fluorescence microscopy. Confocal images at high magnification were acquired with a Leica TCS-4D or Zeiss 510 Meta series confocal (Zeiss laser scanning system; LSM 510) equipped with argon (excitation wavelength 488 nm), HeNe1 (568 nm), and HeNe2 (647 nm) lasers. Then LRAT was pseudocolored green, RPE65 was pseudocolored red, and the nucleus was pseudocolored blue.
Isomerohydrolase Activity Assay. Cells expressing LRAT and RPE65 were lysed in a buffer containing 10 mM BTP (pH 8.0) and 100 mM NaCl. Uninfected cells or cells infected by a control adenovirus expressing GFP (Ad-GFP) were used as negative control. Cell lysates were sonicated on ice for 20 s and used for the activity assay. All-trans [11,12-3H]retinol in ethanol (1 mCi/ml, 52 Ci/mmol, PerkinElmer; 1 Ci = 37 GBq) was dried under argon and resuspended in the same volume of dimethyl formamide (DMF). For each reaction, 2 μl of the nondiluted all-trans [11,12-3H]retinol in DMF and 250 μg of total cell lysate were added into 200 μl of the reaction buffer (10 mM BTP, pH 8.0/100 mM NaCl) containing 0.5% BSA and 25 μM cellular retinaldehyde-binding protein. After 2-h incubation in the dark at 37°C, retinoids generated were extracted by the addition of 300 μl of cold methanol and 300 μl of hexane. The upper organic phase was collected and analyzed by normal phase HPLC coupled to Radiomatic 610TR Flow Scintillation Analyzer (PerkinElmer) as described (24). The identity of each retinoid peak was confirmed by comparison of its retention time with that of known retinoid standards. The activity was calculated from the area of 11-cis [3H]retinol peak using synthetic 11-cis [3H]retinol as a standard for calibration.
Results
Isomerohydrolase Activity of RPE65 Expressed in 293A Cells. RPE65 is abundant in the RPE (11 μg per RPE in bovine), but the isomerohydrolase activity is relatively low compared to the activity of other RPE enzymes such as LRAT and RDH10 (23, 25). Thus, assuming that RPE65 is an enzyme with low specific activity, one explanation for its overall activity is its abundance in the RPE. To obtain a high level of RPE65, we expressed RPE65 under the CMV promoter by using an adenovirus expression vector.
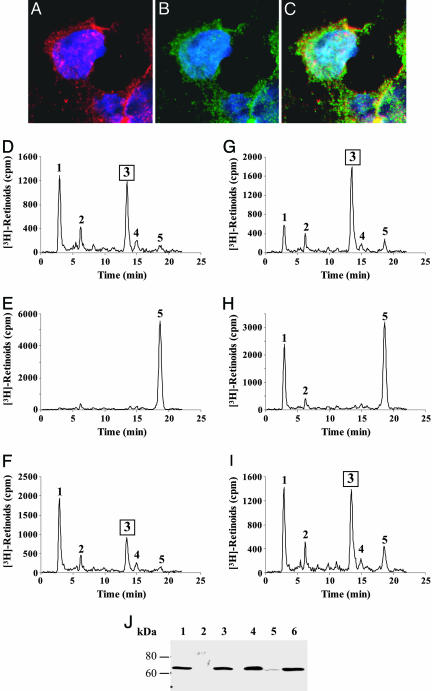
To generate all-trans retinyl ester in the same cells to serve as the in situ substrate for the isomerohydrolase, we expressed both LRAT and RPE65 in QBI-293A cells. The cells were transfected with a LRAT expression plasmid and then infected with Ad-RPE65 at MOI of 20, in triplicate. Twenty-four hours after the infection, recombinant RPE65 was detected at high levels as a single band at ≈65 kDa in cells infected with Ad-RPE65, but not in cells infected with the control virus, Ad-GFP (Fig. 2A and Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Isomerohydrolase activity in QBI-293A cells expressing RPE65 and LRAT. (A) Western blot analysis of total cell lysates. Lane 1, 10 μg of microsomal proteins from bovine RPE (for the RPE65 blot) and 1.25 μg of cell lysates of Sf9 cell expressing LRAT (for the LRAT blot) as positive controls; lanes 2–5, 50 μg of total proteins from 293A cells infected by Ad-GFP (lane 2), transfected with the LRAT plasmid alone (lane 3), infected with Ad-RPE65 alone (lane 4), and transfected with the LRAT plasmid and infected with Ad-RPE65 (lane 5). (B–E) Retinoids generated after incubation of 250 μg of total cellular protein with 0.2 μM all-trans [3H]retinol for 1.5 h. (B) Cells infected by Ad-GFP. (C) Cells infected with Ad-RPE65 alone. (D) Cells transfected with the LRAT plasmid alone. (E) Cells infected by Ad-RPE65 and transfected with LRAT. (F) 11-cis [3H]retinol standard. Peaks 1, retinyl esters; 2, all-trans retinal; 3, 11-cis retinol; 4, 13-cis retinol; 5, all-trans retinol.
To measure the isomerohydrolase activity, equal amounts of cell lysates were used for the isomerohydrolase activity assay in Methods. Lysates from cells infected with Ad-GFP generated low levels of retinyl esters because of endogenous ester synthesizing activities (23) (Fig. 2B). No detectable 11-cis [3H]retinol was produced in the cells infected by Ad-GFP (Fig. 2B). Homogenates from the cells transfected with the LRAT plasmid alone converted almost all of the all-trans [3H]retinol to retinyl ester, but no 11-cis retinol was detected, suggesting that there is no endogenous isomerohydrolase activity in the cell line (Fig. 2D). Cell lysates expressing both RPE65 and LRAT generated a significant amount of 11-cis [3H]retinol, ≈20% of the total [3H]retinoids, in addition to a major retinyl ester peak (Fig. 2E). In the HPLC profile, the retention time of the 11-cis [3H]retinol peak matched that of an 11-cis [3H]retinol standard (Fig. 2F). Interestingly, the cells infected by Ad-RPE65 alone without LRAT expression did not generate detectable 11-cis [3H]retinol from all-trans [3H]retinol (Fig. 2C), although the expression levels of RPE65 were higher than that in cells expressing both the LRAT and RPE65 (Fig. 2A).
Dependence of Isomerohydrolase Activity on RPE65 Expression Levels. To determine the rate of the isomerohydrolase activity of RPE65, the time course of 11-cis [3H]retinol formation was plotted. All-trans [3H]retinol was incubated with cell lysates (0.5 mg of protein) expressing both LRAT and RPE65. The reaction was stopped at varying time points to measure 11-cis [3H]retinol generated. The time course of 11-cis [3H]retinol formation was linear at its initial phase and reached saturation at ≈90 min after the start of the reaction. The initial rate of the reaction was calculated to be 2.9 pmol/min per mg of recombinant RPE65 (Fig. 3A). The time course of 11-cis retinol generation by recombinant RPE65 is similar to that measured in bovine RPE homogenates (Fig. 3B). However, the initial rate of the reaction was 15.1 pmol/min per mg of RPE65 from bovine RPE, which is higher than that of the recombinant protein.
Fig. 3.
Dependence of isomerohydrolase activity on RPE65 expression levels. (A) Time course of 11-cis retinol generation in 293A cell lysates expressing both LRAT and RPE65. (B) Time course of 11-cis retinol generation in bovine RPE homogenates. (C) Dependence of isomerohydrolase activity on MOI of Ad-RPE65. Lysates (250 μg) of 293A cells infected by Ad-RPE65 with increasing MOI and transfected with the same amount of the LRAT plasmid or 33 μg of bovine RPE homogenate were incubated with 0.2 μM all-trans [3H]retinol. In A–C, 11-cis [3H]retinol generated from the reaction were calculated from the area of the 11-cis retinol peak (mean ± SD, n = 3). (D) Western blot analysis of the same lysates used for the activity assay in C. PC, positive control, 10 μg of microsomal proteins from bovine RPE (for the RPE65 blot) and 1.25 μg of cell lysates of Sf9 cell expressing LRAT (for the LRAT blot); NC, negative control (40 μg of total protein from untreated cells); remaining lanes: 40 μg cell lysates from cells transfected with the same amount of LRAT plasmid and infected with Ad-RPE65 at different MOI as indicated. (E) Dependence of isomerohydrolase activity on the RPE65 expression level.
To determine the effect of RPE65 abundance in the cell lysate on isomerohydrolase activity, cells were transfected with the same amount of the LRAT plasmid and then infected with different titers of Ad-RPE65 (MOI from 1 to 40). Cell lysates with equal amounts of total protein (0.25 mg) were used for the isomerohydrolase activity assay. The 11-cis retinol generated in the reaction increased with the MOI (Fig. 3C). Parallel to the increase of isomerohydrolase activity, RPE65 expression levels also increased with the MOI, as shown by Western blot analysis (Fig. 3D). We plotted the amount of 11-cis retinol generated in the reaction against the expression level of RPE65 as determined by densitometry from the Western blots (Fig. 3D) and found a linear dependence (Fig. 3E), indicating that the isomerohydrolase activity depends on RPE65 levels. Variations of LRAT expression levels did not affect the isomerohydrolase activity (Fig. 3 C and D).
Isomerohydrolase Activity of RPE65 Expressed in COS-1 Cells. To exclude the possibility that the isomerohydrolase activity in QBI-293A cells infected by Ad-RPE65 could be a result of an endogenous isomerohydrolase in the cell line, we expressed RPE65 and LRAT in COS-1 cells. COS-1 cells transfected with the equal amount of the LRAT expression plasmid and infected by Ad-RPE65 at MOIs of 40 and 300 generated different RPE65 levels, correlating with MOI (Fig. 4A). Similar to that in QBI-293A cells, lysates from untransfected COS-1 cell or cells expressing either RPE65 alone or LRAT alone did not generate any detectable 11-cis [3H]retinol (Fig. 4 B–D), whereas cells expressing both LRAT and RPE65 had isomerohydrolase activity (Fig. 4 E and F). The level of 11-cis [3H]retinol generated correlated with the expression level of RPE65 in COS-1 cells (Fig. 4 E and F). It should be noted that the MOI required for RPE65 expression in COS-1 cells to show isomerohydrolase activity was higher than that used for QBI-293A cells, probably because adenovirus only replicates in QBI-293A cells (a virus package cell line).
Fig. 4.
Isomerohydrolase activity in COS-1 cells expressing both RPE65 and LRAT. (A) Western blot analysis of COS-1 cells coexpressing LRAT and RPE65. Lanes 1, positive control 10 μg of microsomal proteins from bovine RPE (for the RPE65 blot) and 1.25 μg of cell lysates of Sf9 cells expressing LRAT (for the LRAT blot); 2, negative control (50 μg of total proteins from untreated cells); 3, 50 μg of cell lysate of COS-1 cells transfected with the LRAT expression plasmid and infected with Ad-RPE65 at MOI of 40; 4, 50μg of cell lysate transfected with the same amount of the LRAT plasmid and infected with Ad-RPE65 at MOI of 300. (B–F) Retinoids generated after incubation of 250 μg of COS-1 cell lysates with 0.2 μM all-trans [3H]retinol for 1.5 h: untreated cells (B), cells infected by Ad-RPE65 alone (C), cells transfected by the LRAT plasmid alone (D), cells transfected with the LRAT plasmid and infected by Ad-RPE65 at MOI of 40 (E), and cells transfected with the LRAT plasmid and infected by Ad-RPE65 at MOI 300 (F). Peaks 1, retinyl esters; 2, all-trans retinal; 3, 11-cis retinol; 4, 13-cis retinol; 5, all-trans retinol.
Cofractionation of Isomerohydrolase Activity with LRAT and RPE65. To determine whether the RPE65 colocalized with LRAT, we double-stained QBI-293A cells transfected with the LRAT expression plasmid and infected by Ad-RPE65 by antibodies specific for RPE65 and LRAT. LRAT and RPE65 showed an identical staining pattern in the same cells (Fig. 5 A–C), suggesting the same subcellular localization of RPE65 and LRAT.
Fig. 5.
Colocalization of isomerohydrolase activity with RPE65 and LRAT. (A–C) Double staining of 293A cells expressing RPE65 and LRAT with antibodies specific for RPE65 (A, red) and LRAT (B, green) demonstrated an identical staining pattern of RPE65 and LRAT. (C) Superimposed image of A and B. QBI-293A (D–F) and COS-1 (G–I) cells expressing both RPE65 and LRAT were lysed and centrifuged at 100,000 × g for 1 h. The same amount of proteins (250 μg) from total protein before centrifugation (D and G), the supernatant (E and H), and pellet (F and I) after centrifugation was incubated with 0.2 μM all-trans [3H]retinol in the isomerohydrolase assay buffer for 1.5 h. Peaks were identified as follows: 1, retinyl esters; 2, all-trans retinal; 3, 11-cis retinol; 4, 13-cis retinol; 5, all-trans retinol. (J) Western blot analysis of 40 μg of protein from cell fractionations with antibody for RPE65. Lanes 1, total cell lysate; 2, supernatant from 293A cell after the centrifugation; 3, pellet from 293A cell after the centrifugation; 4, total cell lysate; 5, supernatant from COS-1 cells after the centrifugation; and 6, pellet from COS-1 cells after the centrifugation. MOI for Ad-RPE65 was 40 for 293A cells and 320 for COS-1 cells.
To determine whether the isomerohydrolase activity cofractionates with RPE65, the cells expressing LRAT and RPE65 were lysed and the membrane fraction was separated from the cytosolic proteins by high-speed centrifugation (100,000 × g for 1 h). The activity of the total cell lysate was compared with the cytosolic and membrane fractions. The enzymatic activity of isomerohydrolase was detected only in total cell lysate and the membrane fraction in 293A cells (Fig. 5 D–F and Figs. 8 and 9, which are published as supporting information on the PNAS web site). Correlating with the activity, RPE65 was also detected in the total lysate and the membrane fractions, as shown by Western blot analysis (Fig. 5J). Likewise, RPE65 and the isomerohydrolase activity were also detected in the same subcellular fraction in COS-1 cells. (Fig. 5 G–J).
Discussion
The present study demonstrates that RPE65, when expressed in both 293A and COS-1 cell lines, has a robust isomerohydrolase activity. The isomerohydrolase activity was found to be linearly dependent on RPE65 expression level, as is generally observed for all enzymes (26). Therefore, we conclude that RPE65 is the enzyme responsible for the isomerohydrolase activity. This conclusion that RPE65 is the enzyme does not come in conflict with the observation that RPE65 binds all-trans retinyl ester (17, 18), as it is the substrate of the isomerohydrolase, and enzymes have to bind to their specific substrates (26). The finding that RPE65 is the isomerohydrolase of course explains why the RPE65 gene knockout mice accumulate the substrate of the isomerohydrolase, all-trans retinyl ester, in the RPE.
Our isomerohydrolase activity assays showed that the enzymatic activity of RPE65 requires LRAT coexpression in the same cells (Figs. 2 and 4). This observation is consistent with previous findings that all-trans retinyl esters are the substrate for the isomerohydrolase reaction (24, 27). We propose that LRAT serves as a functional partner of RPE65 by providing the hydrophobic substrate in situ for RPE65. RPE65 is a membrane-associated protein (10, 28). It is likely that its membrane association is necessary for it to obtain hydrophobic all-trans retinyl ester from LRAT through the membrane (Fig. 6). Based on this model, it is important for LRAT and RPE65 to colocalize in the same membrane, as it has been shown that it is almost impossible for a retinyl ester, retinyl palmitate to exchange between different membranes (29). It is not clear whether LRAT and RPE65 have direct protein–protein interactions to form an enzymatic complex or whether they are simply located in the same membrane.
Fig. 6.
Proposed model for functional interactions between LRAT and RPE65. LRAT resides in the membrane via two transmembrane domains. RPE65 is associated with the membrane by three palmitoylated cysteines (indicated by C) (32). LRAT converts all-trans retinol to all-trans-retinyl ester, which is transported to RPE65 through the membrane. RPE65 obtains its substrate, highly hydrophobic all-trans-retinyl ester from the membrane and converted it into 11-cis retinol. at-ROL, all-trans retinol; at-RE, all-trans retinyl ester; 11-cis-ROL; 11-cis retinol.
A conventional test of the validity of the enzymatic function of a protein is that the purified protein shows the enzymatic activity. However, for some enzymes, especially those associated with the membrane, this biochemical approach is not acceptable, because the protein can lose its activity during detergent solubilization and purification. In some cases, membrane-associated enzymes can be reconstituted in liposomes, and their enzymatic activities can be detected. However, it is common that the enzymatic activity cannot be recovered in liposomes. In previous studies, we tested a number of detergents, attempting to solubilize isomerohydrolase in the bovine RPE. All of the detergents inactivated the isomerohydrolase activity. Therefore, it is not surprising that RPE65 purified in detergent solution has no isomerohydrolase activity.
The lack of isomerohydrolase activity in purified RPE65 using the current purification procedures involving detergents raises the question of whether there is an endogenous enzyme in 293A cells that requires the presence of RPE65 for its isomerohydrolase activity. To exclude this possibility, we expressed both RPE65 and LRAT and determined the isomerohydrolase activities in HeLa, a cervical carcinoma cell line, and Hepa G2, a hepatoma cell line, as well as in COS-1 cells. The results show that coexpression of RPE65 and LRAT generated robust isomerohydrolase activities not only in COS-1 cells (Fig. 4), but also in HeLa and Hepa G2 cells (Fig. 10, which is published as supporting information on the PNAS web site). It is unlikely that these cell lines from different origins all contain high levels of an endogenous enzyme that is waiting for RPE65 to show its enzymatic activity.
Isomerohydrolase activity in the visual cycle has been known in the RPE for almost 20 years (30), but the enzyme has not been identified previously, because this enzyme represents one of the most challenging cases for enzymology study as both the substrate and the enzyme are not soluble in water. One major hurdle for identification of the isomerohydrolase is that the enzymatic activity is sensitive to detergent solubilization used during protein purification. Another challenge is the lack of an in vitro activity assay for recombinant protein. Most cultured RPE cell lines do not show any detectable isomerohydrolase activity and also do not express RPE65 at the protein level. The adenovirus-mediated efficient expression and enzymatic activity assay system described here confirms that RPE65 is the enzyme responsible for this isomerization. Identification of RPE65 as the isomerohydrolase filled the gap in our understanding of the visual cycle. However, the precise enzymatic mechanism of the isomerization and hydrolysis by RPE65 remains to be elucidated. The crystal structure of RPE65 has not been revealed. While this paper was in preparation, the crystal structure of carotenoid oxygenase, the first crystallized protein homologous to RPE65, was published (31). Interestingly, the binding of the substrate to the carotenoid oxygenase results in isomerization of its methyl-substituted double bond from trans to cis conformation (31). It is possible that RPE65 uses a similar mechanism for the reaction. The RPE65 and LRAT expression and enzymatic activity assay system provide a useful tool for future mechanistic studies of the isomerohydrolase activity, particularly involving the role of RPE65 mutants in retinal dystrophies.
Supplementary Material
Acknowledgments
We thank Drs. Rosalie K. Crouch and Thomas Ebrey for critical review of the manuscript and helpful comments, Dr. Mas Kono for COS-1 cells, and Dr. John Crabb (Cleveland Clinic Foundation, Cleveland) for the cellular retinaldehyde-binding protein expression system. This study was supported by National Institutes of Health Grants EY12231 and ET015650, a research award from the Juvenile Diabetes Research Foundation, a grant from the American Diabetes Association, and a research grant from Oklahoma Center for the Advancement of Science and Technology and Vision Center of Biomedical Research Excellence (to the Oklahoma University Health Sciences Center).
Author contributions: G.M. and J.-x.M. designed research; G.M., Y.C., Y.T., and B.X.W. performed research; Y.C., Y.T., and B.X.W. contributed new reagents/analytic tools; G.M., Y.C., and Y.T. analyzed data; and G.M. and J.-x.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LRAT, lecithin retinol acyltransferase; RPE, retinal pigment epithelium; Ad-RPE65, adenovirus expressing RPE65; Ad-GFP, adenovirus expressing GFP.
References
- 1.Baylor, D. (1996) Proc. Natl. Acad. Sci. USA 93, 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBee, J. K., Palczewski, K., Baehr, W. & Pepperberg, D. R. (2001) Prog. Retin. Eye Res. 20, 469–529. [DOI] [PubMed] [Google Scholar]
- 3.Rando, R. R. (2001) Chem. Rev. 101, 1881–1896. [DOI] [PubMed] [Google Scholar]
- 4.Saari, J. C., Bredberg, D. L. & Noy, N. (1994) Biochemistry 33, 3106–3112. [DOI] [PubMed] [Google Scholar]
- 5.Crouch, R. K., Chader, G. J., Wiggert, B. & Pepperberg, D. R. (1996) Photochem. Photobiol 64, 613–621. [DOI] [PubMed] [Google Scholar]
- 6.Thompson, D. A. & Gal, A. (2003) Dev. Ophthalmol. 37, 141–154. [DOI] [PubMed] [Google Scholar]
- 7.Deigner, P. S., Law, W. C., Canada, F. J. & Rando, R. R. (1989) Science 244, 968–971. [DOI] [PubMed] [Google Scholar]
- 8.Rando, R. R. (1991) Biochemistry 30, 595–602. [DOI] [PubMed] [Google Scholar]
- 9.Barry, R. J., Canada, F. J. & Rando, R. R. (1989) J. Biol. Chem. 264, 9231–9238. [PubMed] [Google Scholar]
- 10.Hamel, C. P., Tsilou, E., Pfeffer, B. A., Hooks, J. J., Detrick, B. & Redmond, T. M. (1993) J. Biol. Chem. 268, 15751–15757. [PubMed] [Google Scholar]
- 11.Bavik, C. O., Busch, C. & Eriksson, U. (1992) J. Biol. Chem. 267, 23035–23042. [PubMed] [Google Scholar]
- 12.Marlhens, F., Bareil, C., Griffoin, J. M., Zrenner, E., Amalric, P., Eliaou, C., Liu, S. Y., Harris, E., Redmond, T. M., Arnaud, B., Claustres, M. & Hamel, C. P. (1997) Nat. Genet. 17, 139–141. [DOI] [PubMed] [Google Scholar]
- 13.Gu, S. M., Thompson, D. A., Srikumari, C. R., Lorenz, B., Finckh, U., Nicoletti, A., Murthy, K. R., Rathmann, M., Kumaramanickavel, G., Denton, M. J. & Gal, A. (1997) Nat. Genet. 17, 194–197. [DOI] [PubMed] [Google Scholar]
- 14.Aguirre, G. D., Baldwin, V., Pearce-Kelling, S., Narfstrom, K., Ray, K. & Acland, G. M. (1998) Mol. Vis. 4, 23. [PubMed] [Google Scholar]
- 15.Veske, A., Nilsson, S. E., Narfstrom, K. & Gal, A. (1999) Genomics 57, 57–61. [DOI] [PubMed] [Google Scholar]
- 16.Redmond, T. M., Yu, S., Lee, E., Bok, D., Hamasaki, D., Chen, N., Goletz, P., Ma, J. X., Crouch, R. K. & Pfeifer, K. (1998) Nat. Genet. 20, 344–351. [DOI] [PubMed] [Google Scholar]
- 17.Mata, N. L., Moghrabi, W. N., Lee, J. S., Bui, T. V., Radu, R. A., Horwitz, J. & Travis, G. H. (2004) J. Biol. Chem. 279, 635–643. [DOI] [PubMed] [Google Scholar]
- 18.Gollapalli, D. R., Maiti, P. & Rando, R. R. (2003) Biochemistry 42, 11824–11830. [DOI] [PubMed] [Google Scholar]
- 19.von Lintig, J. & Vogt, K. (2000) J. Biol. Chem. 275, 11915–11920. [DOI] [PubMed] [Google Scholar]
- 20.Redmond, T. M., Gentleman, S., Duncan, T., Yu, S., Wiggert, B., Gantt, E. & Cunningham, F. X., Jr. (2001) J. Biol. Chem. 276, 6560–6565. [DOI] [PubMed] [Google Scholar]
- 21.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 22.Ma, J., Zhang, J., Othersen, K. L., Moiseyev, G., Ablonczy, Z., Redmond, T. M., Chen, Y. & Crouch, R. K. (2001) Invest. Ophthalmol. Vis. Sci. 42, 1429–1435. [PubMed] [Google Scholar]
- 23.Ruiz, A., Winston, A., Lim, Y. H., Gilbert, B. A., Rando, R. R. & Bok, D. (1999) J. Biol. Chem. 274, 3834–3841. [DOI] [PubMed] [Google Scholar]
- 24.Moiseyev, G., Crouch, R. K., Goletz, P., Oatis, J., Jr., Redmond, T. M. & Ma, J. X. (2003) Biochemistry 42, 2229–2238. [DOI] [PubMed] [Google Scholar]
- 25.Wu, B. X., Chen, Y., Fan, J., Rohrer, B., Crouch, R. K. & Ma, J. X. (2002) Invest. Ophthalmol. Vis. Sci. 43, 3365–3372. [PubMed] [Google Scholar]
- 26.Fersht, A. (1985) Enzyme Structure and Mechanism (Freeman, New York).
- 27.Gollapalli, D. R. & Rando, R. R. (2003) Biochemistry 42, 5809–5818. [DOI] [PubMed] [Google Scholar]
- 28.Bavik, C. O., Levy, F., Hellman, U., Wernstedt, C. & Eriksson, U. (1993) J. Biol. Chem. 268, 20540–20546. [PubMed] [Google Scholar]
- 29.Rando, R. R. & Bangerter, F. W. (1982) Biochem. Biophys. Res. Commun. 104, 430–436. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein, P. S. & Rando, R. R. (1986) Biochemistry 25, 6473–6478. [DOI] [PubMed] [Google Scholar]
- 31.Kloer, D. P., Ruch, S., Al-Babili, S., Beyer, P. & Schulz, G. E. (2005) Science 308, 267–269. [DOI] [PubMed] [Google Scholar]
- 32.Xue, L., Gollapalli, D. R., Maiti, P., Jahng, W. J. & Rando, R. R. (2004) Cell 117, 761–771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.